SUMMARY
Hundreds of different proteins regulate and implement transcription in Saccharomyces. Yet their inter-relationships have not been investigated on a comprehensive scale. Here we determined the genome-wide binding locations of 200 transcription-related proteins, under normal and acute heat shock conditions. This study distinguishes binding between distal versus proximal promoter regions as well as the 3' ends of genes for nearly all mRNA and tRNA genes. This study reveals 1) a greater diversity and specialization of regulation associated with the SAGA transcription pathway compared to the TFIID pathway, 2) new regulators enriched at tRNA genes, 3) a global co-occupancy network of >20,000 unique regulator combinations that show a high degree of regulatory interconnections among lowly-expressed genes, 4) regulators of the SAGA pathway located largely distal to the core promoter and regulators of the TFIID pathway located proximally, and 5) distinct mobilization of SAGA- versus TFIID-linked regulators during acute heat shock.
Keywords: ChIP-chip, tiling microarrays, transcription initiation, chromatin, Saccharomyces
INTRODUCTION
Eukaryotic transcription is regulated by sequence-specific DNA binding proteins, chromatin regulators, the general transcription machinery, and elongation regulators (Berger, 2000; Li et al., 2007a; Orphanides and Reinberg, 2002; Venters and Pugh, 2009b). Their collective function is to express a subset of genes as dictated by a complex interplay of environmental signals that is only partly understood. Here we report the genome-wide location profiles for 202 representative regulators from diverse stages of transcription under standard conditions and during acute heat stress.
Earlier studies had identified TAF (TFIID)-dependent and TAF-independent genes (Cheng et al., 2002; Kuras et al., 2000; Li et al., 2000). Subsequent genome-wide studies found that ~85-90% of the yeast genome can be generally characterized as being TFIID-dominated (Basehoar et al., 2004; Huisinga and Pugh, 2004), in which such genes are typically TATA-less and utilize TFIID to assemble the transcription pre-initiation complex (PIC). The remaining 10-15% are SAGA-dominated. Such genes typically contain a TATA box and preferentially utilize SAGA rather than TFIID to assemble the transcription machinery. Stress-induced genes tend to reside in the SAGA-dominated class, and are more highly regulated. TFIID-dominated genes tend to be housekeeping genes, many of which are down-regulated in response to stress. Inasmuch as a gene may be involved to varying degrees in both stress and housekeeping responses, differing blends of the two pathways may be used, rather than one or the other exclusively.
Early studies on the genome-wide location of ~100 sequence-specific transcription regulators and their redistribution in response to stress have provided insight into gene regulatory networks (Harbison et al., 2004; Lee et al., 2002). Yet, such regulators represent only part of the transcription control system. We therefore expanded on these earlier studies to include proteins representing nearly all known complexes that regulate chromatin, transcription initiation, and transcription elongation.
We analyzed essentially all mRNA and tRNA genes, interrogated by ChIP-chip with probes placed at distal and proximal regions of the promoter, and near the 3’ ends of genes. In total, >800 ChIP-chip experiments were conducted resulting in ~15 million ChIP measurements. Inasmuch as many proteins involved in transcription elongation may reside in the body of genes, we further examined 20 of these proteins by ChIP-seq, which allow an unbiased evaluation of their genomic binding locations. From this study, a more comprehensive picture of the genome-wide regulatory organization of the genome and its plasticity upon reprogramming was attained. This study may be a useful resource for identifying co-regulatory partners in yeast, and orthologous interactions in other organisms.
RESULTS
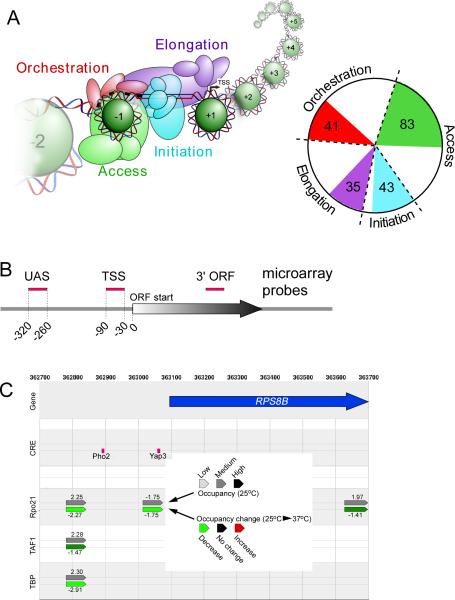
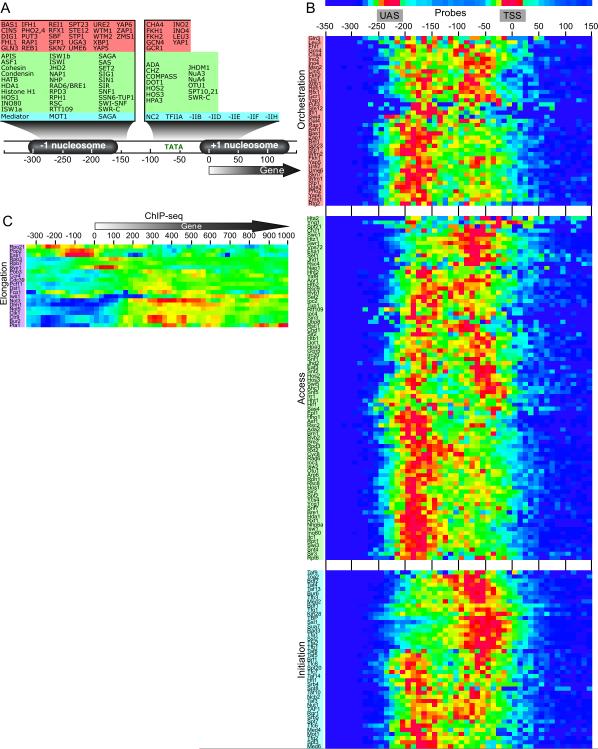
Using Gene Ontology classifications, we identified nearly 400 proteins that potentially regulate transcription. From this list, we categorized proteins according to what protein complex and what stage of the transcription cycle they likely belong. For this purpose, we divided the transcription cycle into four stages (Figure 1A and Table S1): 1) “Orchestration”, representing sequence-specific transcriptional activators and repressors; 2) “Access”, including histones and chromatin regulators; 3) “Initiation”, representing the general transcription factors and their regulators; and 4) “Elongation”, including RNA Polymerase (Pol) II and regulators of transcription elongation and termination.
