Abstract
The inflammasome is a multiprotein complex whose formation is triggered when a NOD-like receptor binds a pathogen ligand, resulting in activated caspase-1, which converts certain interleukins (IL-1β, IL-18, and IL-33) to their active forms. There is currently no information on regulation of this system around the time of birth. We employed transcript profiling of fetal rat intestinal and lung RNA at embryonic days 16 (E16) and 20 (E20) with out-of-sample validation using quantitative RT-PCR. Transcript profiling and quantitative RT-PCR demonstrated that transcripts of core components of the NOD-like receptor Nlrp6 inflammasome (Nlrp6, Pycard, Caspase-1) and one of its substrates, IL-18, were increased at E20 compared with E16 in fetal intestine and not lung. Immunohistochemistry demonstrated increased Pycard in intestinal epithelium. Western blotting demonstrated that IL-18 was undetectable at E16, clearly detectable at E20 in its inactive form, and detectable postnatally in both its inactive and active form. Dramatic upregulation of IL-18 was also observed in the fetal sheep jejunum in late gestation (P = 0.006). Transcription factor binding analysis of the rat array data revealed an overrepresentation of nuclear transcription factor binding sites peroxisome proliferator-activated receptor γ (PPAR-γ) and retinoid X receptor-α and chicken ovalbumin upstream promoter transcription factor 1 in the region 1,000 bp upstream of the transcription start site. Rosiglitazone, a PPAR-γ agonist, more than doubled levels of NLRP6 mRNA in human intestinal epithelial (Caco2) cells. These observations provide the first evidence, to our knowledge, linking activity of PPAR-γ to expression of a NOD-like receptor and adds to a growing body of evidence linking pattern recognition receptors of the innate immune system and intestinal colonization.
Keywords: fetal, pparg, rosiglitazone, microarray, sheep
birth, and the transition from fetal to extrauterine life, involves major adaptation of every organ system of the infant. This is particularly marked in the organs whose primary functions are performed by the placenta in utero, such as the lungs and gastrointestinal tract. As well as taking on new functional roles, both the lungs and intestines are exposed to microorganisms following birth, in contrast to the intrauterine environment, which is sterile. A further complex adaptation of the intestines following birth is to allow colonization by commensal organisms while maintaining defense against pathogens. The intestine has a series of immune defense mechanisms, including the mucosal barrier, passive immunity from the mother, the adaptive immune response, and the innate immune response. The innate immune system consists of multiple elements, including the release of peptides and proteins with antimicrobial activity, such as defensins and cathelicidins (7). It also includes reactive elements, leading to the release of proinflammatory cytokines, which are controlled by the recognition of molecular signatures of microbes by pattern recognition receptors, including Toll-like receptors (TLRs) and NOD-like receptors (NLRs) (25). The TLRs are receptors generally found on the cell membrane and bind a range of microbial ligands (1). Perhaps the best characterized interaction is between lipopolysaccharide (LPS), a component of the cell wall of gram-negative bacteria, and TLR4. Interestingly, TLR4 also recognizes ligands from fungi, parasites, and viruses [see Akira et al. (2) for review]. The NLR family is a collection of pattern recognition receptors that are generally intracellular and consist of four subgroups, in a recently proposed nomenclature (32), on the basis of their amino terminus domain. The current understanding of ligand specificity and postbinding events in all of these have been reviewed elsewhere (4, 9). The Nlrp [Nalp by the old nomenclature (32)] family of proteins has been a focus of particular interest in the last 5 years because they are the key factor regulating assembly of inflammasomes. Inflammasomes are molecular platforms that assemble when a NOD-like receptor protein (Nlrp) binds its ligand. Binding stimulates the formation of Nlrp oligomers, often binding to the caspase-1 adaptor, Pycard (also known as Asc), which then recruits procaspase-1. The end point of the process is generation of activated caspase-1. This in turn can convert inactive precursors of IL-1β, IL-18, and IL-33 to their active forms (26).
We aimed to address a series of hypotheses in relation to preparation of the innate immune system prior to birth. First, we hypothesized that preparation for birth would be associated with upregulation of genes of the innate immune system in both the lungs and the intestines. Second, we hypothesized that the repertoire of innate immune genes upregulated would differ in the two organs, in part because of the specific requirement of the intestines to accommodate commensal colonization. Third, we hypothesized that the coordinated upregulation of these genes would involve “master” regulatory transcription factors. The innate immune system is highly complex and a targeted approach to this hypothesis could easily miss important preparative changes. Transcript profiling by cDNA microarray is a method that allows the expression of thousands of genes to be compared. Therefore, to determine intestinal specific regulation of innate immune genes, we compared the transcript profile in preterm and term rats, in both the lungs and the intestines. Given the possibility for such experiments to yield false positive results, we also performed out-of-sample and out-of-species validation of key findings.
MATERIALS AND METHODS
Animal methods.
All experimental procedures were carried out in accordance with the UK Animals (Scientific Procedures) Act 1986 under an appropriate Home Office License and with approval from the local ethical review process.
Rats.
Twenty time-mated pregnant Wistar rats were euthanized by CO2 overdose at embryonic days 16 (E16; n = 10) and 20 (E20; n = 10), where day 0 was defined as the day of plug and term is 21 days. Pups were delivered by caesarean section or born naturally for postnatal experiments and euthanized by cervical dislocation; tissue including lung and whole intestine was removed under a dissecting microscope. Tissue from multiple siblings was snap frozen in liquid nitrogen prior to storage at −80°C for subsequent RNA or protein analysis. For immunohistochemistry some siblings were fixed by immersion in ice cold neutral buffered formalin for 6 h, then washed twice with PBS, transferred to 70% ethanol and embedded in paraffin wax.
Sheep.
Twenty Welsh Mountain pregnant ewes of known gestational age were kept in individual pens and maintained on 200 g/day concentrates with free access to hay, water, and a saltlick block. Fetuses were delivered for tissue collection by caesarean section under general anesthesia (20 mg/kg sodium pentobarbitone iv) at 100 (n = 5), 114–116 (n = 5), 129–131 (n = 5), and 144–145 (n = 5) days gestation where term is 145 ± 2 days. Immediately following delivery, lambs were euthanized with sodium pentobarbitone (200 mg/kg). During necropsy, samples of fetal jejunum were collected midway between the pyloric sphincter and ileocecal junction. Tissues were frozen in liquid nitrogen and stored at −80°C until analysis.
Microarray analysis.
Transcript profiling using microarray analysis was performed by using lung and intestine RNA from a single animal from each of 10 litters at a given gestational age. Array experiments were performed by the Genomics CoreLab, Cambridge Biomedical Research Centre. Briefly, the total RNA was processed by using Affymetrix one-cycle target labeling protocol (Santa Clara, CA) and hybridized to Affymetrix Rat Genome 230 2.0 GeneChips. Raw data from transcript profiling experiments are available on the Gene Expression Omnibus Database (http://www.ncbi.nlm.nih.gov/geo/), accession ID: GSE16849.
Data were normalized by robust multiarray averaging (RMA) and quantile normalization using LIMMA (27) (http://bioinf.wehi.edu.au/limma/). Normalized transcript abundance data were compared between E16 and E20 by two independent methods: the Cyber-T algorithm (15) and Rank Product Analysis (3). The Cyber-T algorithm is an unpaired t-test, modified by the inclusion of a Bayesian prior based on the variance of other transcripts in the data set. Transcripts that were significantly regulated in both Cyber-T (Bayes P value <0.001, posterior probability of differential expression >0.99) and Rank Product Analysis (P < 0.0001) and showed an absolute fold change of more than five were defined as differentially expressed. Microarray data were annotated by using the NetAffx Analysis Center (Affymetrix) files. To generate the list of up-or downregulated genes only Entrez Genes or UniGene clusters were considered if at least one probe set gave an unambiguous match.
