Abstract
Microtubule-associated protein (MAP) Tau has been reported to be a predictive factor for clinical response to taxanes in metastatic breast cancer. We generated a panel of eight taxane resistant variants from four human breast cancer cell lines (MCF-7, T-47D, MDA-MB-231 and BT-549). Four variants had higher levels of Tau compared to their T47D and MDA-MB-231 parental cells. Using isoform-specific primers, we found that Tau 0N, 1N, 2N, 3R and 4R isoforms are overexpressed in the resistant variants, as is Tau exon 6 but not exons 4A or 8. To determine whether Tau overexpression produces resistance to taxanes, we derived three independent T-47D clones stably over-expressing Tau-3R and Tau-4R isoforms. Tau overexpression did not result in taxane resistance compared to parental cells transfected with vector alone. We then knocked down Tau expression in three cell lines that expressed Tau constitutively (MCF-7 and ZR-75-1 breast cancer cells, and OVCAR-3 ovarian cancer cells). Lentivirus-mediated silencing of Tau expression in MCF-7 and OVCAR-3 cells did not result in increased taxane sensitivity compared with luciferase shRNA-infected cells and uninfected parental cells. Transient silencing using Tau-specific siRNAs also did not alter taxane sensitivity relative to non-targeting controls in both MCF-7 and and ZR-75-1 cells. These results show that neither overexpression nor depletion of Tau modulate cellular sensitivity to taxanes. Although Tau overexpression has been reported to be a predictive marker of taxane resistance, it is not likely to be a direct mechanism of taxane resistance in breast cancer.
Keywords: drug resistance, microtubule associated protein, Tau, breast cancer, taxanes
Introduction
The taxanes, paclitaxel and docetaxel, are an important class of microtubule stabilizing drugs in the treatment of solid tumors, including breast cancer [1]. In breast cancer, taxanes have shown clinical benefit both as adjuvant chemotherapy for localized disease [2, 3] and in the treatment of metastatic disease [4]. However, as is the case for almost all anticancer agents, inherent or acquired taxane resistance greatly limits their clinical utility [5–7].
Resistance to taxanes can involve multiple, diverse mechanisms, including increased drug efflux, altered microtubule dynamics, or impaired cell death signaling. Drug efflux mediated by the overexpression of the multidrug transporter P-glycoprotein (P-gp), the gene product of MDR1 (ABCB1), is an important resistance mechanism for many chemotherapeutic agents, including taxanes [8]. However, the majority of clinically resistant breast cancers are P-gp negative [9], and therefore manifest other mechanisms of resistance to taxanes.
Several lines of evidence suggest that the microtubule-associated protein (MAP) Tau may play a role in modulating response to taxanes. 1) Tau was one of the differentially expressed genes associated with response to preoperative paclitaxel chemotherapy in breast cancer patients [10, 11]. 2) Tau expression across 23 cancer cell lines correlated with IC50 values for paclitaxel [12]. 3) Down regulation of Tau expression in ZR-75-1 and MCF-7 breast cancer cell lines was reported to increase sensitivity to paclitaxel [10]. 4) Reduced Tau expression in gastric cancer was associated with a favorable response to paclitaxel [13].
Tau is a protein enriched in axons of mature and growing neurons whose primary function is to regulate microtubule dynamics [14]. Tau is also found in the distal ends of growing neurons [15, 16], in oligodendrocytes [17, 18] and in muscle [19]. It is encoded by a 100 kb single gene on chromosome 17q21 [20], consisting of 16 exons, of which exons 2, 3, 4A, 6, 8, 10, 13 and 14 are alternatively spliced [19, 21–28]. Alternative splicing of three major exons (2, 3 and 10) results in the expression of six well characterized Tau isoforms in the human brain (Figure 1). Although Tau has been identified as a potential marker of paclitaxel response in breast cancer [10,11], its functional significance and isoform characterization have not been fully investigated.
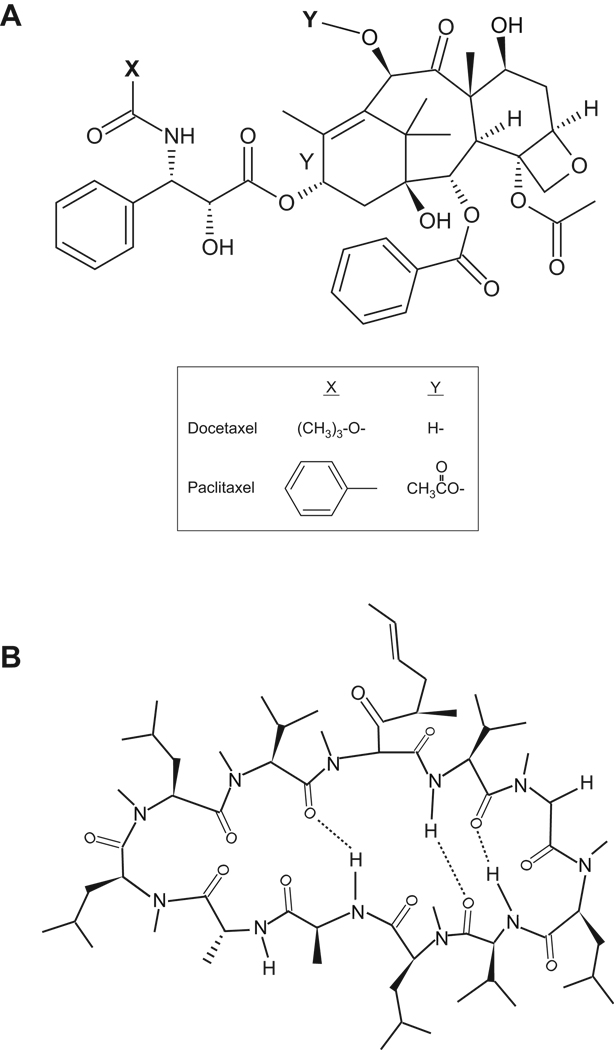
Figure 1. Chemical structures of (A) docetaxel and paclitaxel, and (B) PSC-833.
The aim of this study was to evaluate the functional role of Tau in modulating taxane response and to characterize the expression of Tau isoforms in taxane resistant breast cancer cells. Our results indicate that although Tau isoforms are overexpressed in some taxane-resistant breast cancer cells, neither down regulation nor overexpression of Tau alters cellular sensitivity to taxanes.
Materials and Methods
Materials
Paclitaxel was obtained from the drug repository of the National Cancer Institute (Bethesda, MD), docetaxel from Sanofi Aventis (Bridgewater, NJ) and PSC-833 (PSC) from Novartis (East Hanover, NJ). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Cell lines and cultures
Human cancer cell lines MCF-7, BT-549, MDA-MB-231, T-47D, ZR-75-1, and OVCAR-3 were purchased from and characterized by the American Type Culture Collection (ATCC, Manassas, VA). All cells were cultured in McCoy's 5A media with L-glutamine, supplemented with 10% (v/v) fetal calf serum, penicillin and streptomycin, (all from Invitrogen, Carlsbad, CA). Cells were maintained at 37 °C in an atmosphere containing 5% CO2. T-47D cells transfected with plasmids encoding Tau 3R and Tau 4R isoforms were selected and maintained in complete McCoy5A medium containing 500 µg/ml of G418 (Invitrogen).
Total RNA isolation and cDNA synthesis
Total RNA was extracted using RNeasy Mini Kit (Qiagen, Valencia, CA), resuspended in RNase-free water and stored at −80 °C until analysis. RNA concentration was measured by absorbance reading at 260nm. Total RNA (1–5 µg) was reverse transcribed into cDNA using SuperScript III First-Strand Synthesis Kit with the Oligo(dT)20 primer (both Invitrogen). The final cDNA product was stored at −20 °C until further analysis.