Figure 1. Scope and data display of genome-wide location analysis.
(A) Illustration of the chromatin context of a stereotype promoter. The pie chart depicts the number of regulators analyzed from each stage, out of the total number of potential regulators in those stages (demarcated by dashed lines). Those not covered largely resided within complexes that contain regulators that were covered.
(B) Approximate positions of microarray 60mer probes relative to a gene (arrow). Negative control probes were present at ~300 intergenic regions between two convergently transcribed genes.
(C) Browser display of the occupancy and change in occupancy upon heat shock at the indicated probe locations (pointed bullets), for a selected number of regulators. The upper and lower probe tracks for each regulator reflect occupancy levels at 25°C (grayscale) and changes in occupancy during heat shock (red/green scale), respectively. Any region for the ~200 regulators can be queried at http://atlas.bx.psu.edu.
Occupancy levels were examined at ~20,000 locations across the yeast genome, primarily at canonical locations where upstream activating sequences (UAS), transcriptional start sites (TSS), and the 3' ends of open read frames (ORFs) reside (Figure 1B). Two probes were also located 30 bp and 290 bp upstream of the annotated tRNA transcription start sites. Several hundred negative control probes were placed between two convergently transcribed genes for the purpose of centering the bulk data. Controls and validation of the accuracy and resolution of these arrays was described elsewhere (Venters and Pugh, 2009a). Importantly, the prior study demonstrated that the probes allow for interrogation of entire promoter regions with mapping uncertainties of ±40 bp. Thus, binding to UAS regions can be distinguished from core promoter regions, and promoter regions can be distinguished from the 3’ ends of genes.
Occupancy levels reflect log2 fold over background (hybridization signal of TAP-tagged regulator / hybridization signal for a set of samples lacking a TAP tag), and are reported for individual probes in Table S2 for cells grown at 25°C, and in Table S3 for heat shocked cells. Each normalized log2 dataset was centered by subtracting the median value in 344 control T-T regions, which represent intergenic regions between convergently transcribed genes. Thus, all values are relative to such regions. The occupancy level of each of the ~200 regulators at any gene can also be queried in a browser format at http://atlas.bx.psu.edu (illustrated in Figure 1C). Since a priori knowledge of where a regulator binds within a promoter region (if at all) was not known for all regulators, promoter occupancy levels were taken to be the higher value at either the UAS or TSS probes (reported in Table S4). Based on an error model using an untagged control strain (Li et al., 2008), occupancy values meeting a 5% FDR cut-off were deemed to be significant. A total of 338,704 protein-DNA interactions were determined to be significant (231,704 at mRNA promoters; 99,216 at the 3' end of ORFs; and 7,783 at tRNA genes) for regulators (15% of all possible interactions).
More diverse regulation of SAGA-dominated genes than TFIID-dominated genes
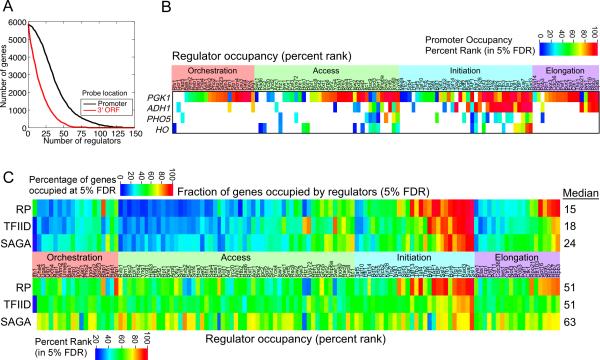
Approximately 93% of the 5,866 mRNA promoter regions had significant occupancy of at least ten regulators (Figure 2A, which includes histones), and ~10% were significantly occupied by at least 75 proteins. As only half of all proteins were examined, we expect the actual number of bound proteins to be approximately twice that. Based upon existing models on transcription initiation it is reasonable to expect at least 75 (or 150) proteins occupying a single promoter, since GTFs, Pol II, Mediator, and TFIID/SAGA are commonly required for transcription, and they alone contribute over half of the proteins. Activators, chromatin regulators, and elongation regulators, many of which are large multisubunit complexes, would make up the remaining. Indeed, a putative gene of unknown function (YGR146C-A) had as many as 145 proteins detected. An important caveat is that these location profiling experiments of cell populations do not distinguish between regulators that simultaneously co-occupy a genomic region from regulators that bind in a mutually exclusive manner or at a different temporal order in the transcription cycle or cell cycle.
Figure 2. Promoter occupancy at 25°C for individual genes and groups of genes.
(A) Number of promoters or 3' ORFs significantly occupied (5% FDR) by the number of regulators indicated along the x-axis.
(B) Regulators significantly bound at the indicated genes are shown as a heatmap. Columns represent regulators. The color scale indicates the occupancy percent rank for those meeting a 5% FDR cutoff. Regulators are grouped by stage. Only those regulators having potentially meaningful occupancy are shown.
(C) Regulator promoter occupancy for genes in the top 10th percentile of transcription frequency (Holstege et al., 1998) are shown, and filtered to be in one of the three following classes: 1) RP genes; 2) TFIID-dominated and TATA-less but not RP genes; 3) SAGA-dominated and TATA-containing (Basehoar et al., 2004; Huisinga and Pugh, 2004). Potentially meaningful occupancy levels are displayed by transcription stage, with the fraction of each gene group occupied also shown. The median values for each class of genes are shown to the right of each row.
Figure 2B displays a heat map representation that quantifies the occupancy levels at selected genes. The number of bound regulators varied widely from gene to gene. For example, two highly transcribed genes from the glycolysis pathway, PGK1 and ADH1 contained 57 and 26 bound complexes (98 and 42 proteins), respectively. Two lowly transcribed genes, HO and PHO5, had fewer regulators bound, and those regulators that were bound had relatively low occupancy levels. Table S5 lists a number of example genes and the complexes present at each.
