Abstract
Processing bodies (P-bodies) contain RNA-protein complexes linked to cytoplasmic RNA decay pathways including mRNA decapping, nonsense-mediated decay and small RNA-mediated decay. Plants deficient in P-body components display severe developmental perturbations, suggesting that these cytoplasmic bodies play important roles in regulating gene expression during plant development. Here, we summarize recent progress in the genetic dissection of P-body components and their roles in translational repression and mRNA decapping.
Introduction
Development is a dynamic process resulting from spatiotemporally controlled gene expression. An increasing body of evidence suggests that post-transcriptional regulation plays important roles in regulating gene expression during development.
The unstable and mobile nature of RNA facilitates both sequence-dependent and independent regulation of gene expression. Cytoplasmic RNA decay serves as the final step to regulate gene expression post-transcriptionally. The 5′ to 3′ degradation of RNAs is a major pathway for cytoplasmic RNA decay, which involves multiple steps including deadenylation, decapping and exonuclease digestion. Among those steps, decapping, the removal of 5′ m7GDP is a rate-limiting step which takes place in P-bodies [1]. Other RNA decay pathways including the 3′ to 5′ decay have been reviewed elsewhere [2–4]. Here, we will focus on recent research relating to P-bodies and their role in plant development.
P-bodies which appear as cytoplasmic foci in eukaryotes cells [5,6] contain RNA-protein particles (RNPs) assembled from RNA and protein components [7]. The evolutionary conservation of P-body proteins in eukaryotes allowed us to identify several Arabidopsis thaliana P-body components by sequence-based genomic surveys.
Protein components of plant P-body
Table 1 shows known and putative P-body components in Arabidopsis homologous to characterized P-body proteins in yeast and human. Plant P-bodies contain VCS, a human Hedls homolog that is not found in yeast, indicating plant P-bodies are structurally closer to human P-bodies than those in yeast. Nevertheless, Arabidopsis DCP1 and DCP2 which mark plant P-bodies well can also localize to yeast P-bodies [8••,9•,10••]. It is not yet clear whether yeast, plant and mammalian P-body structures are truly homologous or are just merely similar.
Table 1. Known homologs of human P-bodies components in Arabidopsis.
| Gene | AGI(AT) | Genetic alleles | Description of alleles | Yeast | Human |
|---|---|---|---|---|---|
| DECAPPING 1 | 1G08370 | dcp1-1;dcp1-2 | Post-embryonic lethal [8••,9•] | DCP1 | DCP1a DCP1b |
| DECAPPING 2 | 5G13570 | dcp2-1;dcp2-2; dcp2-3 | Post-embryonic lethal [8••,9•] | DCP2 | DCP2 |
| tdt1-1;tdt1-2(dcp2-1) | Vascular defects, post-embryonic lethal [12•] | ||||
| VARICOSE | 3G13300 | vcs-1; vcs-2; vcs-3;vcs-4;vcs-5 | Leaf blade and shoot apical meristem defects, temperature sensitive [19] | Hedls Ge-1 |
|
| vcs-6;vcs-7 | Post-embryonic lethal [8••] | ||||
