Abstract
Genome duplication requires accessory helicases to displace proteins ahead of advancing replication forks. Escherichia coli contains three helicases, Rep, UvrD and DinG, that might promote replication of protein-bound DNA. One of these helicases, Rep, also interacts with the replicative helicase DnaB. We demonstrate that Rep is the only putative accessory helicase whose absence results in an increased chromosome duplication time. We show also that the interaction between Rep and DnaB is required for Rep to maintain rapid genome duplication. Furthermore, this Rep–DnaB interaction is critical in minimizing the need for both recombinational processing of blocked replication forks and replisome reassembly, indicating that colocalization of Rep and DnaB minimizes stalling and subsequent inactivation of replication forks. These data indicate that E. coli contains only one helicase that acts as an accessory motor at the fork in wild-type cells, that such an activity is critical for the maintenance of rapid genome duplication and that colocalization with the replisome is crucial for this function. Given that the only other characterized accessory motor, Saccharomyces cerevisiae Rrm3p, associates physically with the replisome, our demonstration of the functional importance of such an association indicates that colocalization may be a conserved feature of accessory replicative motors.
INTRODUCTION
Duplication of DNA requires not only the breakage of hydrogen bonds between the parental strands but also the displacement of proteins bound to the template, both of which are catalysed by the replicative helicase at the fork. This requirement for displacement of proteins ahead of the fork is illustrated by high affinity nucleoprotein complexes such as those associated with transcription, centromeres and inactive replication origins presenting obstacles to movement of the replicative helicase and thus of the fork (1–3). Occasional failure of the replicative helicase to displace such proteins from the template results in fork blockage and the attendant risks of incomplete genome duplication or of genome instability associated with blocked fork repair (4).
Blockage of replication forks by nucleoprotein complexes suggests that recruitment of additional helicases to such forks might facilitate clearance of the block and resumption of replication (5). Rrm3p promotes movement of forks through high affinity non-histone protein–DNA complexes in Saccharomyces cerevisiae, indicating that Rrm3p acts as an accessory replicative helicase (3,6). Lack of Rrm3p is also associated with elevated recombination rates (7), a signature of recombinational repair of blocked forks. However, whether other S. cerevisiae helicases can act in such a capacity is unknown. Indeed, in Escherichia coli multiple helicases can perform this accessory helicase function. Rep and UvrD can each promote movement of E. coli replication forks along protein-bound DNA, a redundancy of function reflected in the inviability on rich medium of cells lacking both Rep and UvrD (8–10). This inviability is associated with nucleoprotein complexes associated with gene expression since RNA polymerase mutations can suppress the rich medium inviability of Δrep ΔuvrD cells (9–11). A third E. coli helicase, DinG, may also promote fork movement along transcribed DNA. DinG is critical in promoting survival of strains bearing chromosomal inversions encompassing highly transcribed rrn operons and is also essential for the survival of Δrep ΔuvrD cells bearing RNA polymerase suppressor mutations (10).
Whether Rep, UvrD and DinG are redundant or whether only one of these helicases is primarily responsible for underpinning fork movement in wild-type cells is unclear. As with ΔdinG cells, Δrep cells are sensitive to inversion of rrn operons but ΔuvrD cells are not (10). Promotion of fork movement along transcribed DNA may be underpinned therefore by Rep and/or DinG rather than UvrD. Mean replication fork speed in vivo may also be reduced 2-fold in the absence of Rep (12), although whether this reduction reflects Rep-directed clearance of proteins ahead of blocked forks or the participation of Rep in PriC-directed replisome reassembly away from oriC is unknown (13). The C-terminus of Rep also interacts physically with the E. coli replicative helicase, DnaB, an interaction that promotes fork movement along protein-bound DNA in vitro and facilitates complementation of Δrep ΔuvrD lethality by plasmid-encoded Rep (9). In contrast, there is no detectable interaction between UvrD and DnaB (9). Thus Rep may be an integral component of the replisome, implying a critical role for Rep in promoting fork movement along protein-bound DNA. Saccharomyces cerevisiae Rrm3p also localizes to replisomes via Pol2p and/or PCNA (14,15), although the functional importance of this colocalization in S. cerevisiae is unknown.