To assess commonalities among similarly regulated genes, we examined regulator occupancy at 124 highly-active (>14 mRNA/hr) ribosomal proteins (RP) genes, which have highly coordinated regulation and are TFIID-dominated (Figure 2C and Table S6). We also examined 190 of the most active TFIID-dominated/TATA-less genes excluding RP genes, and compared them to the 41 most active SAGA-dominated/TATA-containing genes (a transcription frequency cut-off of 14 mRNA/hr was used for both groups). Similar trends were observed at other data thresholds (not shown). The percentage of those genes meeting the 5% FDR threshold for significant occupancy is shown in a heat map representation. In addition, the median occupancy level of each regulator (as a percent rank of those meeting the 5% FDR threshold) is indicated. Percent ranks, which vary between zero and 100, allowed occupancy to be compared (albeit imperfectly) between regulators. Without this scaling, occupancy levels vary widely between regulators due to differences in ChIP efficiency.
More so than other TFIID-dominated/TATA-less genes, we found a higher percentage of the RP genes occupied by the sequence-specific regulators (Orchestration group) Rap1, Ifh1, and Sfp1 confirming previous reports (Marion et al., 2004; Rudra et al., 2005; Yu et al., 2003). The RP genes were comparatively depleted of the histone variant H2A.Z, along with the SWR1 complex, which is responsible for H2A.Z deposition.
SAGA-dominated/TATA-containing genes were occupied by a larger variety of regulators compared to TFIID-dominated/TATA-less genes (Figure 2C, with values reported in Table S4). The median value of the fraction of SAGA-dominated genes occupied by each regulator was 24%, whereas for TFIID-dominated genes the median value was 18%. Moreover, SAGA-dominated genes tended to have higher occupancy levels of bound regulators (12% higher, as a percent rank, than for TFIID-dominated genes).
Table 1 summarizes whether a regulator preferentially occupied active genes dominated by either the SAGA or TFIID pathway. For example, the NuA4 histone acetyltransferase, and TFIID-specific TAFs preferred the TFIID pathway, whereas other chromatin remodeling and histone modification complexes such as RSC, SWI/SNF, RAD6/BRE1, and RTT109 preferred the SAGA pathway. TAFs shared between SAGA and TFIID (Taf5 and Taf10), and the core transcription machinery, displayed no preference. However, certain TBP regulators such as Mot1 and TFIIA, and certain Pol II regulators such as elongation factors and Mediator also tended towards the SAGA pathway. These findings fit well with the notion that SAGA-dominated genes tend to be highly regulated and are more tailored to responding to different environmental cues than the housekeeping and less-variably regulated TFIID class (Basehoar et al., 2004; Huisinga and Pugh, 2004; Tirosh and Barkai, 2008).
Table 1.
Regulators that are biased towards either the TFIID- or SAGA-pathway
| Orchestration | Access | Initiation | Elongation |
|---|---|---|---|
| | |||
| TFIID pathway | |||
| RAP1 | DOT1 | TFIID | ESS1 |
| NuA4 | |||
| SET2 | |||
| SPT10,21 | |||
| SWR1 | |||
| SAGA pathway | |||
|---|---|---|---|
| DIG1 | Cohesin | Mediator | CCR4-NOT |
| FKH1 | COMPASS | MOT1 | PAF1 |
| FKH2 | Condensin | SAGA | SPT6 |
| GCN4 | HATB | TFIIA | THO |
| GCR1 | Histone H1 | ||
| GLN3 | HOS1 | ||
| LEU3 | HOS2 | ||
| SBF | HOS3 | ||
| SKN7 | ISW1b | ||
| STP1 | JHDM1 | ||
| UGA3 | nucleosome | ||
| URE2 | RAD6/BRE1 | ||
| WTM1 | RPD3 | ||
| WTM2 | RSC | ||
| RTT109 | |||
| SAS | |||
| SIG1 | |||
| SNF1 | |||
| TUP1 | |||
| SWI/SNF |
The fraction of SAGA-dominated and TFIID-dominated genes that were significantly occupied by a regulator was determined (Huisinga and Pugh, 2004), and a ratio between the two calculated. Regulators having ratios >1.5 in one direction or the other were listed under the appropriate pathway.
Potential regulatory cross-talk in Pol II and III transcription
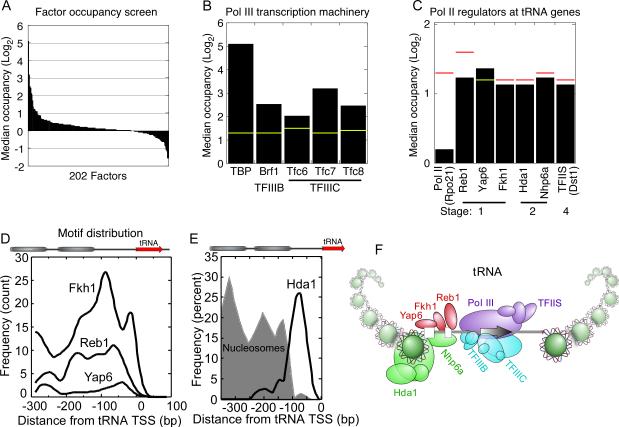
In Saccharomyces, the Pol III transcription machinery transcribes 274 tRNA genes (Dieci et al., 2007). TFIIIC binds within each gene, immediately downstream of the TSS, and then recruits the TBP-containing TFIIIB complex just upstream of the TSS, which then recruits Pol III to the TSS. The core Pol III transcription machinery is largely distinct from that involved in Pol II regulation. Recent reports have found some unexpected regulators at tRNA genes, such as TFIIS and Nhp6a (Braglia et al., 2007; Ghavi-Helm et al., 2008). This led us to consider other regulators that may bind to tRNA genes, and so we screened the 202 datasets for regulators that occupy tRNA genes at 25°C (Figure 3A).
Figure 3. Regulators that occupy tRNA genes.
(A) The median 25°C occupancy for 202 regulators was computed for 274 tRNA genes and displayed as a bar graph. The vast majority of tRNA promoters are >200 bp from a Pol II promoter, and thus would not erroneously pick up signal from nearby genes. Such adjacent genes are also not enriched with these regulators (not shown).
(B, C) Bar graph showing the occupancy level of the Pol III transcription machinery and other high occupancy regulators at tRNA genes.
(D) Frequency distribution plot for the motifs of the three sequence-specific regulator occupying tRNA genes.
(E) Hda1 frequency distribution plot for its interpolated binding location is shown along with nucleosome density (Mavrich et al., 2008) shown in gray fill.
(F) Illustration depicting a composite organization of regulators at tRNA genes. Only the TFIIIB, TFIIIC, and Pol III are like to be present at all genes, with the remaining at a statistically-enriched subset.