Innate immunity gene lists.
Gene ontology (GO) analysis was used to determine whether genes in a given functionally related group were up- or downregulated. Enrichment of GO terms among the significant genes was studied by using FatiGo, part of Babelomics 4.0 suite (18) (http://www.babelomics.org), a two-tailed Fisher exact test was used with statistical significance set at P < 0.05. However, the GO categories “innate immune response” (GO:0045087) or “inflammatory response” (GO:0006954) are not comprehensive and do not include genes with a clear role in defense responses (e.g., several interleukins and pattern recognition receptors). Therefore, we obtained a curated nonredundant list of 5,070 human and mouse innate immune system genes from the Innate Database [InnateDB, URL: http://www.innatedb.ca (16), accessed on 12/06/09]. The rat orthologs of the innate immune system genes were identified by using Biomart (www.ensembl.org), resulting in a list of 4,185 rat Ensembl genes. The Affymetrix array employed in this study included a total of 11,392 Ensembl genes. Of the 4,185 rat Ensembl genes, 2,709 were represented on the array. The significant reduction in numbers is because many rat Entrez Genes, and all UniGene clusters, have no rat Ensembl genes linked to them.
Mapping of transcription factor binding sites.
The Affymetrix probe sets upregulated in the rat intestine were first converted to Ensembl gene IDs by using Biomart. The Ensembl IDs of the mouse and human homologs were also obtained from Biomart by inputting the rat Ensembl gene IDs. To identify overrepresented transcription factor binding sites (TFBSs) the Ensembl gene IDs for rat, mouse or human were subjected to TFBS enrichment analysis using FactorY (4, 10). A 1,000 bp region upstream of the transcription start site (TSS) was analyzed. Both Jaspar (http://www.genereg.net) and Transfac (http://www.gene-regulation.com/) databases were used for searches. FactorY uses hypergeometric distribution to calculate the P value and a false discovery rate (FDR) of 0.05 was used for multitest-correction. Hierarchal clustering analysis of TFBSs was performed using Partek Genomic Suite 6.5 (Partek). TFBSs that occurred in all three species with p < 0.05 were considered significant.
To locate peroxisome proliferator-activated receptor γ-retinoid X receptor-α (PPARG-RXRA) and chicken ovalbumin upstream promoter transcription factor 1 (coup-TF/NR2F1) binding sites 1,000 bp upstream of the TSS, rat, mouse, and human genes were extracted by use of Biomart (Ensembl 58; rat:RGSC3.4, mouse:NCBIM37, human:GRCh37) and aligned by use of ClustalW2 (http://www.ebi.ac.uk/clustalW). Sequences were scanned for TFBSs using matrices from the Jaspar database for PPARG-RXRA (MA0065) and coup-TF (MA0017) by using a threshold score of 80%.
RNA extraction.
Total RNA was purified from rat and sheep frozen tissue by using TRIzol (Invitrogen, Paisley, UK) and treatment with DNaseI (Promega, Southampton, UK) for 30 min at 37°C. The reaction was stopped by addition of stop solution and heating at 65°C for 10 min. RNA was extracted from Caco2 cells using spin columns with on-column DNaseI digestion according to the manufacturer's instructions (Macherey Nagel, Düren, Germany). RNA integrity was confirmed by using RNA 6000 Nano chips on an Agilent 2100 Bioanalyzer (Agilent, Stockport, UK). Only RNA with an optical density 260/280 >1.8 was used for quantitative RT-PCR (qRT-PCR) and RNA with a RNA integrity number greater than 7 was used for array hybridization.
Sheep cDNA cloning.
The sheep genome has not yet fully been sequenced but some sequence information is available in the NCBI expressed sequence tag (EST) database (13). A BLASTN search of the sheep EST database revealed that, for some genes (caspase-1, IL-18, and sucrase-isomaltase), sequences were already available and these sequences were used to design primers for PCR. In the case of pycard, where no sheep EST was available, the human, mouse, rat, and bovine sequences were aligned and degenerate primers were designed to amplify a partial ovine coding sequence. All primers are listed in Table 1. PCR (caspase-1, IL-18, sucrase-isomaltase, and pycard) was performed in 25 μl containing a final concentration of 1.25 mM MgCl2, 200 μm of each dNTPs, 800 nM of each primer pair, and 0.1 units/μl BioTaq DNA polymerase (Bioline, London, UK) and sheep intestinal cDNA as template. The PCR conditions were initial denaturation at 95°C for 5 min followed by 45 cycles of 95°C for 45 s, 55°C for 45 s, 72°C for 90 s, and a final extension step of 72°C for 10 min. In some cases (caspase-1, IL-18, and sucrase-isomaltase) 5% DMSO was added to reduce secondary structures. PCR products were separated on 1.5% agarose gels and bands of the expected size were excised and purified by using the Qiaquick Gel Extraction Kit (Qiagen, Crawley, UK) then cloned into the pGEM-T-Easy plasmid (Promega). For each gene, at least two clones were sequence verified.
Table 1.
Primers for amplification from sheep cDNA of inflammasome related transcripts
| Target Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Pycard | AARAAGTTCAAGMTGAAGCTGCTG | CCTCAGCTCYGYTCCAGGTC |
| IL-18 | ACCGTTCAGATCACGTTTCC | TTCTGGTTTGAACAGTGACCA |
| Caspase1 | GCCGACAAGGTTCTGAAAGA | CCGAATTCCCTGAGACATGA |
| Sucrase Isomaltase | TATGGATGGTCACAGCTGAA | TCTGCAGCAACAATGAGCTT |
qRT-PCR.
Total rat, sheep, or human RNA was reverse transcribed to cDNA by use of Superscript II (Invitrogen) and used as template for qRT-PCR with an ABI Prism 7900HT system (Applied Biosystems, Warrington, UK). For rat transcripts, qRT-PCR was performed by using cDNA derived from 100 ng of RNA as template with Absolute Fast qPCR mix (Abgene, Epsom, UK) and primers and probes for Nlrp6, Pycard, Caspase-1, IL-18, sucrase-isomaltase, and 18S. These primers and probes were predesigned and preoptimized (Applied Biosystems: Rn00592690_m1, Rn00597229_g1, Rn00562724_m1, Rn00564957_m1 and Rn00824548_m1, respectively). All samples were assayed in triplicate. Expression levels were quantified by the standard curve method with normalization to 18S (32a). Sheep sequences were used to design TaqMan probes and primers by use of Applied Biosystems file builder 3.1 (http://marketing.appliedbiosystems.com/mk/get/FB3_login?isource=fr_WWW_Filebuilder_SfweDnload_Filebuilder). Sheep probe and primer sets are listed in Table 2. For sheep qRT-PCR relative quantification was performed against the geometric mean of four housekeeping genes (18S, GAPDH, cyclophilin A, and β-actin) since there was more interanimal variability in the sheep and this method has been reported to be more accurate than any single reference gene or 18S RNA (33).
Table 2.