| DECAPPING 5 | 1G26110 | dcp5-1 | Germination slow, abnormal vein development, defect in mRNA decapping, P-body formation and translational repression [11••] | SCD6 | RAP55 |
| EXORIBONUC LEASE 4 | 1G54490 | xrn4-1 | Elevated transgene silencing [20•] | XRN1 | XRN1 |
| xrn4-4;xrn4-5 | Over-accumulation of specific mRNAs including 3′ products of miRNA-mediated cleavage [21] | ||||
| ein5 | Ethylene insensitive, over-accumulation of EBF1 and EBF2 mRNAs [22,23] elevated small RNA processing [24] | ||||
| DExD/H-BOX HELICASE 1 | 3G61240 | dhh1-1* | DHH1 | Rck/p54 | |
| SUPERSENSITI VE TO ABA AND DROUGHT 1 | 5G48870 | sad1 | Dwarf and small, hypersensitive to ABA and drought [25] | LSM5 | hLSM5 |
| LIKE Sm 4 | 5g27720 | emb1644-1;emb1644-2 | Embryo defective from globular stage (http://www.seedgenes.org) | LSM4 | hLSM4 |
| ARGONAUTE 1 | 1G48410 | ago1-3 | Infertile, defect in gene silencing [26], including translation inhibition [27] | hAGO1 | |
| SILENCING DEFECTIVE 3 | 1G05460 | sde3-1;sde3-2 | Defect in transgene silencing [28] | MOV10 | |
| UP-FRAMESHIFT 1 | 5G47010 | upf1-3;lba1 | Seedling lethality [29,30•] | NAM7 | UPF1 |
| upf1-4;upf1-5 | Leaf and flower abnormality, defect in RNA interference [30•] | ||||
| UP-FRAMESHIFT 2 | 2G39260 | VIGS | Defect in degradation of aberrant spliced RNA [31,32••] | NMD2 | UPF2 |
| UP-FRAMESHIFT 3 | 1G33980 | upf3-1;upf3-2 | Leaf and flower abnormality [30], defect in degradation of aberrant spliced RNA [33] | UPF3 | UPF3 |
| SUPPRESSOR WITH MORPHOGENE TIC EFFECT ON GENITALIA 7 | 5G19400 | VIGS | Defect in degradation of aberrant spl.iced RNA [31,32••] | SMG-7 | |
| CCR4-ASSOCIATED FACTOR1a | 3G44260 | caf1a-1; caf1a-2 | Sensitive to abiotic and biotic stresses [34,35] | POP2 | CAF1 |
| CCR4-ASSOCIATED FACTOR1b | 5G22250 | caf1b-1; caf1b-2; caf1a/caf1 b | |||
| EUKARYOTIC TRANSLATION INITIATION FACTOR 4E | 4G18040 | cum1-1;cum1-2 | Loss-of-susceptibility to potyviruses [36] | CDC33 | eIF4E |
| ISO-EUKARYOTIC TRANSLATION INITIATION FACTOR 4E | 5G35620 | lsp1; ateIF(iso) 4E-1 | Loss-of-susceptibility to potyviruses [37,38] |
Xu and Chua, unpublished
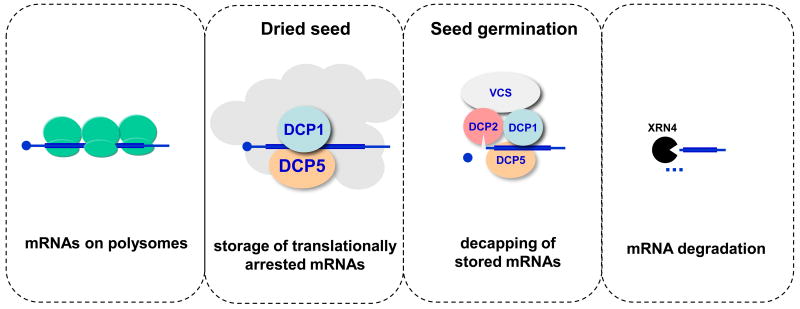
Amongst P-body components investigated, DCP2 is the only one possessing decapping enzymatic activity in vitro [8••]. DCP5 alone has been shown to be a translation suppressor in wheat germ extracts [11••]. Co-localization of DCP1, DCP2, VCS, DCP5, XRN4 and DHH1 suggests plant P-bodies function in both translational repression and mRNA decapping [8••,9•,10••,11••,12•]. We prefer a kinetic model that DCP1, DCP5 and possibly DHH1 are involved in forming RNPs for translational repression of target mRNAs. The untranslated mRNAs then are subjected to decapping by recruiting DCP2 and VCS and the decapped RNAs are subsequently digested by XRN4 (Figure 1) [11••].
Figure 1.
A kinetic model of translational repression and decapping in P-bodies during seed development.
DCP1, DCP5 are involved in forming RNPs for translational repression of target mRNAs. The untranslated mRNAs are then subjected to decapping by recruiting DCP2 and VCS, and the uncapped products are subsequently degraded by XRN4.