In addition to the known Pol III transcription machinery (Figure 3B), a number of Pol II regulators stood out as having significant occupancy at tRNA genes (Figure 3C). The binding sites for these regulators were enriched upstream of tRNA start sites (Figure 3D), and include Fkh1, Reb1, and Yap6. Most of these regulators were also enriched at the RP genes. Their presence at genes that encode protein and RNA structural components of the translation machinery might provide a cross-polymerase mechanism to coordinate translation through the synthesis of ribosomes and tRNAs. Histone deacetylase Hda1 was also enriched at tRNA genes (Figure 3E). Inasmuch as tRNA genes can exert a negative transcriptional effect on adjacent Pol II genes (Hull et al., 1994), deacetylation by Hda1 might generate hypoacetylated regions adjacent to tRNA genes making them refractory to Pol II transcription. Figure 3F illustrates a composite view regarding tRNA transcription regulator assembly for the regulators examined. An important caveat of this composite is that the indicated Pol II regulators may only occupy a small but statistically enriched subset of tRNA genes.
Functionally related regulators tend to work at the same genes
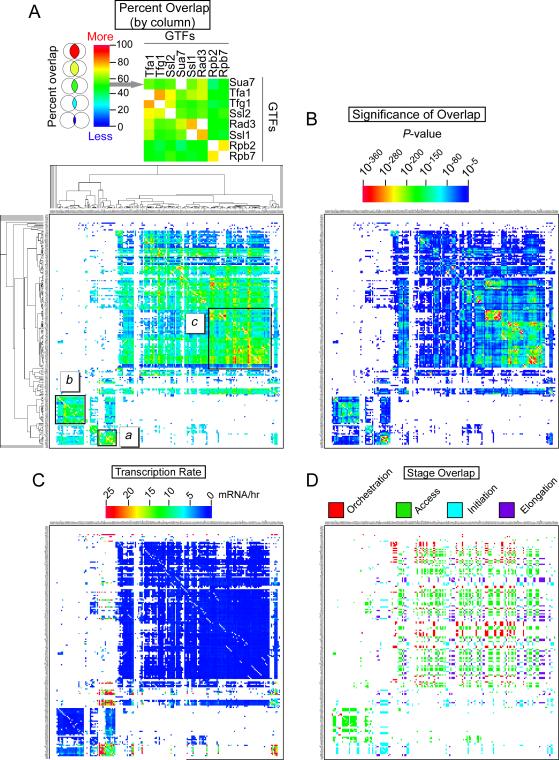
The analysis thus far indicates a preference of some regulatory complexes for distinct PIC assembly pathways. We next address which regulators might be functionally linked to each other by determining their co-occupancy at genes. If the set of regulator-bound genes is defined by a circle, then regulator co-occupancy is defined by the overlap of two circles in a Venn diagram. Figure 4A tabulates the results of >40,000 Venn diagrams (see Figure S1 for a high-resolution version of Figure 4), where the magnitude of percentage overlap is represented as a heatmap color-coded pixel (numerical values are presented in Table S7). Each Venn pair is reported as two reciprocal pixels in a 202 × 202 matrix, reflecting the percentage overlap with respect to the two individual datasets. For example, 26% of all genes bound by Reb1 were also bound by Sua7. However, only 13% of all genes bound by Sua7 were also bound by Reb1. This matrix was then hierarchically clustered to reveal regulators that tend to display similar patterns of co-occupancy. Three clusters were examined further (denoted as a, b, and c in Figure 4A). The significance of each overlap (P-value) is reported in corresponding pixels in Figure 4B. The median transcription frequency for the same sets of genes is reported in Figure 4C. If two co-occupying regulators belonged to the same stage of the transcription cycle as illustrated in Figure 1A, then the corresponding pixel was color-coded as shown in Figure 4D. The purpose for this latter analysis was to assess whether stage-related regulators (represented by clustered pixels) tended to work at the same genes, or whether they were gene-specific.
Figure 4. Regulator co-occupancy matrices.
Shown are promoter co-occupancies and other related properties between 20,301 unique combinations of 202 regulator pairs. (A) Percent overlap for the set of genes co-occupied by each regulator pair is displayed as a heatmap. Each pixel represents the overlap in a Venn diagram between two regulators, heatmap color-coded in accord with the extent of overlap, as indicated. Overlaps that did not meet a significance threshold of P <10-5, or represented a self-by-self comparison were drawn as white pixels. Rows and columns were hierarchically clustered using Cluster (Eisen et al., 1998). See Figure S1 for high resolution images. A blow-up of a portion of cluster a (8 × 8 matrix) is shown in upper portion of the panel.
(B) Statistical significance (P-value determined by the Chi-test) of overlap for the set of genes co-occupied by each regulator pair is displayed as a heatmap. For panels B-D, pixels are in the same position as in A, and were colored white if they were white in panel A.
(C) The corresponding median transcription frequency for the set of genes co-occupied by each regulator pair is displayed as a heat map. Data for mRNA/hr is from (Holstege et al., 1998).
(D) If two compared regulators belonged to the same stage of the transcription cycle (Figure 1A), then the corresponding pixel was color-coded by the stage, as indicated. Otherwise they were colored white.
Cluster a was relatively small and consisted of general transcription regulators (GTFs, in the Initiation class) and Pol II subunits, and as expected, the overlapping sets of genes had high transcription frequencies (Figure 4C). Co-occupancy of these regulators at a common set of genes was verified more directly via standard gene-based clustering (Figure S2A). Other components and regulators of the core transcription machinery occupied many cluster a genes, but also occupied genes having other bound regulators, causing them to cluster apart from the core transcription machinery. The lack of a variety of regulators at cluster a genes indicates that the genes which are most highly occupied by GTFs and most highly transcribed may have fewer regulatory interactions in common than at other genes. If many positive and negative regulators function to antagonize each other, this could result in a rich assortment of co-occupancy that produces a lower net output of transcription. As such, genes with high levels of PICs but few other regulators, may represent an “unattenuated”, and thus highly active, situation.