Correlating probe and primer sets for quantitative RT-PCR of sheep targets
| Target Gene | Probe | Forward Primer | Reverse Primer |
|---|---|---|---|
| Pycard | CTGTCAGGACCTTCCC | CGGGTCACAGTCGTGGAT | CCGCACTGCCTGGTACTG |
| IL-18 | CTGACTGTTCAGATAATG | CCTGTCTTTGAGGATATGCCTGATT | GGTTACAGCCAGACCTCTAGTGA |
| Caspase1 | CACTTCAGGTTCACAGTCTG | CCCACCTGGCAGGAATACTG | CCACTGCTTGGGATTCTTGTCTAAG |
| Sucrase isomaltase | CAGTGTCAACATAAGGTTCC | CCAGCATTTATGGTTACCCCTGTAT | CGAGCATTAGGGACATAACCTTCT |
| GAPDH | CTCCTGCGACTTCAAC | GCTACACTGAGGACCAGGTT | AGCATCGAAGGTAGAAGAGTGAGT |
| Cyclophilin A | CACCCTGGCACATAAA | GGTTCCTGCTTTCACAGAATAATTCC | GTACCATTATGGCGTGTGAAGTCA |
| β-Actin | CATCACGCCCTGGTGCC | CCGTCTTCCCTTCCATCGT | CCCACGTAGGAGTCCTTCTG |
qRT-PCR was performed on cDNA from Caco2 cells with primers and probe sets to detect NLRP6 and retinol binding protein 2 (RBP2) mRNAs (Applied Biosystems: Hs00373246_m1 and Hs00188160_m1, respectively) and normalized to 18S (Hs99999901_s1) by the delta delta cycle threshold method.
Immunohistochemistry.
Paraffin wax-embedded sections were cut and mounted onto Superfrost Plus slides. Sections were dewaxed and hydrated in graded ethanol prior to antigen retrieval by microwaving in 10 mM citrate buffer (pH 6.0) for 10 min. Endogenous peroxidase activity was inactivated with 3% H2O2 in methanol for 20 min. Sections were blocked with 5% goat serum in Tris-buffered saline. Anti-human PYCARD antibody was added at 1:200 (Alexis, Exeter, UK) or anti-mouse IL-18 receptor-α antibody at 1:200 (R&D Systems, Abingdon, UK), and sections were incubated overnight at 4°C. Immunoreactivity was detected by biotinylated secondary antibodies at 1:500 (DAKO, Ely, UK) and avidin biotin complex (Vector Laboratories, Peterborough, UK), and visualized with diaminobenzidine (Sigma, Bookham, UK). Control sections were incubated with rabbit or goat immunoglobulin fraction (DAKO) in place of the primary antibody. Slides were counterstained with hematoxylin, dehydrated, and mounted with DPX mountant.
Western blotting.
Fetal or neonatal rat frozen tissue was homogenized in PTN50 buffer [50 mM Na3PO4, pH 7.4, 50 mM NaCl, 1% Triton X-100] with protease inhibitors (P8340, Sigma). Protein concentration was determined by use of the BCA protein assay reagent kit (Pierce, Cramlington, UK), and 20 μg of protein (per well) was separated by electrophoresis on 18% SDS polyacrylamide gels to enable separation of the 24-kDa inactive and 18-kDa active forms of IL-18. Proteins were transferred to a polyvinylidene difluoride membrane (Invitrogen), blocked with 5% skimmed milk in PBS-Tween-20 (0.05%) for 1 h, and subsequently probed with an antibody to IL-18 (catalog no. sc-6179, Santa Cruz Biotechnology) at 1:1,000 on a shaker at 4°C overnight. Binding was detected by horseradish peroxidase-conjugated rabbit anti-goat secondary antibody at 1:5,000 (DAKO). ECL substrate (Amersham Biosciences, Little Chalfont, UK) was then used to detect binding. Membranes were stripped with Restore Western blot stripping buffer (Pierce) for 15 min at room temperature. To correct for loading variations the membrane was reprobed with a mouse monoclonal antibody to β-actin (AC-15, Ambion, Warrington, UK) at 1:5,000; the secondary antibody was rabbit anti-mouse at 20,000 (DAKO).
Cell culture.
The human epithelial colorectal adenocarcinoma cell line, Caco2, was obtained from the European Collection of Animal Cell Culture and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, penicillin-streptomycin, glutamate, and nonessential amino acids.
Rosiglitazone (Cayman Chemicals, Tallinn, Estonia) was dissolved in ethanol and diluted before use in media; control was vehicle only. Cells were seeded at 1.2 × 105/cm2 and left overnight to settle before addition of rosiglitazone at 1, 10, or 20 μm for 6 h. Cells were then washed with PBS, trypsinized from the well, and snap frozen before RNA extraction and qRT-PCR. Experiments were performed in triplicate on three occasions (n = 9).
Statistics.
Comparison of observed and expected proportions was performed by χ2 test with Yates correction. The association between gestational age and gene expression level was analyzed by the Pearson correlation coefficient. Means were compared by two-tailed Student's t-test. Statistical significance was assumed at P < 0.05.
RESULTS
Transcript profile.
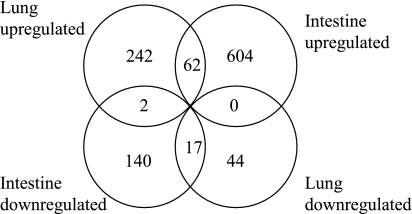
We performed expression array analysis of intestinal samples and lung samples obtained from 10 E16 and 10 E20 rats. A single animal from each litter was used. Rat Affymetrix 230 2.0 arrays have a total of 31,099 probe sets, which map to 28,532 rat genes as defined by Entrez Gene and/or UniGene. In the intestine 769 probe sets (666 genes) were upregulated and 178 probe sets (159 genes) were downregulated greater than fivefold between E16 and E20 embryos (see Supplementary Table S1); 23 genes showed over 100-fold upregulation, including small molecule transporters (Slc2a2, Slc7a9, Slc26a3, Clca3) and lipid binding proteins (Apoa1, Apoa4, Apoc2, Mttp, Fabp2, Rbp2) that play a role in intestinal absorption and transfer. Mucins 3 and 13, as well as a mucin-like protein (Mucdhl), were also upregulated more than 100-fold. In the lung 382 probe sets (306 genes) were upregulated more than fivefold between E16 and E20 and 73 probe sets (61 genes) were downregulated more than fivefold (Supplementary Data S1; the online version of this article contains supplemental data). The largest increase was seen for transcripts encoding surfactant proteins A, B, and D (318-, 132-, and 89-fold, respectively). We then compared gene expression in the lung and the intestine. A Venn diagram representing the intersections between the genes and changes in their expression levels in lung and intestine is shown in Fig. 1.
Fig. 1.
Differentially regulated genes in rat intestine and/or lung. Venn diagram of genes up- or downregulated >5-fold during late gestation in rat fetal intestine and lung.
We assessed the enrichment of GO biological process terms (level 3) of the significantly increased probe sets using FatiGO, Babelomics 4.0. Of the 31,099 probe sets on the array 3,134 had associated GO biological processes. Of the 769 probe sets with significantly increased signal in the rat intestine between E16 and E20, 136 had associated GO biological processes, and of the 382 similarly performing probe sets in the lung, 73 had associated GO biological processes. Overrepresented GO terms in rat intestine and lung are in Supplementary Table S2. In both the intestine and lung two GO terms were significantly overrepresented; “immune response” (P = 0.001 and P = 0.005, respectively) and “antigen processing and presentation” (P < 0.001 and P = 0.041, respectively) and within these categories there were few overlapping genes. In the intestine “cytokine production,” which may also relate to the immune system, was overrepresented (P = 0.019) whereas in the lung “respiratory gaseous exchange” was overrepresented (P = 0.031). Probe sets downregulated in the rat intestine (21 of 178 had associated GO biological processes) showed an overrepresentation of GO terms “developmental maturation” (P = 0.013) and “tissue remodeling” (P = 0.024). The lung had no significantly overrepresented GO terms associated with downregulated probe sets.