Real time PCR
Quantitative real time PCR was carried out in duplicate or triplicate per experiment, with each experiment reproduced multiple times using SYBR Green PCR Master Mix on an ABI 7900HT real-time PCR instrument (Applied Biosystems, Foster City, CA). A PCR reaction mixture of 20 µL contained 10 µL of 2× SYBR Green PCR Master Mix, 3.5 µL of 0.75 µM gene specific forward primer, 3.5 µL of 0.75 µM gene specific reverse primer, and 3 µL cDNA. The amplification program consisted of 1 cycle at 95 °C for 2 min, followed by 40 cycles of 95 °C for 30 sec, annealing temperature at 60 °C for 30 sec and extension temperature at 72 °C for 15 sec. Standard curves for Tau transcripts were generated using plasmids encoding each of the 6 known Tau isoforms and for HPRT1 using a reference cDNA from pooled cell lines. The cycle threshold (Ct) was used to calculate relative amounts of target transcript. Primers for HPRT1 were purchased from RealTimePrimers.com (Elkins Park, PA). Primers for amplification of Tau isoforms were designed using the Primer3 software (Whitehead Institute for Biomedical Research, Cambridge, MA), summarized below:
0N forward: 5’-GCTGGCCTGAAAGCTGAAG-3’
0N reverse: 5’-ATCGCTTCCAGTCCCGTCT-3’
1N forward: 5’-CAACAGCGGAAGCTGAAGAA-3’
1N reverse: 5’-GTGACCAGCAGCTTCGTCTT-3’
2N forward: 5’-ACTCCAACAGCGGAAGATGT-3’
2N reverse: 5’-GTGACCAGCAGCTTCGTCTT-3’
3R forward: 5’-AGGCGGGAAGGTGCAAATA-3’
3R reverse: 5’-GCCACCTCCTGGTTTATGATG-3’
4R forward: 5’-CGGGAAGGTGCAGATAATTAA-3’
4R reverse: 5’-TATTTGCACACTGCCGCCT-3’
End-point PCR
Primers for amplification of Tau exons were designed using the Primer3 software. Primers for the TFRC housekeeping gene were purchased from RealTimePrimers.com. All PCR reactions were performed in a final volume of 10 µL, consisting of 1 µL 10× PCR buffer, 1 µL of RediLoad Dye (Invitrogen), 2.23 µL of each Tau isoform specific primer, 1µL of endogenous control TFRC primer mix, 0.2 µL of dNTPs, 0.3 µL of MgCl2, 2 µL of 1:5 diluted cDNA, and 0.04 µL Platinum Taq. The amplification program consisted of 1 cycle at 95 °C for 2 min, followed by 40 cycles of 95 °C for 30 sec, annealing temperature at 60 °C for 30 sec and extension temperature at 72 °C for 15 sec. The PCR reactions were analyzed by gel electrophoresis and the products were visualized using the SYBR Safe DNA gel stain (Invitrogen). The primer sequences and target regions are summarized below:
0N + exon 6 forward: 5’-GCTGGCCTGAAAGCTGAAG-3’
0N + exon 6 reverse: 5’-GTTCTCAGTGGAGCCGATCT-3’
1N + exon 6 forward: 5’-CAACAGCGGAAGCTGAAGAA-3’
1N + exon 6 reverse: 5’-GGTGCTTCAGGTTCTCAGTGG-3’
2N + exon 6 forward: 5’-ACTCCAACAGCGGAAGATGT-3’
2N + exon 6 reverse: 5’-TTCTCAGTGGAGCCGATCTT-3’
Exons 6 and 8 forward: 5’-CGCATGGTCAGTAAAAGCAA-3’
Exons 6 and 8 reverse: 5’-GTTCTCAGTGGAGCCGATCT-3’
Western blotting
Total protein lysates were isolated from growing cells using 1× RIPA buffer (1% v/v NP-40, 0.5% w/v sodium deoxychlolate, 0.1% w/v SDS in 1× PBS buffer) with freshly added protease inhibitors (PMSF and aprotinin). 10–25 µg of total protein was separated by 4–12% Nu-PAGE gels and transferred onto nitrocellulose membranes using the iBlot transfer system (all Invitrogen). Membranes were blocked overnight at 4 °C in 1× TBS containing 5% (w/v) non-fat milk and 1% (w/v) bovine serum albumin, then incubated for 2 hr at room temperature with the following antibodies: Tau T-1308-1 (rPeptide, Bogart, GA); P-gp (Signet Laboratories, Dedham, MA); MAP4 (BD Transduction Laboratories, Lexington, KY); stathmin-1, class IV tubulin (Abcam, Cambridge, MA); class II and class III tubulin (Covance, Berkeley, CA); class I tubulin, pan alpha- and beta-tubulin (Sigma Aldrich, St Louis, MO). Primary antibodies were recognized by appropriate HRP-conjugated secondary antibodies (GE Healthcare Life Sciences, Piscataway, NJ) and visualized using the enhanced chemiluminescence detection system (ECL, GE Healthcare Life Sciences). Tau protein ladder T-1007 was purchased from rPeptide.
Cell proliferation assays
The surviving fraction of cells exposed to taxanes was determined using a modified clonogenic assay [29, 30]. Briefly, 6,500 cells were seeded in 6-well tissue culture plates (BD Falcon, Franklin Lakes, NJ) and allowed to attach overnight. Cells were exposed to increasing concentrations of taxane (either paclitaxel or docetaxel from 0.1 nM to 1 µM) for 24 hr at which time the medium was aspirated and replaced with drug-free complete media. Cells were incubated for 14 days at 37 °C and 5% CO2, the surviving colonies were stained with 0.4% (w/v) sulforhodamine B (SRB) in 1% (v/v) acetic acid, and colonies greater than 50 cells were counted and expressed as a percentage of an untreated control. Alternatively, a short-term SRB colorimetric cell proliferation assay was used to determine cell survival following drug exposure. In this assay, 8,000 cells were seeded in 96-well tissue culture plates (BD Falcon) and allowed to attach overnight. Drugs at relevant concentrations were added and the plates were incubated for 72 hr, or approximately three cell divisions. Total proteins were fixed in 10% (w/v) TCA overnight, stained with SRB for 1 hr and plates washed thoroughly with 1% (v/v) acetic acid. Protein-bound dye was solubilized in a 10 mM Tris base solution, and plates read in a multi-well spectrophotometer at 570 nm (Molecular Devices, Sunnyvale, CA).
Stable transfection of Tau 3R and Tau 4R isoforms
T-47D cells were seeded in 12-well tissue culture plates (BD Falcon) and allowed to attach overnight to achieve 90% confluency at the time of transfection. 1.6 µg of purified plasmid DNA, encoding either vector alone, EGFP-Tau-3R or EGFP-Tau-4R isoforms, was transfected into cells using Lipofectamine 2000 (Invitrogen) per manufacturer’s protocol. Cells were selected 48 hr post-transfection in media containing 500 µg/ml G418 (Invitrogen) which was replaced every two days. Three independent T-47D clones for each condition (empty vector, Tau-3R and Tau-4R) which survived the G418 selection were grown and used for subsequent experiments. The plasmids were a kind gift of Dr. Kenneth Kosik (University of California, Santa Barbara).
shRNA lentivirus mediated knockdown of Tau and MAP4
Tau and MAP4 gene sequences were entered into the MIT/Harvard Broad Institute RNAi Consortium shRNA Library website (http://www.broad.mit.edu) and the following forward and reverse sequences were synthesized (Integrated DNA Technologies, Inc.)