The Arabidopsis genome also encodes small RNA-mediated decay pathway proteins. Although AGO1 was reported to localize in nuclear dicing bodies [13], the cytoplasmic fraction of AGO1 was found residing in P-bodies [14•,15••]. There is as yet no Arabidopsis homolog of GW182, which connects AGO proteins to mammalian P-bodies. A possible candidate to functionally supplant GW182 is SDE3, which co-localizes with DCP1 in plants (Xu and Chua, unpublished). SDE3 is a MOV10 homolog in plants but contains a GW motif known to mediate interactions between GW182 and AGOs [16]. UPF1 which is a nonsense-mediated decay (NMD) pathway factor also localizes to plant P-bodies (Xu and Chua, unpublished). Although eIF4E marks P-bodies in human and yeast cells [17] it does not localize to plant P-bodies [8••], instead it localizes to plant stress granules (SGs) [10••]. Plant P-bodies share many conserved proteins with yeast and human P-bodies indicating that plant P-bodies execute similar functions, i.e. translational repression and decapping. However, plant P-bodies also contain their own distinct protein components which may form functionally diverged RNPs.
Besides the known homologs (Table 1), other proteins have also been found in plant P-bodies. For example, ISE2 granules represent plant-specific SGs and a small portion of ISE2 has been shown to co-localize with DCP2 [18]. AtTZF1 and hTTP have been reported to co-localize with plant AGO1, DCP2 and XRN4 in P-bodies [15••] and CMV 2b, a viral gene-silencing suppressor, is also associated with P-bodies [14•]. More plant-specific P-body components are expected to emerge with further advances in plant functional genomic studies.
Function implications by phenotypic analysis
Severe single-mutant alleles of DCP1, DCP2, DCP5 and VCS are lethal at the seeding stage with pleiotropic phenotypes including vein, leaf blade and root defects, suggesting the affected proteins are required for post-embryonic development [8••,9•,11••,12•]. The developmental phenotype of mutants defective in P-body component is rather unexpected for three reasons. (1) Among the P-body components only DCP2 has decapping activity in vitro [8••]. The requirement for other components for growth suggests that DCP2 activity in vivo is rate-limiting. The complex formation between DCP2, DCP1, DCP5 and VCS is either indispensible to the mRNA decapping step or DCP1, DCP5 and VCS, may act in steps upstream of decapping. One possibility is that translationally repressed RNP formation is necessary and precedes decapping. Consistent with this notion, DCP5 alone is able to suppress mRNA translation in vitro [11••]. (2) Accumulation of capped mRNAs in decapping-defective mutants results in severe seedling lethality, indicating that these mRNAs are not subject to alternate mechanisms of RNA decay and are able to re-engage with ribosomes for translation. Under this condition, the accumulation of both mRNAs and their products has the same effect as the over-expression of cognate genes. (3) It is not likely that the deadenylation step precedes the decapping step, or the majority of these mRNA substrates are subject to co-translational decapping which may bypass the deadenylation step [39].
Amongst mutants deficient in P-body components the most severe phenotype is found with lsm4; in this mutant, embryos do not develop beyond the globular stage (Table 1). upf1-3 is seedling lethal similar to moderate dcp2 mutants. The developmental defects in mutants of P-body components are pleiotropic, suggesting multi-target effects are common for the affected proteins. Moreover, developmental lethality in P-body mutants suggests that P-body components are required for developmental transition or reprogramming. For example, dcp2-1 seeds are able to germinate but seedling growth ceases with the typical lethal phenotype appearing 6 d after imbibition [8••]. It is likely that P-bodies are required to degrade seed-stored mRNAs, over-accumulation of which would otherwise perturb developmental transition.
RNA substrates of plant P-body
P-bodies contain RNPs assembled from mRNAs and proteins. Although the assembly mechanism of individual RNPs is largely unknown, knowledge of mRNA substrates provides important clues to understanding functions of RNPs or P-bodies in general. As biochemical purification of P-bodies has so far been elusive for unknown reasons, analysis of genetic alleles of specific P-body component has become an important strategy to identify and characterize RNA compositions of plant P-bodies.
