Abstract
Chromosome band 9p24 is frequently amplified in primary mediastinal B-cell lymphoma (PMBL) and Hodgkin lymphoma (HL). To identify oncogenes in this amplicon, we screened an RNA interference library targeting amplicon genes and thereby identified JAK2 and the histone demethylase JMJD2C as essential genes in these lymphomas. Inhibition of JAK2 and JMJD2C cooperated in killing these lymphomas by decreasing tyrosine 41 phosphorylation and increasing lysine 9 trimethylation of histone H3, promoting heterochromatin formation. MYC, a major target of JAK2-mediated histone phosphorylation, was silenced following JAK2 and JMJD2C inhibition, with a corresponding increase in repressive chromatin. Hence, JAK2 and JMJD2C cooperatively remodel the PMBL and HL epigenome, offering a mechanistic rationale for the development of JAK2 and JMJD2C inhibitors in these diseases.
Introduction
Primary mediastinal B-cell lymphoma (PMBL), a subtype of diffuse large B-cell lymphoma (DLBCL), shares clinical, biological and genetic features with Hodgkin lymphoma (HL). PMBL and HL usually occur in young patients, with most PMBLs and over half of HLs involving the mediastinum at presentation. Despite profound histological differences, the malignant cells of PMBL and HL share a characteristic molecular signature, as revealed by gene expression profiling (Rosenwald et al., 2003; Savage et al., 2003). In addition, PMBL and HL share oncogenic mechanisms, including activation of the NF-kB pathway (Lam et al., 2005; Rosenwald et al., 2003; Savage et al., 2003). A recurrent genomic copy number gain in these lymphomas involves a region on chromosome band 9p24, which occurs in ~35–45% of PMBL cases (Joos et al., 1996; Lenz et al., 2008; Meier et al., 2009; Rosenwald et al., 2003) and ~33% of HL cases (Joos et al., 2003; Joos et al., 2000). One gene in this interval is JAK2, which encodes a tyrosine kinase that mediates signaling downstream of several cytokine receptors (Bentz et al., 2001; Joos et al., 2003; Joos et al., 2000; Meier et al., 2009; Rosenwald et al., 2003). Recurrent deletion of SOCS1, an inhibitor of JAK signaling, in PMBL and HL supports a pathogenetic role for JAK2 in these lymphomas (Melzner et al., 2005; Mestre et al., 2005; Mottok et al., 2009; Weniger et al., 2006). The cytokine IL-13 has been proposed as an autocrine stimulus to JAK signaling in HL (Skinnider et al., 2002; Skinnider et al., 2001), but the stimulus activating this pathway in PMBL has not been elucidated (Guiter et al., 2004).
JAK kinases phosphorylate STAT transcription factors, causing their relocation to the nucleus where they activate target genes bearing STAT binding motifs (Ghoreschi et al., 2009). An additional role for JAK signaling in reprogramming chromatin has been revealed by genetic studies in Drosophila (Shi et al., 2006; Shi et al., 2008) and by analysis of histone modifications in mammalian cells (Dawson et al., 2009). Signaling by the Drosophila JAK homologue Hopscotch causes a global decrease in histone H3 lysine 9 methylation and heterochromatin formation (Shi et al., 2006). In human leukemia cells, nuclear JAK2 directly phosphorylates the histone H3 tail on tyrosine 41, thereby blocking recruitment of the heterochromatin protein HP1α (Dawson et al., 2009).
The starting point for the present study was the realization that the recurrent 9p24 amplicon in PMBL and HL does not just involve JAK2 but includes several other genes in the vicinity (Rosenwald et al., 2003). The PDCD1LG2 gene in this interval encodes the negative regulator of T cell activation PD-L2, which blocks signaling from the T cell receptor by engaging the receptor PD-1. Inasmuch as PMBL and HL often originate in the thymus amidst a sea of T cells, overexpression of PD-L2 could plausibly contribute to these malignancies by interdicting immune surveillance.
A putative oncogene in this amplicon is JMJD2C, which encodes a demethylase for trimethylated lysine 9 of histone H3 (H3K9me3) as well as trimethylated lysine 36 of histone H3 (Cloos et al., 2006; Loh et al., 2007; Whetstine et al., 2006; Wissmann et al., 2007). JMJD2C is amplified and overexpressed in esophageal squamous carcinoma, breast cancer, metastatic lung sarcomatoid carcinoma and desmoplastic medulloblastomas (Cloos et al., 2006; Ehrbrecht et al., 2006; Italiano et al., 2006; Liu et al., 2009; Yang et al., 2000) and is involved in a rare translocation in mucosa-associated lymphoid tissue lymphoma (Vinatzer et al., 2008), supporting its oncogenic potential. Moreover, knockdown of JMJD2C in breast, prostate and esophageal cancer cell lines suppresses their proliferation (Cloos et al., 2006; Liu et al., 2009; Wissmann et al., 2007). The mechanism by which JMJD2C is oncogenic is unknown, although it could demethylate chromatin surrounding key oncogenes, thereby activating their transcription.
In the present study, we took an unbiased approach using RNA interference genetic screening to discover the functionally critical genes in the 9p24 amplicon in PMBL and HL, and investigated whether amplicon genes cooperate to sustain the proliferation and survival of these lymphomas.
Results
Functional genomics of the 9p24 amplicon
To explore the extent of the chromosome 9p24 amplicon in PMBL and HL, we analyzed array comparative genomic hybridization (aCGH) data from PMBL patient biopsies (n=31), PMBL cell lines (n=2) and HL cell lines (n=3) (Figure 1A). Gain and/or amplification of sequences on chromosome band 9p24 was detected in 45% of PMBL biopsies but less frequently in the ABC DLBCL subtype (11%) and the GCB DLBCL subtype (7%) (Lenz et al., 2008). Within a ~3.5 megabase minimal common region of copy number gain, 10 genes were upregulated in expression in association with this amplicon (Figure 1B). Because we observed no case that only amplified JAK2, we hypothesized that this region might harbor several oncogenes that confer a selective advantage on PMBL and HL cells.
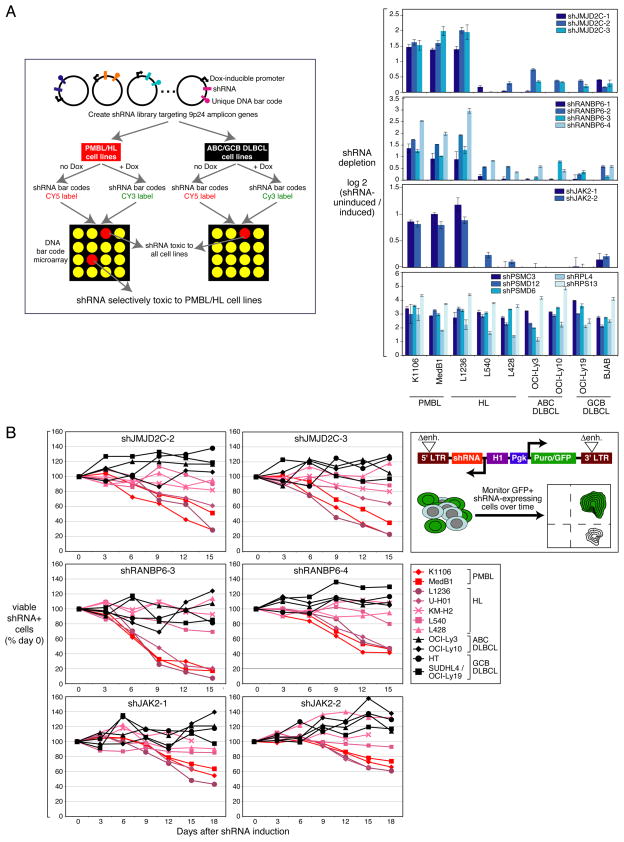
Figure 1. Chromosome 9p24 amplification and an RNAi screen approach.
(A) Array CGH data showing segments of chromosome arm 9p with copy number gains in 18/31 PMBL biopsy samples and in PMBL and HL lines. The box shows the locations of genes in the ~3.5-Mb minimal region of copy number amplification. The 10 genes labeled in red were overexpressed in PMBL cases with the amplicon. (B) Relative expression of 9p24 amplicon genes in cases with and without the amplicon. Shown is the log2-fold overexpression associated with the amplicon along with the t-test p-value for significance. See also Figure S2.
To identify the genes in the 9p24 amplicon that are required for PMBL and HL cell proliferation and survival, we performed a genetic screen utilizing a library of small hairpin RNAs (shRNAs) that mediate RNA interference (Figure 2A). Using a retroviral vector for doxycycline-inducible shRNA expression (Ngo et al., 2006), we created an shRNA library targeting 21 genes in the 9p24 amplicon and, as positive controls, genes encoding proteasome and ribosome subunits, which are essential in all cell types (Figure 1B, See also Table S1).