-
Tau-forward
5’-CCGGCCAGCCTAAGATCATGGTTTACTCGAGTAAACCATGATCTTAGGCTGGTTTTTG-3’
-
Tau-reverse
5’-AATTCAAAAACCAGCCTAAGATCATGGTTTACTCGAGTAAACCATGATCTTAGGCTGG-3’
-
MAP4-forward
5’-CCGGGCCTTCCATCTTACCTTCAAACTCGAGTTTGAAGGTAAGATGGAAGGCTTTTTG-3’
-
MAP4-reverse
5’-AATTCAAAAAGCCTTCCATCTTACCTTCAAACTCGAGTTTGAAGGTAAGATGGAAGGC-3’
The oligonucleotides were resuspended per manufacturer’s instructions and annealed to form a hairpin (referred to as shTau1.1 and shMAP4 4.1) under the following conditions: 94 °C for 5 min, 70 °C for 5 min and drop of 0.1 °C per sec to 4°C. The annealed hairpins (designed with AgeI and EcoRI compatible ends) were ligated into an AgeI/EcoRI digested pLKO.1 vector using T4 ligase protocol per manufacturer’s protocol (Invitrogen). The ligation products were transformed into DH5 alpha competent cells, plasmids were isolated from several single colonies and sequenced. Vectors with confirmed Tau1.1 and MAP4 4.1 hairpin sequence, and pLKO.1 vector containing an shRNA targeting the firefly luciferase gene as a negative control were used for subsequent lentivirus production. Recombinant lentivirus was produced by co-transfecting 293 cells with the pLKO.1/shRNA-luciferase, pLKO.1/shRNA-Tau1.1, or pLKO.1/shRNA-MAP4 4.1 plasmid, along with pMDZG and D8.2 packaging plasmids using a calcium chloride transfection method. Media containing infectious lentiviruses was collected and filtered at 48 hr and 72 hr after transfection. 1,000 – 1,500 MCF-7 or OVCAR-3 cells were plated in 10 cm2 tissue culture dishes (BD Falcon, Franklin Lakes, NJ) and allowed to attach overnight. The media was removed and replaced with 10 mL of lentivirus-containing media plus 10 µL of Polybrene to allow infection of the cells. 48 hr post-infection, the medium was changed to puromycin selection medium (5 µg/mL). The infected cells were maintained in media containing puromycin thereafter.
Transient Tau silencing by siRNA in MCF-7 and ZR-75-1
Five independent Tau-specific siRNAs were designed and synthesized (Qiagen) targeting the following sequences:
MAPT_1: 5’-cgggaaggtgcagataattaa-3’
MAPT_4: 5’-ttaggcaacatccatcataaa-3’
MAPT_5: 5’-ccgccaggagttcgaagtgat-3’
MAPT_7: 5’-ccgcgagaacgccaaagccaa-3’
MAPT_9: 5’-cgggactggaagcgatgacaa-3’
Two siRNAs (5’-aatcacacccaacgtgcagaa-3’, and 5’-aactggcagttctggagcaaa-3’) were previously reported by Rouzier et al [10] and synthesized by Qiagen. In addition, Dharmacon’s siDesign Center (Thermo Scientific, Lafayette, CO) was used to identify one additional siRNA targeting 5’-gggcggagatcg tgtacaa-3’ of the ORF (region 321 to 1379). Qiagen’s AllStars negative siRNA and Dharmacon’s non-targeting siRNA were used as controls for silencing effects. Lipid-mediated siRNA delivery into cells was accomplished using Invitrogen’s RNAiMax transfection reagent. Optimal transfection conditions were determined for MCF-7 and ZR-75-1 cells which included (1) the cell density on the day of transfection, (2) the concentration of siRNA used, (3) the amount of RNAiMax reagent used per condition, and (4) the time course of gene silencing up to 96 hr after the introduction of siRNA.
Results
Development of taxane-resistant breast cancer variants
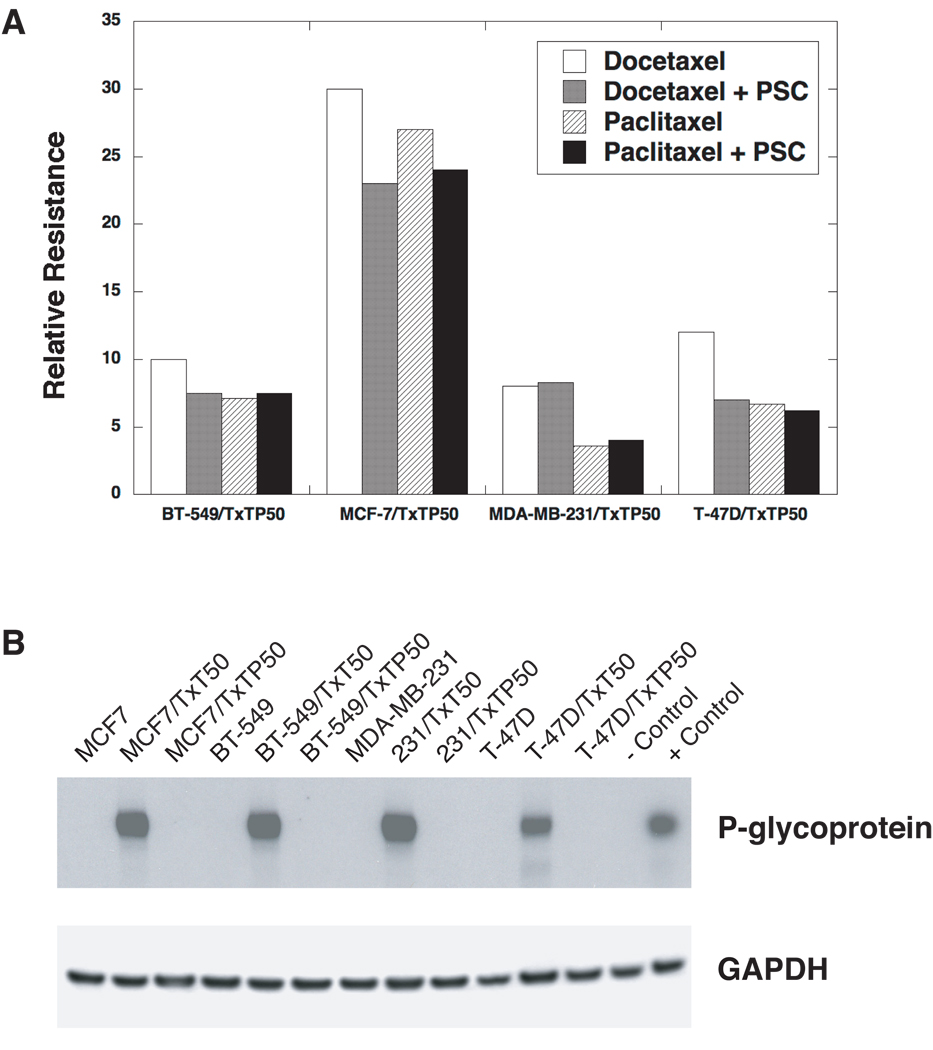
Four breast cancer cell lines (MCF-7, BT-549, MDA-MB-231, and T-47D) were chosen for the selection of taxane resistant variants, based on their constitutive taxane sensitivity and lack of P-gp expression. Stable taxane resistant variants were selected by stepwise exposure to increasing doses of docetaxel (T×T) over several months, with and without the MDR1 inhibitor PSC at 2 µM. The variants are designated according to the name of the parental cell line, the taxane used for selection (T×T for docetaxel), the initial P if the cell line was co-selected with PSC, and the concentration of taxane (e.g., MCF-7/T×TP50 is the MCF-7 variant grown in 50 nM docetaxel plus PSC). The relative taxane resistance of the 4 variants selected with PSC was calculated by dividing the IC50 of the variant by the IC50 of the parental cell line (Figure 2A). The levels of resistance to taxanes ranged from 8 to 30-fold in the PSC co-selected variants compared to parental cells. The four variants co-selected with docetaxel and PSC do not express P-gp protein as assessed by Western blotting (Figure 2B), are negative for MDR1 transcripts by real time PCR, and are not sensitized to taxanes by PSC (data not shown).
Figure 2. Development of taxane resistant breast cancer variants.