GO analysis clearly showed that genes related to the immune system were upregulated in the developing lung and intestine. The immune response, innate immune response, and inflammatory response categories were not well annotated in GO; therefore we assessed whether the lists of up- or downregulated genes that map to Ensembl Gene IDs were enriched for elements of the innate immune system using the InnateDB as our reference dataset (16). We found that of 382 rat Ensembl genes differentially expressed in the intestine, 128 (33.5%) were listed in InnateDB compared with 23.8% (2,709 of 11,392) of all Ensembl genes represented on the array (P < 0.0001). In the lung, 35.1% (66 of 188) of the upregulated rat Ensembl genes were present in InnateDB, which was significantly greater than the proportion in the whole array (P = 0.0003). Table 3 lists genes in the InnateDB list which were upregulated more than fivefold in both lung and intestine. Tables 4 and 5 list genes in the InnateDB list that were specifically upregulated more than 20-fold in the intestines or the lungs, respectively; genes involved in inflammasome formation are in bold.
Table 3.
Genes in the InnateDB significantly upregulated >5-fold in both rat lung and intestine
| Gene Symbol | Gene Title | Intestine FC | Lung FC |
|---|---|---|---|
| Anxa8 | annexin A8 | 7.4 | 66.6 |
| Cd74 | Cd74 molecule, major histocompatibility complex, class II invariant chain | 9.7 | 8.1 |
| Cldn7 | claudin 7 | 25.9 | 5.5 |
| Ctsc | dipeptidyl-peptidase 1 precursor | 14.6 | 7.1 |
| Dpt | dermatopontin precursor | 7.0 | 22.6 |
| Elmo3 | engulfment and cell motility 3 | 21.8 | 5.3 |
| Gpx2 | glutathione peroxidase 2 | 19.5 | 6.0 |
| Hod | HOP homeobox | 7.4 | 12.5 |
| Isg20 | interferon-stimulated exonuclease 20 | 5.4 | 8.5 |
| Itgb6 | integrin β6 | 27.5 | 19.4 |
| LOC691143 | Bq135360 | 23.9 | 37.8 |
| Mgst2 | microsomal glutathione S-transferase 2 | 12.8 | 15.7 |
| Perp | PERP, TP53 apoptosis effector | 16.1 | 11.8 |
| S100 g | S100 calcium binding protein G | 5.7 | 37.6 |
| Tspan8 | tetraspanin 8 | 6.3 | 8.3 |
InnateDB, Innate Database (http://www.innatedb.ca).
Table 4.
Genes in the InnateDB significantly upregulated >20 fold in rat intestine
| Gene Symbol | Gene Title | Intestine FC | Lung FC |
|---|---|---|---|
| Anpep | alanyl (membrane) aminopeptidase | 77.0 | 3.3 |
| Apob | apolipoprotein B (including Ag(x) antigen) | 64.8 | 1.1 |
| Ass | argininosuccinate synthetase 1 | 58.6 | 2.6 |
| Casp1 | caspase-1 | 22.7 | 2.1 |
| Cdh1 | cadherin 1 | 25.3 | 3.7 |
| Cdh17 | cadherin 17 | 62.2 | 1.0 |
| Cx3cl1 | chemokine (C-X3-C motif) ligand 1 | 26.7 | 2.8 |
| Cxcl11 | chemokine (C-X-C motif) ligand 11 | 44.4 | 1.1 |
| Dmbt1 | deleted in malignant brain tumors 1 | 33.8 | −1.0 |
| Elf3 | E74-like factor 3 | 22.8 | 1.8 |
| Entpd2 | ectonucleoside triphosphate diphosphohydrolase 2 | 30.0 | 2.1 |
| Lgals4 | lectin, galactoside-binding, soluble, 4 | 30.2 | −1.2 |
| LOC500292 | cell death-inducing DFFA-like effector c | 46.5 | 1.3 |
| Mamdc4 | MAM domain containing 4 | 56.9 | 1.3 |
| Mucdhl | mucin and cadherin like | 105.7 | 1.2 |
| Nlrp6 | NACHT, LRR and PYD domains-containing protein 6 | 64.3 | −1.3 |
| Nr1 h3 | nuclear receptor subfamily 1, group H, member 3 | 21.9 | 2.5 |
| Pacsin1 | protein kinase C and casein kinase substrate in neurons 1 | 43.1 | −1.0 |
| Pdlim5 | PDZ and LIM domain 5 | 25.0 | 1.4 |
| Plscr1 | phospholipid scramblase 1 | 49.0 | 3.3 |
| Psmb8 | proteasome subunit β type-8 precursor | 38.9 | 2.2 |
| Pycard | PYD and CARD domain containing | 20.9 | 1.4 |
| Q499U7_RAT | RGD1311906 protein | 57.6 | −1.1 |
| Rbp2 | retinol binding protein 2, cellular | 760.7 | −1.0 |
| Rtp4 | receptor transporter protein 4 | 29.3 | 5.5 |
| Slc37a2 | Slc37a2 solute carrier family 37 (glycerol-3-phosphate transporter), member 2 | 20.1 | 1.0 |
| Tacstd1 | tumor-associated calcium signal transducer 1 | 31.4 | 2.8 |
| Tinag | tubulointerstitial nephritis antigen | 42.9 | −1.2 |
Genes involved in inflammasome formation are in bold.
Table 5.
Genes in the InnateDB significantly upregulated >20 fold in rat lung
| Gene Symbol | Gene Title | Intestine FC | Lung FC |
|---|---|---|---|
| Aoc3 | Membrane primary amine oxidase | 4.1 | 24.1 |
| Ces3 | Carboxylesterase 3 precursor | 1.1 | 59.0 |
| Cp | ceruloplasmin | −1.1 | 20.1 |
| Cxcl3 | chemokine (C-X-C motif) ligand 3 | −1.2 | 186.8 |
| Fetub | fetuin B | 1.0 | 21.6 |
| Lamp3 | lysosomal-associated membrane protein 3 | 1.4 | 25.6 |
| LOC691143 | Bq135360 | 23.9 | 37.8 |
| Lyz | lysozyme 2 | 1.6 | 20.9 |
| Muc1 | mucin 1, cell surface associated | 1.0 | 34.4 |
| Scgb1a1 | secretoglobin, family 1A, member 1 (uteroglobin) | 1.1 | 58.9 |
| Scnn1a | sodium channel, nonvoltage-gated, type I, α | 1.2 | 26.9 |
| Sftpb | surfactant protein B | −1.0 | 132.4 |
Validation of changes in inflammasome-related transcripts.
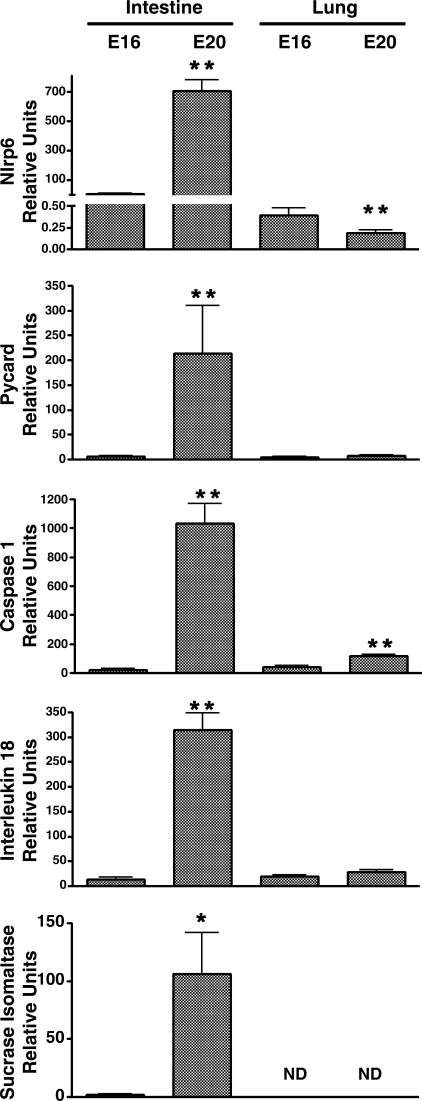
Analysis of the data in Table 4 for functionally related transcripts demonstrated greater than 20-fold upregulation of three transcripts that form the Nlrp6 inflammasome complex (caspase-1, pycard, nlrp6). Moreover, there was a greater than fivefold upregulation of a gene which encodes one of the substrates of the complex IL-18 (Supplementary Table S1). These changes were not observed in the lung. Further analysis therefore focused on this system. Out-of-sample validation was performed by qRT-PCR with intestinal samples obtained from littermates of the animals used for the array analysis. These analyses confirmed the significant upregulation of nlrp6, pycard, caspase-1, and IL-18 mRNAs in the intestinal samples (88-, 216-, 45-, and 22-fold, respectively) (Fig. 2). The magnitude and intestinal specificity of the increase in expression of this system was comparable to that observed for the digestive enzyme sucrase-isomaltase. Nlrp6 and caspase-1 demonstrated small increases in the lung, but the absolute changes were much lower than observed in the intestine (Fig. 2).
