Abstract
Double-stranded DNA breaks (DSBs) induce a phosphorylation-mediated signaling cascade, but the role of phosphatases in this pathway remains unclear. Here we show that human protein phosphatase 4 (PP4) dephosphorylates replication protein A (RPA) subunit RPA2, regulating its role in the DSB response. PP4R2, a regulatory subunit of PP4, mediates the DNA damage-dependent association between RPA2 and the PP4C catalytic subunit. PP4 efficiently dephosphorylates phospho-RPA2 in vitro, and silencing PP4R2 in cells alters the kinetics and pattern of RPA2 phosphorylation. Depletion of PP4R2 impedes homologous recombination (HR) via inefficient loading of the essential HR factor RAD51, causing an extended G2-M checkpoint and hypersensitivity to DNA damage. Cells expressing phosphomimetic RPA2 mutants have a comparable phenotype, suggesting that PP4-mediated dephosphorylation of RPA2 is necessary for an efficient DNA-damage response. These observations provide new insight into the role and regulation of RPA phosphorylation in HR-mediated repair.
The cellular response to DSBs is initiated by the phosphatidylinositol-3 kinase–like (PI3-like) family of kinases that include DNA-dependent protein kinase catalytic subunit (DNA-PKcs), ataxia telangiectasia mutated (ATM) and ATM- and Rad3-related (ATR). A vast network of ~700 mouse and human proteins are phosphorylated by these kinases in response to DNA damage1. The phosphorylated proteins include factors involved in cell-cycle checkpoints and apoptosis as well as bona fide DNA-repair and DNA-replication proteins. However, our understanding of how the DSB signaling network becomes inactivated is limited. It is conceivable that this event involves the dephosphorylation of key factors, with the normal balance of phosphorylation restored by phosphatases.
Recently, we identified a role for PP2A-like phosphatases (PP2AC and PP4C) in the DSB response and observed that they dephosphorylate a key DNA-repair protein, the histone variant H2AX2-4. The members of the PP2A family of phosphatases are found in dimeric or trimeric complexes containing regulatory subunits that confer substrate specificity and tissue- and cell type-specific targeting5,6. PP2AC is a well-characterized phosphatase with a variety of substrates involved in DNA repair, replication and progression of the cell cycle6-8. However, there are few confirmed substrates of PP4C and very limited understanding of its cellular role9. We and others have shown that a PP4C-containing trimeric complex dephosphorylates γ-H2AX generated during DNA replication3,10, with PP4 deficiency specifically affecting the repair of DNA replication–mediated damage3. DSBs induced during DNA replication are typically repaired by HR. Reporter assays show significantly reduced HR-mediated repair in PP4-deficient cells3. Together, these results suggested that factors involved in HR-mediated repair of DSBs are targeted by a human PP4 complex.
Based on these observations, we set out to identify factors involved in HR-mediated repair that are targeted by a PP4 complex in human cell lines. We found that RPA2, the 32-kDa subunit of the RPA heterotrimeric complex, is dephosphorylated by a PP4 complex. RPA is a single-stranded DNA (ssDNA) binding factor that is critical for the ‘three Rs’ of eukaryotic DNA enzymology: replication, recombination and repair11-15. Upon DNA replication, stress or damage, RPA2 undergoes phosphorylation in an ordered and synergistic fashion, with the modification of Ser33 being pivotal in the sequence of phosphorylation events16. Mutation of RPA2 phosphorylation sites causes a defect in the efficiency of DNA repair11,16,17. Because of the importance of RPA2 phosphorylation in DNA repair, we focused on the PP4-mediated dephosphorylation of RPA2 and on studying the functional impact of RPA2 dephosphorylation in human cell lines.
RESULTS
RPA2 interacts with a PP4 complex
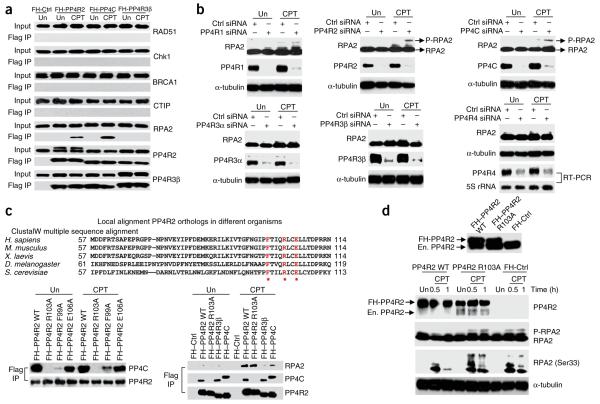
We used a candidate-based approach to investigate the role of PP4 in HR-mediated repair. We probed protein complexes isolated by tandem affinity purification from cells expressing Flag and hemagglutinin (FH)-tagged PP4C, PP4R2 and PP4R3β for established HR-repair proteins, both in the presence or absence of exogenous DNA damage. We found that RPA2 interacts with PP4C and PP4R2 but not with PP4R3β (Fig. 1a). This interaction is DNA damage dependent, as it is detected only in camptothecin (CPT)-treated cells. Immunoprecipitation of endogenous RPA2 in CPT-treated cells also ‘pulled down’ PP4C and PP4R2, further confirming this result (Supplementary Fig. 1). To investigate whether a PP4 complex affects the phosphorylation status of RPA2, we silenced all the known subunits of PP4 and evaluated levels of phosphorylated RPA2. Consistent with the interaction data, silencing PP4C and PP4R2 led to elevated levels of phospho-RPA2 in CPT-treated cells (Fig. 1b). Notably, silencing the other PP4 regulatory subunits did not affect RPA2 phosphorylation. Reducing the levels of other PP2A-like phosphatases, PP2AC and PP6C5, or of the phosphatase Wip1, which is involved in the DNA-damage response18,19, also did not have any effect on RPA2 phosphorylation (Supplementary Fig. 2). PP4C and/or PP4R2 apparently modulate the RPA2 phosphorylation state in response to other types of DNA damage because ionizing radiation induced co-localization of RPA2 and PP4R2 in nuclear foci, and silencing R2 led to elevated levels of phospho-RPA2 in cells treated with ionizing radiation (Supplementary Fig. 3). A PP4 complex including PP4C and PP4R2 dephosphorylates γ-H2AX3,10. Therefore, it is feasible that silencing PP4C and PP4R2 affects RPA2 phosphorylation via H2AX. To address this issue, we silenced PP4R2 in H2AX-depleted cells and assessed the phosphorylation status of RPA2. The absence of H2AX did not alter the impact of PP4R2 on RPA2 phosphorylation (Supplementary Fig. 4). Together, these results suggest that a heterodimeric complex of PP4C and PP4R2 dephosphorylates RPA2.
Figure 1.
