Abstract
Background:
Pancreatic cancer (PC) harbours an activated point mutation (KrasG12D) in the Kras proto-oncogene that has been demonstrated to promote the development of PC.
Methods:
This study was designed to investigate the effect of the oncogenic KrasG12D allele on aggressiveness and metastatic potential of PC cells. We silenced the oncogenic KrasG12D allele expression in CD18/HPAF and ASPC1 cell lines by stable expression of shRNA specific to the KrasG12Dallele.
Results:
The KrasG12D knockdown cells exhibited a significant decrease in motility (P<0.0001), invasion (P<0.0001), anchorage-dependent (P<0.0001) and anchorage-independent growth (P<0.0001), proliferation (P<0.005) and an increase in cell doubling time (P<0.005) in vitro and a decrease in the incidence of metastases upon orthotopic implantation into nude mice. The knockdown of the KrasG12D allele led to a significant increase in the expression of E-cadherin (mRNA and protein) both in vitro and in vivo. This was associated with a decrease in the expression of phoshpo-ERK-1/2, NF-κB and MMP-9, and transcription factors such as δEF1, Snail and ETV4. Furthermore, the expression of several proteins involved in cell survival, invasion and metastasis was decreased in the KrasG12D knockdown cells.
Conclusions:
The results of this study suggest that the KrasG12D allele promotes metastasis in PC cells partly through the downregulation of E-cadherin.
Keywords: activated Kras, invasion, metastasis, motility, pancreatic cancer
Pancreatic cancer (PC) has one of the worst prognoses among all known cancers, with a mortality to incidence ratio of ∼0.83 (Jemal et al, 2009). In the United States, it remains the fourth leading cause of cancer-related deaths with an incidence of ∼12.3 per 100 000 people (Jemal et al, 2009). The median survival of patients with PC is a mere 4.1 months with the overall 5-year survival rate being <5% (Heinemann et al, 2008; Sultana et al, 2008; Jemal et al, 2009). At the time of diagnosis, >85% of patients have metastatic disease, which makes surgical and medical interventions largely ineffective (Matsuno et al, 2004). One of the reasons for the poor outcome of PC is the lack of early detection markers and limited efficacy of existing treatment regimens. Therefore, there is an urgent need to understand the pathogenesis of PC in order to discover early detection marker(s), novel molecular targets and new therapeutic strategies.
Recent advances in molecular genetics have revealed a compendium of genetic lesions associated with the progression and metastasis of PC (Hezel et al, 2006). Of these mutations, Kras is found to be mutated in almost all cases (75–90%) of PC and represents an early event in the development and progression of this malignancy (Almoguera et al, 1988; Shibata et al, 1990; Caldas and Kern, 1995; Dergham et al, 1997; Moskaluk et al, 1997; Wang et al, 2002). Kras is a member of the highly homologous Ras family of proteins and has potent transforming ability (Barbacid, 1987). It is a 21 kDa size monomeric membrane-localised guanine nucleotide (GTP/GDP)-binding protein. A wide variety of extracellular stimuli can activate Kras, and the activated form, in turn, activates a cascade of signals that ultimately regulate cell growth, differentiation (McCormick, 1989) and apoptosis. Mutations in Kras occur most frequently at codon 12 (Gonzalez-Cadavid et al, 1989; Grunewald et al, 1989; Mariyama et al, 1989; Nagata et al, 1990; Shibata et al, 1990; Van Laethem et al, 1995), and less frequently at codons 13 and 61 (Motojima et al, 1991, 1993; Caldas and Kern, 1995). All of these mutations can abolish the intrinsic ability of the Kras protein to hydrolyse GTP, resulting in continuous stimulation of cell proliferation (Barbacid, 1987; Ellis and Clark, 2000).
Recently, several approaches, including short interfering RNAs, antibodies, mutant Kras-specific peptide inhibitors, adenoviruses expressing antisense Kras, dominant-negative Kras, antisense oligonucleotides and different drugs have been investigated for their ability to target the mutant form of Kras (Aoki et al, 1995, 1997; Adjei, 2001). The major drawback of dominant-negative Kras and pharmacological Kras inhibitors is the lack of specificity (Kohl et al, 1994; Feig, 1999; Bolick et al, 2003; de Bono et al, 2003), whereas the antisense oligonucleotides downregulate the wild-type Kras, which is essential for the normal function of all cells in the body. RNA interference (RNAi) has become a novel approach to target the mutant form of this oncogene specifically (Devi, 2006; Gaither and Iourgenko, 2007). Kras is a potent tumour initiator as evidenced by observations that activating mutations in Kras (G12D) are required for the development of pancreatic intraepithelial neoplasms (PanINs), which precede invasive adenocarcinoma (Aguirre et al, 2003). There is also evidence to suggest that several point mutations in codon 12 can result in constitutive activation of Kras (Karapetis et al, 2008). A point mutation (GGT → GAT) resulting in a single amino-acid change from glycine to aspartic acid in codon 12 (KrasG12D) is observed in many cases of PC as well as in many PC cell lines (Hohne et al, 1992). Various studies have shown the role of mutant KrasG12D in enhanced cell proliferation and transformation of normal pancreatic epithelial cells (Hingorani et al, 2003; Tuveson et al, 2004); however, its function in the late stage of PC progression remains unknown.
The objective of this study was to investigate the role of mutant KrasG12D allele in PC by knockdown of this allele in the highly metastatic PC cell lines CD18/HPAF and ASPC1, followed by examination of the effects on cellular functions (through in vitro and in vivo functional studies) and intracellular signalling cascades. Altogether, our data indicate that silencing of KrasG12D causes a significant reduction in the motility, invasion and metastatic potential of PC cells. This is done through an upregulation of E-cadherin and downregulation of Snail, δEF1 and ETV4 transcription factors, and signalling pathways such as Akt, FAK and ERK1/2.
Materials and methods
Cell culture, plasmid construction and transfection
CD18/HPAF, Capan-1, ASPC-1 cells were cultured in DMEM, whereas BXPC-3 cells were grown in RPMI and HPDE cells in Keratinocyte media, respectively, supplemented with 10% fetal calf serum and antibiotics (penicillin and streptomycin 100 μg ml–1). The pSUPER.retro.puro vector was digested with BglII and HindIII restriction enzymes and dephosphorylated with calf intestinal alkaline phosphatase (CIAP). Two complementary oligonucleotides, 5′-GATCCCCGTTGGAGCTGATGGCGTAGTTCAAGAGACTACGCCATCAGCTCCAACTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGTTGGAGCTGATGGCGTAGTCTCTTGAACTACGCCATCAGCTCCAACGGG 3′, corresponding to the mutant KrasG12D gene with BglII and HindIII sites were synthesised, annealed, phosphorylated and ligated into the digested pSUPER vector (Restriction sites at 3′ and 5′ ends are underlined while the sequence of the hairpin loop sequence is indicated by underline italics). The presence of the insert was confirmed by sequencing and digestion with EcoRI and HindIII restriction enzymes. The shRNA construct (pSUPER-KrasG12D) was transfected in the phoenix cells, a packaging cell line that produces high-viral titer in culture using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA). At the same time, the phoenix cells were also transfected with pSUPER vectors bearing scramble oligonucleotide sequence. After 24hr, CD18/HPAF PC cells were seeded in 6-well plates at 5 × 104 cells per well and grown to 60% confluence in DMEM without serum medium. The media supernatant was collected from phoenix cells after 48 and 72hr post-transfection and the viral supernatant was used to infect the sub-confluent cultures of CD18/HPAF PC cells after addition of 4mg mL−1 polybrene. Pooled populations of stable oncogenic Kras knockdown and control (ShRNA, Scramble) cells were selected by puromycin (5 μg ml–1) containing 10% DMEM medium. The ASPC1 and BXPC3 cells were transiently knocked down for oncogenic shKras allele by using the aforementioned vector constructs (pSUPER-KrasG12D and Scramble vectors). The protein was isolated from ASPC1-shKras as well as scramble controls after 48 h of transient transfection and it was analysed for downstream signalling molecules.