Fig. 2.
Quantitative RT-PCR (qRT-PCR) analysis of inflammasome assembly mRNAs in the rat. Nlrp6, Pycard, Caspase-1, and substrate IL-18 were normalized to 18S. Sucrase-isomaltase was used to demonstrate maturation of the intestine and was not detectable (ND) in the lungs. Bars represent means ± SE. Independent t-test P values: *P < 0.02, **P < 0.001 for 16 (E16) vs. 20 (E20) days gestation.
Analysis of transcripts in fetal sheep tissues.
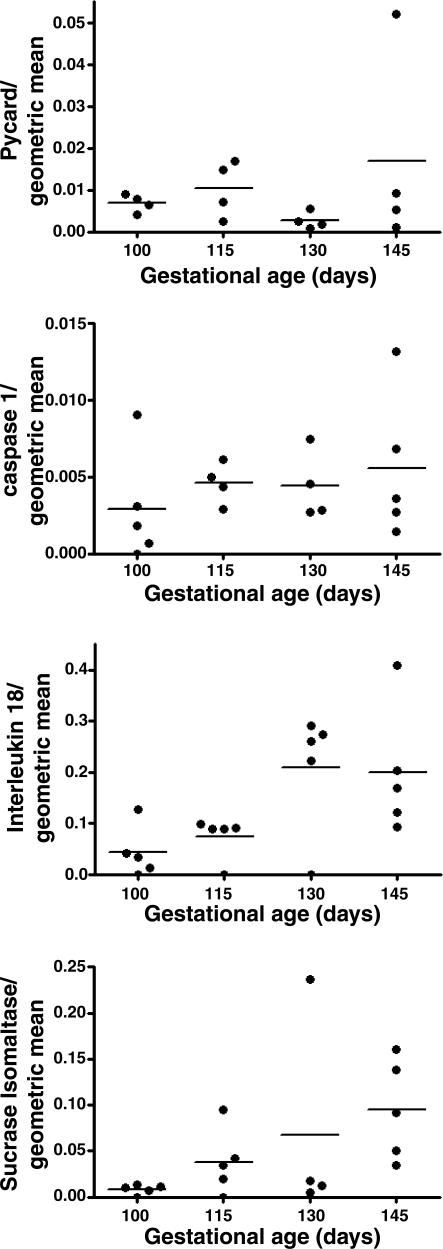
For validation in a second species qRT-PCR was performed by using cDNA from sheep jejunum at various gestational ages (100, 115, 130, and 145 days). There was considerable interanimal variability in the sheep specimens, and no clear pattern between levels of expression of pycard or caspase-1 and gestational age was found (Fig. 3). However, there was a striking increase in the mRNA encoding IL-18 in late gestation (4.6-fold, P = 0.006), the magnitude of which was comparable to that observed for the intestinal digestive enzyme, sucrase-isomaltase (11.6-fold, P = 0.048).
Fig. 3.
qRT-PCR analysis of inflammasome assembly mRNAs of sheep jejunum. Pycard, Caspase-1, and substrate IL-18 and sucrase-isomaltase expression levels are normalized to the geometric mean of 4 housekeeping genes (18S, GAPDH, cyclophilin A, and β-actin). Plots show the mean at each gestational age (line) and individual values. Correlation between gestational age and gene expression for Pycard (P = 0.56), Caspase-1 (P = 0.42), IL-18 (P = 0.006), and Sucrase-isomaltase (P = 0.048) were determined by the Pearson correlation coefficient.
Analysis at the protein level.
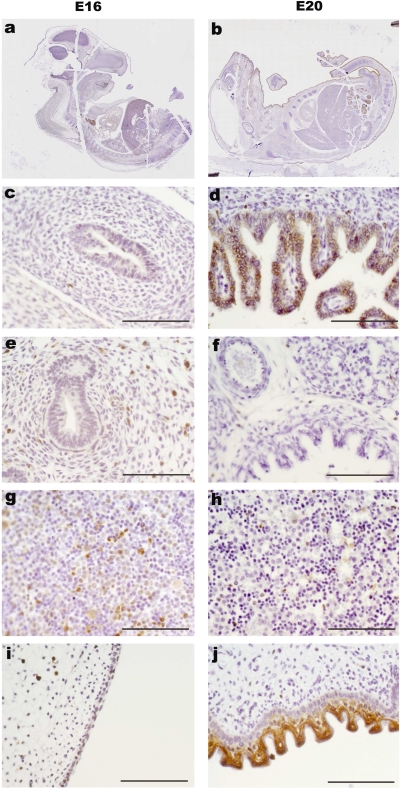
Pycard, the central adaptor protein of the inflammasome, was at the lower level of limit of detection by immunohistochemistry in the intestine of E16 embryos but was clearly detectable in the intestinal epithelium in E20 whole mount embryos. Pycard was also detectable in the liver and skin at E20 (Fig. 4) and remained clearly detectable in the intestinal epithelium from animals at postnatal days 1, 2, and 5 (data not shown). The IL-18 receptor subunit α was not detectable in the intestine of E16 embryos but was expressed on the apical surface of intestinal epithelial cells at E20 and also on cells within the intestinal villi (Fig. 5).
Fig. 4.
Pycard protein localization in fetal rat. Immunohistochemistry with antibody against PYCARD at 16 days gestation (a, c, e, g, i) and 20 days gestation (b, d, f, h, j) of rat embryos. Higher magnification of intestine (c and d), lung (e and f), liver (g and h), and skin (i and j). Scale bar = 100 μm.
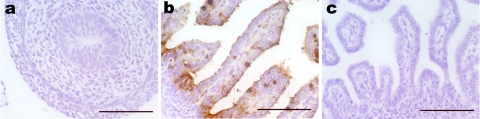
Fig. 5.
IL-18 receptor-α protein expression in rat intestine. Immunohistochemistry of the rat intestine with an antibody against murine IL-18Rα at E16 (a) or E20 (b) and negative control for E20 (c). Scale bar = 100 μm.
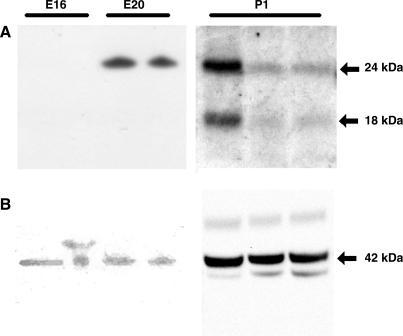
Western blotting using an antibody specific for IL-18 demonstrated no apparent signal at E16 (Fig. 6). A single band with an estimated molecular weight of 24-kDa was observed at E20. Samples from three animals obtained on the first day of life demonstrated bands at both 24-kDa (inactive precursor of IL-18) and 18-kDa (active form).