Seed storage proteins (SSP) mRNAs are P-body substrates in dried seed and during germination. In dcp1-1, dcp1-2 and dcp5-1, SSP mRNAs accumulate greatly at 6 d after germination [11••,12•]. Similarly, SSP protein levels are increased in these mutants, suggesting that P-bodies target these mRNAs for both translational repression and decapping. Dried seeds accumulate high levels of DCP1 and DCP5 but not DCP2, which is consistent with the storage of SSP mRNAs in seeds without degradation. After germination, DCP1 and DCP5 levels decline along with increase in DCP2 levels and decapping and degradation of SSP mRNAs. In dried seed, SSP mRNAs are stored and translationally arrested but become degraded during germination. These observations are in line with the kinetic model suggesting that translation repression and RNP formation precedes the decapping step. Decapping, therefore, is crucial to the developmental transition from a quiescent state and to rapid post-embryonic growth. Besides SSP mRNAs, many unstable mRNAs [40] including EXPL1 and SEN1 are also stabilized in decapping mutants indicating that a broad spectrum of mRNAs are subjected to translational repression and decapping [8••,11••].
EBF1 and EBF2 mRNAs are likely the direct targets of EIN5/XRN4. In ein5 mutant, EBF1 and EBF2 mRNAs are highly expressed, resulting in reduced levels of EIN3 [22,23]. Although its enzyme activity has not yet been directly demonstrated XRN4 is presumably an exoribonuclease preferring RNA substrates with 5′-monophosphate.
Assuming this activity of XRN4, the increased levels of EBF1/2 mRNAs in xrn4 mutant are likely to be uncapped. It is not clear how uncapped mRNAs are able to re-engage with ribosomes for translation. The other characterized targets of XRN4 includes 3′ products of miRNA cleavage [21], transcripts of the STM-GR transgene [20•] and also 130 endogenous transcripts determined by microarray analysis [24]. Another microarray experiment on uncapped RNAs demonstrates the presence of diverse uncapped RNAs in flowers [46]. Further work is needed to understand how uncapped mRNAs are channeled to cap-insensitive translation or are directly degraded or serve as substrates for dsRNA production.
Several lines of evidence support the view that P-body formation is likely not required for degradation of mRNAs targeted by miRNA, siRNA and ta-siRNA. (1) Although dcp5-1 mutant displays reduced formation of P-bodies it does not significantly accumulate miRNA targets [11••]. (2) In contrast to mammalian systems, the majority of miRNAs in plants show complete or near complete sequence complementarity to their mRNA targets, and 3′ cleavage products are usually readily detectable. (3) RNA interference has been successfully used in plants to achieve degradation/silencing of specific mRNAs in a sequence-dependent manner [41]. Notably, VCS has been found to be required for miRNA-guided translational repression but had no effect on mRNA levels of genes like SPL3, SCL6-IV and CIP4 [27]. It is not clear if the translational-arrested miRNA targets are stored in P-bodies and if P-bodies are required for this miRNA-guided translational repression. Levels of several miRNAs are reduced in decapping mutants suggesting a possible connection between miRNA biogenesis and plant P-bodies (Xu and Chua, unpublished).
Non-sense mediated decay (NMD) is a eukaryotic mRNA quality-control mechanism for elimination of aberrant mRNAs containing a premature termination codon (PTC) to prevent the production of truncated proteins. Although mRNA targets responsible for the developmental defect of upf 1 and upf 3 mutants have not been fully identified, several aberrant mRNAs containing PTC have been found to over-accumulate, i.e. SMD-7 and ATGRP7 transcripts [31,42]. A genome-wide survey revealed that expression of not only protein-coding transcripts but also many non-coding RNAs (ncRNAs) is upregulated in both upf 1 and upf 3 mutants [32••]. Typical ncRNAs include natural antisense transcripts, and transcripts of transposable elements and pseudogenes, all of which contain no or short ORFs and share the same features with canonical PTC-containing NMD substrates. Interestingly, UPF1, which is required for degradation of aberrant RNAs, appears to play an important role in RNA interference [30•].