Quantitative real-time PCR
Total RNA was isolated and the cDNA was synthesised by reverse transcription as described previously (Moniaux et al, 2008). The real-time primers for KrasG12D were designed as described by Gupta et al (2005) by keeping the mutation at the 3′ end of the forward primer and an additional base mutation was also included before the Kras mutation in order to amplify the Kras mutant allele selectively (Gupta et al, 2005). For all other genes, the primers were designed using Primer 3 software (Supplementary Table 1). Real-time PCR was performed on Roche 480 Real-Time PCR System (Indianapolis, IN, USA). Real-time PCR reactions were performed in triplicate and template controls (NTCs) were run for each assay under the same conditions. PCR was then performed in 10 μl reaction containing 5 μl 2 × SBYR green Master Mix, 3.2 μl of autoclaved nuclease free water, 1 μl diluted RT product (1 : 10) and 0.4 μl each of forward and reverse primers (5 pmol) for Kras mutation (F-5′-ACTTGTGGTAGTTGGAGCAGA-3′ and R-5′-TTGGATCATATTCGTCCACAA-3′). The cycling conditions comprised: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and by 58 °C for 1 min. Gene expression levels were normalised to the level of β-actin expression, which we have shown to be unresponsive to Kras mutation, and were reported relative to mutant Kras expression level in the scramble RNA-transfected cells.
Immunoblot analysis
Immunoblot analysis was done as described previously (Moniaux et al, 2008). The primary antibodies for the activated form and total FAK, Kras, cyclins D1, E and A and NF-κB were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA), cMyc and p27Kip1 from Epitomics (Burlingame, CA, USA), activated and total Akt and ERK, Caspase-3 and Cleaved-Caspase-9 from Cell Signaling (Danvers, MA, USA), matrix metalloproteinase-9 (MMP-9) and E-cadherin were gifts from Dr Rakesh Singh and β-actin was from Sigma Aldrich (St Louis, MO, USA).
Growth kinetics, clonogenicity and apoptosis assays
Cells (1.0 × 104 cells per 3 ml of medium containing 1.0% FBS) were seeded in six-well plates and allowed to grow for different time intervals. The growth of the cells was monitored by counting the number of viable cells on a Vi-CELL (Boulevard, CA, USA) counter every day for 8 days. The cell population doubling time (Td) was calculated during the exponential growth phase (96–144 h) using the following formula: Td = 0.693,t/log ,(Nt/N0), where t is the time difference (in h), Nt is the cell number at time t (144 h) and N0 is the cell number at the initial time (96 h) (Zhang et al, 2002). To assess clonogenic potential, the cells were trypsinised and either plated in 0.3% agarose with a 0.5% agarose underlay (1 × 103 cells per well in 24-well plate) for assessment of anchorage-independent growth or on plastic coated petri dishes for anchorage-dependent respectively. The number of foci >100 μm was counted after 14 days. Apoptosis was measured by Annexin V FITC staining as described previously (Chaturvedi et al, 2007). This assay is based on the principle that during the process of apoptosis, phosphatidyl serine (normally localised on the inner leaflet of the plasma membrane) is flipped out. Annexin V has a high affinity for phosphatidyl serine and binds to it. However, at this stage (called early apoptosis), the cell membrane is still intact and hence propidium iodide (PI) is excluded from these cells. Therefore, early apoptotic cells are defined by a positive staining for Annexin V and a negative staining for PI.
As apoptosis progresses, the cell membrane permeability increases, leading to increased entry of PI that binds to the DNA. Hence, cells during the later stage of apoptosis are positive for both Annexin V and PI. A similar process also takes place during necrosis, making it impossible to distinguish late apoptosis from necrosis with this assay.
Tumourigenicity assay
Subconfluent cultures of CD18/HPAF-derived clones were trypsinised and washed with phosphate-buffered saline. Cell viability was determined by Trypan blue staining and single-cell suspensions of >90% viability was used for the orthotopic injections. The cells were resuspended in a normal saline (NS) solution at a concentration of 5 × 104 cells per 50 μl. Immunodeficient mice were purchased from the Animal Production Area of the National Cancer Institute-Frederick Cancer Research and Development Center (Frederick, MD, USA). The mice were treated in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines. The orthotopic implantation was performed as previously described (Choudhury et al, 2004). All mice were killed after 21 days of implantation. The presence of metastatic lesions in different organs was determined thorough gross inspection and histological analysis. Pancreatic tumours were excised, weighed and measured.
Motility and invasion assay
For motility assays, 1 × 106 cells suspended in serum-free medium were plated in the top chamber of polyethylene teraphthalate membranes (six-well insert, pore size 8 μm) (Becton Dickinson, Franklin Lakes, NJ, USA). Then, 2 ml of 10% serum-containing medium was added to the lower chamber of the well and the cells were allowed to migrate for 22 h under chemotactic drive. After incubation, the cells that did not migrate through the pores in the membrane were removed by scraping the membrane with a cotton swab. The migrated cells on the lower side of the membrane were stained with Diff-Quick cell stain kit (Dade-Behring Inc., Newark, DE, USA) and photographed in 10 random fields of viewed at × 100 magnification. Cell numbers were counted and expressed as the average number of cells per field of view. For invasion assay, cells (1 × 106) were seeded on Matrigel-coated membrane inserts (BD Biosciences, Bedford, MA, USA). The bottom chamber contained 2.0 ml of serum-supplemented medium as a chemoattractant. After incubation for 22 h at 37 °C, the cells that had invaded through the Matrigel-coated membrane were fixed and stained using a Diff-Quick reagent kit. After air drying the membrane, the cells were counted at a magnification of × 10 in 10 random fields of view under a microscope. Three independent experiments were done in each case. The data were represented as the average of the three independent experiments with the standard error of mean (s.e.m.).
Oligonucleotide array gene expression analysis
Human oligonucleotide array containing probes for 39 200 genes was constructed at the Microarray Core Facility of University of Nebraska Medical Center. Total RNA was isolated from shK-ras and K-ras scramble transfected CD18/HPAF cells by Qiagen RNEasy kit (Qiagen Sciences, Valencia, CA, USA) according to the manufacturer's directions. The procedure for the microarray hybridisation and the subsequent analysis has been previously described by us (Chaturvedi et al, 2007).
Statistical analysis
For analysis of microarray data, a gene chip containing 39 200 genes was used. The data were normalised using BRB Array Tools. Random-variance paired t-tests were used to determine which genes are differentially expressed between tumour samples and the normal samples. The random-variance paired t-test allows for sharing information among genes about variation without assuming that all genes have the same variance, which gives a more accurate estimate of the variability when sample sizes are small (2). A significance level of 0.001 was selected to help limit the false discovery rate (FDR) due to multiple comparisons. The FDR was limited to <10%. Parametric data were compared using the two-tailed Student's t-test, whereas nonparametric data were analysed using a two-way ANOVA or χ2 test. Data were analysed using the Medcalc for Windows version 9·6·4·0 software (MedCalc Software, Broekstraat, Mariakerke, Belgium). A P-value of <0.05 was considered significant.