(A) The relative resistance of four breast cancer variants co-selected by stepwise exposure to docetaxel (T×T), with and without the P-gp inhibitor PSC-833 (PSC). The highest T×T concentration used in the step-wise selections was 50 nM. Hence, the BT-549 variant selected with T×T at 50 nM drug concentration and PSC-833 at 2 µM is designated “BT-549/T×TP50”. The resistance levels are expressed as the ratios of the IC50’s of the variants to the parental cell lines respectively. These IC50 values were determined by a 72 hour SRB assay. (B) P-glycoprotein expression in parental and taxane resistant variants. The cells were selected with T×T, with and without PSC (designated as “P”). Protein extracts were prepared from the parental and taxane resistant cell lines, resolved by PAGE, and transferred to nitrocellulose membranes. The membranes were probed with an antibody against P-gp (C219) and an anti-GAPDH antibody was used as a loading control. All of the docetaxel alone selections resulted in the activation of the MDR1 gene and these variants were positive for P-gp expression. All of the variants co-selected with PSC were P-gp negative and drug accumulation assays determined that the taxane resistance observed was non-transporter mediated (data not shown). The doxorubicin-selected human uterine sarcoma MES-SA/D×5 variant was used as a positive control for P-gp expression.
All four resistant variants selected with docetaxel alone expressed high levels of P-gp (Figure 2B), and the taxane resistance in these variants was completely reversed by PSC. Substantially higher levels of resistance to taxanes were noted in the P-gp expressing variants selected with a docetaxel alone, ranging from 110 to 1,000-fold (data not shown).
[3H]-docetaxel intracellular accumulation was similar in the resistant variants and parental controls, indicating that the resistance observed in the variants is not related to a taxane transporter (data not shown)
Expression of Tau protein in taxane resistant variants
To evaluate Tau protein expression in the four parental lines and eight taxane resistant variants, whole cell lysates were analyzed by Western blotting (Figure 3A). We used an antibody that recognizes all Tau isoforms, and confirmed the identity of the isoforms (Figure 3A, lane 13) by using a commercially available Tau protein ladder composed of the 6 known Tau isoforms described in Figure 1. The results show that MCF-7 cells express Tau isoforms at similar levels in both the parental and taxane resistant variants, and BT-549 cells do not express Tau in either parental or taxane resistant variants. T-47D and MDA-MB-231 cells show elevated levels of all Tau isoforms in the taxane resistant variants compared to parental cell lines. The overexpression of Tau in MDA-MB-231/T×TP50 and T-47D/T×TP50 variants suggests that Tau levels may be associated with taxane resistance in breast cancer.
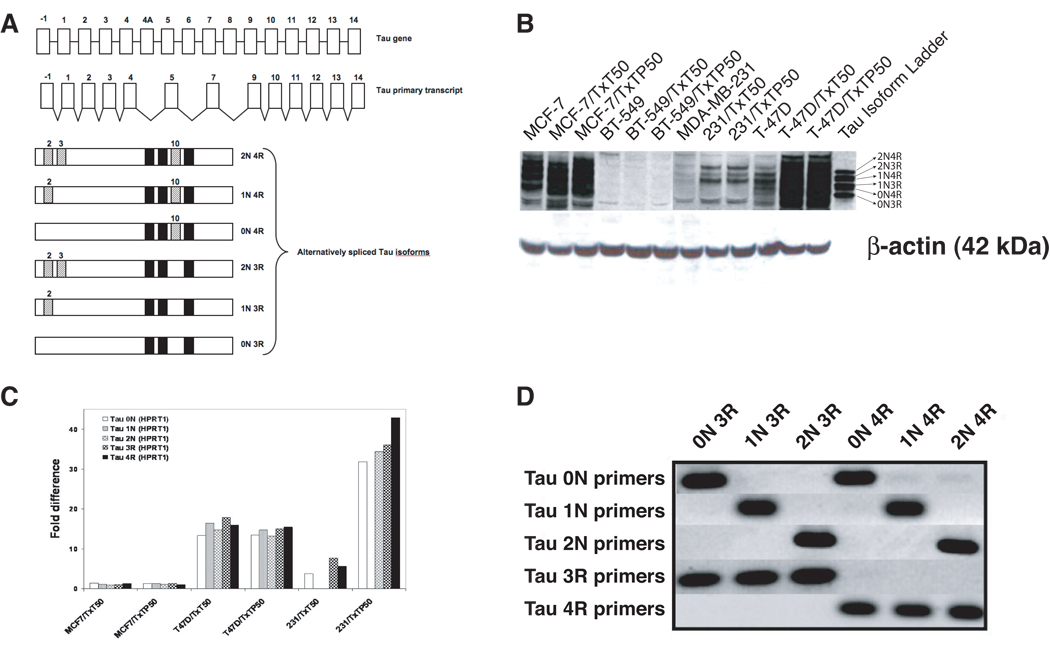
Figure 3. Schematic diagram of the Tau gene, primary transcript and alternatively spliced isoforms, and expression of Tau isoforms in taxane resistant breast cancer variants.
(A) The human Tau gene encodes 16 exons but the human brain primary transcript lacks exons 4A, 6 and 8. Exons 2, 3 and 10 are alternatively spliced, giving rise to six major isoforms. The inclusion or exclusion of exon 10 determines whether the isoform will be a 4R-Tau (with exon 10) or a 3R-Tau (without exon 10). The inclusion or exclusion of exons 2 and 3 determines whether the isoform will be 0N-Tau (without exons 2 and 3), 1N-Tau (with exon 2 but without exon 3), or 2N-Tau (with both exons 2 and 3). Expression of Tau was assessed in the panel of breast cancer variants (B) at the protein level using Western blot analysis with an antibody recognizing all Tau isoforms; and (C) at the mRNA level using quantitative real time PCR analysis with Tau isoform-specific primers. The ratio of Tau isoforms was calculated between taxane resistant variants and parental cells after normalization to HPRT1 internal control. The presented data is representative of at least three independent experiments. (D) Specificity of primer oligonucleotides was confirmed in a PCR reaction using six plasmids, each encoding a specific Tau isoform.
Expression of Tau mRNA isoforms in taxane resistant variants
Although several studies have identified Tau as a potential marker of taxane response, expression of specific Tau isoforms in breast cancer has not yet been investigated. To evaluate the levels of each Tau isoform at the mRNA level using quantitative real time PCR assay, we designed primers that would selectively detect the following Tau isoforms: 0N Tau (no exons 2 and 3), 1N Tau (exon 2 but no exon 3), 2N Tau (both exons 2 and 3), 3R Tau (no exon 10) and 4R Tau (exon 10). In order to validate the isoform specificity of the primers, we used plasmids encoding each of the Tau isoforms as templates. Figure 3C shows that each of the primer pairs amplified only the targeted isoforms without significant cross-reactivity. Using the validated primers, we then measured the levels of each Tau isoform in the parental and taxane resistant variants by quantitative real time PCR. The standard curves for each Tau isoform were generated using serially diluted plasmids encoding a specific isoform. The amount of each isoform expression was calculated from the standard curve and normalized to the expression of HPRT1. The normalized quantity of each isoform in taxane resistant variants was then divided by the quantity measured in parental cells to calculate fold difference. A value of 1 indicates no difference in mRNA expression, a value greater than 1 indicates an increase in expression in taxane-resistant variants, and no value indicates that expression was not detectable in either the parental cells or the variants. Real time PCR assays were conducted in duplicate or triplicate for each sample on different cDNA preparations. Figure 3B shows that the expression levels of all Tau isoforms in T-47D and MDA-MB-231 taxane resistant variants are much higher compared to parental cells, consistent with Western blot results in Figure 2B.