A PP4 complex interacts with RPA2 in the context of DNA damage, and silencing the PP4 complex enhances RPA2 phosphorylation. (a) Immunoprecipitation from HeLa S3 cells where the indicated FH-tagged PP4 subunits were stably expressed. Anti-Flag antibody was used for immunoprecipitation from untreated (Un) or CPT-treated cells and the western blot probed with indicated antibodies. (b) Silencing PP4R2 and PP4C enhances phospho-RPA2 levels. All known PP4 subunits in U2OS cells were depleted by siRNA transfection, and the phosphorylation of RPA2 was checked by immunoblotting after CPT treatment. Owing to lack of a commercially available antibody, the expression of the newly discovered PP4R4 (ref. 43) subunit was determined by RT-PCR. α-tubulin served as loading control. (c) PP4R2 mediates the interaction of PP4C with RPA2. Top, alignment of the N-terminal region of PP4R2 across different organisms. The highlighted and starred residues represent conserved residues which were consequently mutated to alanine for study. Bottom, HeLa S3 cells stably expressing FH-tagged wild-type (WT) or mutant PP4R2 proteins were subjected to immunoprecipitation after CPT treatment. Immunoprecipitation with FH-PP4C– and FH-PP4R3β–expressing cells were performed as additional controls. (d) Interaction of PP4C and PP4R2 is necessary for dephosphorylation of RPA2. Endogenous PP4R2 was depleted in HeLa S3 cells expressing either FH–PP4R2 WT or FH–PP4R2 R103A by siRNAs targeting the 3′ UTR of PP4R2. Endogenous (En.) and FH-PP4R2 can be distinguished by mobility shift on immunoblot (upper panel). The phosphorylation of RPA2 was observed at indicated times after CPT treatment. Antibody against a RPA2 epitope with phosphorylated Ser33 was used for the immunoblot. α-tubulin served as loading control.
PP4R2 mediates the interaction of PP4C and RPA2
We have previously shown that PP4C–PP4R2–PP4R3β forms a heterotrimeric complex involved in the DSB response. However, several studies have shown that PP4C and PP4R2 form a heterodimeric complex in vivo and in vitro, which then recruits PP4R3α or R3β9,20. Because our results suggest that only a heterodimeric complex of PP4C–PP4R2 affects RPA2, we wanted to confirmed that PP4C and PP4R2 interact efficiently in the absence of PP4R3β (Supplementary Fig. 5) and PP4R3α (data not shown). It is possible, however, that PP4R2 and PP4C both regulate RPA2 phosphorylation independent of each other, and not as a PP4 complex. To address this issue, we isolated an R2 mutant that did not interact with PP4C but retained the capacity to interact with RPA2. Our rationale was that, if PP4R2 mediates dephosphorylation of RPA2 by recruiting PP4C, cells expressing these PP4R2 mutants should have elevated levels of phospho-RPA2. We generated PP4R2 mutants based on species conservation (Fig. 1c, top) and observed that an arginine-to-alanine mutation of residue 103 (R103A) abolished the interaction of PP4R2 and PP4C (Fig. 1c, bottom left), without altering the DNA damage–dependent interaction of PP4R2 and RPA2 (Fig. 1c, bottom right). We replaced endogenous PP4R2 with FH-tagged wild type (WT) or R103A mutant using siRNAs targeting the 3′ untranslated region (UTR) of PP4R2 and then evaluated the phosphorylation status of RPA2. Consistent with our hypothesis, CPT-treated cells expressing the R103A mutation had elevated levels of phospho-RPA2 and resembled the PP4R2-silenced cells (Fig. 1d). We further established the existence of this heightened RPA2 phosphorylation by probing for the phospho-Ser33 residue of RPA2. This observation clearly shows that PP4R2 mediates RPA2 dephosphorylation by recruiting PP4C. This result allowed us to investigate the impact of PP4 phosphatase complex on RPA2 phosphorylation by silencing PP4R2 and avoid the possible pleiotropic effects of silencing the catalytic subunit PP4C.
PP4C efficiently dephosphorylates RPA2 in vitro
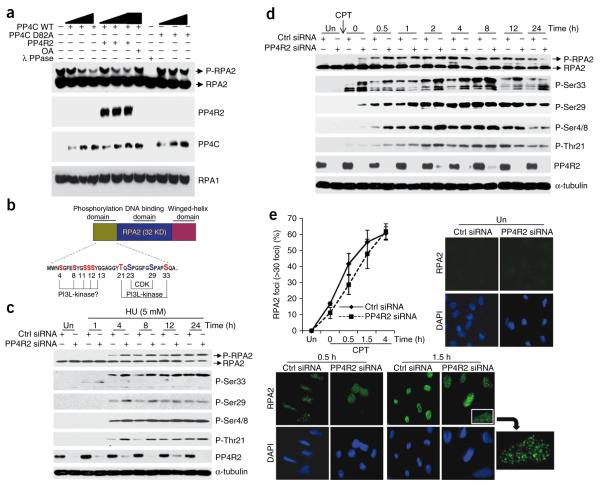
To determine whether PP4C can dephosphorylate phospho-RPA2 directly, we used the baculoviral system to make recombinant PP4C and PP4R2 (Supplementary Fig. 6). Based on conservation with catalytic subunit of PP2A21, we made mutant PP4C (PP4C D82A), which is expected to be catalytically inactive (Supplementary Fig. 7). We immunopurified endogenous phospho-RPA2 from CPT-treated cells and performed dephosphorylation assays (Fig. 2a). PP4C dephosphorylated phospho-RPA2 in a dose-dependent manner. Notably, the presence of PP4R2 did not influence the dephosphorylation reaction, suggesting that its role in cells is restricted to the recruitment of PP4C to substrates such as RPA2. Consistent with the predicted active site21 and the biochemistry of the catalytic subunit5, this reaction was inhibited by a mutation in PP4C and by okadaic acid (Fig. 2a). λ-phosphatase, which is insensitive to okadaic acid, served as a control.
Figure 2.
PP4 dephosphorylates RPA2 in vitro and influences the kinetics and pattern of RPA2 phosphorylation in cells. (a) PP4 dephosphorylates RPA2 in vitro. Wild-type PP4C, mutant PP4C (PP4C D82A) and PP4R2 were purified using the baculoviral system and were serially diluted in the phosphatase reaction. PP4C dephosphorylates phospho-RPA2 in a dose-dependent manner. Phosphatase reactions were probed with indicated antibodies. RPA1 serves as a loading control. OA, okadaic acid. (b) Schematic representation of RPA2 with an expanded view of the N-terminal phosphorylation domain. Serine and threonine residues highlighted with red are confirmed or potential DNA damage–responsive PI3-like kinase sites, and blue represents the cell cycle–dependent CDK sites. (c,d) Time course and pattern of RPA2 phosphorylation in PP4R2-depleted U2OS cells after hydroxyurea (HU; c) or CPT (d) treatment using antibodies against specific RPA2 phosphoresidues; untreated (Un) cells serve as controls. In c, cells were incubated in media containing 5 mM hydroxyurea for indicated time periods. In d, cells after CPT treatment were washed and incubated for indicated time points. Elevated levels of hyperphosphorylated RPA2 were detected in PP4R2-silenced cells, a difference found to be more pronounced at early times after DNA damage. (e) Delayed RPA2 focus formation after CPT treatment in PP4R2-depleted U2OS cells. Cells were stained for RPA2 and 4′,6-diamidino-2-phenylindole (DAPI), and images were captured by fluorescence microscopy. The RPA2 focus–positive cells (>30 foci) were quantified manually by comparison with DAPI images (~300 cells total). A magnified image of a RPA2 focus–positive cell is shown.