Results
Targeting of mutant KrasG12D allele by stable expression of KrasG12D shRNA leads to decreased oncogenic Kras expression
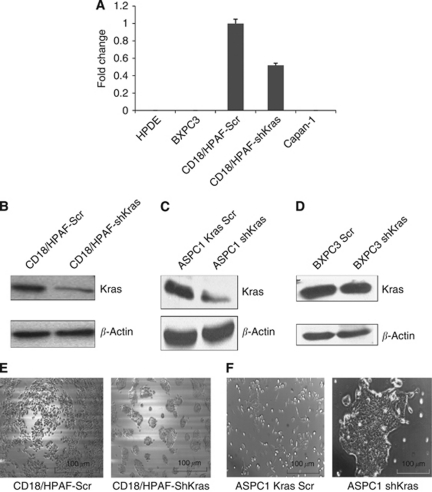
To target the KrasG12D mutant allele, siRNA oligos were designed that were 64 nucleotides long and covered with a point mutation in codon 12 (G → D) of the Kras gene. They were cloned into the pSUPER RETRO mammalian expression vector. Similarly, a scramble expression vector construct (pSUPER Kras-Scr) was made using scramble shRNA oligonucleotides. The resultant constructs (pSUPER-shKrasG12D and pSUPER Kras-Scr) were transfected into CD18/HPAF pancreatic adenocarcinoma cells. Pooled populations of CD18/HPAF-shKrasG12D and CD18/HPAF-Kras-Scr were selected for puromycin resistance. The effective inhibition of the mutant KrasG12D allele was determined by real-time PCR using a primer set that selectively amplifies the mutated Kras allele but not the wild-type (WT) allele. RNAs isolated from BXPC3, HPDE and Capan-1 cells were used as a control as it is known that BXPC3 and HPDE cells express only the WT alleles and Capan-1 has a G12V mutation instead of the G12D mutation. The CD18/HPAF-shKras cells had a significantly decreased expression of KrasG12D mRNA compared with the CD18/HPAF-Kras-Scr cells (Figure 1A). Furthermore, the expression of total Kras protein was reduced in the CD18/HPAF-shKrasG12D cells compared with Kras-Scr-transfected cells (Figure 1B). Similar results were observed with transient knockdown of the oncogenic Kras allele in ASPC1 cells (Figure 1C).
Figure 1.
Strategy for shRNA-mediated sequence-specific silencing of oncogenic Kras in CD18/HPAF cells. (A) Real-time PCR analysis using primers that specifically amplify mutated codon 12 but not wild-type allele. CD18/HPAF-shKrasG12D pooled population cells show a 60–70% decrease in the expression of mutant Kras (G → D) allele at the mRNA level compared with the control population. No amplification was observed in BXPC3, HPDE and Capan-1 cells, which are negative for the K-rasG12D mutant allele. (B, C) Western blot analysis of the CD18/HPAF (or ASPC-1) shKrasG12D, CD18/HPAF (or ASPC-1) Scr pooled population shows significant inhibition in total Kras protein expression, respectively. (D) Specificity of Kras shRNAs for KrasG12D allele: BXPC3 cells transiently transfected with pSUPER retro shKras or Kras-Scr revealed no significant difference in total levels of Kras protein by immunoblotting. β-Actin was served as a loading control. (E, F) A morphological comparison between the CD18/HPAF (or shASPC-1) shKrasG12D and CD18/HPAF (or ASPC-1) Kras-Scr cells. The Kras knockdown CD18/HPAF (or shASPC-1) cells grow as aggregates compared with the scramble cells.
In order to confirm the specificity of the oligos, we transiently transfected BXPC3 cells (KrasG12D negative) with pSUPER-shKrasG12D. The pSUPER-Kras-Scr vectors revealed no significant decrease in Kras total protein between the BXPC3 Scr and BXPC3 shKras (Figure 1D).
Silencing of mutant KrasG12D allele leads to altered morphology, decreased growth rate and reduced clonogenicity of pancreatic cancer cells
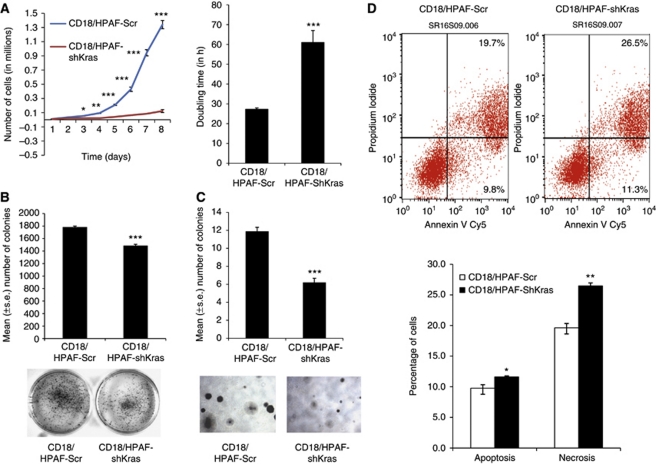
The morphology and growth rates of the pooled populations of CD18/HPAF-shKras and CD18/HPAF-Kras-Scr cells were monitored after inhibiting oncogenic Kras (mutant allele) in tumour cells. The CD18/HPAF-shKras cells showed a tendency to grow as clumps when compared with the CD18/HPAF-Kras-Scr cells (Figure 1E). Similar growth pattern was observed with silencing of mutant KrasG12D allele in ASPC1 cells (Figure 1F). A growth curve was plotted to determine the effect of Kras knockdown on the cell doubling time. Calculation of population doubling time during the exponential phase (96–144 h) demonstrated a significant (P<0.0045) increase in cell doubling time in the CD18/HPAF-shKras cells (61.0 h) compared with scramble siRNA-transfected cells (27.0 h; Figure 2A). Furthermore, on the last day (day 8), there was nearly a 90% reduction in the number of cells in the CD18/HPAF-shKras group when compared with the CD18/HPAF-Kras-Scr group (Figure 2A).
Figure 2.
(A) Growth kinetics of shKras pooled population and vector control (scramble) cells. A total of 10 000 cells were plated in six-well plates in DMEM media containing 1.0% FBS. Cells were counted every 24 h for 8 days and a growth curve was plotted. The shKras clones showed reduced cell growth compared with the vector control cells. Doubling time of these cells was calculated at 96–144 h; the shKras clone showed a higher doubling time compared with the vector control cells (*P<0.05, **P<0.001, ***P<0.0001). (B, C) Anchorage-dependent (B) and -independent (C) analysis for short-term colony formation in CD18/HPAF-shKrasG12D and scramble cells. Statistical analysis revealed a significant variation (anchorage-independent (***P<5.002E–08) and anchorage-dependent conditions (***P<0.0001)) between the two populations. (D) Annexin V and propidium iodide staining analysis by flow cytometry to identify apoptosis in CD18/HPAF-shKrasG12D and scramble cells. The statistical analysis showed significant variation in necrotic cells (**P=0.002) and apoptotic cells (*P=0.05).
The effect of silencing oncogenic KrasG12D expression on the clonogenic properties of CD18/HPAF cells was studied in anchorage-independent and anchorage-dependent conditions. We observed that the CD18/HPAF-shKrasG12D cells had a significantly reduced ability to divide as evidenced by the reduction in the number of colonies formed in both anchorage-independent (P<0.0001) and anchorage-dependent conditions (P<0.0001; Figures 2B and C). Analysis of PI- and Annexin V-positive cells by flow cytometry indicated a significant increase in the number of apoptotic cells (P<0.05; with Annexin V-positive but PI-negative staining) and late apoptotic/necrotic cells (P<0.002; with Annexin V-positive and PI-positive staining) in CD18/HPAF-shKras cells when compared with CD18/HPAF-Kras-Scr cells (Figure 2D).