Expression of Tau exons 4A, 6 and 8 in taxane resistant variants
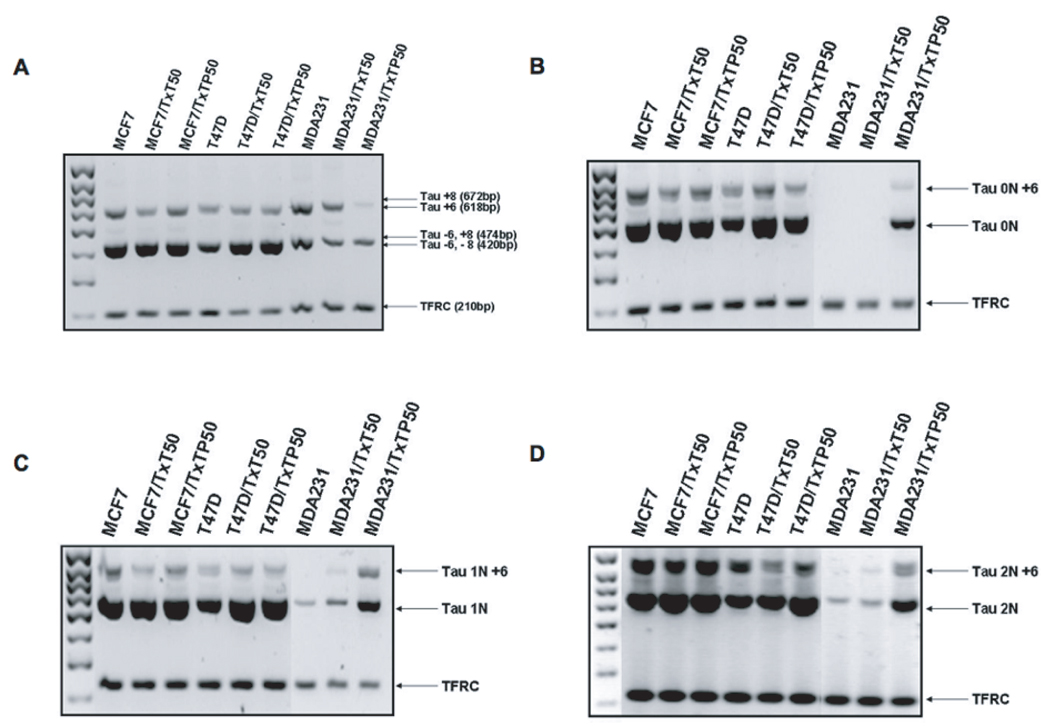
The Tau primary transcript contains 16 exons, with exons 4A, 6 and 8 absent in the human brain mRNA [31]. Exons 4A and 6 have been found in peripheral tissues whereas exon 8 has never been described in human tissues [14, 31]. To test whether our breast cancer variants express exons 4A, 6 and 8, we performed a PCR reaction with a set of primers designed to span exons 4 and 5, exons 5 and 9, and exons 7 and 9. As a control, we co-amplified the TFRC housekeeping gene in each of the reactions. We showed that exon 4A and exon 8 are not expressed in breast cancer cells (data not shown), however, PCR amplification spanning exons 5 and 9 (Figure 4A) yielded products of 420 bp (Tau transcripts lacking exon 6 and 8) and 618 bp (Tau transcripts containing exon 6 but lacking exon 8). We further evaluated whether exon 6 is expressed in 0N, 1N and 2N Tau isoforms by designing forward primers specific to 0N, 1N and 2N and reverse primers specific to exon 7. Figure 4B-D shows that exon 6 is indeed expressed in all three isoforms.
Figure 4. Expression of Tau exons 4A, 6 and 8.
Total RNA was extracted from each cell line, reverse transcribed to cDNA and amplified by polymerase chain reaction to test for the presence of (A) exon 6 and 8 using primers spanning exons 5 and 9; (B) exon 6 in Tau 0N isoforms; (C) exon 6 in Tau 1N isoforms; (D) exon 6 in Tau 2N isoforms. Each PCR reaction co-amplified housekeeping gene TFRC as an internal and loading control. The arrows point to all possible products given the particular primer pair.
Taxane sensitivity of cells overexpressing Tau
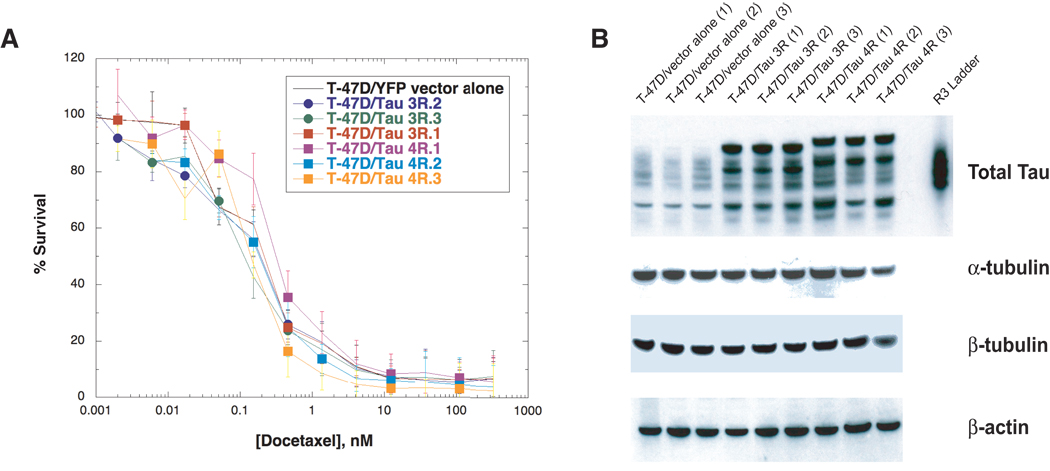
To determine whether Tau overexpression modulates response to taxanes, we derived three independent T-47D clones stably overexpressing Tau 3R isoforms and three clones overexpressing Tau 4R isoforms. Figure 5A shows that Tau overexpression did not result in altered sensitivity to docetaxel compared with the T-47D cells transfected with empty vector alone. Additional experiments confirmed this finding in cells exposed to another taxane, paclitaxel, under identical conditions (data not shown). Furthermore, Tau overexpression did not alter the sensitivity to tubulin depolymerizing agents such as the vinca alkaloids, vinblastine and vincristine, and colchicine (data not shown). Tau overexpression was demonstrated in the transfected cells by Western blotting (Figure 5B). We performed unpaired t-testing of the vector alone clones and the 3R and 4R expression clones in figure 5. There was no significant difference between vector and 4R clones (P = 0.2), and there was increased rather than decreased taxane sensitivity in the 3R clones (P = 0.01).
Figure 5. Survival assay of T-47D cells overexpressing Tau after docetaxel exposure.
(A) SRB assays were used to determine cell survival of T-47D cells stably transfected with Tau-3R (three clones), Tau-4R (three clones) and vector alone (three clones) after drug exposure as described in Materials and Methods. Briefly, 8,000 cells were seeded in 96-well tissue culture plates, allowed to attach overnight followed by addition of drug at increasing concentrations for 72 hr. Total proteins were fixed and stained with SRB, plates were washed thoroughly and read in a multi-well spectrophotometer at 570 nm. (B) Each clone was assessed for protein expression of Tau, β-tubulin and α-tubulin by Western blot analysis.
We hypothesized that perhaps the absence of observed taxane resistance may be due to upregulation of tubulin which would allow the formation of more microtubules, and thus make more binding sites available for the drug. However, the levels of tubulin proteins were not changed between the parental and stably transfected cells (data not shown).