Silencing PP4R2 impacts the kinetics of RPA2 phosphorylation
The N terminus of RPA2 is a flexible domain containing roughly nine sites that undergo both stress- and cell cycle–dependent phosphorylation by both PI3-like kinases and the cyclin-dependent kinases (CDKs) (reviewed in refs. 11,14; Fig. 2b). We used phosphospecific antibodies to examine both the modification pattern and the kinetics of phosphorylation at five of the RPA2 sites in PP4R2-silenced cells after treatment with CPT or the dNTP synthesis inhibitor hydroxyurea. We determined that robust RPA2 phosphorylation was detectable after 4 h in 5 mM hydroxyurea, with phosphorylation gradually increasing thereafter (Supplementary Fig. 8). The phosphorylation levels of all four phosphoresidues that are targeted by the PI3-like kinases (specifically, Ser33, Thr21 and Ser4 and Ser8 (Ser4/8)) were distinctly elevated at all indicated time points in PP4R2-silenced cells (Fig. 2c). The CDK target Ser29 also had a relatively higher level of phosphorylation after 4 and 8 h of hydroxyurea treatment.
To focus on the repair process, we repeated the experiment using an initial 30-min treatment with CPT and then followed the kinetics of RPA2 phosphorylation for 24 h. In control cells, Ser33 phosphorylation was detectable within 1 h, peaked between 4 and 8 h and diminished by 12 h (Fig. 2d). However, in the absence of PP4R2, phospho-Ser33 was distinctly increased at all times and remained elevated even 12 h after removing CPT. The effect of PP4R2 deficiency on different RPA2 phosphoresidues was detectable at varying degrees in the early time points, particularly at early times (0–2 h) after removal of CPT. Together, these results clearly show that, in response to DNA damage, the PP4R2–PP4C complex regulates the kinetics of RPA2 phosphorylation, possibly in conjunction with the PI3-like kinases.
In response to both CPT and hydroxyurea, recruitment of RPA2 to DNA-repair foci occurs before hyperphosphorylation22,23. Although constitutively hyperphosphorylated RPA2 can be recruited to DNA-repair foci24,25, it is unclear whether the phosphorylation affects the kinetics or efficiency of RPA2-focus formation. We therefore investigated the kinetics of RPA2-focus formation in PP4R2-silenced cells using immunofluorescence microscopy. To eliminate non–chromatin bound RPA2, we extracted the cells with nonionic detergent before formaldehyde fixation. We found that early in the DNA-damage response (0.5 h and 1.5 h after CPT treatment), there were significantly (P < 0.031) fewer RPA-positive foci in cells lacking PP4R2, but by 4 h this difference disappeared (Fig. 2e). This observation suggests that the increase of hyperphosphorylated RPA2 in PP4R2-silenced cells may delay the formation of RPA foci and potentially affects the DNA-damage response.
Dephosphorylation of RPA2 affects post-damage DNA synthesis
To elucidate the functional significance of RPA hyperphosphorylation, mutant forms of RPA designed to mimic the hyperphosphorylated protein, with aspartate substituted for phosphorylatable RPA2 residues, have been successfully used24,25. Phosphomimetic RPA mutants are efficiently incorporated in the RPA complex and do not affect normal cell division24,25. These mutants selectively prevent the association of RPA with replication centers but not repair foci24,25. Similarly, others have found that ATR-dependent phosphorylation of RPA inhibits DNA synthesis following UV irradiation25. Also, inhibition of radioresistant DNA synthesis by RPA appears to be mediated by Mre11 (ref. 25). These results are consistent with the reduced affinity of hyperphosphorylated RPA for DNA polymerase α-primase26,27.
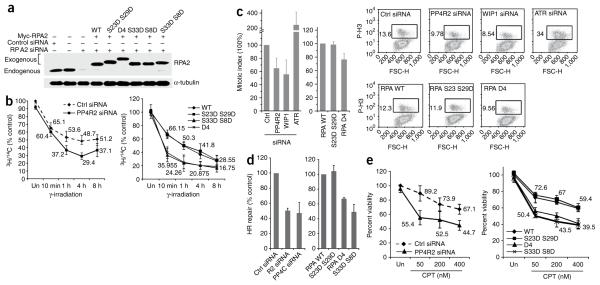
To confirm these results and further establish that the effect of PP4R2 on DNA synthesis is due to hyperphosphorylated RPA2, we used the RPA2 replacement strategy24. In cells ectopically expressing Myc-tagged RPA2 (WT and phosphomimic mutants), the endogenous RPA2 is depleted using siRNAs to the 3′ UTR, effectively replacing endogenous RPA2 with the ectopic version. We expressed either RPA2 WT or the different RPA2 mutants individually or in combination (RPA2 S23D S29D, RPA2 S33D S8D, RPA2 S33D, RPA2 S8D and RPA2 S23D S29D S33D S8D (referred to as RPA2 D4); Fig. 3a). To investigate whether the elevated levels of phospho-RPA2 in PP4R2-deficient cells had any impact on post-damage DNA synthesis, we measured the [3H]thymidine incorporation at different time points after ionizing radiation. There was significantly less DNA synthesis in PP4R2-silenced cells from 1 to 8 h after ionizing radiation (Fig. 3b, top). Only cells expressing the RPA2 S33D S8D (P < 0.029) and RPA2 D4 mutant (P < 0.012), and not the RPA2 S23D S29D mutant, had significantly impaired DNA synthesis after ionizing radiation (Fig. 3b bottom). This is consistent with the observation that residues Ser33 and Ser8 are phosphorylated by the PI3-like kinases in response to DNA damage17,25,28,29, whereas Ser23 and Ser29 phosphorylation is largely cell-cycle regulated16,26,30. Cells expressing either RPA2 S33D or RPA2 S8D also have reduced DNA synthesis after DNA damage (Supplementary Fig. 9a). Notably, the decrease in [3H]thymidine incorporation at different times after ionizing radiation in RPA2 S33D S8D– and RPA2 D4–expressing cells follow the same trend as the PP4R2-silenced cells. However, silencing PP4R2, or transient expression of RPA2 mutants, does not alter normal cell-cycle progression or DNA synthesis in undamaged cells (Supplementary Fig. 10). These data suggest that PP4 complex–mediated dephosphorylation of RPA2 is necessary to modulate the inhibition of DNA synthesis after DNA damage.
Figure 3.