Figure 2. Identification of shRNAs selectively toxic to PMBL and HL cells.
(A) Left panel: Schematic of the shRNA library screen to identify the genes on chromosome band 9p24 essential for PMBL and HL cell proliferation and/or survival. Each shRNA vector contains a unique barcode sequence that allows its abundance in a population of transduced cells to be measured by a microarray. In cell lines transduced with the library, shRNAs can be induced by doxycycline (dox). The toxic effect of an shRNA is measured by the relative abundance of each bar code in shRNA-induced vs. uninduced cells. See text and methods for details. Right panel: shRNAs targeting JMJD2C, RANBP6 and JAK2 that were selectively toxic to PMBL and HL cells are displayed. Positive control shRNAs are shown in the bottom panel. Data, expressed as log2-fold changes in shRNA barcode depletion from the shRNA-induced culture, represent means ± SD of 4 parallel infections. (B) Validation of shRNA toxicity. Positive shRNAs (Figure 2A) were expressed using a retroviral vector that co-expressed GFP and were screened for toxicity as indicated in the top right panel. Live shRNA-expressing, GFP+ cells were monitored by flow cytometry over time following shRNA induction. Data were normalized based on cells transduced with a negative control shRNA targeting luciferase. See also Figure S1, Table S1.
We screened this library in a pooled fashion, searching for shRNA vectors that decreased tumor cell proliferation and/or survival over 21 days in culture in shRNA-induced cells relative to uninduced cells (Ngo et al., 2006). We tested 2 PMBL and 3 HL lines with the 9p24 amplicon as well as 2 ABC DLBCL and 2 GCB DLBCL lines without the amplicon. As expected, each of the control shRNAs targeting proteasome and ribosome subunits was similarly toxic to all lines (Figure 2A, bottom panel). To identify essential amplicon genes, we focused on shRNAs that were toxic for PMBL and HL lines but not for control DLBCL lines and we required that at least two distinct shRNAs targeting the same gene had the same toxicity spectrum. By these criteria, we identified 3 candidate genes whose knockdown was toxic for PMBL and HL cells: JAK2, JMJD2C and RANBP6, encoding a paralogue of RANBP1 with no known function (Figure 2A). shRNAs targeting these genes were strongly toxic for 2 PMBL lines and 1 HL line with the 9p24 amplicon, but not for 2 other HL lines or for the ABC and GCB DLBCL lines. We confirmed that these shRNAs decreased expression of their targets as expected (Figures S1A, B, C). The specificity of the shRNAs targeting JAK2, JMJD2C or RANBP6 was further demonstrated by the ability of their corresponding cDNAs to rescue PMBL cells from their toxicity (Figure S1D).
JAK2, JMJD2C and RANBP6 were each strong candidate oncogenes since they were included in the minimal region of gain/amplification in PMBL (Figure 1A) and since their mRNA levels were correlated with DNA copy number increases (Figures 1B, See also Figure S2). To validate the RNAi screening results, we cloned shRNAs from the library into a retroviral vector that co-expresses green fluorescent protein (GFP), allowing us to gauge the toxicity of an shRNA by the percentage of GFP+ cells over time (Figure 2B). For JAK2, JMJD2C and RANBP6, two different shRNAs displayed a strong time-dependent toxicity for the two PMBL lines and the L1236 HL line, in accord with the RNAi screening, but had no effect on a variety of ABC and GCB DLBCL lines (Figure 2B). In addition, these shRNAs were toxic for another HL line with the 9p24 amplicon, U-H01 (Mader et al., 2007), but had little if any toxicity to the L540, KM-H2, and L428 HL lines, despite the fact that they also bear this amplicon (Figure 1A) (Joos et al., 2003). In the case of L540 and KM-H2, the ineffectiveness of these individual shRNAs can be traced to functional redundancy of cancer amplicon genes (see below).
Analysis of apoptosis and the cell cycle by flow cytometry revealed that JAK2 knockdown induced apoptosis but did not inhibit proliferation (Figure S1E, F). Conversely, JMJD2C and RANBP6 knockdown caused a 10%–15% increase in G1 phase of the cell cycle and a ~10% decrease in S phase after 6 days but did not induce apoptosis (Figure S1G, H and data not shown). Hence, JMJD2C, RANBP6 and JAK2 are differentially required for the proliferation and survival of PMBL and HL lines with the 9p24 amplicon but are not essential genes in other DLBCL subtypes.
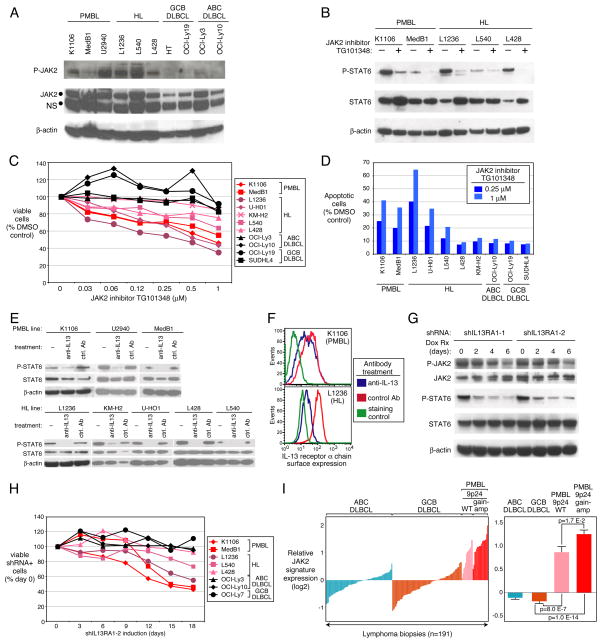
Autocrine activation of JAK2
JAK2 protein was highly expressed in PMBL and HL lines with JAK2 amplification, and JAK2 phosphorylation was detected exclusively in these cells (Figure 3A, Figure S3A). To test the requirement for JAK2 kinase activity, we treated lymphoma lines with a selective JAK2 inhibitor, TG101348 (Geron et al., 2008; Lasho et al., 2008; Wernig et al., 2008). TG101348 reduced STAT6 phosphorylation in PMBL and HL lines (Figure 3B), decreased viable cells in a dose-dependent fashion (Figure 3C) and induced apoptosis in the same PMBL and HL lines that were sensitive to JAK2 knockdown (Figure 3D). Like the JAK2 shRNA, TG101348 did not block cell cycle progression (Figure S1H). Similar effects on cell viability and STAT6 signaling were obtained with another JAK2 inhibitor, AZD1480 (Hedvat et al., 2009) (Figure S3B, C).
Figure 3. Active JAK2-STAT6 required for survival of PMBL and HL cells.
(A) Immunoblotting for total and phosphorylated JAK2 protein in the indicated lines. NS: non-specific band. (B) JAK2 kinase inhibition reduces STAT6 phosphorylation. Cells were treated with 2 μM TG101348 for 2 h. (C) Cell viability by MTT assay after 72 h treatment of the indicated lines with TG101348. (D) Flow cytometric analysis of apoptotic cells. The percentage of activated caspase 3-positive cells was calculated relative to cells treated with the DMSO carrier alone. (E–F) The indicated lines were treated with anti-IL-13 antibody (20 μg/ml) or an isotype control antibody for 18 h and analyzed by immunoblotting for total and phosphorylated STAT6 (E) or by flow cytometry for IL13RA1 expression (F). (G) Immunoblotting for the indicated proteins in K1106 cells expressing IL13RA1 shRNAs. (H) Cell survival assay in the indicated lines expressing the IL13RA1-2 shRNA. (I) Left: Average expression of 55 JAK2-regulated genes that are highly expressed in PMBL (Figure S3G) in primary biopsies of the indicated lymphoma categories. Right: Average JAK2 signature expression within each lymphoma category with p-values indicating significantly higher levels in PMBL than in GCB DLBCL. Error bars depict SEM. See also Figure S3.
Antibody inhibition of IL-13 decreased STAT6 phosphorylation in all 5 HL lines and in all 3 PMBL lines (Figure 3E). Interestingly, anti-IL-13 also reduced the cell surface expression of the IL-13 receptor α chain (IL13Rα) in both cell types as did treatment with the JAK2 inhibitor TG101348 (Figure 3F, See also Figure S3D), suggested that IL-13 secretion initiates a positive feedback loop that enhances IL-13 receptor expression and signaling in PMBL and HL cells. We generated two shRNAs that knocked down IL13Rα expression, reduced downstream signaling in PMBL cells, and were selectively toxic to PMBL and HL cells (Figure 3G, H, See also Figure S3E, F). We conclude that amplification and overexpression of JAK2 cooperates with autocrine IL-13 signaling to promote the survival of PMBL and HL cells.