As P-bodies contain many RNPs functional in regulating plant development it is not surprising that they may be involved in pathogen infection and defense. In yeast, brome mosaic virus (BMV) is localized to P-bodies and viral RNA-dependent RNA polymerase binds to the yeast lsm1p [43•]. More important, decapping complex factors including Lsm1p-7p/Pat1p/Dhh1p have been reported to be necessary for BMV replication in yeast [44•]. These results obtained with a heterozygous host suggest that BMV may likely replicate in plant P-bodies as well.
P-body formation
Franks and Lykke-Anderson proposed a kinetic model for P-body formation in which the formation of translationally repressed RNPs precedes the mRNA decapping step [45]. DCP5 is a translation repressor and does not stimulate DCP2 activity in vitro. However, P-body formation is reduced in the dcp5-1 mutant [10••], suggesting DCP5 is required for P-body formation. DCP1, DCP5 and possibly DHH1 are involved in forming RNPs for translational repression of target mRNAs. The untranslated mRNAs then are subjected to decapping by recruiting DCP2 and VCS and the uncapped products are subsequently degraded by XRN4 (Figure 1).
There is evidence that P-body formation is dispensable to active decapping, since DCP1 marked P-bodies are still detected in plants lacking DCP2 [11••]. P-bodies devoid of DCP2 most likely contain translationally repressed RNPs. The multi-steps between translation and mRNA decapping facilitate regulation of the decapping step. During stress signaling-pathways are found to regulate RNP formation through modifying DCP1 (Xu and Chua, unpublished). Therefore, translatability of a specific group of mRNAs is regulated by responses to environmental clues before being committed to the irreversible and rate-limiting step of decapping.
Conclusions
Plant P-bodies share many protein components with yeast and mammalian P-bodies, but are more closely related to mammalian P-bodies. However, plant P-bodies also contain specific components. The phenotype of mutants and specific substrates found in P-bodies suggest that their function are developmental rather than housekeeping. The most conserved functions of P-bodies are translational repression and mRNA decapping. P-bodies are more likely a compartment rather than a single RNP complex. Diversity and specificity uncovered from the characterization of many P-body-related RNA substrates indicate that translationally repressed RNPs from different RNA decay pathways may localize to this compartment. A major future challenge will be to understand how this diversity and specificity are achieved and genetic dissection using Arabidopsis to discover more P-body components and RNPs will certainly be an important approach.
Research Highlights.
Developmental defects in mutants of P-body components are pleiotropic, suggesting multi-target effects are common for the affected proteins.
Developmental lethality in P-body mutants suggests that P-body components are required for developmental transition or reprogramming.
A kinetic model of translational repression and decapping in P-bodies during seed development.
Acknowledgments
We apologize to those colleagues whose work could not be cited because of space limitations. This work is supported by NIH grant GM44640 to N.-H. C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest published within the period of review have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 2.Belostotsky DA. State of decay: an update on plant mRNA turnover. Curr Top Microbiol Immunol. 2008;326:179–199. doi: 10.1007/978-3-540-76776-3_10. [DOI] [PubMed] [Google Scholar]
- 3.Bailey-Serres J, Sorenson R, Juntawong P. Getting the message across: cytoplasmic ribonucleoprotein complexes. Trends Plant Sci. 2009;14:443–453. doi: 10.1016/j.tplants.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Belostotsky DA, Sieburth LE. Kill the messenger: mRNA decay and plant development. Curr Opin Plant Biol. 2009;12:96–102. doi: 10.1016/j.pbi.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulkarni M, Ozgur S, Stoecklin G. On track with P-bodies. Biochem Soc Trans. 2010;38:242–251. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]
- 8.Xu J, Yang JY, Niu QW, Chua NH. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 2006;18:3386–3398. doi: 10.1105/tpc.106.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The first report on plant P-bodies and mRNA decapping complex. Using an in vitro decapping assay, the authors confirmed that DCP2 but not other components is a decapping enzyme, indicating the involvement of multiple steps in mRNA decapping. Genetic analysis highlighted the requirement for decapping complex in postembryonic development.