Hyperphosphorylation of RPA2 affects HR-mediated repair of DSBs and the DNA-damage response. (a) Endogenous RPA2 was replaced with phosphomimetic mutants. Myc-tagged RPA2, wild-type (WT) and indicated RPA2 mutants were expressed in cells depleted of endogenous RPA2. α-tubulin served as loading control. (b) Radioresistant DNA synthesis is inhibited by hyperphosphorylated RPA2. Following silencing of PP4R2 (left) or replacement of endogenous RPA2 with phosphomimetic RPA2 mutants (right), U2OS cells were incubated with 14C, exposed to γ-radiation (undamaged controls, Un), and treated with 3H for indicated time periods. (c) Hyperphosphorylation of RPA2 affects the G2-M checkpoint. U2OS cells similar to those described in b were irradiated and released in medium containing nocodazole, and mitotic cells evaluated by flow cytometry. ATR- and WIP1-silenced cells served as controls. Representative flow cytometry images are shown on the right, and the results from three independent experiments are graphically represented on the left. (d) Measurement of HR-mediated repair of an I-SceI–induced DSB. U2OS cells carrying a single copy of the recombination substrate were transfected with control siRNA, PP4R2 siRNA or PP4C siRNA (left). In a parallel experiment, endogenous RPA2 was replaced with RPA2 WT or indicated mutants. The I-SceI expression plasmid was transfected, and green fluorescent protein–positive cells were measured by flow cytometry. (e) Hyperphosphorylation of RPA2 influences sensitivity to DNA damage. Cells depleted of PP4R2 or expressing RPA2- WT or indicated mutants were incubated with CPT at different concentration.
Dephosphorylation of RPA2 influences the G2-M checkpoint
PP4C has been implicated in regulating the G2-M checkpoint10, but whether hyperphosphorylated RPA2 contributes to the defect in PP4C-deficient cells is not clear. To assess the impact of PP4-RPA interaction on the G2-M checkpoint, PP4R2-silenced cells or cells expressing the RPA2 phosphomimic mutants were exposed to ionizing radiation and released in medium containing nocodazole. We determined the mitotic index of these cells after 24 h by analyzing expression of phospho-H3. Wip1- and ATR-silenced cells served as controls. Wip1 is a serine/threonine phosphatase that dephosphorylates Chk1 (ref. 18), and Wip1-deficient cells have a prolonged G2-M checkpoint and a relatively lower proportion of cells in mitosis following DNA damage18. ATR silencing abrogates the G2-M checkpoint, allowing cells with damaged DNA to enter mitosis31,32. We found that PP4R2-silenced cells (P < 0.0067) had an extended checkpoint, with a ~35% reduction of the number of cells in mitosis after ionizing radiation (Fig. 3c). Cells expressing RPA2 WT or RPA2 S23D S29D mutant had a G2-M checkpoint similar to that of control cells, whereas the phenotype of cells expressing RPA2 D4 (P < 0.024). was similar to that of PP4R2-silenced cells (Fig. 3c). These results indicate that PP4C–PP4R2 complex–mediated dephosphorylation of RPA2 facilitates release from a DNA damage–induced G2-M checkpoint.
RPA2 phosphorylation status affects HR-mediated DSB repair
Although RPA2 phosphorylation is induced in response to DSBs, the functional significance of this phosphorylation in DSB repair remains unclear. RPA acts in HR-mediated repair of DSBs33. To test the role of RPA phosphorylation on HR, we expressed the rare-cutting I-SceI endonuclease in U2OS cells containing a single, stably integrated copy of the artificial recombination substrate DR-GFP with an I-SceI site34.
This system permits quantification of HR efficiency through assay of the fraction of cells expressing GFP and was previously used to show that RPA-deficient cells have diminished HR-mediated repair35. We confirmed our earlier observation that HR is significantly reduced in PP4C-silenced cells3 and determined that knockdown of PP4R2 also suppressed HR (Fig. 3d, left). To address whether PP4R2-mediated dephosphorylation of RPA2 had any impact on the efficiency of DSB-induced HR, we expressed I-SceI in cells replaced with the RPA2 variants. Cells expressing RPA2 S33D S8D (P < 0.011) and RPA2 D4 (P < 0.037) had a significantly lower HR efficiency than cells expressing RPA2 WT or RPA2 S23D S29D (Fig. 3d, right). Again, the cells expressing either RPA2 S33D or RPA2 S8D also had reduced HR efficiency (Supplementary Fig. 9). To further confirm the impact of phosphorylated RPA2 on DSB repair, we measured DSBs using single-cell gel electrophoresis (neutral comet assay) in cells expressing RPA2 WT or the phosphomimetic mutants. CPT treatment induces DSBs visible by increased DNA mobility, or ‘comet tails’. Based on the comet moments, which quantify the extent of DNA damage, we determined that significantly more unresolved DNA damage was present in cells expressing RPA2 D4 (P < 0.034), and in PP4R2-silenced (P < 0.015) cells than in controls (Supplementary Fig. 11). Defects in the efficiency of DSB repair would be expected to be biologically relevant, and indeed PP4R2-deficient cells (P < 0.0195) and cells expressing RPA2 S33D S8D (P < 0.024) and RPA2 D4 (P < 0.0278) had lower viability than control cells at all tested doses of CPT (Fig. 3e). Together, these results show that absence of a PP4C–PP4R2 complex leads to elevated levels of hyperphosphorylated RPA2, which impedes HR-mediated repair of DSBs and sensitizes cells to DNA-damaging agents.
RPA2-focus formation is regulated by hyperphosphorylation
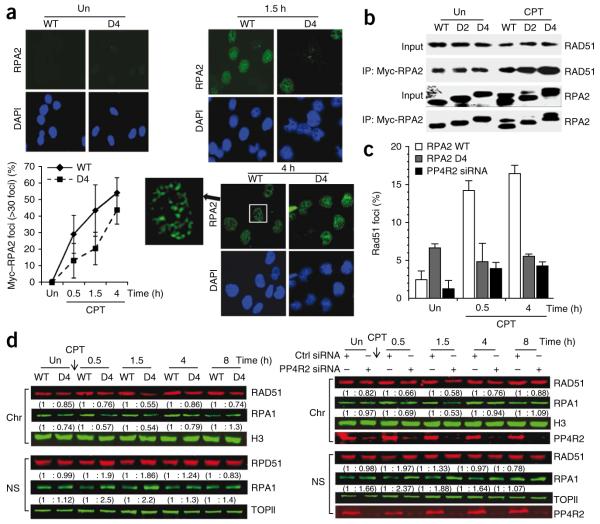
One of the early steps in HR-mediated repair of DSBs is RPA binding to the ssDNA generated at the break by resection. RPA must be rapidly loaded on the ssDNA, preventing formation of secondary structures; this step is critical for efficient HR33,36,37. Interestingly, in response to DNA damage, recruitment of RPA to DNA-repair foci precedes detection of the hyperphosphorylated form of RPA2 (refs. 22,23), and our results (Fig. 2e) suggest that premature hyperphosphorylation of RPA2 delays focus formation. We speculated that DNA damage–dependent formation of RPA2 foci is delayed in cells expressing the phosphomimetic RPA2 mutants, impeding efficient HR. To test this idea, we examined the kinetics of RPA2 foci formation in U2OS cells expressing RPA2 D4. We pre-extracted cells to remove soluble RPA2 before fixation and immunostaining. Whereas more than 30% of cells expressing RPA2-WT had clear RPA2 foci within 0.5 h after CPT treatment, cells expressing RPA2 D4 showed a significant (P < 0.0085) delay in RPA2-focus formation during the initial 2-h incubation after CPT (Fig. 4a). This result suggests that hyperphosphorylated RPA2 detected early in the DSB response in PP4R2-silenced cells impairs the rapid recruitment of RPA to DSB-induced foci, thereby reducing the efficiency of HR.