Fig. 6.
IL-18 protein expression in rat intestine and its activation. Western blot showing interleukin-18 expression at E16, E20, and postnatal day 1 (P1) (A) and β-actin loading control (B).
Mapping of transcription factor binding sites.
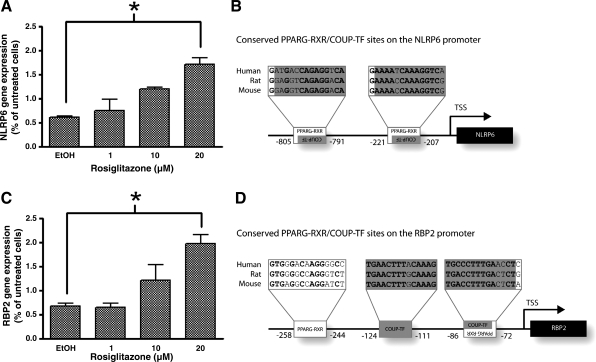
Six Jaspar and seven Transfac motifs were significantly overrepresented in the 1,000 bp upstream of the transcription start sites of genes upregulated in rat fetal intestine between E16 and E20 in all three species (rat, mouse, and human) (Table 6). Hierarchical clustering revealed that the TFBSs PPARG-RXRA and coup-TF commonly occurred together. Nineteen genes upregulated at E20 contained PPARG-RXRA and coup-TF binding sites (Table 7). These genes had various functions including protection against oxidative stress (PON3, GDA), roles in digestion or metabolism (CYP2D6, MEP1A, NAALADL1, RBP2, SLC5A11), and also immunity (CD74, NLRP6). NLRP6 contained both these motifs, is involved in the innate immune response, and was therefore selected for further investigation. Alignment of the promoter regions of human, rat, and mouse Nlrp6 revealed two potential PPARG-RXRA binding sites that were conserved across all three species. Both PPARG-RXRA sites also contained a coup-TF binding site (human nt −221 to −207 and −805 to −791 from TSS) (Fig. 7B). The promoter regions for the positive control gene, retinol binding protein (RBP2), were also aligned and predicted to contain two PPARG-RXRA binding sites (−72 to −86 and −258 to −244) and a further coup-TF binding site (−124 to −111) (Fig. 7D).
Table 6.
TFBSs significantly overrepresented in intestinal upregulated genes of rat, mouse and human determined by Factor Y analysis
| P Value | |||||
|---|---|---|---|---|---|
| Accession Number | Motif Name | Also Known As | Human | Rat | Mouse |
| Jaspar | |||||
| MA0017 | COUP-TF | NR2F1 | 0.0000 | 0.0008 | 0.0000 |
| MA0056 | MZF_1-4 | MZF1_1-4 | 0.0001 | 0.0467 | 0.0132 |
| MA0103 | δEF1 | ZEB1 | 0.0080 | 0.0005 | 0.0081 |
| MA0061 | NF-κB | NF-κB | 0.0126 | 0.0198 | 0.0001 |
| MA0065 | PPARG-RXRA | PPAR-γ-RXRal | 0.0147 | 0.0011 | 0.0084 |
| MA0105 | p50 | NFKB1 | 0.0430 | 0.0269 | 0.0001 |
| Transfac | |||||
| V$HNF4_01_B | F4 | 0.0000 | 0.0000 | 0.0000 | |
| V$HNF4_01 | HNF4 | 0.0000 | 0.0005 | 0.0000 | |
| V$GC_01 | GC | 0.0000 | 0.0006 | 0.0000 | |
| V$COUP_01 | COUP | 0.0000 | 0.0018 | 0.0000 | |
| V$SP1_01 | SP1 | 0.0000 | 0.0158 | 0.0000 | |
| V$HEN1_02 | HEN1 | 0.0089 | 0.0082 | 0.0497 | |
| V$NFKAPPAB_01 | NFKAPPAB | 0.0280 | 0.0448 | 0.0000 | |
Table 7.
Genes significantly upregulated >5 fold in the rat intestine containing PPARG-RXRA and COUP-TF motifs
| Gene Symbol | Gene Title |
|---|---|
| CD74 | CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
| CREB3L3 | cAMP responsive element binding protein 3-like 3 |
| COL17A1 | collagen, type XVII, α1 |
| CYP2D6 | cytochrome P-450, family 2, subfamily D, polypeptide 6 |
| PON3 | paraoxonase 3 |
| FRK | fyn-related kinase |
| MEP1A | meprin 1α |
| RBP2 | retinol binding protein 2, cellular |
| TNNT2 | troponin T2, cardiac |
| GDA | guanine deaminase |
| CCDC93 | coiled-coil domain containing 93 |
| C19orf66 | chromosome 19 open reading frame 66 |
| TAOK3 | TAO kinase 3 |
| SLC5A11 | solute carrier family 5 (sodium/glucose cotransporter), member 11 |
| FAM151A | family with sequence similarity 151, member A |
| TMEM79 | transmembrane protein 79 |
| NAALADL1 | N-acetylated α-linked acidic dipeptidase-like 1 |
| NLRP6 | NLR family, pyrin domain containing 6 |
| F2 | coagulation factor II |
Inflammasome-related-gene NLRP6 is shown in bold.
Fig. 7.
NLRP6 and RBP2 mRNA expression in rosiglitazone treated Caco2 cells. qRT-PCR analysis of NLRP6 (A) and RBP2 (C) mRNA in Caco2 cells treated with rosiglitazone for 6 h as a percentage of untreated cells (normalized to 18S). Asterisks denote significant increase in mRNA after rosiglitazone treatment (P < 0.05). Alignments of NLRP6 (B) and RBP2 (D) regions upstream of transcription start sites (TSSs) showing the locations of PPARG-RXRA and coup-TF binding sites. Nucleotide locations relate to the human sequence.
Regulation of NLRP6 in vitro.
Caco2 cells showed a significant increase in NLRP6 mRNA that was dose dependent in response to the PPAR-γ agonist rosiglitazone (Fig. 7A). RBP2, which is known to be regulated by PPARs in Caco2 cells (29), was also increased significantly (Fig. 7C). Treatment of Caco2 cells with rosiglitazone (20 μm) for 6 h significantly increased NLRP6 and RBP2 mRNA above that of vehicle control (P = 0.023 and 0.025, respectively).
DISCUSSION
The key findings of the present analysis are that we observed substantial upregulation of transcripts encoding elements of the Nlrp6 inflammasome (nlrp6, pycard, and caspase-1) and its substrate, IL-18, in the late-gestation fetal rat intestine. The observed transcript changes in vivo were not due to an influx of immune cells, sine we localized one of the key proteins, Pycard, to the intestinal epithelium and staining with antibodies against lymphocyte-specific markers CD3, CD79a, and CD43 did not show an increase in immune cells in the intestine of 1-day-old rat pups (mouse spleen was used as a positive control and showed specific staining; data not shown). We also used this physiological upregulation of an inflammasome to study the regulation of gene expression, using bioinformatic analysis of TFBSs in the regions upstream of the TSSs of differentially expressed genes. We found that the promoter regions of genes upregulated in the late gestation intestine, including Nlrp6, were more likely to contain binding sites for PPARG-RXRA. Furthermore, we observed upregulation of Nlrp6 when we incubated a human intestinal epithelial cell line, Caco2, with medium containing the PPAR-γ agonist (rosiglitazone).