Oncogenic Kras knockdown results in an inhibition of cell motility and invasion
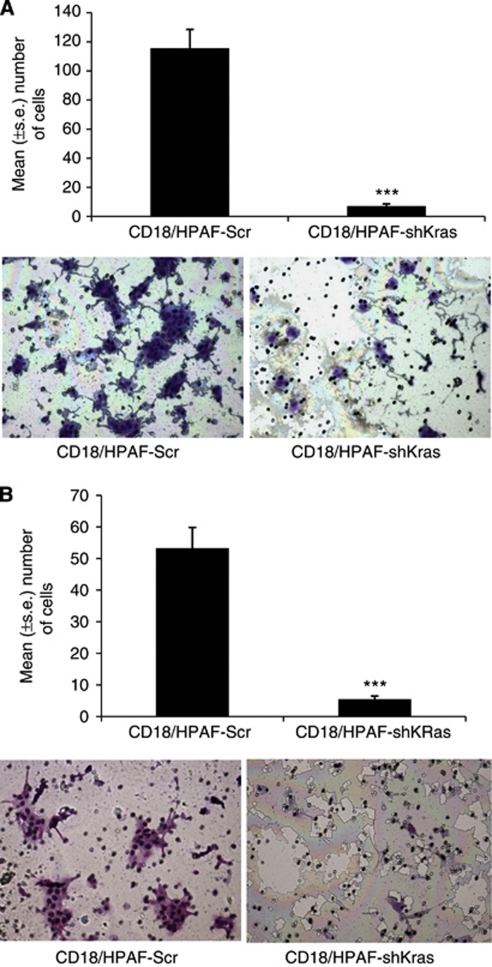
Several studies have reported that invasive and metastatic properties of tumour cells are partly influenced by their phenotypic characteristics such as motility and invasion. Silencing of oncogenic KrasG12D leads to a significant (P<0.0001) reduction in cellular motility and invasive ability (∼6- and 10-fold, respectively) in CD18/HPAF cells (Figures 3A and B).
Figure 3.
Effect of oncogenic K-ras on cell motility and invasive behaviour of CD18/HPAF cells. Cells (1 × 106) were seeded on non-coated or Matrigel-coated membranes for motility (A) and invasion (B) assays, respectively, and incubated for 24 h. Medium containing 10% fetal bovine serum in the lower chamber was used as a chemoattractant. The cells that migrated through the membrane were stained and photographed under bright-field microscopy (magnification × 10). The number of cells that migrated through the membrane was determined by averaging 10 random fields of view. The data are expressed as the number of cells per field of view and is the average of three independent experiments. Error bars indicate s.e. of the average (***P<0.0001). The total numbers of CD18/HPAF-shKrasG12D cells that migrated and invaded were reduced by six- and ten-fold respectively as compared with the vector control cells.
Selective inhibition of oncogenic KrasG12D in pancreatic cancer cells results in the suppression of tumourigenicity and metastasis
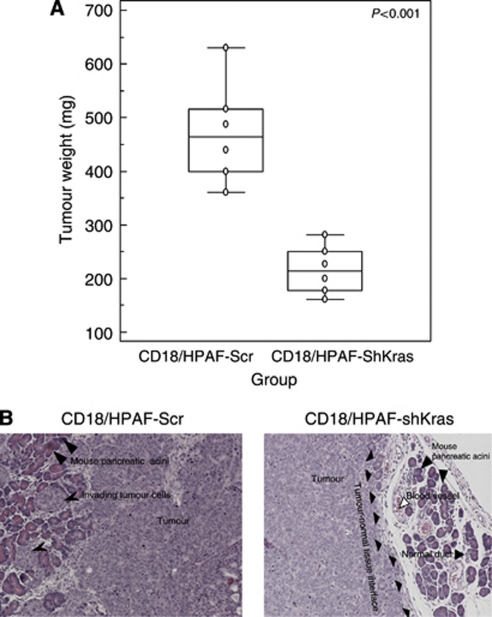
To examine the effect of oncogenic Kras knockdown in vivo, a pooled population of CD18/HPAF-shKrasG12D and CD18/HPAF-Kras-Scr cells was orthotopically implanted into the pancreas of nude mice. The animals were killed at 21 days post-implantation and the pancreatic tumours were removed and weighed. We carried out haematoxylin and eosin staining (Figures 4A and B). Liver, lung, diaphragm, intestine, kidney and mesenteric lymph nodes were examined for the presence of metastatic lesions. A primary pancreatic tumour and metastatic lesions in the spleen and on the intestinal wall were found in all the mice implanted with CD18/HPAF-Kras-Scr cells. In this group, some animals also had metastasis in the liver (n=2) and/or kidney (n=1). In contrast, animals injected with CD18/HPAF-shKras cells had significantly smaller tumours (P<0.001), and had fewer or no metastatic lesions (Table 1).
Figure 4.
Effect of oncogenic Kras knockdown on size of orthotopically grown primary tumours. (A) The mean weight of tumours formed by CD18/HPAF-shKras cells was significantly less than that formed by the scramble cells (P<0.001). (B) Haematoxylin and eosin-stained sections of orthotopic tumours demonstrating the aggressive invasion of the normal pancreatic tissue by the CD18/HPAF-Kras-Scr cells (left). In comparison, CD18/HPAF-shKrasG12D cells showed significantly decreased invasiveness (as evidenced from a clear demarcation between the edge of the tumour and the healthy tissue) (right).
Table 1. Incidence of metastases developed by orthotopic implantation of pooled populations of CD18/HPAF Scr cells and shKras clones in immunodeficient mice.
| Cell type | Spleen | Liver | Peritoneum | Mesenteric lymph nodes | Kidney | Intestinal wall |
|---|---|---|---|---|---|---|
| CD18/HPAF scramble | 4/6 (67%) | 2/6 (33%) | 4/6 (67%) | 4/6 (67%) | 1/6 (17%) | 5/6 (83%) |
| CD18/HPAF shK-ras | 0/6 (0%) | 0/6 (0%) | 0/6 (0%) | 1/6 (17%) | 0/6 (0%) | 0/6 (0%) |
| P-value (χ2 test) | 0.06 | 0.45 | 0.06 | 0.24 | 1.0 | 0.01 |
Effect of KrasG12D silencing on downstream signalling
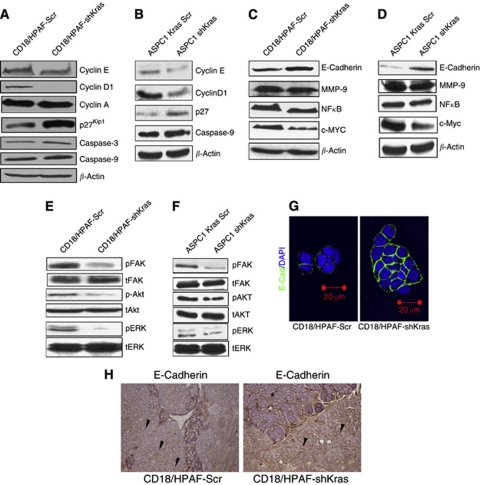
Kras mutation has previously been reported to be associated with the upregulation of cyclins D and E and downregulation of p27kip1 (Fan and Bertino, 1997). As shown in Figures 5A and B, the sequence-specific knockdown of the activated Kras allele led to a decreased expression of cyclins D1 and E in the CD18/HPAF-shKras and ASPC1-shKras cells in comparison with the Kras-Scr-transfected cells, whereas no change was observed in cyclin A levels. Similarly, the expression of p27kip1, caspase-3 and cleaved caspase-9 was also increased in shKrasG12D transfected cells compared with the Kras-Scr-transfected CD18/HPAF and ASPC1 cells. Inhibition of the KrasG12D allele expression resulted in a significant decrease in the activation of downstream signalling molecules, including phospho-ERK1/2, phospho-Akt and phospho-FAK, MMP-9, c-Myc, NF-κB in the shKras-transfected cells compared with Kras-Scr-transfected cells (Figures 5C–F). The level of total ERK-1/2, Akt and total FAK, however, remained unchanged. Furthermore, immunoblot analysis also revealed an increased expression of E-cadherin in the CD18/HPAF-shKras and ASPC1-shKras cells compared with the scrambled population (Figures 5C and D). In agreement with these results, immunofluorescence analysis also revealed an increase in E-cadherin expression and membrane localisation in CD18/HPAF-shKras-transfected cells when compared with CD18/HPAF-Kras-Scr knockdown cells (Figure 5G). Furthermore, immunohistochemical analysis of the primary orthotopic tumour sections (from the orthotopically implanted mice) revealed an increased E-cadherin expression in the CD18/HPAF-shKras tumours compared with the scramble vector-transfected cells (Figure 5H).