Taxane sensitivity of cells depleted in Tau
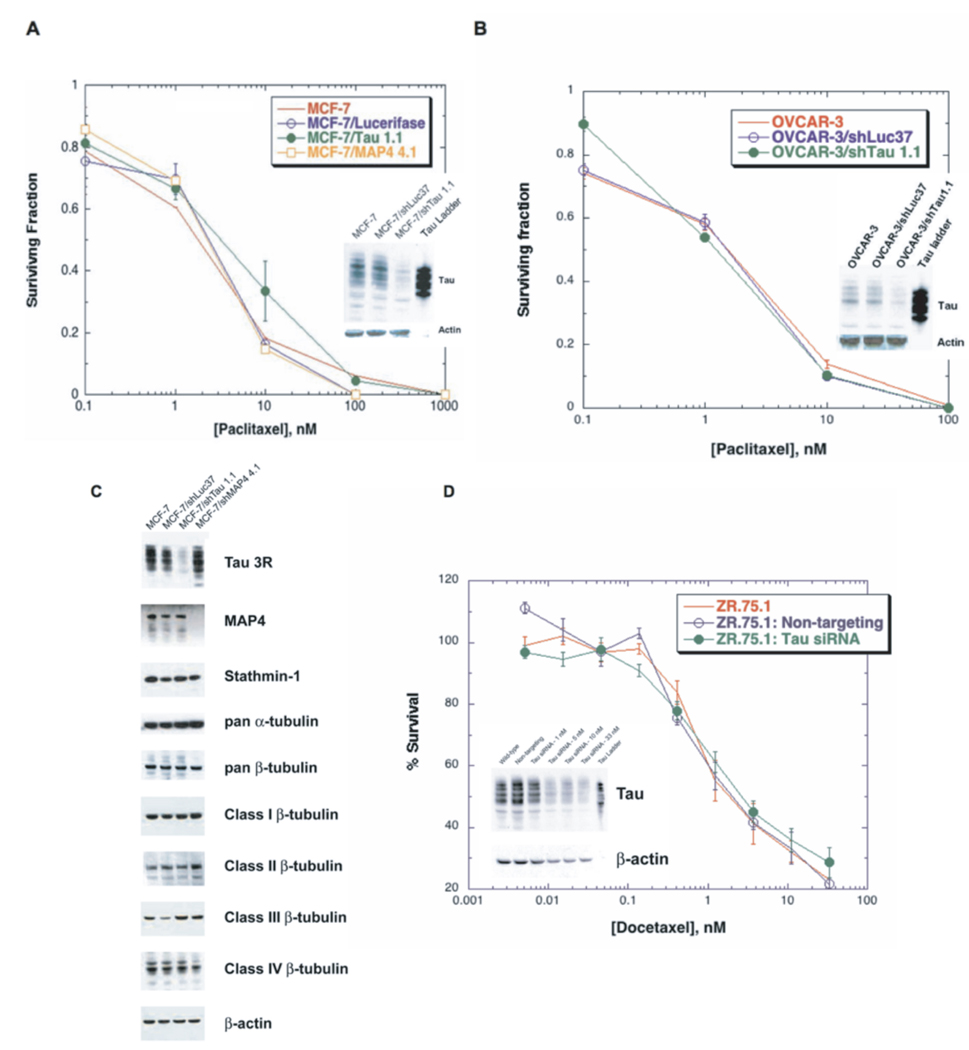
To further investigate whether Tau plays a functional role in modulating response to taxanes, we knocked down Tau expression in MCF-7 parental breast cancer cells and assessed survival after exposure to taxanes. We chose MCF-7 due to the high Tau expression in parental cells, and applied the lentivirus-mediated shRNA approach for Tau silencing. The Tau targeting shRNA (shTau1.1) or control luciferase shRNA (shLuc37) were cloned into a pLKO.1 vector, lentivirus-containing media preparation, infection and selection process were performed as described in Materials and Methods. Following selection, the infected MCF-7 cells were exposed to escalating doses of paclitaxel for 24 hr and cellular survival was assessed using clonogenic assays. Colonies greater than 50 cells were counted and expressed as a percentage of an untreated control. Figure 6A shows that Tau knockdown did not result in increased sensitivity to paclitaxel compared to the luciferase shRNA-infected cells and uninfected parental cells. To confirm that Tau was indeed silenced in the infected cells, its expression was assessed by a Western blot (Figure 6A inset). SRB cytotoxicity assays following 72 hr drug incubations confirmed these clonogenic assay results.
Figure 6. Sensitivity assays in Tau-silenced cells following taxane exposure.
Clonogenic survival assays after exposure to paclitaxel were done in (A) MCF-7 cells infected with Tau, luciferase or MAP4 shRNA, and (B) OVCAR-3 cells infected with Tau or luciferase shRNA. Cells were seeded in 6-well tissue culture plates, allowed to attach overnight, exposed to various concentrations of paclitaxel for 24 hr in the presence of 2 µM PSC, and incubated in drug free medium for 14 days. The surviving cells were stained with sulforhodamine B (SRB) and colonies greater than 50 cells were counted and expressed as a percentage of an untreated control. Tau knockdown was confirmed by Western blot analysis (inset in panels A and B). Results were confirmed using an SRB colorimetric assay following a 72 hr taxane incubation. (C) Protein extracts from cells knocked down in Tau and MAP4 were probed for microtubule associated proteins and tubulin isoforms. Actin was used as a loading control. (D) Treatment with a Tau-specific siRNA from Dharmacon resulted in >90% silencing in ZR-75-1 breast cancer cells. Docetaxel sensitivity was not altered in response in Tau knockdown (33 nM siRNA) relative to wild-type and non-targeting controls as determined by an SRB assay. Similar results were obtained in the MCF-7 cell line under identical conditions (data not shown).
Tau belongs to the same family of proteins as MAP4; therefore, we sought to determine whether silencing of MAP4 would result in altered taxane sensitivity. Lentivirus mediated down regulation of MAP4 in MCF-7 cells also did not alter taxane sensitivity (Figure 6A). We hypothesized that perhaps down regulation of Tau is compensated by changes in expression of other microtubule associated proteins or tubulin proteins. We examined the expression of MAP4, stathmin-1, and tubulin isoforms by Western blot and did not show any significant changes in expression (Figure 6C). To validate the finding that silencing of Tau does not sensitize cells to taxanes, we also silenced Tau in the OVCAR-3 ovarian cancer cell line (Figure 6B). The results again showed that down regulation of Tau did not result in increased sensitivity to taxanes compared to luciferase shRNA-infected cells and parental cells.
In order to further test the effects of silencing Tau on taxane sensitivity, we also used a transient siRNA approach in MCF-7 and ZR-75-1 cells. We achieved approximately 90% Tau silencing in MCF-7 cells relative to untreated and non-targeting controls using both a Fast-Forward protocol (introducing the siRNA-lipid complexes immediately after plating cells) and a traditional approach where complexes are added once cells have attached. This effect was maintained for 96 hr as determined by immunoblotting, and total levels of α- and β-tubulin levels were not altered in Tau-silenced cells (data not shown). The Dharmacon-designed Tau-specific siRNA was very effective in the ZR-75-1 cell line, resulting in >90% silencing (Figure 6D). Transient silencing did not alter taxane sensitivity in either cell line. Sensitivity to docetaxel as determined by SRB assays was not altered in ZR-75-1 cells with silenced Tau, compared to untransfected and non-targeting controls, Figure 6D. This lack of effect was substantiated using clonogenic assays following exposure to either paclitaxel or docetaxel (data not shown).
Discussion
Based on the role of Tau in microtubule stabilization, it has been hypothesized that Tau competes with taxanes for microtubule binding sites. In this model, elevated Tau expression leads to decreased taxane binding to microtubules and in turn, decreased taxane efficacy and suboptimal clinical response to taxane-based chemotherapy regimens. Previously published studies have reported that Tau is one of the genes that discriminate between breast cancer cases with pathologic complete response (pCR) and those with residual disease following paclitaxel-based chemotherapy [10]. However, while Tau-mediated modulation of taxane response was an intriguing hypothesis, it remained unclear whether Tau overexpression represented a true mechanism of taxane resistance or was a marker of another biological phenomenon. In addition, the role of particular Tau isoforms was poorly understood. In this study, we sought to address these questions by studying the functional role of Tau in modulating taxane response by both gene silencing and overexpression, and characterizing the expression of the various Tau isoforms in a panel of taxane resistant breast cancer cells.