We observed that IL-18 was absent in the intestines of preterm animals, was expressed wholly in its precursor form in the intestines of fetal rats at term, and was present in both its inactive and activated form in the neonate (Fig. 6). It is plausible that the generation of active IL-18 in the neonatal intestines was due to assembly of the inflammasome, given the changes observed in late gestation. IL-18 activates similar signaling pathways to IL-1 (22). IL-18 binds to its receptor, IL-18Rα, which then recruits IL-18Rβ to form a heterodimer. This activation initiates a signaling cascade involving MyD88, TRAF6, and IRAK (31). The IL-18Rα, which is responsible for binding active IL-18, was present on the intestinal epithelium, suggesting a potential autocrine role for active IL-18 during the perinatal period.
We also determined whether the same changes were evident in late pregnancy in the sheep fetus. This was technically problematic for a number of reasons. First, there is no commercially available microarray for the sheep at present. Hence, we had to use a more targeted approach. Second, sheep are much more expensive than rats and we were limited to five animals in each group. Third, the interanimal variability was much greater, sheep being an outbred species. Finally, since this was a secondary analysis of specimens collected for another study, we had ovine samples from the small intestine only. Hence, we were unable to determine whether any differences between the rat and sheep were explained by the fact that the rat samples included both small and large intestine. We did not observe any change in the key inflammasome-related transcripts, Pycard and Caspase-1, in fetal sheep small intestine in late gestation. This may reflect different biological mechanisms controlling the intestinal innate immune system in rats and sheep, or it could reflect the technical limitations discussed above. However, despite the limitations of these experiments, we observed a dramatic upregulation of IL-18 mRNA in the fetal sheep jejunum in late gestation. Indeed, the magnitude of upregulation of this cytokine message with advancing gestational age was comparable to upregulation of the transcript encoding a key digestive enzyme (sucrase-isomaltase). Hence, these experiments demonstrate that IL-18 mRNA is profoundly upregulated in the intestine in preparation for birth, a phenomenon that is conserved in phylogenetically distant mammalian fetuses.
It has previously been shown that the majority of IL-18 in the adult rat and mouse intestine is produced in epithelial cells rather than immune cells (11). Moreover, active IL-18 has not been observed in the normal adult intestine and exists as the precursor 24-kDa form (19, 24). We observed readily detectable levels of IL-18 in its activated 18-kDa form in the neonatal rat, and this may indicate that there is transient activation of IL-18 in early neonatal life. A subset of lymphocytes, the intraepithelial lymphocytes, express the IL-18 receptor and have been shown to proliferate in response to IL-18 in combination with other interleukins (23). Therefore, the activation of IL-18 observed in the early neonatal period may contribute to the homing of lymphocytes to the intestinal mucosa. There are data that suggest dysregulation of IL-18 may be of importance in intestinal pathology in the neonatal period. Neonatal necrotizing enterocolitis (NEC) is a life-threatening complication associated with preterm birth (5). The risk of the condition increases linearly with the degree of prematurity, indicating that it is likely related to functional immaturity of the intestines. A rat model of NEC (created by enteral feeding with milk substitute, asphyxia, and cold stress) demonstrated elevated expression of IL-18 (12). IL-18 was localized in the enterocytes and the degree of inflammation was correlated with IL-18 gene expression (11). Moreover, when the same procedure was employed in IL-18 null mice, the incidence and severity of NEC was reduced compared with wild-type animals (12). We hypothesize that during the perinatal period activation of the inflammasome, and production of active IL-18, is a normal physiological process and this differs from diseased states such as NEC in which there is excessive IL-18 activation. The precise regulation of the level, location, and timing of IL-18 activation is likely to be complex. Other studies have indicated anti-inflammatory effects of inflammasomes. NLRP3−/− animals had altered intestinal microbiota and were more susceptible to dextran sodium sulfate-induced colitis, implying a role for the inflammasome in maintenance of intestinal homeostasis in physiological and pathological conditions (14). The stimulus that activates Nlrp6 (either pathogen or stress related) is unknown and, although Nlrp6 null mice have been generated, they are not yet fully characterized (21). Given the substantial upregulation of Nlrp6 mRNA observed in the present study, identification of the ligand for Nlrp6 could shed light on adaptation of the intestine to exposure to microbes in the neonatal period.
The PPARG-RXRA binding motif was overrepresented in the promoter regions of genes upregulated in the intestine of E20 rats including NLRP6. In a human intestinal cell line, NLRP6 was also upregulated in vitro by rosiglitazone, a PPAR-γ agonist. This is the first description of a regulatory mechanism for any Nlrp and will provide a manipulatable system to investigate NOD-like receptors further. It is noteworthy that upregulation of NLRP6 by rosiglitazone occurred in undifferentiated Caco2 cells which model the more immature enterocyte and are therefore a better model of the perinatal intestine. PPAR-γ is known to be involved in intestinal homeostasis (8) and administration of rosiglitazone to rodents has been shown to reduce the severity of a model of intestinal colitis (28). These findings suggest that modulators of the PPAR-γ system may have potential as a treatment for neonatal NEC and that further studies in animal models of this condition are warranted.
Array analysis revealed that expression profiles in both the lung and intestine change in late fetal development. GO analysis revealed that both the intestine and lung upregulated transcripts relating to immune response and antigen processing and presentation; upon closer inspection the upregulated genes within these categories differed between the two organs. Our array analysis also revealed that interferon regulatory factor (IRF) mRNAs 1, 6, and 7 were significantly upregulated in the intestine. The IRFs have been identified as components of the signaling cascade associated with detection of viruses by TLRs (30). There is experimental evidence for a role of IRFs in intestinal function. Irf1 knockout mice exhibited an apparently normal intestinal phenotype in steady state but, when challenged with experimental colitis, demonstrated higher levels of inflammation and crypt distortion (17). Previous studies have demonstrated that Irf7 was differentially expressed in the intestines of mice kept in germ-free vs. specific pathogen-free environment (20), suggesting a role for Irf7 in adaptation to intestinal colonization. Collectively, these observations indicate that better understanding of preparation for birth in the fetal intestines may yield insights into the physiological control of colonization by and tolerance of intestinal commensal organisms, as well as the causes and potential treatment of intestinal complications of preterm birth.
In conclusion, we have demonstrated upregulation of IL-18 in the fetal rat and sheep intestine in late gestation that does not occur in the fetal lung. In the rat, there is parallel upregulation of the transcripts encoding three components of the NLRP6 inflammasome, a molecular platform that assembles on binding a microbial ligand and produces activated IL-18 from its inactive precursor. Genes upregulated in the intestine, including Nlrp6, had an overrepresentation of PPAR-γ binding sites in their promoter regions. NLRP6 could also be regulated in Caco2 cells by rosiglitazone. Consistent with a role for this system in the physiological regulation of intestinal colonization following birth, IL-18 was observed in its activated form in neonatal, but not late fetal life.
GRANTS
This work was supported by the Beebe Fund of Cambridge University, the Biotechnology and Biological Sciences Research Council, and the National Institute of Health Research Cambridge Comprehensive Biomedical Research Centre.
Supplementary Material
ACKNOWLEDGMENTS
Present address of R. D. Catalano: MRC Human Reproductive Sciences Unit, The Queen's Medical Research Institute, Edinburgh, EH16 4TJ, United Kingdom.