Figure 4.
Premature formation of hyperphosphorylated RPA2 impedes recruitment of RPA and RAD51 to chromatinized DNA damage–induced foci. (a) RPA2 focus formation after CPT or mock (represented as Un) treatment of U2OS cells expressing RPA2 WT or RPA2 D4. Cells were stained for RPA2 and DAPI, and images were captured by fluorescence microscopy. The RPA2 focus–positive cells (>30 foci) were quantified manually by comparison with DAPI images (~300 cells total). Representative images are shown on the left. (b) Interaction of Myc-tagged RPA2 WT, RPA2 D2 or RPA2 D4 with RAD51 after CPT treatment. Cells were subjected to immunoprecipitation after CPT treatment using an anti-Myc and probed for RAD51. (c) Reduced nuclear staining of RAD51 in damaged cells lacking PP4R2 or replaced with RPA2 D4. Following replacement of endogenous RPA2 with phosphomimetic RPA2 mutants or depletion of PP4R2, U2OS cells were mock- or CPT-treated. Cells were then extracted to remove soluble RAD51, stained with DAPI and anti-RAD51, and imaged by epifluorescence microscopy using identical exposure times. RAD51 nuclear staining was quantified and plotted. (d) Nuclear localization of RAD51 is altered by hyperphosphorylated RPA2. RAD51 localization after CPT treatment of U2OS cells, either where endogenous RPA2 was replaced by RPA2 WT or RPA2 D4 (left) or where PP4R2 was silenced (right). Nuclei were biochemically fractionated, and nuclear soluble (NS) and chromatin-bound (Chr) fractions were probed for RAD51. Topoisomerase II (TOPII) and histone H3 (H3) was probed for loading and fractionation controls, respectively. The relative amounts of RAD51 and RPA1 are shown in parentheses.
RAD51 localization is influenced by RPA2 hyperphosphorylation
It is also possible that hyperphosphorylated RPA2 impairs HR by causing defects in the loading of other DSB repair factors. Several DSB-repair factors, including RAD51, preferentially interact with the hyperphosphorylated form of RPA2 after UV or CPT treatment38. In conjunction with a recombination mediator, such as BRCA2 in mammalian cells33, RPA is dislodged concomitant with RAD51 binding to generate a recombinogenic RAD51 ssDNA filament33. We speculated that hyperphosphorylated RPA2 generated in an inappropriate location (nuclear soluble fraction) and time (early in the DSB response) in PP4-deficient cells would sequester RAD51, thereby preventing its recruitment to repair foci. To test this hypothesis, we first confirmed that RAD51 preferentially co-immunoprecipitates with RPA2 D4 (Fig. 4b). We then evaluated the effect of silencing PP4R2 or replacing endogenous RPA2 with either RPA2 WT or RPA2 D4 on formation of detergent-resistant RAD51 nuclear staining. CPT treatment caused a substantial increase in nuclear RAD51 staining only in cells replaced with RPA2 WT. In contrast, only background levels of nuclear RAD51 cells were detected in mock-treated cells or in CPT-treated cells deficient in PP4R2 or replaced with RPA2 D4 (Fig. 4c; representative images are shown in Supplementary Fig. 12). To further establish that the decrease of RAD51 foci in PP4R2-deficient cells and cells expressing RPA2 D4 is due to inappropriate localization of RAD51, we fractionated these cells and evaluated the amount of RAD51 and RPA1 in chromatin and soluble nuclear fractions. To accurately quantify the amount of RAD51 and RPA1 in the immunoblots, we used the Odyssey Infrared Imaging System. After CPT treatment, there was a detectable increase in soluble nuclear RAD51 and RPA1 and a parallel decrease in chromatin-bound RAD51 and RPA1, both in cells expressing RPA2 D4 (Fig. 4d, left) and also in PP4R2-silenced cells (Fig. 4d, right). These results strongly indicate that PP4-mediated dephosphorylation of RPA2 is necessary for efficient recruitment of the RPA complex and factors that associate with hyperphosphorylated RPA2, such as RAD51, to chromatin in response to DNA damage.
DISCUSSION
RPA2 phosphorylation is an integral component of the DSB response. ATR and DNA-PKcs phosphorylate RPA2 in response to variety of DNA-damaging agents, including CPT, hydroxyurea and UV16,22,25. In this study, we identify a heterodimeric phosphatase complex (PP4C–PP4R2) that specifically dephosphorylates RPA2 and regulates its role in the DNA-damage response. Absence of PP4C or PP4R2 results in a detectable increase in hyperphosphorylated RPA2. Specifically, Ser33 of RPA2, which is critical in the cooperative phosphorylation of other RPA2 residues16, had the largest increase. Silencing PP4C does not affect ATR activity3, and inhibiting PP2A-like phosphatases (PP2A, PP4 and PP6) diminishes the enzymatic activity of DNA-PKc2,39. Therefore, the elevated levels of hyperphosphorylated RPA2 induced by CPT or hydroxyurea in PP4-silenced cells is not due to constitutive activation of these kinases.
We show that there is DNA damage–dependent association of PP4C and RPA2 that can be disrupted by a single mutation in the targeting subunit PP4R2. PP4C dephosphorylated phospho-RPA2 in vitro in a dose-dependent manner, and mutations in the ‘active site’ of PP4C abolished this activity. Also, the impact of PP4C–PP4R2 on RPA2 is independent of H2AX. Finally, by expressing specific RPA2 phosphomimetic mutants, we can recapitulate the effect of silencing PP4R2 on the DNA-damage response to a considerable degree, further strengthening the notion of a direct impact of the PP4 complex on RPA2 phosphorylation. Although we cannot formally exclude the possibility that PP4C regulates RPA2 phosphorylation indirectly via some other factors, together these results suggest that PP4C directly dephosphorylates RPA2.