- 9.Iwasaki S, Takeda A, Motose H, Watanabe Y. Characterization of Arabidopsis decapping proteins AtDCP1 and AtDCP2, which are essential for post-embryonic development. FEBS Lett. 2007;581:2455–2459. doi: 10.1016/j.febslet.2007.04.051. [DOI] [PubMed] [Google Scholar]; • Independent study on DCP1 and DCP2 concludes that these two proteins are essential for post-embryonic development.
- 10.Weber C, Nover L, Fauth M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 2008;56:517–530. doi: 10.1111/j.1365-313X.2008.03623.x. [DOI] [PubMed] [Google Scholar]; •• The authors provided evidence that XRN4 localizes to P-bodies. The comparison of P-bodies, stress granules and heat stress granules revealed that heat stress granules are distinct from plant stress granules.
- 11.Xu J, Chua NH. Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell. 2009;21:3270–3279. doi: 10.1105/tpc.109.070078. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This study identified DCP5 as another P-body component. DCP5 is a translation repressor required for P-body formation and postembryonic development. Seed storage protein mRNAs were suggested in this study to be P-body substrates.
- 12.Goeres DC, Van Norman JM, Zhang W, Fauver NA, Spencer ML, Sieburth LE. Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell. 2007;19:1549–1564. doi: 10.1105/tpc.106.047621. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Genetic screen for vein development mutants identified TDT which encodes DCP2.
- 13.Fang Y, Spector DL. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol. 2007;17:818–823. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The study demonstrated that plant P-body contains viral proteins against AGO1 slicer functions.
- 15.Pomeranz MC, Hah C, Lin PC, Kang SG, Finer JJ, Blackshear PJ, Jang JC. The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol. 2010;152:151–165. doi: 10.1104/pp.109.145656. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The study identified AtTZF1 as a P-body component suggesting that plant P-bodies may contain RNPs linked to the ARE-mediated RNA decay pathway.
- 16.Karlowski WM, Zielezinski A, Carrere J, Pontier D, Lagrange T, Cooke R. Genome-wide computational identification of WG/GW Argonaute-binding proteins in Arabidopsis. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brengues M, Parker R. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2592–2602. doi: 10.1091/mbc.E06-12-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi K, Otegui MS, Krishnakumar S, Mindrinos M, Zambryski P. INCREASED SIZE EXCLUSION LIMIT 2 encodes a putative DEVH box RNA helicase involved in plasmodesmata function during Arabidopsis embryogenesis. Plant Cell. 2007;19:1885–1897. doi: 10.1105/tpc.106.045666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deyholos MK, Cavaness GF, Hall B, King E, Punwani J, Van Norman J, Sieburth LE. VARICOSE, a WD-domain protein, is required for leaf blade development. Development. 2003;130:6577–6588. doi: 10.1242/dev.00909. [DOI] [PubMed] [Google Scholar]
- 20.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]; • The first report demonstrating that XRN4 is an endogenous silencing suppressor. Together with Ref. [30•], these studies highlight a link between mRNA turnover and RNA interference.
- 21.Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell. 2004;15:173–183. doi: 10.1016/j.molcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Olmedo G, Guo H, Gregory BD, Nourizadeh SD, Aguilar-Henonin L, Li H, An F, Guzman P, Ecker JR. ETHYLENE-INSENSITIVE5 encodes a 5′-->3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc Natl Acad Sci U S A. 2006;103:13286–13293. doi: 10.1073/pnas.0605528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potuschak T, Vansiri A, Binder BM, Lechner E, Vierstra RD, Genschik P. The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell. 2006;18:3047–3057. doi: 10.1105/tpc.106.046508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gregory BD, O'Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu JK. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell. 2001;1:771–781. doi: 10.1016/s1534-5807(01)00087-9. [DOI] [PubMed] [Google Scholar]
- 26.Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci U S A. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 28.Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoine M, Nishii T, Nakamura K. Arabidopsis UPF1 RNA helicase for nonsense-mediated mRNA decay is involved in seed size control and is essential for growth. Plant Cell Physiol. 2006;47:572–580. doi: 10.1093/pcp/pcj035. [DOI] [PubMed] [Google Scholar]
- 30.Arciga-Reyes L, Wootton L, Kieffer M, Davies B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 2006;47:480–489. doi: 10.1111/j.1365-313X.2006.02802.x. [DOI] [PubMed] [Google Scholar]; • See annotation to Ref. [20•].