REFERENCES
- 1. Abreu MT, Fukata M, Arditi M. TLR signaling in the gut in health and disease. J Immunol 174: 4453–4460, 2005 [DOI] [PubMed] [Google Scholar]
- 2. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell 124: 783–801, 2006 [DOI] [PubMed] [Google Scholar]
- 3. Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92, 2004 [DOI] [PubMed] [Google Scholar]
- 4. Chamaillard M, Girardin SE, Viala J, Philpott DJ. Nods, Nalps and Naip: intracellular regulators of bacterial-induced inflammation. Cell Microbiol 5: 581–592, 2003 [DOI] [PubMed] [Google Scholar]
- 5. Chandler JC, Hebra A. Necrotizing enterocolitis in infants with very low birth weight. Semin Pediatr Surg 9: 63–72, 2000 [DOI] [PubMed] [Google Scholar]
- 7. Dann SM, Eckmann L. Innate immune defenses in the intestinal tract. Curr Opin Gastroenterol 23: 115–120, 2007 [DOI] [PubMed] [Google Scholar]
- 8. Dubuquoy L, Rousseaux C, Thuru X, Peyrin-Biroulet L, Romano O, Chavatte P, Chamaillard M, Desreumaux P. PPARgamma as a new therapeutic target in inflammatory bowel diseases. Gut 55: 1341–1349, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol 7: 1250–1257, 2006 [DOI] [PubMed] [Google Scholar]
- 10. Guruceaga E, Segura V, Corrales FJ, Rubio A. FactorY, a bioinformatic resource for genome-wide promoter analysis. Comput Biol Med 39: 385–387, 2009 [DOI] [PubMed] [Google Scholar]
- 11. Halpern MD, Holubec H, Dominguez JA, Williams CS, Meza YG, McWilliam DL, Payne CM, McCuskey RS, Besselsen DG, Dvorak B. Up-regulation of IL-18 and IL-12 in the ileum of neonatal rats with necrotizing enterocolitis. Pediatr Res 51: 733–739, 2002 [DOI] [PubMed] [Google Scholar]
- 12. Halpern MD, Khailova L, Molla-Hosseini D, Arganbright K, Reynolds C, Yajima M, Hoshiba J, Dvorak B. Decreased development of necrotizing enterocolitis in IL-18-deficient mice. Am J Physiol Gastrointest Liver Physiol 294: G20–G26, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hecht J, Kuhl H, Haas SA, Bauer S, Poustka AJ, Lienau J, Schell H, Stiege AC, Seitz V, Reinhardt R, Duda GN, Mundlos S, Robinson PN. Gene identification and analysis of transcripts differentially regulated in fracture healing by EST sequencing in the domestic sheep. BMC Genomics 7: 172, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hirota SA, Ng J, Lueng A, Khajah M, Parhar K, Li Y, Lam V, Potentier MS, Ng K, Bawa M, McCafferty DM, Rioux KP, Ghosh S, Xavier RJ, Colgan SP, Tschopp J, Muruve D, Macdonald JA, Beck PL. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm Bowel Dis 2010. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Long AD, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J Biol Chem 276: 19937–19944, 2001 [DOI] [PubMed] [Google Scholar]
- 16. Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, Gardy JL, Roche FM, Chan TH, Shah N, Lo R, Naseer M, Que J, Yau M, Acab M, Tulpan D, Whiteside MD, Chikatamarla A, Mah B, Munzner T, Hokamp K, Hancock RE, Brinkman FS. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol Syst Biol 4: 218, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mannick EE, Cote RL, Schurr JR, Krowicka HS, Sloop GD, Zapata-Velandia A, Correa H, Ruiz B, Horswell R, Lentz JJ, Byrne P, Gastanaduy MM, Hornick CA, Liu Z. Altered phenotype of dextran sulfate sodium colitis in interferon regulatory factor-1 knock-out mice. J Gastroenterol Hepatol 20: 371–380, 2005 [DOI] [PubMed] [Google Scholar]
- 18. Medina I, Carbonell J, Pulido L, Madeira SC, Goetz S, Conesa A, Tarraga J, Pascual-Montano A, Nogales-Cadenas R, Santoyo J, Garcia F, Marba M, Montaner D, Dopazo J. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res 38 Suppl: W210–W213, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Monteleone G, Trapasso F, Parrello T, Biancone L, Stella A, Iuliano R, Luzza F, Fusco A, Pallone F. Bioactive IL-18 expression is up-regulated in Crohn's disease. J Immunol 163: 143–147, 1999 [PubMed] [Google Scholar]
- 20. Munakata K, Yamamoto M, Anjiki N, Nishiyama M, Imamura S, Iizuka S, Takashima K, Ishige A, Hioki K, Ohnishi Y, Watanabe K. Importance of the interferon-alpha system in murine large intestine indicated by microarray analysis of commensal bacteria-induced immunological changes. BMC Genomics 9: 192, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ, Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 452: 103–107, 2008 [DOI] [PubMed] [Google Scholar]
- 22. Netea MG, Simon A, van de Veerdonk F, Kullberg BJ, Van der Meer JW, Joosten LA. IL-1beta processing in host defense: beyond the inflammasomes. PLoS Pathog 6: e1000661, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Okazawa A, Kanai T, Nakamaru K, Sato T, Inoue N, Ogata H, Iwao Y, Ikeda M, Kawamura T, Makita S, Uraushihara K, Okamoto R, Yamazaki M, Kurimoto M, Ishii H, Watanabe M, Hibi T. Human intestinal epithelial cell-derived interleukin (IL)-18, along with IL-2, IL-7 and IL-15, is a potent synergistic factor for the proliferation of intraepithelial lymphocytes. Clin Exp Immunol 136: 269–276, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pizarro TT, Michie MH, Bentz M, Woraratanadharm J, Smith MF, Jr, Foley E, Moskaluk CA, Bickston SJ, Cominelli F. IL-18, a novel immunoregulatory cytokine, is up-regulated in Crohn's disease: expression and localization in intestinal mucosal cells. J Immunol 162: 6829–6835, 1999 [PubMed] [Google Scholar]
- 25. Sanderson IR, Walker WA. TLRs in the Gut I. The role of TLRs/Nods in intestinal development and homeostasis. Am J Physiol Gastrointest Liver Physiol 292: G6–G10, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schroder K, Tschopp J. The inflammasomes. Cell 140: 821–832, 2010 [DOI] [PubMed] [Google Scholar]
- 27. Smyth G. Limma: linear models for microarray data. In: Bioinformatics and Computational Biology Solutions using R and Bioconductor, edited by Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W. New York: Springer, 2005, p. 397–420 [Google Scholar]
- 28. Su CG, Wen X, Bailey ST, Jiang W, Rangwala SM, Keilbaugh SA, Flanigan A, Murthy S, Lazar MA, Wu GD. A novel therapy for colitis utilizing PPAR-gamma ligands to inhibit the epithelial inflammatory response. J Clin Invest 104: 383–389, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suruga K, Mochizuki K, Suzuki R, Goda T, Takase S. Regulation of cellular retinol-binding protein type II gene expression by arachidonic acid analogue and 9-cis retinoic acid in caco-2 cells. Eur J Biochem 262: 70–78, 1999 [DOI] [PubMed] [Google Scholar]
- 30. Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol 26: 535–584, 2008 [DOI] [PubMed] [Google Scholar]
- 31. Thomassen E, Bird TA, Renshaw BR, Kennedy MK, Sims JE. Binding of interleukin-18 to the interleukin-1 receptor homologous receptor IL-1Rrp1 leads to activation of signaling pathways similar to those used by interleukin-1. J Interferon Cytokine Res 18: 1077–1088, 1998 [DOI] [PubMed] [Google Scholar]
- 32. Ting JP, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, Flavell RA, Girardin SE, Godzik A, Harton JA, Hoffman HM, Hugot JP, Inohara N, Mackenzie A, Maltais LJ, Nunez G, Ogura Y, Otten LA, Philpott D, Reed JC, Reith W, Schreiber S, Steimle V, Ward PA. The NLR gene family: a standard nomenclature. Immunity 28: 285–287, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a. User Bulletin #2 ABIPRISM 7700 Sequence Detection System. http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_040980.pdf. [11/06/2009.]
- 33. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.