In cells exposed to hydroxyurea for 24 h, PP2A has been recently reported to dephosphorylate RPA2 (ref. 40). Interestingly, hydroxyurea does not directly cause DNA damage but rather impedes DNA synthesis. There are no detectable DNA lesions in cells treated with up to 500 μM hydroxyurea for 4 h41. Treatment for 18 h, however, leads to an inconsistently significant increase in DNA breaks, accompanied with severe cell-cycle abnormalities, cytotoxic effects (reduced population doubling and reduced mitotic index) and increased frequencies of cells with chromosomal aberrations41. Therefore, prolonged exposure to hydroxyurea causes a global stress response in cells, and the physiological relevance of RPA phosphorylation in this scenario remains unclear. Also, the primary observations in this study were derived by inhibiting or silencing the catalytic subunit of PP2A, which has pleiotropic effects on a variety of cellular processes, including DNA replication. Depleting or inhibiting PP2AC impedes the initiation of DNA replication42. Therefore, it remains unclear whether PP2A has any direct role in dephosphorylating RPA2, or the impaired DNA synthesis in PP2AC-silenced cells cause RPA hyperphosphorylation. However, it is feasible that, similar to the PI3-like kinases, multiple phosphatases work in combination to regulate RPA2 phosphorylation.
Why is it important to dephosphorylate RPA2? Consistent with earlier reports24,25, we find that hyperphosphorylated RPA2 impedes DNA replication. Dephosphorylation of RPA2 is therefore necessary for the resumption of post-damage DNA synthesis, and this in turn allows the cell to resume cycling. In addition, premature formation of hyperphosphorylated RPA2 impedes HR-mediated repair of DSBs and enhances sensitivity to DNA-damaging agents. We provide a mechanistic explanation for this observation. For efficient HR-mediated repair, RPA needs to be loaded rapidly on ssDNA generated at the DSB site33. We find that there is delayed formation of chromatinized RPA2 foci in PP4-silenced cells or in cells expressing RPA2 phosphomimic mutants. This is consistent with in vitro studies showing that hyperphosphorylated RPA2 competes with ssDNA to bind the basic DNA binding domain of RPA1, impeding the DNA-binding ability of the RPA complex13,38. Alternatively, increased association of hyperphosphorylated RPA2 with DNA repair factors38 before focus formation may impede the process. Moreover, we find that the hyperphosphorylated RPA2 retained in the soluble nuclear fraction sequesters RAD51, preventing its recruitment to DSB sites and further impairing the DSB repair process. We speculate that, early in the DSB response, the PI3-like kinases phosphorylate all nuclear proteins that have consensus phosphorylation sites and are in the vicinity of the DSB. RPA2 is prematurely phosphorylated in this initial signaling cascade but is immediately dephosphorylated by PP4 to facilitate its role in the DNA-repair process. Future studies better defining the kinetics and biochemistry of PP4-mediated dephosphorylation of RPA2 will elucidate the roles of RPA and PP4 in DNA repair.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Michael and members of the Chowdhury and Borowiec laboratories for useful discussions. This work was supported by the Joint Center for Radiation Therapy and a Barr Award (D.C.), US National Institutes of Health grant GM083185 and an Exceptional Project Award Grant from the Breast Cancer Alliance (J.A.B.).
Appendix
ONLINE METHODS
Cell culture, antibodies and reagents
We grew HeLa S3, U2OS and U2OS–DR-GFP cells in DMEM supplemented with 10% (v/v) FBS. Antibodies used were against RAD51 (Santa Cruz), Chk1 (Cell Signaling), BRCA1 (Calbiochem), CTIP (Santa Cruz), RPA2 (NeoMarker; Cell Signaling), PP4R1 (Bethyl), PP4R2 (Bethyl), PP4R3α (Bethyl), PP4R3β (Bethyl), PP4C (Bethyl), WIP1 (Bethyl), PP2AC (Upstate Biotech), PP6C (Sigma), topoisomerase II (Cell Signaling), origin recognition complex 2 (Abcam), phospho-RPA2 (Ser33) (Bethyl), phospho-RPA2 (Ser4/8) (Bethyl), phospho-RPA2 (Thr21) (Abcam), phospho-RPA2 (Ser29), α-tubulin (Sigma), histone H3 (Cell Signaling), phospho–histone H3 (Upstate) and c-Myc (Santa Cruz). We obtained from Sigma-Aldrich camptothecin (CPT) and hydroxyurea. We purchased okadaic acid from Calbiochem.
Plasmids
We constructed phosphomimetic mutants (RPA2 S33D, RPA2 S8D, RPA2 S33D S8D), PP4C mutant (PP4C D82A) and PP4R2 mutants (PP4R2 F99A, PP4R2 R103A, PP4R2 E106A) by QuikChange II XL site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. We used the following primers: for RPA2 S33D-F, 5′-TGGATCGCCCGCACCTGATCAAGCCGAAAAGAAATCAAG-3′; for RPA2 S33D-R, 5′-CTTGATTTCTTTTCGGCTTGATCAGGTGCGGGCGATCCA-3′; for RPA2 S8D-F, 5′-GTGGAACAGTGGATTCGAAGACTATGGCAGCTCCTCATAC-3′; for RPA2 S8D-R, 5′-GTATGAGGAGCTGCCATAGTCTTCGAATCCACTGTTCCAC-3′; for PP4C D82A-F, 5′-CCTCTTCATGGGGGCCTTTGTGGACCGTG-3′; for PP4C D82A-R, 5′-CACGGTCCACAAAGGCCCCCATGAAGAGG-3′; for PP4R2 F99A-F, 5′-GTCACTGGATTTAATGGTATCCCTGCTACTATTCAGCGACTATGTGAATT-3′; for PP4R2 F99A-R, 5′-AATTCACATAGTCGCTGAATAGTAGCAGGGATACCATTAAATCCAGTGAC-3′; for PP4R2 R103A-F, 5′-AATGGTATCCCTTTTACTATTCAGGCACTATGTGAATTGTTAACAGATCC-3′; for PP4R2 R103A-R, 5′-GGATCTGTTAACAATTCACATAGTGCCTGAATAGTAAAAGGGATACCATT-3′; for PP4R2 E106A-F, 5′-TTACTATTCAGCGACTATGTGCATTGTTAACAGATCCAAGGAG-3′; for PP4R2 E106A-R, 5′-CTCCTTGGATCTGTTAACAATGCACATAGTCGCTGAATAGTAA-3′.
siRNAs and transfection
We transfected siRNA duplexes (Dharmacon and Invitrogen) using Lipofectamine 2000 (Invitrogen). The PP4, PP2AC and PP6C siRNAs were described previously2,3,44; the other siRNAs were as follows: WIP1, siRNA #1 sense 5′-GGGUCUUCCUAGCACAUCAUU-3′, antisense 5′-UGAUGUGCUAGGAAGACCCGU-3′; RPA2, siRNA #1 (3′-UTR target) sense 5′-AACCUAGUUUCACAAUCUGUU-3′, antisense 5′-CAGAUUGUGAAACUAGGUUUU-3′.