Figure 5.
Western blot analysis comparing the expression of key molecules involved in cell proliferation (c-myc, cyclins A, D1, E and p27), apoptosis (caspase-3 and 9), metastasis and invasion (E-cadherin, MMP-9, NF-κB, phospho and total FAK, phosphor and total Akt and phosphor and total ERK-1/2) in CD18/HPAF (A, C, E) and ASPC-1 (B, D, F) Scr vs shKrasG12D cells. (H) Immunohistochemical analysis of primary tumours developed following orthotopic implantation of CD18/HPAF-shKrasG12D and scramble cells in immunodeficient mice. Tumour sections were stained for E-cadherin using specific anti-mouse monoclonal antibody (original magnification × 100). Tissue derived from CD18/HPAF-shKrasG12D tumours showed a stronger membrane staining for E-cadherin (arrowheads). (G) Confocal analysis showed increased E-cadherin expression on the cell membrane in CD18/HPAF-shKrasG12D cells compared with scramble cells (*Normal mouse pancreatic tissue, primary tumour with E-cadherin expression).
Alteration in signalling pathways because of knockdown of mutant Kras allele in CD18/HPAF cells
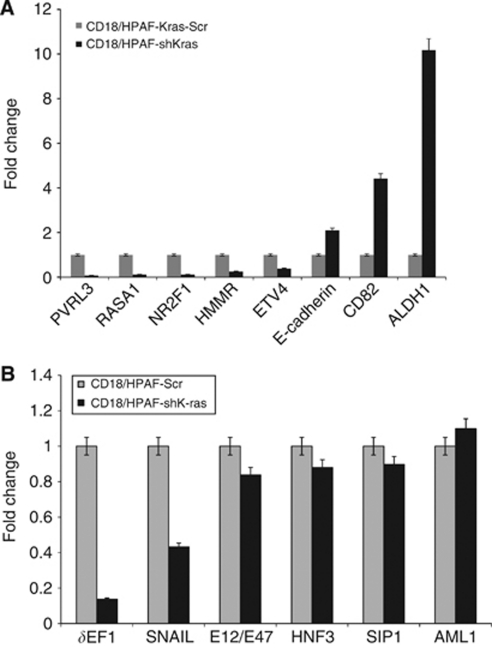
In order to identify the pathways dysregulated in PC cells because of the knockdown of the mutant Kras allele, we compared the gene expression profiles of CD18/HPAF-shKras and scramble cells by global microarray analysis. The microarray analysis revealed that many genes were significantly up- or down-regulated more than two-fold in CD18/HPAF-shKras cells compared with the scrambled cells (Supplementary Tables 2 and 3). Notably, the functional classes of genes affected by Kras silencing included tumour suppressors (HMMR, CAV1 and BHLHE41), cell adhesion molecules (CDH1, LGALS4 and PVRL3), genes regulating cellular motility and invasion (ETV4, NT5E and ALDH1A1), cell growth (GCNT3), cell cycle (HPGD, CDKN1A and CAV1), metastasis (CD82) and signal transduction (TM4SF4 and NR2F1). Out of these pathways, the pathway modulated by the transcription factor ETV4, SNAIL and δEF1 appeared to be highly perturbed. Using the Ingenuity Pathway Analysis (Ingenuity Systems, Mountain View, CA, USA) software, the differentially expressed genes were grouped into several gene networks (Supplementary Figure 1). Some of the differentially expressed genes were validated by real-time PCR (Figure 6A). The real-time PCR also showed a reduced expression of SNAIL and δEF1 transcription factors in Kras knockdown cells compared with control cells (Figure 6B). The gene ontology-based clustering analysis revealed that many of the genes differentially regulated upon silencing of Kras were involved in cell adhesion and metastasis. In summary, the microarray analysis suggests that Kras signalling is important in the process of PC metastasis and may crosstalk with other signalling pathways.
Figure 6.
Quantitative real-time PCR analysis. (A) The expression of selected upregulated and downregulated genes (identified by global microarray analysis) was validated by Q-RT–PCR using specific primers. (B) Real-time PCR analysis of the expression of transcription factors known to downregulate E-cadherin expression.
Discussion
Cancer development involves a multistep process in which tumour cells acquire various genetic and epigenetic changes to grow and metastasise to distant organs. Investigation of the molecular genetics of pancreatic adenocarcinoma has revealed a specific pattern of genetic lesions that occur during the initiation and progression of PC (Aguirre et al, 2003; Hezel et al, 2006). Out of these, mutations in the Kras gene are reported to be an early event, being observed in virtually all cases of PC (75–95% Almoguera et al, 1988; Wang et al, 2002). These mutations in Kras have an important role in the initiation and progression of PC. During the later part of the disease, other genetic and epigenetic alterations occur in EGFR, HER2, p16Ink4a, p53, Smad/DPC4, and other genes (Aguirre et al, 2003; Hezel et al, 2006) that facilitate the development of pancreatic adenocarcinoma and, subsequently, its metastasis (Aguirre et al, 2003; Hingorani et al, 2005; Bardeesy et al, 2006; Hezel et al, 2006). The dominant nature of the mutant Kras allele results in the cells exhibiting a transformed ability even when a single allele of mutant Kras is expressed. Consequently, inhibition of the oncogenic Kras allele expression in human cancers is a promising approach for tumour-specific gene therapy (Friday and Adjei, 2005). Previous studies have shown that KrasG-12D has lower intrinsic GTPase activity than WT Kras. Furthermore, this mutant is also insensitive to p-120-GAP (Bollag et al, 1996), leading to constitutive activation of Ras-mediated downstream signalling pathways in cells expressing the mutant Kras allele. Therefore, studies on the molecular and cellular functions associated with this mutant hold paramount importance for therapeutic purposes.
In this study, the expression of the mutated KrasG12D allele was selectively inhibited by shRNA, specifically targeting the mutant allele in CD18/HPAF and ASPC-1 PC cells. Subsequently, we studied the effect of KrasG12D silencing on the function of PC cells in vitro (motility, invasion and clonogenicity) and in vivo (tumourigenesis and metastasis) and its impact on downstream signalling pathways by using pooled populations. In the case of CD18/HPAF cells, suppression of the oncogenic KrasG12D allele led to a significant reduction in their tumourigenic and metastatic potential in vivo. Microarray analysis identified several genes associated with cell growth, proliferation and metastasis that were significantly altered in the CD18/HPAF-KrasG12D knockdown cells.
The stable expression of a shRNA targeting the mutant KrasG12D allele led to a decreased expression of the mutant allele at the mRNA level in CD18/HPAF-shKras cells (Figure 1A). The total Kras protein expression was also significantly decreased in the CD18/HPAF-shKras ASPC1-shKras cells (Figures 1B and C). These results suggest that siRNAs can serve as powerful tools for sequence-specific inhibition of an oncogenic mutant allele. Our study also corroborates previous studies in human (Capan-1 KrasG12V) and murine (C26 colorectal cells, KrasG12D) cells, wherein the stable knockdown of mutant Kras allele resulted in a reduced expression of both the oncogenic and total Kras protein and mRNA levels (Brummelkamp et al, 2002; Smakman et al, 2005).