We next used both the JAK2 shRNA and TG101348 to identify genes regulated by JAK2 signaling in K1106 PMBL cells (Figure S3G). Remarkably, this JAK2-regulated gene signature accounted for roughly one sixth (55/341; 16%) of the genes that were more highly expressed in primary PMBL tumors than in GCB DLBCL tumors (Figure S3G, lanes 1–2), a highly significant overlap (p=2.4 E-36). Most of these genes were more highly expressed in PMBL cases with the 9p24 amplicon than in cases with wild type 9p24 (p=0.017), indicating that this genetic abnormality has a broad influence on the signaling output of the JAK2 pathway (Figure 3I; See also Figure S3G, lanes 3–6). Of note, PMBL cases with wild type 9p24 copy number still had higher expression of these JAK2-regulated genes than did GCB DLBCLs (p=8.0 E-7), indicating that JAK2 signaling imparts a pervasive phenotype in a majority of PMBL tumors that is augmented by the 9p24 amplicon. Moreover, a majority of these JAK2-regulated genes were also more highly expressed in HL lines than in GCB DLBCL lines (Figure S3G, lanes 7–8), demonstrating that JAK2 signaling significantly shapes the biology of HL as well.
Functional cooperation between JAK2 and JMJD2C
Since both JAK2 and JMJD2C can modify the genome epigenetically, we focused our subsequent work on the mechanism by which these two amplicon genes might jointly alter PMBL and HL biology. Our interest in JAK2 and JMJD2C was further stimulated by a tissue microarray analysis that demonstrated high expression of these proteins in 70% and 38% of PMBL biopsies, respectively, but in significantly fewer biopsies of other DLBCL subtypes (Figure S4).
We investigated whether JAK2 and JMJD2C might cooperatively sustain the survival and proliferation of PMBL and HL cells. To test this, we infected a population of cells with vectors expressing an shRNA targeting JMJD2C or a control shRNA together with GFP, and treated the cells with various concentrations of the JAK2 inhibitor TG101348. The equal exposure of both shRNA-transduced and non-transduced cells to the JAK2 inhibitor allowed us to compare the effects of JAK2 inhibition in the two populations and observe a cooperative effect of JAK2 inhibition and JMJD2C knockdown. Knockdown of JMJD2C alone was toxic for the K1106 PMBL line and the UH-01 HL line, but treatment with the JAK2 inhibitor increased the loss of shJMJD2C-transduced cells in a dose-dependent manner. By contrast, expression of a control shRNA did not alter the sensitivity of lymphoma cells to TG101348 (Figure 4A). In these experiments in which JAK2 and JMJD2C were simultaneously inhibited, the effect of JMJD2C knockdown was still restricted to cell cycle blockade while JAK2 inhibition primarily induced apoptosis (Figure S1G, H). Hence, the cooperative toxicity of JAK2 and JMJD2C inactivation stems from their dual inhibition of two primary oncogenic processes, proliferation and survival. Of particular interest were the HL lines L540 and KM-H2, in which JMJD2C knockdown alone was not toxic but did sensitize the cells to the JAK2 inhibitor (Figure 4A). This result suggests that JAK2 signaling and JMJD2C may affect the same regulatory pathway in these cells in a partially redundant fashion. The L428 HL line was not affected by combined inhibition of both of these factors, indicating either that there is further functional redundancy in this cell line for other amplicon genes (e.g. RANBP6) or that other survival pathways that are active in this line play a dominant role, such as NF-kB (Emmerich et al., 1999). The functional cooperation of the JAK2 inhibitor with JMJD2C knockdown was not observed in the control GCB DLBCL line SUDHL4 (Figure 4A).
Figure 4. Cooperative toxicity of JAK2 inhibition and JMJD2C knockdown.
(A) The indicated lines were transduced with vectors expressing a JMJD2C shRNA or a control luciferase shRNA. Cells were treated with a range of concentrations of TG101348 (red lines) or the DMSO carrier alone (blue lines) and simultaneously induced for shRNA expression using doxycycline. As controls (green lines), cells were treated with the DMSO carrier and were not induced for shRNA expression. shRNA-expressing cells were monitored by flow cytometry for a co-expressed GFP marker, as described in Figure 2B. The ratio of live GFP-positive cells to GFP-negative cells for each time point was normalized to the day 0 value. (B) Cooperative effect of shRNAs targeting JAK2 and JMJD2C. See text for details. See also Figure S4.
To confirm the cooperation between JAK2 and JMJD2C, we examined the effect of shRNA-mediated knockdown of these two genes, either alone or in combination (Figure 4B). Cells were first transduced with a vector expressing a JAK2 shRNA or with an empty vector control and selected for stable retroviral integration. These two cell populations were then transduced with vectors expressing GFP together with a JMJD2C shRNA or a control shRNA. We monitored the fraction of GFP+ cells over time following shJMJD2C induction and compared the stable pools expressing the JAK2 shRNA or the control shRNA. As single agents, the JAK2 and JMJD2C shRNAs were toxic for K1106 PMBL and L1236 HL cells but not for control GCB DLBCL cells, as expected (Figure 4B). The toxicity of JMJD2C knockdown was increased with dual knockdown of JAK2, confirming the functional cooperation between these two factors. Also of note, combined knockdown of JAK2 and JMJD2C was toxic to L540 HL cells despite the fact that these cells were not sensitive to knockdown of either gene alone, again suggesting that JAK2 and JMJD2C may redundantly regulate the same pathway in these cells.
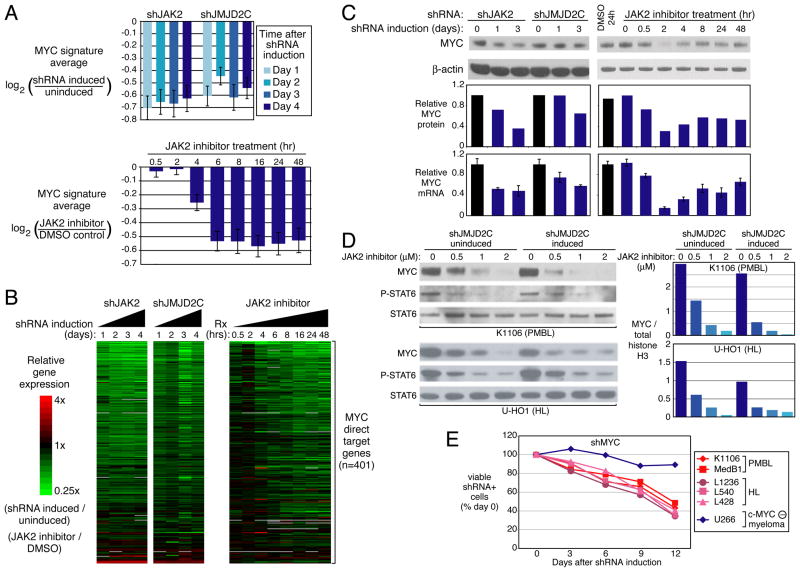
Activation of the MYC transcriptional network by JAK2 and JMJD2C
To investigate the molecular mechanisms of JAK2/JMJD2C cooperation, we profiled gene expression in K1106 PMBL cells following knockdown of these two genes. We identified a set of genes that were downregulated both by JAK2 inhibition and by JMJD2C inhibition, and compared this gene list to a database of previously characterized gene expression signatures (Shaffer et al., 2006). We observed a striking overlap between the genes downmodulated by these treatments and signatures of MYC target genes (Bild et al., 2006; Zeller et al., 2006) (Table S2A). A signature of genes that are activated by MYC overexpression (Bild et al., 2006) was downmodulated in expression when JAK2 or JMJD2C were inhibited (Figure 5A, See also Table S2B), as was a set of genes that MYC directly binds and positively regulates in B cells (Zeller et al., 2006) (Figure 5B, See also Figure S5).
Figure 5. The MYC target gene network depends on JAK2 and JMJD2C in PMBL and HL.