To replace endogenous RPA2 with WT or phosphomimetic mutants, we co-transfected RPA2, RPA2 siRNA (3′ UTR) and plasmid using Lipofectamine 2000 (Invitrogen). For HR assay, we transfected U2OS–DR-GFP cells transfected with RPA2 plasmids and used blasticidin S (InvivoGen) selection (5 μg ml−1) with RPA2 siRNA (3′ UTR) to deplete endogenous RPA2.
Protein purification from Sf 9 insect cells and in vitro enzymatic analysis
We purified PP4C and PP4R2 proteins using the Bac-to-Bac Baculovirus Expression System according to the manufacturer’s manual. Briefly, we introduced pFastBac HT-A plasmid containing cDNA of PP4C or PP4R2 into DH10Bac E. coli. We selected and sequenced positive clones. We purified bacmids transposed with pFast-Bac HT-A and introduced them into Sf9 insect cell by transfection using Cellfectin Reagent (Invitrogen). After 72 h post-transfection, we used viral soup containing recombinant baculoviral particles for infection of Sf9 cells (250 ml, 1 × 106 cells per ml). We lysed cells with buffer containing 150 mM NaCl, 50 mM Tris, pH 7.5, 1% (v/v) nonyl phenoxylpolyethoxylethanol (NP-40), 0.1% (w/v) SDS, 20 mM imidazole and protease inhibitor cocktail, and conjugated them with nickel–nitrilotriacetic acid agarose for 2 h. We eluted protein with different amounts of imidazole. We dialyzed indicated fractions in phosphatase reaction buffer. For the dephosphorylation assay, we prepared endogenous phospho-RPA2 by immunoprecipitation with anti-RPA2 from CPT-treated U2OS cells. We performed phosphatase reactions as described3. We resolved reaction mixtures by 12% (v/v) SDS-PAGE and determined relative phosphatase activity by loss of phospho-RPA2 immunoreactivity.
Immunofluorescence
We treated U2OS cells (2 × 105) with 0.5 μM CPT for indicated times. For detection of RPA and RAD51 foci, we first extracted cells with cytoskeletal (CSK) buffer containing 0.5% (v/v) Triton X-100 for 5 min on ice, fixed them with 4% (v/v) paraformaldehyde for 30 min, and permeabilized and immunostained them in 1 × permeabilization/wash (P/W) buffer (BD Biosciences) containing 5% (v/v) donkey serum. Secondary antibodies were Alexa594-conjugated donkey anti–mouse IgG and Alexa488-conjugated donkey anti–rabbit Ig (Molecular Probes). We acquired images with epifluorescent illumination on a Zeiss microscope and analyzed them with the US National Institutes of Health Image J program (http://rsweb.nih.gov/ij/index.html).
Co-immunoprecipitation
We extracted cell lysates from HeLa S3 or U2OS in buffer containing 50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 5 mM EDTA, 0.5% (v/v) NP-40 and protease inhibitor cocktail (Roche). We incubated anti–Flag-agarose (Sigma) or anti–c-Myc with lysate at 4 °C for 16 h. We used protein A/G PLUS agarose (Santa Cruz) to pull down immunocomplexes. We washed precipitates three times with 50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 5 mM EDTA, and 0.5% (v/v) NP-40. We resolved the immunoprecipitated proteins by 4–15% SDS-PAGE and analyzed them by immunoblot.
Chromatin fractionation and western blot by Odyssey Infrared Imaging System
We performed chromatin fractionation as described45. We quantified each fraction for equal loading using NanoDrop 1000 (Thermo Scientific). We performed immunoblotting using the Odyssey Infrared Imaging System. After primary antibody incubation, we incubated blots with goat anti–mouse IRDye 800CW or goat anti–rabbit IRDye 680(LI-COR) for 1 h and scanned them using LI-COR instrument. We quantified images by Odyssey V3.0 software (http://biosupport.licor.com/index.jsp?m=Proteomics&menu=Software&spec=Odyssey_Software).
Radioresistant DNA synthesis
We transfected U2OS cells (0.6 × 105 cells) with siRNA by RNAiMAX (Invitrogen) per manufacturer’s instructions. After 24 h, we incubated cells with 0.025 μCi ml−1 of [14C]thymidine (PerkinElmer) for 24 h, and then treated them with CPT (0.5 μM for 30 min) and washed. To label nascent DNA, we added 0.5 μCi ml−1 of [3H]thymidine (PerkinElmer) for 10 min at indicated times after CPT treatment. We mixed cell lysate with EcoscintH (National Diagnostics) and we measured incorporation of 14C and 3H labels into DNA using a Beckman scintillation counter.
G2-M checkpoint assay
We irradiated (5 Gy) siRNA-transfected U2OS cells and incubated them in medium containing 100 ng ml−1 nocodazole (Sigma) for 24 h. We fixed cells in 4% (v/v) formaldehyde and permeabilized them in cold (−20 °C) 90% (v/v) methanol. We stained ~0.5 × 105 cells with phospho–histone H3 (1:100) for 1 h at 25 °C and then with Alexa488-conjugated donkey anti–rabbit Ig for 1 h at room temperature, followed by propidium iodide (PI/RNase staining buffer, BD Biosciences) staining. We performed flow cytometry and analysis using FloJo (http://www.flowjo.com/home/tutorial.html).
HR Assay
We performed HR assay as described3.
Cell viability assay
We seeded siRNA-U2OS cells (3 × 103 per 200 μl) into octuplicate microtiter wells, incubated them overnight, and then treated them with CPT for 48 h. We measured viability as described3,46.
Single-cell gel electrophoresis (comet) assay
BrdU incorporation assay and cell cycle
We labeled U2OS cells lacking PP4R2 or expressing RPA2 mutants with 10 μM BrdU (6 μl ml−1 of 5 mg ml−1 stock) for 10 min and then fixed them with 70% (v/v) ethanol. After incubation with anti-BrdU (Invitrogen) for 30 min, we added as secondary antibody Alexa Fluor 488–conjugated F(ab’)2 fragment of goat anti–mouse IgG (H+L) (Molecular Probes). We monitored cellular fluorescence with flow cytometry. In a parallel experiment, we stained cells with PI/RNase staining buffer (BD Biosciences) for cell cycle analysis by flow cytometry. We used FloJo software for analysis.