Cluster b was enriched with regulators in the Access class (Figure 4A), including chromatin remodelers, chromatin modifying regulators, and chromatin binding proteins. Interestingly, in contrast to cluster a, cluster b tended to be lowly transcribed (Figure 4C), and contained generally repressive chromatin remodelers and histone deacetylases, such as Isw1a/b, Isw2, Cyc8/Ssn6, Hos1, and Hda1 (Figure 4D). The Xbp1 and Rfx1/Crt1 sequence-specific repressors were also enriched in cluster b, suggesting that they may be functionally linked to these chromatin regulators. Thus, many chromatin regulators tend to occupy the same set of lowly expressed genes (also verified by gene-based clustering in Figure S2B). The expression of such genes might be rate-limited by the combined repressive action of these chromatin regulators. We observed a substantial intersection of the regulators in clusters a and b, and such genes tended to have an intermediate level of transcription (Figure 4A and C). We interpret this overlap to mean that many genes may have both repressive and activating proteins bound, although not temporally resolved, with the net output being an intermediate level of transcription.
Cluster c had a large membership of regulators from all stages of the transcription cycle (Figure 4D), and the corresponding genes tended to be lowly expressed (Figure 4C). We interpret the large cluster size to reflect a tendency of a large number of positive and negative regulators at all stages of the transcription cycle to work at many of the same genes, to produce a relatively low transcriptional output. This contrasts to cluster a, where apparently unbridled PICs produce high transcriptional outputs.
The co-occupancy data are displayed as a Cytoscape network in Figure S3A (Shannon et al., 2003), and those with stronger co-occupancies in Figure S3B. Many of the sequence-specific regulators (“Orchestration” class shown in red) were located towards the periphery of the network, which likely reflects their gene-selective behavior, and thus had less overlap with other regulators. Ume6, Hho1, and Asf1 stood out as having a high degree of co-occupancy, and this was verified by standard gene-based clustering (Figure S3C, cluster 2). Ume6 is a key regulator of early meiotic genes by repressing transcription during vegetative growth (Strich et al., 1994). Asf1 is involved in chromatin assembly/disassembly during DNA replication and transcription (Schwabish and Struhl, 2006; Tyler et al., 1999). The presence of Asf1 at Ume6-repressed genes is consistent with the role Asf1 in establishing silenced chromatin (Krawitz et al., 2002), perhaps in helping assemble the repressive linker histone H1 (Hho1).
Further exploration of the occupancy data is presented in Figure S4 and Table S7, wherein we report on the overlapping membership (measured as –log10 P-value) between regulator-occupied genes and genes that are in the upper or lower tenth percentile of a measured genome-wide property available in the public domain (e.g. expression changes in mutants, motif enrichment, and regulator occupancy levels). Over 2,300 relationships are presented for each regulator. The dataset provides a rich resource of empirical information about a regulator that is essentially untapped here.
Spatially distinct promoter organization of regulators associated with the SAGA and TFIID pathways
Understanding how the diverse repertoire of transcription regulators is organized in promoter regions can provide insight into the interplay of the transcription machinery and chromatin. Thus with microarray probes situated at the distal (“UAS”) and proximal (“TSS”) ends of each promoter (230 bp center-to-center distance), we interpolated the location of each significantly bound regulator, based upon an occupancy level weighting of each probe location (see Methods). The distributions of all bound locations relative to the nearest transcriptional start site were then plotted for each regulator as a heat map (Figure 5A and Table S8). The upper panel provides a summary list of those complexes that bind the distal versus core promoter region. Regulatory proteins appeared to separate out into distal and proximal promoter binding, with the dividing line around -120 relative to the TSS. Inasmuch as high density ChIP-chip microarrays produced similar genome-wide maps as the low density arrays described here (Figure S5), it is likely that the distal and proximal occupancy domains were not an artifact of the microarray platform. As further support, a previous study using site-specific DNA cleavage probes tethered to the HIS4 promoter qualitatively agrees with the interpolated distribution profiles for Pol II and the GTFs (Miller and Hahn, 2006). Nonetheless there are caveats. A regulator might not have a consensus location that resides between the two probes, or a regulator may not have any consensus location (relative to the TSS). In addition, patterns may be biased to reflect the most highly occupied locations.
Figure 5. Composite distribution of regulators across promoter regions.
(A) Complexes particularly enriched in the distal (<-120bp) versus proximal (>-120bp) promoter region based on panel B are listed. The consensus locations for the TATA box, the “-1” and “+1” nucleosomes are shown (Basehoar et al., 2004; Mavrich et al., 2008). The frequency distribution mode around the TSS for individual subunits shown in panel B defined the consensus binding location for each complex. If two or more subunits were available for a complex, then the modes for all subunits were averaged.
(B) In the heat map, the ChIP-chip binding locations for each regulator (one regulator per line) were interpolated between the UAS and TSS probe at each gene, binned, and aligned by the TSS. Heatmap color was used to indicate bin counts. Data were filtered to have at least twofold occupancy levels. The upper-most track indicates the distribution of microarray probes.
(C) For elongation regulators, ChIP-seq data were used to generate a heatmap display for the composite frequency distributions relative to the TSS. The ChIP-seq data for each regulator were binned in 25 bp intervals and normalized to an untagged control. Log2 values were centered so that the lowest quartile value was the same in all datasets. Genes with >2-fold normalized occupancy were plotted. See also Figures S5-6.
Complexes favoring the SAGA pathway (Table 1 and Figure 5) tended to reside in the distal promoter region where SAGA resides. For example, Mot1, Mediator, Histone H1, HOS1, RPD3, SSN6-TUP1, and SWI/SNF preferentially occupied the upstream promoter region. The SAGA pathway involves substantial chromatin regulation. Nearly three-quarters of the chromatin regulators (29 of 41 complexes) we examined tended to occupy the region closer to where the -1 nucleosome resides rather than where the +1 nucleosome resides, which implicates substantial regulation of the -1 nucleosome. In contrast, Nua4, SWR1, and SPT10,21, which are more enriched at TFIID-dominated genes tended to reside at core promoter along with TFIID or in the adjacent downstream region near the +1 nucleosome. Taken together, the spatial arrangement of SAGA versus TFIID suggests that these complementary PIC assembly pathways may have distinct spatial organizations at promoters.
Since the caveats associated with location mapping in promoter regions may be particularly problematic with elongation regulators, we mapped the locations for 20 regulators in the Elongation class by ChIP-seq (Figure 5C and Figure S6). Pol II and the Pop2 subunit of CCR4-NOT were enriched across the promoter. Ess1, which is a prolyl isomerase that acts on the Pol II C-terminal heptad repeat, and Bye1 which has genetic interactions with Ess1 (Wu et al., 2003) were enriched within 100 bp of the TSS, suggesting that they might enter the elongation complex at the promoter region. Most other complexes, including components of the CCR-NOT, PAF, CTK1, and FACT complexes were enriched from 200-600 bp downstream of the TSS, possibly reflecting entry into the elongation complex as Pol II transitions from a serine-5 phosphorylated state to serine-2 phosphorylation. These results are in line with related studies reported elsewhere (Mayer et al., 2010; Rahl et al., 2010). Some of these regulators were also detected in the promoter region, which could reflect lower levels of interactions or fewer genes having those interactions.