(A) Average expression levels of genes belonging to a set of MYC targets (Bild et al., 2006) following knockdown of JAK2 or JMJD2C in K1106 PMBL cells, or treatment with the JAK2 inhibitor TG101348 (2 μM). Expression is illustrated relative to cells without shRNA induction or relative to DMSO-treated cells, as indicated. Error bars depict S.E.M. (B) Relative expression levels of 401 direct MYC target genes (Zeller et al., 2006) in K1106 cells, as treated in Figure 5A, depicted according to the color scale shown. (C) MYC protein levels by immunoblotting and MYC mRNA levels by QPCR (normalized to beta-2-microglobulin mRNA levels) in K1106 cells after JAK2 or JMJD2C knockdown, or after treatment with 2 μM TG101348. mRNA data represent mean ± SD (n=3). (D) Cooperative downregulation of MYC protein by JAK2 and JMJD2C inhibition. K1106 PMBL or U-H01 HL cells were induced for expression of an shRNA targeting JMJD2C for 2 days, or were uninduced. Cells were treated with the indicated concentrations of TG101348 for 2 h and analyzed by immunoblotting as indicated (left). Densitometric analysis of MYC protein levels relative to total histone H3 levels in the indicated samples (right). (E) Knockdown of MYC is toxic to PMBL and HL lines. The U266 myeloma line, which expresses L-MYC but not c-MYC, served as a negative control. See also Figure S5, Table S2.
In keeping with this observation, MYC mRNA and protein expression levels were reduced after induction of these same shRNAs and after JAK2 inhibition (Figure 5C). Interestingly, MYC downregulation by the JAK2 inhibitor was dynamic, reaching a nadir at 2 h and partially recovering at later time points (Figure 5C), suggesting the possibility of homeostatic regulation of MYC levels under these conditions. Of note, combined blockade of JAK2 and JMJD2C reduced MYC protein levels more than blockade of either regulator alone, in both K1106 PMBL cells and in U-H01 HL cells (Figure 5D).
We next examined the dependence of PMBL and HL lines on MYC using a previously validated shRNA targeting MYC (Shaffer et al., 2008). Knockdown of MYC proved toxic to all lines except for the myeloma U266, which expresses L-myc rather than c-myc (encoded by MYC) (Figure 5E). Expression of the MYC shRNA increased cell apoptosis but had little effect on cell proliferation (Figure S1E, F). The toxic effect of the MYC shRNA could be reversed by ectopic expression of a MYC cDNA (Shaffer et al., 2008) and data not shown). We conclude that MYC and its transcriptional network is an important aspect of JAK2 and JMJD2C regulation that is required for the survival of PMBL and HL cells. However, MYC is not the only important downstream target of JAK2 and JMJD2C in these lymphomas since MYC overexpression did not rescue PMBL cells from the toxic effect of JAK2 or JMJD2C knockdown (data not shown).
Cooperative epigenetic modulation by JAK2 and JMJD2C
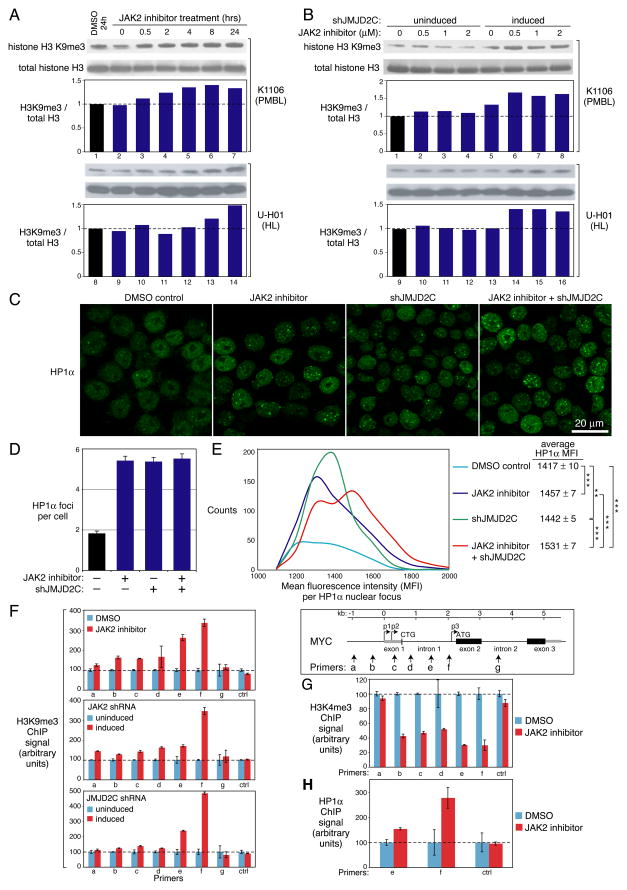
JMJD2C is a demethylase for H3K9me3 (Cloos et al., 2006; Loh et al., 2007; Wissmann et al., 2007), a histone mark that is recognized by the chromo domain of HP1α (Bannister et al., 2001). HP1α uses its chromo shadow domain to bind to a second region on the histone H3 tail surrounding tyrosine 41, and phosphorylation of this residue by nuclear JAK2 prevents this interaction (Dawson et al., 2009). Hence JMJD2C and JAK2 inhibit HP1α recruitment and heterochromatin formation by distinct mechanisms, suggesting the possibility that JAK2 and JMJD2C might collaborate in modifying the epigenome of PMBL and HL cells. Upon treatment of K1106 PMBL or U-H01 cells with the JAK2 inhibitor TG101348, we observed a time-dependent increase in total cellular H3K9me3 levels by immunoblotting, suggesting that JAK2 signaling counteracts heterochromatin formation in these lymphoma cells (Figure 6A).
Figure 6. JAK2 and JMJD2C modulate the epigenome of PMBL and HL.
(A) The K1106 PMBL and U-H01 HL lines were treated with the 2 μM TG101348 for the indicated times before immunobloting for H3K9me3 or total histone H3. Relative protein levels were quantitated by densitometry. (B) K1106 or U-H01 cells transduced with JMJD2C shRNAs (shJMJD2C-3 and shJMJD2C-4) were induced for shRNA expression for 2 days, or left uninduced, before treatment with the indicated concentrations of TG101348 for 2h. H3K9me3 and total histone H3 levels were determined by immunoblotting and quantitated by densitometry. The experiment was repeated 3 times with similar results. (C) Representative images of HP1α staining in K1106 cells that were induced to express a JMJD2C shRNA, treated with TG101348, or both. (D) The number of HP1α foci per nucleus was quantitated under the indicated conditions. Data represent mean ± SEM. (E) Histogram of mean fluorescence intensity data from a confocal imaging analysis of HP1α nuclear foci in K1106 cells treated as indicated. ***: p < 0.01; **: p < 0.05, calculated by a two-tailed t-test. (F–H) ChIP analysis of the MYC locus following JMJD2C knockdown or JAK2 inhibition for H3K9me3 (F), H3K4me3 (G) and HP-1α (H). shRNAs targeting JAK2 or JMJD2C were induced in K1106 cells for 2 days, or cells were treated with 2 μM TG101348 for 2 h. Primers from the MYC locus are shown at the upper right. The location of 3 MYC transcriptional start sites (p1, p2, p3) and 2 translation initiation codons (CTG, ATG) are shown. The median value of the signals was first normalized to the input DNA signal. The signals in the shRNA-induced and TG101348-treated samples were further normalized to those from shRNA-uninduced and DMSO-treated samples, respectively. Error bars depict SEM (n=3). See also Figure S6.
To examine the cooperative effects of JAK2 inhibition and JMJD2C knockdown, we treated K1106 and U-H01 cells with low concentrations of the JAK2 inhibitor for a short period of time (2 hrs), with and without JMJD2C knockdown. At this time point, the JAK2 inhibitor and the JMJD2C shRNA had little impact on their own, but the JAK2 inhibitor clearly increased H3K9me3 levels in cells in which JMJD2C had been silenced, demonstrating their cooperative effect on chromatin structure in these lymphoma cells (Figure 6B).
Since the JAK2 inhibitor TG101348 induces cell apoptosis, we examined whether an increase in H3K9me3 is a general feature of apoptosis. We induced apoptosis in K1106 PMBL cells with the topoisomerase II inhibitor VP16, and chose a dose (1.25 μM) that yielded apoptosis comparable to that achieved with 2 μM TG101348 (Figure S6A). VP16-induced apoptosis was not associated with any increase in H3K9me3 over a 24 hour period. Since knockdown of JMJD2C blocks proliferation, we additionally examined whether cell cycle inhibition generally increased H3K9me3 levels. Treatment of K1106 PMBL cells with a specific CDK inhibitor, PD0332991, caused proliferation arrest but did not increase H3K9 trimethylation (Figure S6B). We conclude that the rise in H3K9me3 associated with JAK2 and JMJD2C inhibition in PMBL and HL cells is not an indirect consequence of either apoptosis or cell cycle blockade.