- 31.Kerenyi Z, Merai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 2008;27:1585–1595. doi: 10.1038/emboj.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, Morosawa T, Tanaka M, Kaminuma E, Mochizuki Y, Matsushima A, et al. Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:2453–2458. doi: 10.1073/pnas.0808902106. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The authors provided a genome-wide view of putative targets for NMD. The study also extended our understanding that NMD targets include not only protein-coding transcripts but also ncRNAs.
- 33.Hori K, Watanabe Y. UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 2005;43:530–540. doi: 10.1111/j.1365-313X.2005.02473.x. [DOI] [PubMed] [Google Scholar]
- 34.Liang W, Li C, Liu F, Jiang H, Li S, Sun J, Wu X. The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res. 2009;19:307–316. doi: 10.1038/cr.2008.317. [DOI] [PubMed] [Google Scholar]
- 35.Walley JW, Kelley DR, Nestorova G, Hirschberg DL, Dehesh K. Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol. 2010;152:866–875. doi: 10.1104/pp.109.149005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato M, Nakahara K, Yoshii M, Ishikawa M, Uyeda I. Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. FEBS Lett. 2005;579:1167–1171. doi: 10.1016/j.febslet.2004.12.086. [DOI] [PubMed] [Google Scholar]
- 37.Duprat A, Caranta C, Revers F, Menand B, Browning KS, Robaglia C. The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 2002;32:927–934. doi: 10.1046/j.1365-313x.2002.01481.x. [DOI] [PubMed] [Google Scholar]
- 38.Lellis AD, Kasschau KD, Whitham SA, Carrington JC. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr Biol. 2002;12:1046–1051. doi: 10.1016/s0960-9822(02)00898-9. [DOI] [PubMed] [Google Scholar]
- 39.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narsai R, Howell KA, Millar AH, O'Toole N, Small I, Whelan J. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell. 2007;19:3418–3436. doi: 10.1105/tpc.107.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGinnis K, Chandler V, Cone K, Kaeppler H, Kaeppler S, Kerschen A, Pikaard C, Richards E, Sidorenko L, Smith T, et al. Transgene-induced RNA interference as a tool for plant functional genomics. Methods Enzymol. 2005;392:1–24. doi: 10.1016/S0076-6879(04)92001-0. [DOI] [PubMed] [Google Scholar]
- 42.Schoning JC, Streitner C, Page DR, Hennig S, Uchida K, Wolf E, Furuya M, Staiger D. Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 2007;52:1119–1130. doi: 10.1111/j.1365-313X.2007.03302.x. [DOI] [PubMed] [Google Scholar]
- 43.Beckham CJ, Light HR, Nissan TA, Ahlquist P, Parker R, Noueiry A. Interactions between brome mosaic virus RNAs and cytoplasmic processing bodies. J Virol. 2007;81:9759–9768. doi: 10.1128/JVI.00844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The study found a specific interaction between LSM1 and viral RNA-dependent RNA polymerase. Together with Ref. [44•], these studies strongly suggest that plant P-bodies are likely to be virus replication sites.
- 44.Mas A, Alves-Rodrigues I, Noueiry A, Ahlquist P, Diez J. Host deadenylation-dependent mRNA decapping factors are required for a key step in brome mosaic virus RNA replication. J Virol. 2006;80:246–251. doi: 10.1128/JVI.80.1.246-251.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; • See annotation of Ref. [43•].
- 45.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiao Y, Riechmann JL, Meyerowitz EM. Transcriptome-wide analysis of uncapped mRNAs in Arabidopsis reveals regulation of mRNA degradation. Plant Cell. 2008;20:2571–2585. doi: 10.1105/tpc.108.062786. [DOI] [PMC free article] [PubMed] [Google Scholar]