Mobilization of the transcription machinery in response to acute heat shock differs in the SAGA vs TFIID pathways
In yeast the common environmental stress response, including the heat shock response (abrupt change from 25°C to 37°C), involves transient down-regulation of ~600 genes and up-regulation of ~300 genes within 15 minutes of the stress (Causton et al., 2001). While the promoter association and dissociation of a number of regulators in response to heat shock have been determined (Harbison et al., 2004; Lee et al., 2004; Zanton and Pugh, 2004), no study has comprehensively examined the genome-wide change in occupancy of the vast majority of regulatory complexes.
Changes in occupancy upon heat shock, were examined at promoter regions and the 3' end of ORFs (rows in Figure 6A-B, respectively, and Table S9), and were K-means clustered into seven groups. Within each region, regulators were separated by their classification into transcriptional stages (Figure 1A). Within each stage, individual regulators (columns) were hierarchically clustered. A list of complexes that increase/decrease in occupancy at heat shock induced/repressed genes can be found in Table S10.
Figure 6. Assembly and disassembly of the transcription machinery in response to heat shock.
Log2 ratios of 37°C/25°C occupancy changes for promoter regions that increased (red), decreased (green), did not change (black), or did not have data (black), the latter comprising <10% of the data. Rows representing promoter regions (A) were clustered alongside 3' ORF regions (B) by K-means (K = 8), and are ordered identically in the two panels. The data were filtered to include promoters having >75% data present and >3-fold change in occupancy in at least 7 datasets, which resulted in 934 genes. Columns represent ChIP-chip data for a given regulator, and were clustered hierarchically by stage. For clarity, only a subset of the 200 regulators are shown; the cluster of all regulators is shown in Figure S7. The changes in gene expression during heat shock are aligned to the right (Zanton and Pugh, 2004). The average transcription rate at 25°C (Holstege et al., 1998) for each cluster are indicated on the right of the panel.
Two clusters of heat shock induced genes were evident. Modestly induced genes (cluster 1 in Figure 6A) tended to preferentially accumulate TFIID, SWR1, and RSC, which may reflect increased activity of housekeeping genes. Shifting cultures to 37°C actually increases growth rates, and this likely requires increased activity of housekeeping genes. Accordingly, cluster 1 genes are moderately transcribed at 25°C (4 mRNA/hr, Figure 6A far right panel). The most highly induced genes (cluster 2), which are enriched with SAGA-dominated stress-induced genes, saw the largest increase in occupancy of regulators such as SAGA, SWI/SNF, INO80, NuA4, Mediator, GTFs, NC2, Cyc8/Tup1, Pol II, Iws1/Spt6, FACT, Pcf11, COMPASS, Rad6/Bre1, and Set2. Some of these regulators act negatively, which may reflect their involvement in quenching the transient heat shock response. In addition, heat-shock enrichment of the elongation regulatory complexes were most evident in the body of the gene (Figure 6B), which is consistent with their role in events that are linked to transcription elongation and mRNA transport.
One group of genes (cluster 3) stood out because Pol II and a variety of elongation and termination regulators (Iws1/Spt6, FACT, Pcf11, Rtt103, Spt2, and Bye1) displayed a strong increase in occupancy in the promoter region but not in the 3’ ORF region. These genes were lowly transcribed at 25°C (3 mRNA/hr) and remained unchanged during the heat shock response. Furthermore, the enrichment pattern for two negative regulators of transcription elongation, Bye1 and Spt2 (Kruger and Herskowitz, 1991; Roeder et al., 1985; Wu et al., 2003), mirrored the Pol II pattern, suggesting that these regulators may be involved in generating an elongation-refractory Pol II at the 5’ ends of these genes.
Cluster 4 stood out as having increased occupancy levels of the GTFs and Pol II at the 3’ end of the genes, but with little detectable RNA output. The basis for this needs to be further investigated, although it could reflect heat-shock induced production of unstable antisense transcription, as one potential example.
Occupancy changes at heat shock repressed genes (clusters 5-8 tended to involve many of the same regulators as seen at induced genes, but in the opposite direction. However, the repressed genes of cluster 5 also acquired the Xbp1 repressor, three histone deacetylases (Rpd3, Hda1, and Hos1), the Jhd1 demethylase, and Isw1a/b in the promoter region. This cluster was enriched with ribosome biosynthesis and rRNA maturation genes, and these genes are highly transcribed at 25°C (85 mRNA/hr). Thus the transient shut down of ribosome biosynthesis that accompanies heat shock appears to be an active shut-down by an influx of co-repressor complexes, rather than simply a loss of the transcription machinery.
DISCUSSION
Saccharomyces utilizes two complementary strategies to regulate its mRNA genes (Basehoar et al., 2004; Bhaumik and Green, 2002; Cheng et al., 2002; Huisinga and Pugh, 2004; Kuras et al., 2000). Here we provide a description of the extent to which transcription proteins participate in TFIID and SAGA pathways for PIC assembly. The namesake for these pathways is not intended to imply a disproportionate importance of these components compared to others. The TFIID pathway serves the vast majority of genes by providing a relatively steady production of mRNA that may be an inherent property of genes. The variety of regulators occupying TFIID-dominated genes is rather limited. In contrast, the SAGA pathway provides a greater dynamic range of mRNA output, and thus is more highly regulated. The SAGA pathway may be best suited to respond to major cellular reprogramming events ranging from acute stress responses to cellular differentiation. Importantly, a gene may utilize both pathways to varying extents to achieve an appropriate level of regulation. Here, we identify which complexes tend to be connected to which pathway in yeast cells grown in rich media.
We surmise the following framework of regulators specific for each pathway, based upon current and previous functional and genome-wide occupancy studies. Since the occupancy data presented here do not provide information on the order of assembly, we utilize prevailing models, for example (Sikorski and Buratowski, 2009), on the order of events in the transcription cycle to place regulators into context within each pathway. The genome-wide location analyses presented here on hundreds of proteins are consistent with the specificity of the assembly pathways described below.