The influence of JAK2 and JMJD2C on H3K9 methylation prompted us to study whether these factors globally alter heterochromatin content in these lymphomas. HP1α is a marker of heterochromatin that can be quantitatively assessed by immunofluorescence (Dawson et al., 2009). Treatment with the JAK2 inhibitor TG101348 or knockdown of JMJD2C increased the number of HP1α foci per nucleus (Figure 6C, D), and the intensity of the HP1α foci increased under both conditions (Figure 6C, E). When JAK2 and JMJD2C were simultaneously inhibited, the HP1α intensity increased substantially, with a new population of high-intensity HP1α foci clearly indicated by the shoulder on the HP1α intensity histogram (Figure 6E). In cells expressing a control shRNA, TG101348 did not produce this new population of high-intensity HP1α foci (data not shown). We conclude that JAK2 and JMJD2C cooperatively suppress heterochromatin formation in PMBL cells.
The concerted effect of JAK2 and JMJD2C on MYC expression (Figure 5D) raised the possibility that the chromatin structure of the MYC locus might be affected by these regulators. We investigated H3K9me3 at the MYC locus by chromatin immunoprecipitation (ChIP). Several pairs of primers for quantitative PCR (QPCR) were designed to span most MYC regions required for transcriptional regulation (Wierstra and Alves, 2008) (Figure 6F, right panel). The JAK2 inhibitor TG101348 increased H3K9me3 localization to all MYC regions examined except intron 2, a region without major transcriptional regulatory elements (Wierstra and Alves, 2008), and these changes were echoed in cells in which JAK2 was silenced by RNA interference (Figure 6F, upper and middle left panels). The changes in H3K9me3 localization were most pronounced in intron 1, where a minor transcription start site (p3) resides just upstream of the major translation start site of MYC (Wierstra and Alves, 2008) (Figure 6F, right panel). Similar increases in H3K9me3 localization at the MYC locus occurred upon JMJD2C knockdown (Figure 6F, lower left panel). Together, these results suggest that JAK2 and JMJD2C inhibition cause the MYC locus to adopt a repressive heterochromatic structure. In keeping with this model, a marker of active chromatin, histone H3 lysine 4 trimethylation, was diminished at the MYC locus by treatment with the JAK2 inhibitor (Figure 6G). Moreover, JAK2 inhibition increased recruitment of the heterochromatin protein HP1α to the MYC locus, as would be predicted by the increase in H3K9me3, which is bound by HP1α (Figure 6H). Thus, MYC adopts a repressive chromatin structure upon silencing of JAK2 or JMJD2C, in keeping with its decreased expression under these conditions.
Epigenetic modulation by JAK2 phosphorylation of histone H3 tyrosine 41
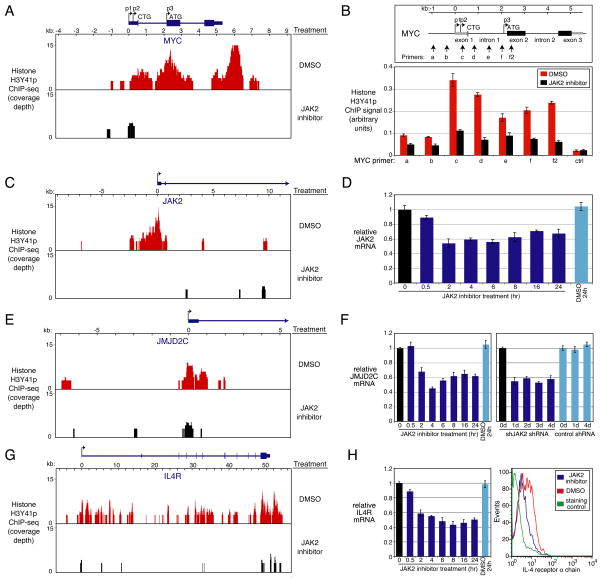
Recent evidence suggests that JAK2 can modify the epigenome in mammalian cells by phosphorylating tyrosine 41 of the histone H3 tail (H3Y41p), thereby diminishing the recruitment of HP-1α (Dawson et al., 2009). We localized H3Y41 phosphorylation across the genome by ChIP followed by high-throughput DNA sequencing (ChIP-Seq), comparing K1106 PMBL cells treated with the JAK2 inhibitor TG101348 with control cells treated with the vehicle DMSO. Overall, we identified 9,087 H3Y41 peaks in the combined data set, 65% of which were in the vicinity of a protein-coding gene either within the body of the gene (72%) or in the promoter region within 2 kilobases of the transcriptional start site (28%). For 2,140 genes, H3Y41p marks were more prominent in the control cells than in cells treated with the JAK2 inhibitor and consequently we will refer to these as JAK2 direct target genes (Table S3). As in leukemias with mutant JAK2 isoforms (Dawson et al., 2009), LMO2 was a JAK2 direct target gene in PMBL (Figure S7A). Among 341 genes that were more highly expressed in PMBL than GCB DLBCL tumors, over one fifth (n=75; 22%) were JAK2 direct target genes, a highly significant overlap (p=4.1E-22; Figure S7B). These genes include PDCD1L2 and CD274, which encode the T cell inhibitory ligands PD-L2 and PD-L1 that are hallmarks of PMBL (Rosenwald et al., 2003). Likewise, among 914 genes that were downregulated upon JAK2 inhibition in PMBL cells, nearly one quarter (n=216; 23.6%) were JAK2 direct target genes, again highly significant (p=2.1E-71; Figure S7C). By contrast, among 416 genes that were upregulated following JAK2 inhibitor treatment (data not shown), fewer than one tenth (n=41; 9.8%) were JAK2 direct target genes, little more than expected by chance. We conclude that JAK2 modifies the chromatin surrounding a substantial subset (~8%) of all protein-coding genes in PMBL cells and that these JAK2 direct targets are enriched for genes that are transcriptionally activated by JAK2 signaling in these lymphomas.
The MYC locus had especially notable H3Y41p peaks that were significantly diminished upon JAK2 inhibitor treatment (p<0.001; Figure 7A). A prominent H3Y41p peak spanning the MYC intron 1 – exon 2 boundary overlapped the region that was modified by H3K9me3 and HP-1α upon JAK2 inhibition (Figure 6F); JAK2-induced phosphorylation of this region was confirmed by QPCR (Figure 7B). These observations support the notion that dysregulated MYC expression in PMBL results from epigenetic changes at the MYC locus initiated by JAK2 phosphorylation of nucleosomes.
Figure 7. JAK2-mediated histone H3Y41 phosphorylation in PMBL and HL.
(A) H3Y41p at the MYC locus. Shown are H3Y41p ChIP-seq histograms in K1106 PMBL cells treated with 2 μM TG101348 for 4h or with DMSO as a negative control. (B) ChIP analysis of H3Y41p at the MYC locus with and without treatment with TG101348 (2 μM) for 4h. QPCR was performed using the indicated primers from MYC and negative control primers from the ubiquitin B promoter. The median value of H3Y41p signals was normalized to the input DNA signal. Error bars denote SEM (n=3). (C) H3Y41p at the JAK2 locus, as in Figure 7A. (D) QPCR analysis of JAK2 mRNA levels in K1106 cells treated with 2 μM TG101348 for the indicated times or with DMSO. Error bars depict SEM (n=3). (E) H3Y41p at the JMJD2C locus, as in Figure 7A. (F) QPCR analysis of JMJD2C mRNA levels. K1106 cells were treated as in Figure 7D (left) or induced to express shJAK2-1 or a negative control shRNA for the indicated days (right). Error bars depict SEM (n=3). (G) H3Y41p at the IL4R locus, as in Figure 7A. (H) QPCR analysis of IL4R mRNA levels in K1106 cells, as treated in Figure 7D (left), and FACS analysis of the IL4Rα on the surface of K1106 cells following treatment with 2 μM TG101348 for 18h or with DMSO (right). Error bars depict SEM (n=3). See also Figure S7, Table S3.
Also notable were H3Y41p peaks at both the JAK2 and JMJD2C loci (Figure 7C, E), which were confirmed by QPCR (Figure S7D). Upon treatment of K1106 PMBL cells with the JAK2 inhibitor TG101348, JAK2 mRNA levels decreased, suggesting that JAK2 signaling creates a feed-forward loop that enhances its own expression (Figure 7D). Similarly, TG101348 treatment or shRNA-mediated knockdown of JAK2 decreased JMJD2C mRNA levels (Figure 7F), revealing another mechanism by which JAK2 and JMJD2C act cooperatively in PMBL.