Tau is encoded by a single-copy gene which produces three transcripts of 2, 6 and 9 kb. These transcripts are differentially expressed and localized depending on the cell type and stage of maturation, and produce multiple alternatively spliced isoforms. Taking into account all of the splice sites, Tau can hypothetically produce at least 30 variants by splicing alone, excluding further post-translational modifications of each isoform [19]. The probe sets used in gene expression studies that originally identified Tau as a potential modulator of taxane response targeted Tau domains shared by all isoforms. We sought to characterize the expression of each of the six known major Tau splice variants in our taxane resistant breast cancer variants (Figure 1). Our results demonstrate that T-47D and MDA-MB-231 taxane resistant variants express higher levels of all Tau isoforms compared to parental cells both at the protein level (Figure 2A) as well as the mRNA level (Figure 2B).
To further characterize the expression of Tau isoforms, we sought to determine whether the breast cancer variants express exons 4A, 6 and 8, which are typically absent in the human brain. Interestingly, we showed that exon 6, but not exons 4A and 8, is expressed in 0N, 1N and 2N Tau isoforms in breast cancer cells (Figure 4). Exon 6 is an alternatively spliced cassette whose expression profile differs from that of other regulated exons, implying the existence of distinct regulatory factors. Inclusion of the proline rich exon 6 results in a Tau protein containing a more rigid and extended hinge region, which might alter microtubule spacing [32]. Exon 6 may be involved in modulating the dynamicity or extent of the microtubule network. However, it is not clear whether its inclusion in the breast cancer variants has a functional significance. The lack of expression of exon 8 (data not shown) is consistent with the finding that no human tissue has yet been demonstrated to express exon 8. Exon 4A is the longest exon and is exclusively present in the 9kb Tau transcript. We have shown that exon 4A is expressed in skeletal muscle and heart but not other types of tissues, including our breast cancer variants (data not shown).
Because Tau overexpression was found in four of our breast cancer taxane resistant variants, we sought to ascertain the functional role of Tau via knockdown and overexpression experiments. It should be noted that two of these variants also expressed P-gp, and that taxane resistance in the P-gp positive variants was completely reversed by PSC. Since the MCF-7 parental cell line expressed relatively high baseline levels of Tau, we used it for the stable shRNA mediated Tau silencing experiments. Our results demonstrated that the depletion of Tau did not alter cellular response to taxanes (Figure 6A). We also knocked down Tau in the human ovarian carcinoma OVCAR-3 cell line, which expresses moderate baseline levels of Tau. We again showed that although Tau was successfully depleted (Figure 6B), the sensitivity to taxanes was not affected. To confirm our findings, we used eight independent siRNAs including two siRNAs previously published by Rouzier et al. [10]. Although we observed >90% Tau silencing, taxane sensitivity was not altered relative to wild-type and non-targeting controls in both the MCF-7 and ZR-75-1 breast cancer cells (Figure 6D). This is in contrast to other reports, that down regulation of Tau by transient siRNA transfection conferred sensitivity to taxanes in these two cell lines [10, 12].
There are important differences between our set of experiments and those reported by Rouzier et al. [10] and Wagner et al. [12]. First, we used both an shRNA approach via lentiviral delivery, which allowed us to select cells with stable Tau knockdown, as well as a transient siRNA transfection method. Second, our drug exposure was for 72 hr for the SRB cell proliferation assays rather than 48 hr, allowing cells to undergo at least 3 cell divisions in the presence of drug. Third, we confirmed our results with a clonogenic assay to assess cell survival after drug exposure rather than the less reliable tetrazolium based assay or CellTiter-Glo luminescent cell viability assays used in the prior studies.
Tau belongs to the same family of proteins as MAP4; therefore, we hypothesized that perhaps down regulation of Tau is compensated by changes in expression of other microtubule associated proteins or tubulin proteins. We examined the expression of MAP4, stathmin-1, and tubulin isoforms by Western blot and did not show any significant changes in expression (Figure 6C). We also measured the kinetics of taxane-driven tubulin polymerization in MCF-7 cells knocked down in Tau, demonstrating no difference in comparison to parental or luciferase shRNA controls (data not shown). Altogether, these results show that depletion of Tau does not confer taxane sensitivity, and suggest that Tau overexpression in taxane resistant breast cancer clinical specimens may be an epiphenomenon associated with another taxane resistance mechanism(s).
To further evaluate a possible causal relationship between Tau overexpression and taxane resistance, we developed three T-47D clones that stably express Tau-3R, three clones stably expressing Tau-4R, and three clones stably expressing vector alone. We hypothesized that if Tau indeed competes with taxanes for binding to microtubules, then cell lines overexpressing Tau should show resistance to taxanes. Our results show that the overexpression of Tau did not result in resistance to taxanes (Figure 6A). In fact, some of the clones overexpressing Tau showed slightly more sensitivity to taxanes compared to vector alone.
Tau expression is reported to be regulated by estrogen. The Tau gene contains an imperfect ER response element upstream of its promoter, and Tau expression has been induced in an estrogen-dependent fashion [33, 34]. Tau levels were substantially higher in our T-47D/T×TP50 and MDA-31/T×TP50 variants, as measured by quantitative real time PCR, while estrogen receptor levels did not significantly change from parental cells (data not shown). It was previously suggested that high levels of Tau may be a potential marker of sensitivity to anti-estrogen therapy whereas low levels of Tau may be a potential marker of sensitivity to paclitaxel therapy [35]. Our findings indicate that upregulation of Tau in taxane resistant variants is not dependent upon upregulation of estrogen receptor.
A recently published, extensive analysis of Tau expression in the NSABP-B28 study confirmed that high Tau expression was associated with estrogen receptor (ER) expression in breast cancers. However, there was no association of Tau expression and clinical benefit from paclitaxel chemotherapy in either ER positive or negative patients (36). Thus, Tau expression does not appear to be a useful biomarker for clinical sensitivity to taxanes in breast cancers.
In summary, in this study we show that modulation of Tau expression (knockdown and overexpression) does not result in altered cellular responses to taxanes in several breast cancer cell lines and one ovarian cancer cell line. These findings indicate that Tau expression is not a mechanism of resistance to taxanes.
Acknowledgments
We thank Dr. Stuart Feinstein for plasmids encoding each of the six major Tau isoforms, Dr. Kenneth Kosik for the EGFP-Tau-3R and EGFP-Tau-4R plasmids, and Sanofi-Aventis and Novartis Pharmaceuticals for the drugs docetaxel and PSC.
Grant Support: Supported by NIH Grant R01 CA 114037 from the U. S. Public Health Service (B. I. Sikic), a grant from Sanofi-Aventis (B. I. Sikic), Postdoctoral Fellowship Award #13FB-0144 from the California Breast Cancer Research Program (T. Spicakova) and an American Society of Clinical Oncology Young Investigator Award (M. M. O’Brien).