It is now clearly established that activating mutations in Kras are found in the majority of PCs and that these mutations, along with other genetic lesions, contribute to the increased aggressiveness of the tumour (Almoguera et al, 1988; Dergham et al, 1997; Wang et al, 2002; Aguirre et al, 2003; Hezel et al, 2006). Therefore, the Kras has emerged as an attractive target for the therapy of PC (Friday and Adjei, 2005). In this study, the sequence-specific inhibition of mutant KrasG12D allele resulted in a significant decrease in tumour cell growth and an altered morphology and clonogenic ability (Figures 1E and 1F and 2A–C). This was expected because of the established role of Kras in cell proliferation and cell survival (Fan and Bertino, 1997; Hingorani et al, 2003; Tuveson et al, 2004). Activated Kras regulates the cell cycle through activation of ERK and Akt signalling pathways (Wang et al, 2009) and induction of cyclin D1, a protein important for progression from the G1 to the S phase (Stacey, 2003). Furthermore, it also has an antiapoptotic effect mediated by activation of the PI3-K/Akt pathway (Downward, 2004). In our study, inhibition of mutant Kras resulted in suppression of the ERK and Akt pathways (Figures 5E and F). Similarly, there was a significant decrease in cellular levels of c-Myc, NF-κB, cyclins D1 and E and an increase in caspase-3, 9 and p27kip1 levels in the CD18/HPAF-shKras and ASPC1-shKras cells (Figures 5A–D). This result suggests that the constitutively active Kras allele is important for survival, proliferative ability, motility and invasiveness of PC cells. Thus, the KrasG12D allele appears to have an important role in regulating key cellular processes in PC cells, reinforcing its importance as a target for anticancer therapy.
Tumour metastasis comprises a series of distinct and sequential steps, involving the growth of the tumour locally, invasion by transmigration through basement membrane and nontumor host tissue, intravasation into blood vessels, dissemination and survival in the bloodstream and finally extravasation and re-establishment at distant sites (Chambers et al, 2002). It requires a series of cellular processes to occur, including phenotypic changes, loss of the cell–cell and cell–extracellular matrix (ECM) interactions and increased cellular motility. In this study, we observed a decrease in cell motility and invasion in the Kras knockdown cells (pooled population) associated with decreased activation of the Akt pathway (Figures 3A, B, 5E, F). Cell motility has a key role in tumour cell invasion into the surrounding non-tumour tissues and is a major determinant of the aggressive nature of a tumour cell. The Ras pathway has also been implicated in cytoskeletal rearrangements, altering the expression of integrins and cell migration through activation of the PI3-K/Akt pathway (Potempa and Ridley, 1998; Okudela et al, 2004; Fleming et al, 2005).
Inhibition of Krasval12 by shRNA has been previously shown to decrease tumourigenicity in Capan-1 cells upon subcutaneous implantation (Brummelkamp et al, 2002) and in murine C26 colorectal cancer cells in vitro. The Kras knockdown cells formed fewer tumours and did not cause morbidity (Smakman et al, 2005). We also noted that inhibition of KrasG12D resulted in a significant decrease in the tumourigenic and metastatic potential of CD18/HPAF (Figure 4A and Table 1). However, there was no decrease in the incidence of tumours with 100% of the animals injected with the CD18/HPAF-shKrasG12D cells forming tumours. The possible explanations for this observation include the presence of heterogeneous cells in the pooled clonal population and the partial compensation of the anti-tumourigenic effect of KrasG12D knockdown by other unknown signalling pathways. Although these earlier studies indicated that oncogenic Kras has a key role in the proliferation of PC cells, its role in regulating metastasis of PC has remained largely unexplored. In order to determine the role of activated Kras in metastasis, we utilised an orthotopic model that revealed a significant inhibition of metastasis upon selective silencing of KrasG12D (Table 1). Our results suggest that the constitutively active form of Kras not only regulates tumour cell growth, but it also has an important role in modulating the invasive nature of the malignant cells.
Tumour cell invasion through the ECM and tissue barriers requires the combined effects of increased cell motility and proteolytic degradation. We observed decreased levels of activated FAK (pY925) in the CD18/HPAF-shKras and ASPC1-shKras cells when compared with that in the scramble cells (Figures 5E and F). The decreased activation of FAK may be responsible for the reduced motility observed in the CD18/HPAF-shKras cells compared with scramble cells similar to that reported in an earlier study (Sieg et al, 2000).
Additionally, the phenotypic changes associated with epithelial mesenchymal transition (EMT) include both an increased cellular motility (Thiery, 2002) and an increased production of ECM-degrading enzymes, accompanied by disruption of E-cadherin-mediated cell–cell adhesion (Takeichi, 1995; Christofori and Semb, 1999). The function of epithelial E-cadherin is altered in most epithelial tumours and can be disrupted by various genetic and epigenetic mechanisms, including modulation by signalling molecules. Loss of E-cadherin activates signals that promote tumour cell migration, invasion and dissemination (Thiery, 2002). The increased expression of E-cadherin in the Kras knockdown cells (Figures 5C, D, G, H and 6A, and Supplementary Figure 1) seems to suggest that oncogenic KrasG12D can inhibit E-cadherin function partly by suppressing its expression. Furthermore, the expression of MMP-9, a key mediator of the invasive property of malignant cells (Fridman et al, 2003), was also decreased upon silencing of the oncogenic Kras allele. Kras-mediated ERK activation is known to induce MMP-9 that, in turn, causes cleavage of E-cadherin leading to the disruption of cell–cell contacts (Wang et al, 2009). Our results suggest that a MMP-9-mediated decrease in E-cadherin expression may contribute to the highly metastatic property of CD18/HPAF cells. This suggests that activated Kras contributes to the metastatic nature of CD18/HPAF cells.
To elucidate the global cellular pathways that are altered in PC cells upon downregulation of the mutant Kras allele, we conducted microarray analysis and observed that most of the genes differentially expressed in the Kras-Scr compared with the shKras cells were associated with cell proliferation, motility and metastatic behaviour of tumour cells (Figure 6A, Supplementary Tables 2 and 3). Of those genes that were altered because of silencing of oncogenic Kras, alteration in the expression of the transcription factor Snail supports Kras-mediated E-cadherin regulation. Snail has been shown to promote tumour cell invasion by either inducing the transcription of MMP-9 (Jorda et al, 2005) or suppression of transcription of E-cadherin (Huber et al, 2005). Recently, a report showed that δEF-1/ZEB1 binds to the promoter of the E-cadherin and represses its expression (Eger et al, 2005). In the present study, real-time PCR analysis showed that the expression of both SNAIL and δEF1/ZEB1 was decreased in the CD18/HPAF-shKras cells (Figure 6B). Altogether, these results, including the increase in E-cadherin, point to SNAIL and δEF1/ZEB1-mediated regulation of E-cadherin as a target of Kras in PC cells. In addition, the microarray analysis identified several differential genes associated with cell proliferation, motility and metastatic behaviour of tumour cells (Supplementary Tables 2 and 3 and Supplementary Figure 1). Taken together, the results of our study provide an insight into the diverse pathways altered in PC cells upon the sequence specific inhibition of the mutant Kras allele.
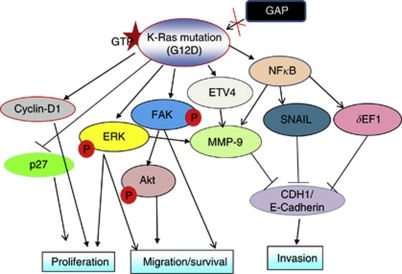
This study was aimed at understanding the pathological role of the highly oncogenic KrasG12D allele in PC. The specific silencing of the oncogenic Ras allele downregulated multiple signalling pathways that are involved in promoting cell proliferation, inhibiting apoptosis, breaking cell–cell contacts and regulating expression of protease like MMP-9. The observed inhibition of cell proliferation in the Kras knockdown cells may be mediated through the inhibition of the MAPK pathway, whereas the increase in the metastatic property of KrasG12D expressing cells might be at least partly because of the activation of FAK and a reduction in the expression of E-cadherin (Figure 7). Finally, our studies demonstrated that PC cells harbouring the KrasG12D mutation are dependent on Ras signalling and suggest that shRNA-mediated gene silencing could be an effective approach for selective inhibition of activated Kras. The results of our study could be useful to target novel proteins downstream of activated Kras in order to disrupt Ras-mediated oncogenic signalling pathways.