Another JAK2 direct target gene, IL4R, encodes the IL-4 receptor α chain (IL4Rα), which is an integral component of the IL-13 receptor that increases its affinity for IL-13 by 2–3 orders of magnitude (Andrews et al., 2002; LaPorte et al., 2008) (Figure 7G). H3Y41 phosphorylation of the IL4R locus was confirmed by ChIP (Figure S7D), and JAK2 inhibitor treatment of PMBL cells decreased IL4R mRNA and protein levels (Figure 7H). These data suggest that JAK2-mediated epigenetic modification creates another positive autoregulatory loop that could augment the autocrine IL-13 signaling that is characteristic of PMBL and HL (Figure 3E, F, G, H).
Discussion
Cancer genome copy number changes are opportunistic, preferentially altering chromosomal regions that provide the greatest selective advantage for the malignant clone. This principle is exemplified by a recurrent chromosome amplicon in PMBL and HL that does not focus on a single gene but rather on a several megabase region on chromosome band 9p24. Using a functional genomics screen, we discovered that three amplicon genes – JAK2, JMJD2C, and RANBP6 – are required for the proliferation and survival of lymphoma lines bearing this amplicon. These genes are not essential to human cells in general since lymphoma lines lacking this amplicon were not dependent upon these genes. It thus appears that amplification of this genomic region creates a simultaneous addiction to these three genes. In some lines, inactivation of any one of these genes was toxic. In others, the simultaneous inactivation of JAK2 and JMJD2C was required to efficiently kill the cells. Our results thus demonstrate that a cancer amplicon can harbor more than one “driver” gene, and suggest that functional genomics will be required to gain a full understanding of the multiple addictions created by amplicons. This understanding may in turn lead to the rational combination of therapeutic agents targeting these addictions.
Although JAK2 is amplified in both PMBL and HL, mutations such as those in myeloproliferative disorders have not been found in these lymphoma types (Melzner et al., 2006; Wu et al., 2009). Rather, our data suggest that wild type JAK2 is activated by autocrine IL-13 signaling in these lymphomas and that the 9p24 amplicon increases signal strength through this pathway. STAT6 activation was blocked in all PMBL and HL lines treated with an anti-IL-13 antibody, and IL13Rα knockdown had a similar effect. IL-13 signaling in PMBL and HL cells up-regulated expression of IL13Rα, thereby creating a positive feed-forward loop. Perhaps as a result, expression of IL13RA1 mRNA is a hallmark of PMBL and HL that distinguishes them from other lymphoma types (Rosenwald et al., 2003; Savage et al., 2003). Moreover, IL4R is a direct target of JAK2 histone phosphorylation in PMBL, leading to increased expression of IL4Rα, a subunit of the IL-13 receptor that significantly increases its affinity for IL-13.
Remarkably, one sixth of the genes that are characteristically expressed in PMBL tumors relative to GCB DLBCL tumors were activated by JAK2 signaling in a PMBL line. These JAK2-regulated genes were more highly expressed in PMBL tumors even in the absence of the 9p24 amplicon, suggesting that autocrine IL-13 signaling and JAK2 activation takes place in the absence of JAK2 amplification. However, the 9p24 amplicon further increased expression of these JAK2-regulated genes suggesting that one or more genes within the 9p24 amplicon augment the signaling output of the JAK2 pathway. Thus, JAK2 signaling has a defining influence on the biology of this lymphoma subtype that is aided and abetted by the 9p24 amplicon.
The cooperation between JAK2 and the histone demethylase JMJD2C suggests that JAK2 mediates its oncogenic effect in PMBL and HL by modulating the epigenome. Classically, JAK signaling mediates its biological effects by phosphorylating STAT transcription factors that then transactivate target genes bearing STAT binding motifs (Ghoreschi et al., 2009). This signaling pathway undoubtedly plays a role in modulating the gene expression profile of PMBL and HL cells. However, of the genes that were most downmodulated in expression upon JAK2 inhibition in PMBL (n=1701) and HL (n=1027), only 2.5% contain canonical STAT6 binding sites in their regulatory regions (data not shown). Thus, much of the biology of PMBL and HL cells that is controlled by JAK2 is likely to come from other regulatory mechanisms. Studies in Drosophila (Shi et al., 2006; Shi et al., 2008) and human leukemia (Dawson et al., 2009) have highlighted the ability of JAK signaling to globally decrease heterochromatin formation. In our study, JAK2 cooperated with the histone demethylase JMJD2C in several assays, suggesting that epigenetic modulation by JAK2 is a key aspect of its oncogenic action in lymphomas bearing the 9p24 amplicon. Specifically, inhibition of JAK2 and JMJD2C cooperatively killed PMBL and HL lines, increased genome-wide histone H3K9me3 levels, and promoted heterochromatin formation. Moreover, inhibition of JAK2 and JMJD2C cooperated to repress MYC expression, which was associated with remodeling of the MYC locus by two hallmarks of heterochromatin, H3K9me3 and HP1α recruitment.
Heterochromatin has been conceptually subdivided into stable “constitutive” heterochromatin and dynamic “facultative” heterochromatin (reviewed in (Trojer and Reinberg, 2007)). The local epigenetic modification that we observed at the MYC locus is most reminiscent of the facultative heterochromatin state, such as is mediated by the Rb protein, which represses the S-phase gene cyclin E during G1 phase by recruiting a histone H3K9 methyltransferase, leading to HP1 recruitment (Nielsen et al., 2001). On the other hand, JAK2 and JMJD2C inhibition was associated with a microscopically discernable increase in HP1α-associated nuclear speckles. Previous work has shown that the chromatin in these nuclear domains recruits HP1α by possessing H3K9me3 marks and lacking H3Y41 phosphorylation (Dawson et al., 2009). These nuclear domains may represent the formation of stable foci of constitutive heterochromatin or alternatively may represent the reversible recruitment of genes such as MYC to nuclear regions where gene silencing occurs.
Our working model of the epigenetic cooperation between JAK2 and JMJD2C is shown in Figure 8. Both regulators control recruitment of the heterochromatin protein HP1α to histone tails, but by different mechanisms. HP1α uses its chromo domain to bind histone H3K9me3 (Bannister et al., 2001; Lachner et al., 2001), and demethylation of this residue by JMJD2C removes this HP1α binding site (Cloos et al., 2006; Loh et al., 2007; Whetstine et al., 2006; Wissmann et al., 2007). HP1α uses its chromo shadow domain to bind to a second region of the histone H3 tail centered around tyrosine 41, and phosphorylation of this residue by nuclear JAK2 blocks this binding (Dawson et al., 2009). Because the chromo domain and the chromo shadow domain are linked in the same polypeptide, the simultaneous interaction with these two regions of the histone H3 tail would be expected to cooperatively increase HP1α binding avidity. Of note, HP1α also interacts with SUV39H1 (Yamamoto and Sonoda, 2003) and SETDB1 (Verschure et al., 2005), which are H3K9 methylases. SUV39H1 methyltransferase activity is required for the spreading of heterochromatin and the recruitment of HP1α. On a nucleosome lacking H3K9 methylation and H3Y41 phosphorylation, HP1α might initially bind through its chromo shadow domain to the histone H3 tail near tyrosine 41, thereby recruiting SUV39H1/SETDB1 to methylate lysine 9 and facilitate HP1α binding through its chromo domain. The 9p24 amplicon appears to engage both JAK2 signaling and JMJD2C to decrease HP1α deposition genome-wide, thereby promoting an active chromatin configuration surrounding functionally critical genes, such as MYC. JAK2-mediated H3Y41 phosphorylation sets up several positive feedback loops by targeting JMJD2C and JAK2 itself, as well as IL4R, which encodes IL4Rα, a subunit of the IL-13 receptor.
Figure 8. Model of cooperative modulation of the cancer epigenome by JAK2 and JMJD2C.
Autocrine signaling by IL-13 activates JAK2 kinase in PMBL and HL. Amplification of a genomic region on chromosome band 9p24 increases the abundance of JAK2 and JMJD2C, both of which modify histone H3 tails and impede the recruitment of the heterochromatin protein HP1α. The active chromatin that results facilitates the expression of genes such as MYC, JAK2, JMJD2C, and IL4R. See text for details.
Our findings have several implications for the development of new therapeutic modalities for PMBL and HL. Despite the fact that current chemotherapy regimens for HL are quite effective, they fail to cure roughly 20% of patients with advanced stage HL (Diehl et al., 2003) and 25% of patients with PMBL (Zinzani et al., 2009). Moreover, PMBL and HL tumors in the mediastinum are often irradiated, causing later sequelae such as coronary artery disease. Inhibitors of JAK2 signaling are just entering the clinic and are beginning to show activity in myelofibrosis associated with activating JAK2 mutations (Santos et al., 2010). The JAK2 pathway is an attractive therapeutic target in PMBL and HL based on the genetic and functional evidence in the present study along with previous work implicating SOCS1 inactivation in PMBL and HL (Melzner et al., 2005; Mestre et al., 2005; Mottok et al., 2009; Weniger et al., 2006) and autocrine IL-13 signaling in HL (Skinnider et al., 2002; Skinnider et al., 2001). Together, these considerations support the further development of JAK2 inhibitors as potential therapeutic agents in these lymphomas.