In the TFIID-dominated pathway, aggregate available evidence supports the idea that Rap1 (as one clear example of a sequence-specific activator in this pathway) recruits NuA4 and TFIID to promoters (Garbett et al., 2007; Reid et al., 2000). NuA4 acetylates histones H4 and H2A.Z, particularly at the +1 nucleosome located at the downstream edge of the promoter (Allard et al., 1999; Keogh et al., 2006; Li et al., 2007a). These acetylation sites dock the bromodomain-containing regulator Bdf1, which interacts with the SWR1 complex to promote and maintain H2A.Z deposition and acetylation (Jacobson et al., 2000; Kobor et al., 2004; Krogan et al., 2003). Bdf1 might also facilitate TFIID-promoter interactions, since it copurifies with TFIID (Matangkasombut et al., 2000). A TFIID-based core PIC then assembles, involving TBP, TFIIB, -E, -F, -H, and Pol II. Continuing along the TFIID pathway, polymerase-associated Ctk1 phosphorylates serine-2 of the Pol II C-terminal heptad repeats (Sterner et al., 1995). Phosphoserine-2 helps recruits Set2 to methylate H3K36 in the body of the gene (Li et al., 2003), which allows the RPD3-S complex to remove activating histone acetylation marks and help suppress internal initiation (Li et al., 2007b).
When cells are shifted from 25°C to 37°C, the observed relocation of regulators provides additional insight into the TFIID pathway. For example, a number of sequence-specific negative regulators (e.g., Rfx1/Crt1, Xbp1, and others) move from heat-shock activated genes to heat shock repressed genes, but they largely stay with genes regulated by the TFIID pathway. Others such as those involved in removing active histone marks and setting up repressive chromatin tend to accumulate at the heat shock repressed genes.
In contrast to the TFIID pathway, the SAGA pathway appears to have many more points of regulation. The utilization of a greater variety and gene selectivity of sequence-specific regulators provides one means of integrating many different stress and/or cell type specific signaling events. The SAGA-dominated genes display more heterogeneity in chromatin organization (Albert et al., 2007), which suggests that nucleosomes have gene-specific and position-specific roles in regulating these genes. In support of this notion, ATP-dependent chromatin remodeling complexes and histone deacetylases were particularly enriched at SAGA-dominated genes. Indeed many other types of chromatin regulators, including those that write, read, and remove histone marks were also enriched.
Positioning of the -1 and +1 nucleosomes could play major roles in limiting promoter access to the GTFs at SAGA-dominated promoters. Within this context, why SAGA delivers TBP to such promoters and utilizes a TATA box for TBP, as opposed to TFIID that delivers TBP to TATA-less promoters (or at least without apparent regard for TATA) is currently unclear. Perhaps the TATA box provides added TBP-promoter stability in light of encroaching nucleosomes, while at the same time leaving TBP accessible to its many negative regulators such as Mot1, which favors the SAGA pathway. In the SAGA pathway, TBP is stabilized against these regulators by TFIIA (reviewed by Lee and Young, 1998; Pugh, 2000). In the TFIID pathway, TAFs may prevent negative regulators from accessing TBP. Events surrounding the early stages of transcription, such as COMPASS-directed H3K4 trimethylation of the nucleosomes at the 5' ends of genes and originating through Rad6/Bre1-directed H2B ubiquitylation (Sun and Allis, 2002), show a preference for the SAGA pathway.
The enrichment of elongation regulators at SAGA-dominated genes further fits with the highly regulated status and high dynamic range of these genes. In particular, FACT and Spt6/Iws1 allow Pol II to move through chromatin more rapidly by being highly efficient at dismantling nucleosomes in the path of Pol II (Fleming et al., 2008; Kaplan et al., 2003). Finally, transcripts from SAGA-dominated genes appear to have a more direct route to nuclear pores, in that such genes tend to be enriched with the THO/TREX complex which is nuclear pore-associated and linked to nucleo-cytoplasmic trafficking of mRNAs (Strasser et al., 2002).
While the genome-wide location analysis indicates that more regulatory proteins at various stages of the transcription cycle tend to be linked to the SAGA pathway rather than the TFIID pathway, the fact that genes utilize both pathways to varying extents suggests that components of the two pathways could cross-regulate each other. The data presented here provide a snapshot of how regulatory complexes that operate at various stages of the transcription cycle may be integrated across the yeast genome. Certainly, much more can be mined from the data. The combined datasets may be of value in assessing direct versus indirect effects in expression profiling studies, and in serving as a platform to develop new questions such as the relationship between genomic binding locations and function. Importantly, the comprehensiveness of the analyses should provide guidance in exploring co-regulation in more complex organisms. Future work will be aimed at achieving higher resolution maps of regulator binding locations in an effort to define the precise organizational structure at promoters. In addition, it will be important to discern how occupancy of each regulator depends on other regulatory components.
EXPERIMENTAL PROCEDURES
Yeast strains and epitope tagging
Strains used for this study were obtained from the Yeast TAP-Fusion Library (Open Biosystems), in which tagged proteins were immunoprecipitated with IgG antibodies. In the ChIP-chip experiments, BY4741 was used as an untagged Mock IP control for occupancy normalization. The identity of TAP-tagged stains were confirmed by size-matching from Western blot analysis and/or by validating microarray data with published data sets (not shown).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as described previously (Venters and Pugh, 2009a; Zanton and Pugh, 2006). Briefly, each strain (500 ml) was grown at 25°C to mid log phase (A600 = 0.8) in 500 ml YPD. For heat shock experiments, each culture was rapidly shifted to 37°C at mid log phase by the addition of hot media, and incubation continued at 37°C for 15 minutes. Cells were then fixed by the addition of formaldehyde to a final concentration of 1% for 15 minutes at 25°C, and quenched for 5 minutes with glycine. For both normal and heat shock conditions, cultures were diluted two fold with the same volume of temperature-adjusted distilled water just prior to addition of formaldehyde so as to achieve a media temperature of 25°C. The harvested cells were lysed with glass beads, and the chromatin pellet washed and sonicated. Sheared chromatin was immunoprecipitated with IgG-sepharose. This ChIP enriched DNA was amplified by ligation-mediated PCR as described elsewhere (Harbison et al., 2004), and 75-300 bp LM-PCR amplified fragments were gel purified according to the manufacturer's protocol (Qiagen). The gel-purified DNA yield was measured using the Nanodrop ND-1000 spectrophotometer.