Figure 7.
Proposed model for Kras-mediated signalling events that promote progression and metastasis of pancreatic cancer. A glycine (G) to aspartate (D) mutation in the Kras protein makes it constitutively active by making it insensitive to inactivation by GTPase-activating proteins (GAPs). The constitutively active Kras (KrasG12D) can then activate several pathways involved in cell proliferation, migration, invasion and metastasis of PC cells. The activated Kras upregulates cyclin D1 and downregulates the expression of p27, leading to cell proliferation. Mutant Kras-mediated activation of extracellular signal-regulated kinase (ERK) can activate downstream signalling pathways that promote cell proliferation, survival, migration and metastasis. Likewise, Kras-mediated activation of focal adhesion kinase (FAK) leads to the activation of Akt, and thus promotes cell migration and survival. The activated FAK can also directly regulate cell motility. Kras-mediated upregulation of NF-κB leads to an upregulation of the transcription factors SNAIL and δEF1 that, in turn, repress the expression of E-cadherin, a key molecule regulating cell migration/invasiveness. At the same time, it also upregulates ETV4 (a transcription factor) that, in turn, promotes the expression of matrix metalloproteinase-9 (MMP-9), a gelatinase that promotes cancer cell invasion.
Acknowledgments
We thank Ms Kristi L Berger for editing the paper. We acknowledge the invaluable technical support from Mr Erik Moore and Ms Kavita Mallya. We also thank Janice A Taylor and James R Talaska of the confocal laser scanning microscope core facility at the UNMC for their support. This work was supported in part by the grants from the National Institutes of Health (CA RO1 CA78590, UO1 CA111294, RO1 CA133774, RO1 CA131944 and P50 CA 127297).
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
The authors declare no conflict of interest.
Supplementary Material
References
- Adjei AA (2001) Blocking oncogenic Ras signaling for cancer therapy. J Natl Cancer Inst 93(14): 1062–1074 [DOI] [PubMed] [Google Scholar]
- Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA (2003) Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev 17(24): 3112–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M (1988) Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 53(4): 549–554 [DOI] [PubMed] [Google Scholar]
- Aoki K, Yoshida T, Matsumoto N, Ide H, Sugimura T, Terada M (1997) Suppression of Ki-ras p21 levels leading to growth inhibition of pancreatic cancer cell lines with Ki-ras mutation but not those without Ki-ras mutation. Mol Carcinog 20(2): 251–258 [PubMed] [Google Scholar]
- Aoki K, Yoshida T, Sugimura T, Terada M (1995) Liposome-mediated in vivo gene transfer of antisense K-ras construct inhibits pancreatic tumor dissemination in the murine peritoneal cavity. Cancer Res 55(17): 3810–3816 [PubMed] [Google Scholar]
- Barbacid M (1987) ras genes. Annu Rev Biochem 56: 779–827 [DOI] [PubMed] [Google Scholar]
- Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, Hanahan D, Redston MS, Chin L, DePinho RA (2006) Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci USA 103(15): 5947–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolick SC, Landowski TH, Boulware D, Oshiro MM, Ohkanda J, Hamilton AD, Sebti SM, Dalton WS (2003) The farnesyl transferase inhibitor, FTI-277, inhibits growth and induces apoptosis in drug-resistant myeloma tumor cells. Leukemia 17(2): 451–457 [DOI] [PubMed] [Google Scholar]
- Bollag G, Adler F, elMasry N, McCabe PC, Conner Jr E, Thompson P, McCormick F, Shannon K (1996) Biochemical characterization of a novel KRAS insertion mutation from a human leukemia. J Biol Chem 271(51): 32491–32494 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2(3): 243–247 [DOI] [PubMed] [Google Scholar]
- Caldas C, Kern SE (1995) K-ras mutation and pancreatic adenocarcinoma. Int J Pancreatol 18(1): 1–6 [DOI] [PubMed] [Google Scholar]
- Chambers AF, Groom AC, MacDonald IC (2002) Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2(8): 563–572 [DOI] [PubMed] [Google Scholar]
- Chaturvedi P, Singh AP, Moniaux N, Senapati S, Chakraborty S, Meza JL, Batra SK (2007) MUC4 mucin potentiates pancreatic tumor cell proliferation, survival, and invasive properties and interferes with its interaction to extracellular matrix proteins. Mol Cancer Res 5(4): 309–320 [DOI] [PubMed] [Google Scholar]
- Choudhury A, Moniaux N, Ulrich AB, Schmied BM, Standop J, Pour PM, Gendler SJ, Hollingsworth MA, Aubert JP, Batra SK (2004) MUC4 mucin expression in human pancreatic tumours is affected by organ environment: the possible role of TGFbeta2. Br J Cancer 90(3): 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofori G, Semb H (1999) The role of the cell-adhesion molecule E-cadherin as a tumour-suppressor gene. Trends Biochem Sci 24(2): 73–76 [DOI] [PubMed] [Google Scholar]
- de Bono JS, Tolcher AW, Rowinsky EK (2003) Farnesyltransferase inhibitors and their potential in the treatment of breast carcinoma. Semin Oncol 30(5 Suppl 16): 79–92 [DOI] [PubMed] [Google Scholar]
- Dergham ST, Dugan MC, Kucway R, Du W, Kamarauskiene DS, Vaitkevicius VK, Crissman JD, Sarkar FH (1997) Prevalence and clinical significance of combined K-ras mutation and p53 aberration in pancreatic adenocarcinoma. Int J Pancreatol 21(2): 127–143 [DOI] [PubMed] [Google Scholar]
- Devi GR (2006) siRNA-based approaches in cancer therapy. Cancer Gene Ther 13(9): 819–829 [DOI] [PubMed] [Google Scholar]
- Downward J (2004) PI 3-kinase, Akt and cell survival. Semin Cell Dev Biol 15(2): 177–182 [DOI] [PubMed] [Google Scholar]
- Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R (2005) DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 24(14): 2375–2385 [DOI] [PubMed] [Google Scholar]
- Ellis CA, Clark G (2000) The importance of being K-Ras. Cell Signal 12(7): 425–434 [DOI] [PubMed] [Google Scholar]
- Fan J, Bertino JR (1997) K-ras modulates the cell cycle via both positive and negative regulatory pathways. Oncogene 14(21): 2595–2607 [DOI] [PubMed] [Google Scholar]
- Feig LA (1999) Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat Cell Biol 1(2): E25–E27 [DOI] [PubMed] [Google Scholar]
- Fleming JB, Shen GL, Holloway SE, Davis M, Brekken RA (2005) Molecular consequences of silencing mutant K-ras in pancreatic cancer cells: justification for K-ras-directed therapy. Mol Cancer Res 3(7): 413–423 [DOI] [PubMed] [Google Scholar]
- Friday BB, Adjei AA (2005) K-ras as a target for cancer therapy. Biochim Biophys Acta 1756(2): 127–144 [DOI] [PubMed] [Google Scholar]
- Fridman R, Toth M, Chvyrkova I, Meroueh SO, Mobashery S (2003) Cell surface association of matrix metalloproteinase-9 (gelatinase B). Cancer Metastasis Rev 22(2–3): 153–166 [DOI] [PubMed] [Google Scholar]
- Gaither A, Iourgenko V (2007) RNA interference technologies and their use in cancer research. Curr Opin Oncol 19(1): 50–54 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Cadavid NF, Zhou D, Battifora H, Bar-Eli M, Cline MJ (1989) Direct sequencing analysis of exon 1 of the c-K-ras gene shows a low frequency of mutations in human pancreatic adenocarcinomas. Oncogene 4(9): 1137–1140 [PubMed] [Google Scholar]
- Grunewald K, Lyons J, Frohlich A, Feichtinger H, Weger RA, Schwab G, Janssen JW, Bartram CR (1989) High frequency of Ki-ras codon 12 mutations in pancreatic adenocarcinomas. Int J Cancer 43(6): 1037–1041 [DOI] [PubMed] [Google Scholar]