Because of the functional redundancy between JAK2 and JMJD2C in some lymphomas with the 9p24 amplicon, it is likely that successful therapy of some cases might require simultaneous inhibition of both enzymes. For example, some HL lines (KM-H2 and L540) showed little or no response to JAK2 inhibition or JMJD2C inhibition as single interventions, but were killed when JAK2 and JMJD2C were simultaneously inhibited. JMJD2C is a potentially druggable enzyme that is an attractive therapeutic target because of its involvement in PMBL and HL. Moreover, JMJD2C is a potentially interesting target in other cancers such as esophageal carcinoma, which can amplify JMJD2C and depend upon JMJD2C for proliferation (Cloos et al., 2006; Yang et al., 2000), and prostate cancer, which can rely upon JMJD2C for androgen-dependent proliferation (Wissmann et al., 2007). It is important to emphasize that JMJD2C is not required by all cells for proliferation and survival, potentially opening a therapeutic window for cancer treatment. The development of JMJD2C-directed therapeutics may be especially attractive in PMBL and HL as they may have cooperative activity with JAK2-directed agents that are already in clinical trials.
Materials and methods
Array CGH
Array CGH and gene-expression profiling of DLBCL biopsies were described (Lenz et al., 2008). The National Cancer Institute Institutional Review Board has approved the study and written informed consent has been obtained.
Gene expression
Gene expression profiling was performed using Agilent 4X44K whole genome arrays (Shaffer et al., 2008). Quantitative RT-PCR was performed using Applied Biosystems probes.
The JAK2 signaling signature (Figure 3I, S10) was defined as those genes (n=1035) that were decreased in abundance by 0.5 log2 in at least 2/4 time points following knockdown of JAK2 and that were decreased by 0.5 log2 in at least 4/8 time points following treatment with TG101348.
Apoptosis assays and flow cytometric analysis
Apoptosis assays were performed by flow cytometric analysis of caspase 3 activity with a FITC-VAD-FMK kit (Promega). Cells grown in 96-well plates were treated with TG101348 for 72 h. FITC-VAD-FMK was added to the cells (10 μM) for 20 min at 37°C and cells were harvested and washed with PBS before FACS.
ChIP-Seq for H3Y41 phosphorylation
Chromatin immunoprecitipated with anti-H3Y41p antibody was used to construct ChIP-Seq libraries (Illumina), selecting genomic fragments ~350 bp in size. Single-end 36 bp sequence tags were obtained using libraries from K1106 PMBL cells treated with 2μM TG101348 or DMSO for 4h. H3Y41p peaks were defined for the combined data set and peaks with significantly fewer sequence tags from the TG101348-treated data were identified.
See additional methods in the Supplemental Experimental Procedures.
Significance.
We show that the selective advantage of an amplicon for a malignant clone can be due to the cooperative oncogenic effects of multiple genes within the amplicon. Two genes in the 9p24 amplicon of PMBL and HL, JAK2 and JMJD2C, cooperate to modify the epigenome of these lymphomas, thereby promoting proliferation and survival. JAK2 signaling, fostered by an autocrine IL-13 loop and the 9p24 amplicon, appears to be a pervasive feature among PMBL tumors. JAK2 inhibitors emerge from this work as promising therapeutic agents that may have activity against a majority of PMBL and HL tumors. JMJD2C inhibitors should also be developed since they might synergize with JAK2 inhibitors in the treatment of these lymphomas.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and an NCI SPECS grant (UO1-CA 114778). L.R. is the recipient of a CJ Martin Fellowship from the National Health and Medical Research Council (NHMRC) of Australia. G.L. was also supported by a research grant from the German Research Foundation (DFG). C.S. is funded by a fellowship award from the Cancer Research Society of Canada. R.D.G. is funded by a Terry Fox Foundation award (019001) and a Canadian Institutes for Health Research grant (178536). We thank Dennis Huszar of AstraZeneca for providing AZD1480.
Footnotes
Accession number
Array data are at http://www.ncbi.nlm.nih.gov/geo/ (accession # GSE20988).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrews AL, Holloway JW, Puddicombe SM, Holgate ST, Davies DE. Kinetic analysis of the interleukin-13 receptor complex. J Biol Chem. 2002;277:46073–46078. doi: 10.1074/jbc.M209560200. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bentz M, Barth TF, Bruderlein S, Bock D, Schwerer MJ, Baudis M, Joos S, Viardot A, Feller AC, Muller-Hermelink HK, et al. Gain of chromosome arm 9p is characteristic of primary mediastinal B-cell lymphoma (MBL): comprehensive molecular cytogenetic analysis and presentation of a novel MBL cell line. Genes Chromosomes Cancer. 2001;30:393–401. doi: 10.1002/1098-2264(2001)9999:9999<::aid-gcc1105>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl V, Franklin J, Pfreundschuh M, Lathan B, Paulus U, Hasenclever D, Tesch H, Herrmann R, Dorken B, Muller-Hermelink HK, et al. Standard and increased-dose BEACOPP chemotherapy compared with COPP-ABVD for advanced Hodgkin’s disease. N Engl J Med. 2003;348:2386–2395. doi: 10.1056/NEJMoa022473. [DOI] [PubMed] [Google Scholar]
- Ehrbrecht A, Muller U, Wolter M, Hoischen A, Koch A, Radlwimmer B, Actor B, Mincheva A, Pietsch T, Lichter P, et al. Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components. J Pathol. 2006;208:554–563. doi: 10.1002/path.1925. [DOI] [PubMed] [Google Scholar]
- Emmerich F, Meiser M, Hummel M, Demel G, Foss HD, Jundt F, Mathas S, Krappmann D, Scheidereit C, Stein H, Dorken B. Overexpression of I kappa B alpha without inhibition of NF-kappaB activity and mutations in the I kappa B alpha gene in reed-sternberg cells. Blood. 1999;94:3129–3134. [PubMed] [Google Scholar]
- Geron I, Abrahamsson AE, Barroga CF, Kavalerchik E, Gotlib J, Hood JD, Durocher J, Mak CC, Noronha G, Soll RM, et al. Selective inhibition of JAK2-driven erythroid differentiation of polycythemia vera progenitors. Cancer Cell. 2008;13:321–330. doi: 10.1016/j.ccr.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Ghoreschi K, Laurence A, O’Shea JJ. Janus kinases in immune cell signaling. Immunol Rev. 2009;228:273–287. doi: 10.1111/j.1600-065X.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiter C, Dusanter-Fourt I, Copie-Bergman C, Boulland ML, Le Gouvello S, Gaulard P, Leroy K, Castellano F. Constitutive STAT6 activation in primary mediastinal large B-cell lymphoma. Blood. 2004;104:543–549. doi: 10.1182/blood-2003-10-3545. [DOI] [PubMed] [Google Scholar]
- Hedvat M, Huszar D, Herrmann A, Gozgit JM, Schroeder A, Sheehy A, Buettner R, Proia D, Kowolik CM, Xin H, et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16:487–497. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano A, Attias R, Aurias A, Perot G, Burel-Vandenbos F, Otto J, Venissac N, Pedeutour F. Molecular cytogenetic characterization of a metastatic lung sarcomatoid carcinoma: 9p23 neocentromere and 9p23-p24 amplification including JAK2 and JMJD2C. Cancer Genet Cytogenet. 2006;167:122–130. doi: 10.1016/j.cancergencyto.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Joos S, Granzow M, Holtgreve-Grez H, Siebert R, Harder L, Martin-Subero JI, Wolf J, Adamowicz M, Barth TF, Lichter P, Jauch A. Hodgkin’s lymphoma cell lines are characterized by frequent aberrations on chromosomes 2p and 9p including REL and JAK2. Int J Cancer. 2003;103:489–495. doi: 10.1002/ijc.10845. [DOI] [PubMed] [Google Scholar]
- Joos S, Kupper M, Ohl S, von Bonin F, Mechtersheimer G, Bentz M, Marynen P, Moller P, Pfreundschuh M, Trumper L, Lichter P. Genomic imbalances including amplification of the tyrosine kinase gene JAK2 in CD30+ Hodgkin cells. Cancer Res. 2000;60:549–552. [PubMed] [Google Scholar]