DNA labeling and microarray hybridization
DNA labeling and hybridization to DNA microarrays were performed as described previously (Zanton and Pugh, 2004). For the custom tiling microarrays, 100 ng of gel purified LM-PCR ChIP enriched DNA was amplified by 15 PCR cycles, coupled to Cy3 or Cy5 fluorescent dyes, and subsequently cohybridized to the arrays. For each ChIP sample, at least two biological replicates were performed employing a dye swap for each replicate.
Low-density tiling microarray design
Oligonucleotide probes (60 bp) were designed and synthesized by Operon Biotechnologies, and were primarily against regions -320 to -260 and -90 to -30 from the start of each open reading frame in the S. cerevisiae genome, with positional adjustments made to keep a uniform Tm. Additional genic oligos comprised the Yeast Genome Oligo Set Version 1.1 (Operon Biotechnologies). An additional 1,205 probes were synthesized against non-promoter intergenic regions, and in snRNA and tRNA promoters. Each microarray consisted of >21,000 oligonucleotide probes spotted onto amino-silane treated glass slides.
ChIP-seq sample preparation and analysis
ChIP-enriched DNA samples were prepared for the Illumina Genome Analyzer platform according to the manufacturer's instructions. The sequencing data was normalized using an untagged control (BY4741) and centered to the lowest quartile of reads in a 25 bp window.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Pugh laboratory and the Center for Gene Regulation for many helpful discussions. We thank Istvan Albert for bioinformatics support, and Ben Pugh for conducting strain validation tests. This work was supported by NIH grant ES013768. This project is funded, in part, under a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
ChIP-chip and ChIP-seq data are available at ArrayExpress under accession numbers E-MTAB-311 and E-MTAB-440, respectively.
SUPPLEMENTAL DATA
Supplemental data include microarray data analysis methods, seven figures, and ten tables and can be found with this article online at www.cell.com/molecular-cell/.
REFERENCES
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. Embo J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Berger SL. Gene regulation. Local or global? Nature. 2000;408:412–413. 415. doi: 10.1038/35044160. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol Cell Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braglia P, Dugas SL, Donze D, Dieci G. Requirement of Nhp6 proteins for transcription of a subset of tRNA genes and heterochromatin barrier function in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:1545–1557. doi: 10.1128/MCB.00773-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Molecular biology of the cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JX, Floer M, Ononaji P, Bryant G, Ptashne M. Responses of four yeast genes to changes in the transcriptional machinery are determined by their promoters. Curr Biol. 2002;12:1828–1832. doi: 10.1016/s0960-9822(02)01257-5. [DOI] [PubMed] [Google Scholar]
- Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Garbett KA, Tripathi MK, Cencki B, Layer JH, Weil PA. Yeast TFIID serves as a coactivator for Rap1p by direct protein-protein interaction. Mol Cell Biol. 2007;27:297–311. doi: 10.1128/MCB.01558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Michaut M, Acker J, Aude JC, Thuriaux P, Werner M, Soutourina J. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev. 2008;22:1934–1947. doi: 10.1101/gad.471908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Hull MW, Erickson J, Johnston M, Engelke DR. tRNA genes as transcriptional repressor elements. Mol Cell Biol. 1994;14:1266–1277. doi: 10.1128/mcb.14.2.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS biology. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawitz DC, Kama T, Kaufman PD. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol Cell Biol. 2002;22:614–625. doi: 10.1128/MCB.22.2.614-625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Kruger W, Herskowitz I. A negative regulator of HO transcription, SIN1 (SPT2), is a nonspecific DNA-binding protein related to HMG1. Mol Cell Biol. 1991;11:4135–4146. doi: 10.1128/mcb.11.8.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Kosa P, Mencia M, Struhl K. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nature genetics. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Lee TI, Young RA. Regulation of gene expression by TBP-associated proteins. Genes Dev. 1998;12:1398–1408. doi: 10.1101/gad.12.10.1398. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007a;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007b;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. The Journal of biological chemistry. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- Li XY, Bhaumik SR, Green MR. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS biology. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, O'Shea EK. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci U S A. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matangkasombut O, Buratowski RM, Swilling NW, Buratowski S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome research. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nature structural & molecular biology. 2010 doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- Miller G, Hahn S. A DNA-tethered cleavage probe reveals the path for promoter DNA in the yeast preinitiation complex. Nature structural & molecular biology. 2006;13:603–610. doi: 10.1038/nsmb1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Pugh BF. Control of gene expression through regulation of the TATA-binding protein. Gene. 2000;255:1–14. doi: 10.1016/s0378-1119(00)00288-2. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JL, Iyer VR, Brown PO, Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- Roeder GS, Beard C, Smith M, Keranen S. Isolation and characterization of the SPT2 gene, a negative regulator of Ty-controlled yeast gene expression. Mol Cell Biol. 1985;5:1543–1553. doi: 10.1128/mcb.5.7.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Zhao Y, Warner JR. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. Embo J. 2005;24:533–542. doi: 10.1038/sj.emboj.7600553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski TW, Buratowski S. The basal initiation machinery: beyond the general transcription factors. Current opinion in cell biology. 2009;21:344–351. doi: 10.1016/j.ceb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Lee JM, Hardin SE, Greenleaf AL. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol Cell Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Strich R, Surosky RT, Steber C, Dubois E, Messenguy F, Esposito RE. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome research. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome research. 2009a;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Crit Rev Biochem Mol Biol. 2009b;44:117–141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Rossettini A, Hanes S. The ESS1 prolyl isomerase and its suppressor BYE1 interact with RNA Pol II to inhibit transcription elongation in Saccharomyces cerevisiae. Genetics. 2003;165:1687. doi: 10.1093/genetics/165.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Qiu R, Foland TB, Griesen D, Galloway CS, Chiu YH, Sandmeier J, Broach JR, Bi X. Rap1p and other transcriptional regulators can function in defining distinct domains of gene expression. Nucleic acids research. 2003;31:1224–1233. doi: 10.1093/nar/gkg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanton SJ, Pugh BF. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc Natl Acad Sci U S A. 2004;101:16843–16848. doi: 10.1073/pnas.0404988101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanton SJ, Pugh BF. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev. 2006;20:2250–2265. doi: 10.1101/gad.1437506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.