- Gupta M, Yates CR, Meibohm B (2005) SYBR Green-based real-time PCR allelic discrimination assay for beta2-adrenergic receptor polymorphisms. Anal Biochem 344(2): 292–294 [DOI] [PubMed] [Google Scholar]
- Heinemann V, Boeck S, Hinke A, Labianca R, Louvet C (2008) Meta-analysis of randomized trials: evaluation of benefit from gemcitabine-based combination chemotherapy applied in advanced pancreatic cancer. BMC Cancer 8: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, DePinho RA (2006) Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 20(10): 1218–1249 [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA (2003) Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4(6): 437–450 [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA (2005) Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7(5): 469–483 [DOI] [PubMed] [Google Scholar]
- Hohne MW, Halatsch ME, Kahl GF, Weinel RJ (1992) Frequent loss of expression of the potential tumor suppressor gene DCC in ductal pancreatic adenocarcinoma. Cancer Res 52(9): 2616–2619 [PubMed] [Google Scholar]
- Huber MA, Kraut N, Beug H (2005) Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol 17(5): 548–558 [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ (2009) Cancer statistics, 2009. CA Cancer J Clin 59(4): 225–249 [DOI] [PubMed] [Google Scholar]
- Jorda M, Olmeda D, Vinyals A, Valero E, Cubillo E, Llorens A, Cano A, Fabra A (2005) Upregulation of MMP-9 in MDCK epithelial cell line in response to expression of the Snail transcription factor. J Cell Sci 118(Pt 15): 3371–3385 [DOI] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR (2008) K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359(17): 1757–1765 [DOI] [PubMed] [Google Scholar]
- Kohl NE, Wilson FR, Mosser SD, Giuliani E, deSolms SJ, Conner MW, Anthony NJ, Holtz WJ, Gomez RP, Lee TJ, Smith RL, Graham SL, Hartman GD, Gibbs JB, Oliff A (1994) Protein farnesyltransferase inhibitors block the growth of ras-dependent tumors in nude mice. Proc Natl Acad Sci USA 91(19): 9141–9145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariyama M, Kishi K, Nakamura K, Obata H, Nishimura S (1989) Frequency and types of point mutation at the 12th codon of the c-Ki-ras gene found in pancreatic cancers from Japanese patients. Jpn J Cancer Res 80(7): 622–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno S, Egawa S, Fukuyama S, Motoi F, Sunamura M, Isaji S, Imaizumi T, Okada S, Kato H, Suda K, Nakao A, Hiraoka T, Hosotani R, Takeda K (2004) Pancreatic Cancer Registry in Japan: 20 years of experience. Pancreas 28(3): 219–230 [DOI] [PubMed] [Google Scholar]
- McCormick F (1989) ras GTPase activating protein: signal transmitter and signal terminator. Cell 56(1): 5–8 [DOI] [PubMed] [Google Scholar]
- Moniaux N, Chakraborty S, Yalniz M, Gonzalez J, Shostrom VK, Standop J, Lele SM, Ouellette M, Pour PM, Sasson AR, Brand RE, Hollingsworth MA, Jain M, Batra SK (2008) Early diagnosis of pancreatic cancer: neutrophil gelatinase-associated lipocalin as a marker of pancreatic intraepithelial neoplasia. Br J Cancer 98(9): 1540–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskaluk CA, Hruban RH, Kern SE (1997) p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res 57(11): 2140–2143 [PubMed] [Google Scholar]
- Motojima K, Tsunoda T, Kanematsu T, Nagata Y, Urano T, Shiku H (1991) Distinguishing pancreatic carcinoma from other periampullary carcinomas by analysis of mutations in the Kirsten-ras oncogene. Ann Surg 214(6): 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motojima K, Urano T, Nagata Y, Shiku H, Tsurifune T, Kanematsu T (1993) Detection of point mutations in the Kirsten-ras oncogene provides evidence for the multicentricity of pancreatic carcinoma. Ann Surg 217(2): 138–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata Y, Abe M, Motoshima K, Nakayama E, Shiku H (1990) Frequent glycine-to-aspartic acid mutations at codon 12 of c-Ki-ras gene in human pancreatic cancer in Japanese. Jpn J Cancer Res 81(2): 135–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okudela K, Hayashi H, Ito T, Yazawa T, Suzuki T, Nakane Y, Sato H, Ishi H, KeQin X, Masuda A, Takahashi T, Kitamura H (2004) K-ras gene mutation enhances motility of immortalized airway cells and lung adenocarcinoma cells via Akt activation: possible contribution to non-invasive expansion of lung adenocarcinoma. Am J Pathol 164(1): 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potempa S, Ridley AJ (1998) Activation of both MAP kinase and phosphatidylinositide 3-kinase by Ras is required for hepatocyte growth factor/scatter factor-induced adherens junction disassembly. Mol Biol Cell 9(8): 2185–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata D, Almoguera C, Forrester K, Dunitz J, Martin SE, Cosgrove MM, Perucho M, Arnheim N (1990) Detection of c-K-ras mutations in fine needle aspirates from human pancreatic adenocarcinomas. Cancer Res 50(4): 1279–1283 [PubMed] [Google Scholar]
- Sieg DJ, Hauck CR, Ilic D, Klingbeil CK, Schaefer E, Damsky CH, Schlaepfer DD (2000) FAK integrates growth-factor and integrin signals to promote cell migration. Nat Cell Biol 2(5): 249–256 [DOI] [PubMed] [Google Scholar]
- Smakman N, Veenendaal LM, van DP, Bos R, Offringa R, Borel RI, Kranenburg O (2005) Dual effect of Kras(D12) knockdown on tumorigenesis: increased immune-mediated tumor clearance and abrogation of tumor malignancy. Oncogene 24(56): 8338–8342 [DOI] [PubMed] [Google Scholar]
- Stacey DW (2003) Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr Opin Cell Biol 15(2): 158–163 [DOI] [PubMed] [Google Scholar]
- Sultana A, Tudur SC, Cunningham D, Starling N, Neoptolemos JP, Ghaneh P (2008) Meta-analyses of chemotherapy for locally advanced and metastatic pancreatic cancer: results of secondary end points analyses. Br J Cancer 99(1): 6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M (1995) Morphogenetic roles of classic cadherins. Curr Opin Cell Biol 7(5): 619–627 [DOI] [PubMed] [Google Scholar]
- Thiery JP (2002) Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2(6): 442–454 [DOI] [PubMed] [Google Scholar]
- Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D, Hingorani SR, Zaks T, King C, Jacobetz MA, Wang L, Bronson RT, Orkin SH, DePinho RA, Jacks T (2004) Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell 5(4): 375–387 [DOI] [PubMed] [Google Scholar]
- Van Laethem JL, Vertongen P, Deviere J, Van RJ, Rickaert F, Cremer M, Robberecht P (1995) Detection of c-Ki-ras gene codon 12 mutations from pancreatic duct brushings in the diagnosis of pancreatic tumours. Gut 36(5): 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Lian ST, Chen YF, Yang YC, Chen LT, Lee KT, Huang TJ, Lin SR (2002) Unique K-ras mutational pattern in pancreatic adenocarcinoma from Taiwanese patients. Cancer Lett 180(2): 153–158 [DOI] [PubMed] [Google Scholar]
- Wang XQ, Li H, Van PV, Winn RA, Heasley LE, Nemenoff RA (2009) Oncogenic K-Ras regulates proliferation and cell junctions in lung epithelial cells through induction of cyclooxygenase-2 and activation of metalloproteinase-9. Mol Biol Cell 20(3): 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhao W, Zhang HJ, Domann FE, Oberley LW (2002) Overexpression of copper zinc superoxide dismutase suppresses human glioma cell growth. Cancer Res 62(4): 1205–1212 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.