Abbreviations used
- MAP
microtubule-associated protein
- P-gp
P-glycoprotein
- PSC
the P-gp inhibitor PSC-833
- SRB
sulforhodamine B
References
- 1.Ring AE, Ellis PA. Taxanes in the treatment of early breast cancer. Cancer Treat Rev. 2005;31:618–627. doi: 10.1016/j.ctrv.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Bria E, Nistico C, Cuppone F, et al. Benefit of taxanes as adjuvant chemotherapy for early breast cancer: pooled analysis of 15,500 patients. Cancer. 2006;106:2337–2344. doi: 10.1002/cncr.21886. [DOI] [PubMed] [Google Scholar]
- 3.Henderson IC, Berry DA, Demetri GD, et al. Improved outcomes from adding sequential Paclitaxel but not from escalating Doxorubicin dose in an adjuvant chemotherapy regimen for patients with node-positive primary breast cancer. J Clin Oncol. 2003;21:976–983. doi: 10.1200/JCO.2003.02.063. [DOI] [PubMed] [Google Scholar]
- 4.Ghersi D, Wilcken N, Simes RJ. A systematic review of taxane-containing regimens for metastatic breast cancer. Br J Cancer. 2005;93:293–301. doi: 10.1038/sj.bjc.6602680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orr GA, Verdier-Pinard P, McDaid H, Horwitz SB. Mechanisms of Taxol resistance related to microtubules. Oncogene. 2003;22:7280–7295. doi: 10.1038/sj.onc.1206934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGrogan BT, Gilmartin B, Carney DN, McCann A. Taxanes, microtubules and chemoresistant breast cancer. Biochim Biophys Acta. 2008;1785:96–132. doi: 10.1016/j.bbcan.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Cortes J, Baselga J. Targeting the microtubules in breast cancer beyond taxanes: the epothilones. Oncologist. 2007;12:271–280. doi: 10.1634/theoncologist.12-3-271. [DOI] [PubMed] [Google Scholar]
- 8.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 9.Trock BJ, Leonessa F, Clarke R. Multidrug resistance in breast cancer: a meta-analysis of MDR1/gp170 expression and its possible functional significance. J Natl Cancer Inst. 1997;89:917–931. doi: 10.1093/jnci/89.13.917. [DOI] [PubMed] [Google Scholar]
- 10.Rouzier R, Rajan R, Wagner P, et al. Microtubule-associated protein tau: a marker of paclitaxel sensitivity in breast cancer. Proc Natl Acad Sci USA. 2005;102:8315–8320. doi: 10.1073/pnas.0408974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hess KR, Anderson K, Symmans WF, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 12.Wagner P, Wang B, Clark E, et al. Microtubule Associated Protein (MAP)-Tau: a novel mediator of paclitaxel sensitivity in vitro and in vivo. Cell Cycle. 2005;4:1149–1152. doi: 10.4161/cc.4.9.2038. [DOI] [PubMed] [Google Scholar]
- 13.Mimori K, Sadanaga N, Yoshikawa Y, et al. Reduced tau expression in gastric cancer can identify candidates for successful Paclitaxel treatment. Br J Cancer. 2006;94:1894–1897. doi: 10.1038/sj.bjc.6603182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreadis A. Tau gene alternative splicing: expression patterns, regulation and modulation of function in normal brain and neurodegenerative diseases. Biochim Biophys Acta. 2005;1739:91–103. doi: 10.1016/j.bbadis.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Black MM, Slaughter T, Moshiach S, Obrocka M, Fischer I. Tau is enriched on dynamic microtubules in the distal region of growing axons. J Neurosci. 1996;16:3601–3619. doi: 10.1523/JNEUROSCI.16-11-03601.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiTella M, Feiguin F, Morfini G, Caceres A. Microfilament-associated growth cone component depends upon Tau for its intracellular localization. Cell Motil Cytoskeleton. 1994;29:117–130. doi: 10.1002/cm.970290204. [DOI] [PubMed] [Google Scholar]
- 17.Gorath M, Stahnke T, Mronga T, Goldbaum O, Richter-Landsberg C. Developmental changes of tau protein and mRNA in cultured rat brain oligodendrocytes. Glia. 2001;36:89–101. doi: 10.1002/glia.1098. [DOI] [PubMed] [Google Scholar]
- 18.LoPresti P, Szuchet S, Papasozomenos SC, Zinkowski RP, Binder LI. Functional implications for the microtubule-associated protein tau: localization in oligodendrocytes. Proc Natl Acad Sci USA. 1995;92:10369–10373. doi: 10.1073/pnas.92.22.10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei ML, Andreadis A. Splicing of a regulated exon reveals additional complexity in the axonal microtubule-associated protein tau. J Neurochem. 1998;70:1346–1356. doi: 10.1046/j.1471-4159.1998.70041346.x. [DOI] [PubMed] [Google Scholar]
- 20.Neve RL, Harris P, Kosik KS, Kurnit DM, Donlon TA. Identification of cDNA clones for the human microtubule-associated protein tau and chromosomal localization of the genes for tau and microtubule-associated protein 2. Brain Res. 1986;387:271–280. doi: 10.1016/0169-328x(86)90033-1. [DOI] [PubMed] [Google Scholar]
- 21.Gao QS, Memmott J, Lafyatis R, et al. Complex regulation of tau exon 10, whose missplicing causes frontotemporal dementia. J Neurochem. 2000;74:490–500. doi: 10.1046/j.1471-4159.2000.740490.x. [DOI] [PubMed] [Google Scholar]
- 22.Himmler A. Structure of the bovine tau gene: alternatively spliced transcripts generate a protein family. Mol Cell Biol. 1989;9:1389–1396. doi: 10.1128/mcb.9.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Couchie D, CMavilia C, Georgieff IS, et al. Primary structure of high molecular weight tau present in the peripheral nervous system. Proc Natl Acad Sci USA. 1992;89:4378–4381. doi: 10.1073/pnas.89.10.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goedert M, Spillantini MG, Jakes R, Rutherford D, Crowther RA. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer's disease. Neuron. 1989;3:519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 25.Goedert M, Spillantini MG, Crowther RA. Cloning of a big tau microtubule-associated protein characteristic of the peripheral nervous system. Proc Natl Acad Sci USA. 1992;89:1983–1987. doi: 10.1073/pnas.89.5.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Himmler AD, Drechsel D, Kirschner MW, Martin DW., Jr Tau consists of a set of proteins with repeated C-terminal microtubule-binding domains and variable N-terminal domains. Mol Cell Biol. 1989;9:1381–1388. doi: 10.1128/mcb.9.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosik KS, Orecchio LD, Bakalis S, Neve RL. Developmentally regulated expression of specific tau sequences. Neuron. 1989;2:1389–1397. doi: 10.1016/0896-6273(89)90077-9. [DOI] [PubMed] [Google Scholar]
- 28.Lee G, Cowan N, Kirschner M. The primary structure and heterogeneity of tau protein from mouse brain. Science. 1988;239:285–288. doi: 10.1126/science.3122323. [DOI] [PubMed] [Google Scholar]
- 29.Pauwels B, Korst AE, de Pooter CM, et al. Comparison of the sulforhodamine B assay and the clonogenic assay for in vitro chemoradiation studies. Cancer Chemother Pharmacol. 2003;51:221–226. doi: 10.1007/s00280-002-0557-9. [DOI] [PubMed] [Google Scholar]
- 30.Leonard CE, Chan DC, Chou TC, Kumar R, Bunn PA. Paclitaxel enhances in vitro radiosensitivity of squamous carcinoma cell lines of the head and neck. Cancer Res. 1996;56:5198–5204. [PubMed] [Google Scholar]
- 31.Buee L, Bussiere T, Buee-Scherrer V, Delacourte A, Hof PR. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res Brain Res Rev. 2000;33:95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Tse SW, Andreadis A. Tau exon 6 is regulated by an intricate interplay of trans factors and cis elements, including multiple branch points. J Neurochem. 2007;100:437–445. doi: 10.1111/j.1471-4159.2006.04252.x. [DOI] [PubMed] [Google Scholar]
- 33.Matsuno A, Takekoshi S, Sanno N, et al. Modulation of protein kinases and microtubule-associated proteins and changes in ultrastructure in female rat pituitary cells: effects of estrogen and bromocriptine. J Histochem Cytochem. 1997;45:805–813. doi: 10.1177/002215549704500605. [DOI] [PubMed] [Google Scholar]
- 34.Ferreira A, Caceres A. Estrogen-enhanced neurite growth: evidence for a selective induction of Tau and stable microtubules. J Neurosci. 1991;11:392–400. doi: 10.1523/JNEUROSCI.11-02-00392.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andre F, Hatzis C, Anderson K, et al. Microtubule-associated protein-tau is a bifunctional predictor of endocrine sensitivity and chemotherapy resistance in estrogen receptor-positive breast cancer. Clin Cancer Res. 2007;13:2061–2067. doi: 10.1158/1078-0432.CCR-06-2078. [DOI] [PubMed] [Google Scholar]
- 36.Pusztai L, Jeojg JH, Gong Y, et al. Evaluation of microtubule-associated protein-Tau expression as a prognostic and predictive marker in the NSABP-B 28 randomized clinical trial. J Clin Oncol. 2009;27:4287–4292. doi: 10.1200/JCO.2008.21.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]