- 44.Pandey AV, Mellon SH, Miller WL. Protein phosphatase 2A and phosphoprotein SET regulate androgen production by P450c17. J. Biol. Chem. 2003;278:2837–2844. doi: 10.1074/jbc.M209527200. [DOI] [PubMed] [Google Scholar]
- 45.Xu X, Stern DF. NFBD1/KIAA0170 is a chromatin-associated protein involved in DNA damage signaling pathways. J. Biol. Chem. 2003;278:8795–8803. doi: 10.1074/jbc.M211392200. [DOI] [PubMed] [Google Scholar]
- 46.Lal A, et al. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 2009;16:492–498. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 2.Chowdhury D, et al. γ-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Chowdhury D, et al. A PP4-phosphatase complex dephosphorylates γ-H2AX generated during DNA replication. Mol. Cell. 2008;31:33–46. doi: 10.1016/j.molcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keogh MC, et al. A phosphatase complex that dephosphorylates γ-H2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 5.Honkanen RE, Golden T. Regulators of serine/threonine protein phosphatases at the dawn of a clinical era? Curr. Med. Chem. 2002;9:2055–2075. doi: 10.2174/0929867023368836. [DOI] [PubMed] [Google Scholar]
- 6.Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell. 2009;33:537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 7.Janssens V, Goris J, Van Hoof C. PP2A: the expected tumor suppressor. Curr. Opin. Genet. Dev. 2005;15:34–41. doi: 10.1016/j.gde.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Janssens V, Longin S, Goris J. PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail) Trends Biochem. Sci. 2008;33:113–121. doi: 10.1016/j.tibs.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Hastie CJ, Carnegie GK, Morrice N, Cohen PT. A novel 50 kDa protein forms complexes with protein phosphatase 4 and is located at centrosomal microtubule organizing centres. Biochem. J. 2000;347:845–855. [PMC free article] [PubMed] [Google Scholar]
- 10.Nakada S, Chen GI, Gingras AC, Durocher D. PP4 is a γ-H2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep. 2008;9:1019–1026. doi: 10.1038/embor.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anantha RW, Borowiec JA. Mitotic crisis: the unmasking of a novel role for RPA. Cell Cycle. 2009;8:357–361. doi: 10.4161/cc.8.3.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell. Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst.) 2004;3:1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 15.Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 16.Anantha RW, Vassin VM, Borowiec JA. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J. Biol. Chem. 2007;282:35910–35923. doi: 10.1074/jbc.M704645200. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, et al. Replication protein A2 phosphorylation after DNA damage by the coordinated action of ataxia telangiectasia-mutated and DNA-dependent protein kinase. Cancer Res. 2001;61:8554–8563. [PubMed] [Google Scholar]
- 18.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19:1162–1174. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia Y, Ongusaha P, Lee SW, Liou YC. Loss of Wip1 sensitizes cells to stress- and DNA damage-induced apoptosis. J. Biol. Chem. 2009;284:17428–17437. doi: 10.1074/jbc.M109.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gingras AC, et al. A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol. Cell. Proteomics. 2005;4:1725–1740. doi: 10.1074/mcp.M500231-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y, Chen Y, Zhang P, Jeffrey PD, Shi Y. Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated τ dephosphorylation. Mol. Cell. 2008;31:873–885. doi: 10.1016/j.molcel.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakasai R, et al. Differential involvement of phosphatidylinositol 3-kinase-related protein kinases in hyperphosphorylation of replication protein A2 in response to replication-mediated DNA double-strand breaks. Genes Cells. 2006;11:237–246. doi: 10.1111/j.1365-2443.2006.00942.x. [DOI] [PubMed] [Google Scholar]
- 23.Cruet-Hennequart S, Glynn MT, Murillo LS, Coyne S, Carty MP. Enhanced DNA-PK-mediated RPA2 hyperphosphorylation in DNA polymerase eta-deficient human cells treated with cisplatin and oxaliplatin. DNA Repair (Amst.) 2008;7:582–596. doi: 10.1016/j.dnarep.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 24.Vassin VM, Wold MS, Borowiec JA. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol. Cell. Biol. 2004;24:1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olson E, Nievera CJ, Klimovich V, Fanning E, Wu X. RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. J. Biol. Chem. 2006;281:39517–39533. doi: 10.1074/jbc.M605121200. [DOI] [PubMed] [Google Scholar]
- 26.Oakley GG, et al. RPA phosphorylation in mitosis alters DNA binding and protein-protein interactions. Biochemistry. 2003;42:3255–3264. doi: 10.1021/bi026377u. [DOI] [PubMed] [Google Scholar]
- 27.Patrick SM, Oakley GG, Dixon K, Turchi JJ. DNA damage induced hyperphosphorylation of replication protein A. 2 Characterization of DNA binding activity, protein interactions, and activity in DNA replication and repair. Biochemistry. 2005;44:8438–8448. doi: 10.1021/bi048057b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zernik-Kobak M, Vasunia K, Connelly M, Anderson CW, Dixon K. Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J. Biol. Chem. 1997;272:23896–23904. doi: 10.1074/jbc.272.38.23896. [DOI] [PubMed] [Google Scholar]
- 29.Shao RG, et al. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA: DNA-PK complexes. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anantha RW, Sokolova E, Borowiec JA. RPA phosphorylation facilitates mitotic exit in response to mitotic DNA damage. Proc. Natl. Acad. Sci. USA. 2008;105:12903–12908. doi: 10.1073/pnas.0803001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim SH, Holway AH, Wolff S, Dillin A, Michael WM. SMK-1/PPH-4.1-mediated silencing of the CHK-1 response to DNA damage in early C. elegans embryos. J. Cell Biol. 2007;179:41–52. doi: 10.1083/jcb.200705182. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 34.Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sleeth KM, et al. RPA mediates recombination repair during replication stress and is displaced from DNA by checkpoint signalling in human cells. J. Mol. Biol. 2007;373:38–47. doi: 10.1016/j.jmb.2007.07.068. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Haber JE. Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol. 2004;2:E21. doi: 10.1371/journal.pbio.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- 38.Wu X, Yang Z, Liu Y, Zou Y. Preferential localization of hyperphosphorylated replication protein A to double-strand break repair and checkpoint complexes upon DNA damage. Biochem. J. 2005;391:473–480. doi: 10.1042/BJ20050379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Douglas P, Moorhead GB, Ye R, Lees-Miller SP. Protein phosphatases regulate DNA-dependent protein kinase activity. J. Biol. Chem. 2001;276:18992–18998. doi: 10.1074/jbc.M011703200. [DOI] [PubMed] [Google Scholar]
- 40.Feng J, et al. PP2A-dependent dephosphorylation of replication protein A (RPA) is required for the repair of DNA breaks induced by replication stress. Mol. Cell. Biol. 2009;29:5696–5709. doi: 10.1128/MCB.00191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Speit G, Schutz P. The effect of inhibited replication on DNA migration in the comet assay in relation to cytotoxicity and clastogenicity. Mutat. Res. 2008;655:22–27. doi: 10.1016/j.mrgentox.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 42.Petersen P, et al. Protein phosphatase 2A antagonizes ATM and ATR in a Cdk2- and Cdc7-independent DNA damage checkpoint. Mol. Cell. Biol. 2006;26:1997–2011. doi: 10.1128/MCB.26.5.1997-2011.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen GI, et al. PP4R4/KIAA1622 forms a novel stable cytosolic complex with phosphoprotein phosphatase 4. J. Biol. Chem. 2008;283:29273–29284. doi: 10.1074/jbc.M803443200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.