- Joos S, Otano-Joos MI, Ziegler S, Bruderlein S, du Manoir S, Bentz M, Moller P, Lichter P. Primary mediastinal (thymic) B-cell lymphoma is characterized by gains of chromosomal material including 9p and amplification of the REL gene. Blood. 1996;87:1571–1578. [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lam LT, Davis RE, Pierce J, Hepperle M, Xu Y, Hottelet M, Nong Y, Wen D, Adams J, Dang L, Staudt LM. Small molecule inhibitors of IkappaB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clin Cancer Res. 2005;11:28–40. [PubMed] [Google Scholar]
- LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, Keegan AD, Garcia KC. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132:259–272. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasho TL, Tefferi A, Hood JD, Verstovsek S, Gilliland DG, Pardanani A. TG101348, a JAK2-selective antagonist, inhibits primary hematopoietic cells derived from myeloproliferative disorder patients with JAK2V617F, MPLW515K or JAK2 exon 12 mutations as well as mutation negative patients. Leukemia. 2008;22:1790–1792. doi: 10.1038/leu.2008.56. [DOI] [PubMed] [Google Scholar]
- Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, Carty S, Lam LT, Shaffer AL, Xiao W, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A. 2008;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Bollig-Fischer A, Kreike B, van de Vijver MJ, Abrams J, Ethier SP, Yang ZQ. Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene. 2009;28:4491–4500. doi: 10.1038/onc.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader A, Bruderlein S, Wegener S, Melzner I, Popov S, Muller-Hermelink HK, Barth TF, Viardot A, Moller P. U-HO1, a new cell line derived from a primary refractory classical Hodgkin lymphoma. Cytogenet Genome Res. 2007;119:204–210. doi: 10.1159/000112062. [DOI] [PubMed] [Google Scholar]
- Meier C, Hoeller S, Bourgau C, Hirschmann P, Schwaller J, Went P, Pileri SA, Reiter A, Dirnhofer S, Tzankov A. Recurrent numerical aberrations of JAK2 and deregulation of the JAK2-STAT cascade in lymphomas. Mod Pathol. 2009;22:476–487. doi: 10.1038/modpathol.2008.207. [DOI] [PubMed] [Google Scholar]
- Melzner I, Bucur AJ, Bruderlein S, Dorsch K, Hasel C, Barth TF, Leithauser F, Moller P. Biallelic mutation of SOCS-1 impairs JAK2 degradation and sustains phospho-JAK2 action in the MedB-1 mediastinal lymphoma line. Blood. 2005;105:2535–2542. doi: 10.1182/blood-2004-09-3701. [DOI] [PubMed] [Google Scholar]
- Melzner I, Weniger MA, Menz CK, Moller P. Absence of the JAK2 V617F activating mutation in classical Hodgkin lymphoma and primary mediastinal B-cell lymphoma. Leukemia. 2006;20:157–158. doi: 10.1038/sj.leu.2404036. [DOI] [PubMed] [Google Scholar]
- Mestre C, Rubio-Moscardo F, Rosenwald A, Climent J, Dyer MJ, Staudt L, Pinkel D, Siebert R, Martinez-Climent JA. Homozygous deletion of SOCS1 in primary mediastinal B-cell lymphoma detected by CGH to BAC microarrays. Leukemia. 2005;19:1082–1084. doi: 10.1038/sj.leu.2403741. [DOI] [PubMed] [Google Scholar]
- Mottok A, Renne C, Seifert M, Oppermann E, Bechstein W, Hansmann ML, Kuppers R, Brauninger A. Inactivating SOCS1 mutations are caused by aberrant somatic hypermutation and restricted to a subset of B-cell lymphoma entities. Blood. 2009;114:4503–4506. doi: 10.1182/blood-2009-06-225839. [DOI] [PubMed] [Google Scholar]
- Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, Staudt LM. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O’Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, Chan WC, Zhao T, Haioun C, Greiner TC, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med. 2003;198:851–862. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos FP, Kantarjian HM, Jain N, Manshouri T, Thomas DA, Garcia-Manero G, Kennedy D, Estrov Z, Cortes J, Verstovsek S. Phase 2 study of CEP-701, an orally available JAK2 inhibitor, in patients with primary or post-polycythemia vera/essential thrombocythemia myelofibrosis. Blood. 2010;115:1131–1136. doi: 10.1182/blood-2009-10-246363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage KJ, Monti S, Kutok JL, Cattoretti G, Neuberg D, De Leval L, Kurtin P, Dal Cin P, Ladd C, Feuerhake F, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102:3871–3879. doi: 10.1182/blood-2003-06-1841. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Wright G, Yang L, Powell J, Ngo V, Lamy L, Lam LT, Davis RE, Staudt LM. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- Shi S, Calhoun HC, Xia F, Li J, Le L, Li WX. JAK signaling globally counteracts heterochromatic gene silencing. Nat Genet. 2006;38:1071–1076. doi: 10.1038/ng1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Larson K, Guo D, Lim SJ, Dutta P, Yan SJ, Li WX. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol. 2008;10:489–496. doi: 10.1038/ncb1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinnider BF, Elia AJ, Gascoyne RD, Patterson B, Trumper L, Kapp U, Mak TW. Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood. 2002;99:618–626. doi: 10.1182/blood.v99.2.618. [DOI] [PubMed] [Google Scholar]
- Skinnider BF, Elia AJ, Gascoyne RD, Trumper LH, von Bonin F, Kapp U, Patterson B, Snow BE, Mak TW. Interleukin 13 and interleukin 13 receptor are frequently expressed by Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood. 2001;97:250–255. doi: 10.1182/blood.v97.1.250. [DOI] [PubMed] [Google Scholar]
- Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Verschure PJ, van der Kraan I, de Leeuw W, van der Vlag J, Carpenter AE, Belmont AS, van Driel R. In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol Cell Biol. 2005;25:4552–4564. doi: 10.1128/MCB.25.11.4552-4564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinatzer U, Gollinger M, Mullauer L, Raderer M, Chott A, Streubel B. Mucosa-associated lymphoid tissue lymphoma: novel translocations including rearrangements of ODZ2, JMJD2C, and CNN3. Clin Cancer Res. 2008;14:6426–6431. doi: 10.1158/1078-0432.CCR-08-0702. [DOI] [PubMed] [Google Scholar]
- Weniger MA, Melzner I, Menz CK, Wegener S, Bucur AJ, Dorsch K, Mattfeldt T, Barth TF, Moller P. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene. 2006;25:2679–2684. doi: 10.1038/sj.onc.1209151. [DOI] [PubMed] [Google Scholar]
- Wernig G, Kharas MG, Okabe R, Moore SA, Leeman DS, Cullen DE, Gozo M, McDowell EP, Levine RL, Doukas J, et al. Efficacy of TG101348, a selective JAK2 inhibitor, in treatment of a murine model of JAK2V617F-induced polycythemia vera. Cancer Cell. 2008;13:311–320. doi: 10.1016/j.ccr.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Wierstra I, Alves J. The c-myc promoter: still MysterY and challenge. Adv Cancer Res. 2008;99:113–333. doi: 10.1016/S0065-230X(07)99004-1. [DOI] [PubMed] [Google Scholar]
- Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- Wu D, Dutra B, Lindeman N, Takahashi H, Takeyama K, Harris NL, Pinkus GS, Longtine J, Shipp M, Kutok JL. No evidence for the JAK2 (V617F) or JAK2 exon 12 mutations in primary mediastinal large B-cell lymphoma. Diagn Mol Pathol. 2009;18:144–149. doi: 10.1097/PDM.0b013e3181855c7f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sonoda M. Self-interaction of heterochromatin protein 1 is required for direct binding to histone methyltransferase, SUV39H1. Biochem Biophys Res Commun. 2003;301:287–292. doi: 10.1016/s0006-291x(02)03021-8. [DOI] [PubMed] [Google Scholar]
- Yang ZQ, Imoto I, Fukuda Y, Pimkhaokham A, Shimada Y, Imamura M, Sugano S, Nakamura Y, Inazawa J. Identification of a novel gene, GASC1, within an amplicon at 9p23-24 frequently detected in esophageal cancer cell lines. Cancer Res. 2000;60:4735–4739. [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzani PL, Stefoni V, Finolezzi E, Brusamolino E, Cabras MG, Chiappella A, Salvi F, Rossi A, Broccoli A, Martelli M. Rituximab combined with MACOP-B or VACOP-B and radiation therapy in primary mediastinal large B-cell lymphoma: a retrospective study. Clin Lymphoma Myeloma. 2009;9:381–385. doi: 10.3816/CLM.2009.n.074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.