Here we show that the optimal rate of genome duplication in E. coli requires a single accessory helicase, Rep. We show also that interaction between Rep and DnaB is critical for the maintenance of rapid genome duplication and that this interaction reduces the need both for blocked fork processing by recombination enzymes and for replisome reassembly. However, whilst physical interaction with the replisome is critical for the function of Rep in vivo, interaction with Rep has no allosteric effect on the integral speed of the replisome. The only function of the Rep–DnaB interaction is to increase therefore the local concentration of an accessory motor at the fork. Our demonstration of the functional importance of interaction of an accessory helicase with the replisome in E. coli implies that this colocalization may be a key signature of such helicases.
MATERIALS AND METHODS
Strains and plasmids
Strains are listed in Supplementary Table S1. A rep allele bearing a deletion of the final 33 codons, repΔC33, was generated using the primers MKG64 (TTTTTGCTGGAGCTGCCGCAGGATGATCTGATTTGGTAAGTGTAGGCTGGAGCTGCTTC) and MKG63 (CCGGATGCGATGCTGACGCATCTTTTCCGGCCTTGACATATGAATATCCTCCTTAC) to amplify the kanamycin resistance cassette from pKD4 (16). The resulting PCR product was then inserted into the chromosome of DY330 via λRed integration (17) to generate repΔC33 linked to a kanamycin resistance gene immediately downstream of the repΔC33 stop codon. An otherwise isogenic rep+ strain was generated in a similar manner except that MKG62 (CTGAAAGCGATGATGGCGGCAAAACGAGGGAAATAAGTGTAGGCTGGAGCTGCTTC) was used in combination with MKG63. Both alleles were sequenced to ensure accurate integration and to confirm the absence of unwanted mutations in either rep allele.
pAM374, 403 and 407 are pRC7 derivatives encoding priA, rep and uvrD, respectively (9,18).
Flow cytometry
For measurement of chromosome duplication time in dnaA46 and dnaA5 strains, the indicated strains were grown in 5 ml of LB at 30°C with shaking until an A650 of 0.3 was reached. Cultures were then transferred to a 42°C shaking waterbath for 2 h. 5 ml of LB pre-cooled to 18°C was then added directly to the 42°C cultures to alter the culture temperature rapidly to 30°C and the mixture then placed in a 30°C shaking waterbath for 10 min. This 10-min period at the permissive temperature was the minimum needed to allow the majority of cells to initiate chromosome replication. 10 ml of LB preheated to 55°C was then added to the 30°C cultures, achieving a temperature of 42°C rapidly, and the mixture placed into a 42°C shaking waterbath for a further 70 min. Culture samples were removed immediately prior to the temperature downshift to 30°C and designated as time zero. Samples were then removed immediately prior to the temperature upshift to 42°C and at 10-min intervals thereafter. Culture samples were fixed in methanol and DNA stained with Sytox Green (Invitrogen). Flow cytometry was performed on a FACSCalibur Benchtop Cytometry Analyser (Becton Dickinson).
Estimation of intracellular Rep concentrations
To estimate the number of Rep molecules in wild-type cells (TB28), cells were grown in 10 ml of LB at 37°C with shaking until an A650 of 0.4 was reached. Serial dilutions were plated onto LB agar and the number of viable cells per ml estimated after incubation overnight at 37°C. The remaining 10 ml was chilled on ice and cells harvested by centrifugation at 4°C. Cell pellets were resuspended in 400 µl of 50 mM Tris.HCl (pH 8.4), 150 mM NaCl, 0.2 mg/ml lysozyme and 20 mM EDTA and incubated on ice for 10 min before addition of Brij 58 to a final concentration of 0.1%. Incubation was continued on ice for another 20 min. Protein concentration was determined using Bio-Rad protein assay reagent to give an estimate of the total protein content per ml of culture. Combining this value with the number of viable cells per millilitre of culture was used to estimate the number of viable cells required to obtain 10 µg of total cell protein. 10 µg of extract was then loaded onto a 8% SDS polyacrylamide gel along with 0.5, 1, 2.5, 5 and 10 ng of Rep, purified as described (9). After electrophoresis at 180 V for 80 min in an Invitrogen XCell Surelock gel tank, proteins were transferred onto Hybond-P (GE Healthcare) in transfer buffer (0.48 mM Tris base, 3.84 mM glycine and 20% methanol). The filter was then blocked overnight at 4°C in 25 ml of PBS/T (100 mM NaCl, 80 mM disodium hydrogen phosphate, 20 mM sodium dihydrogen phosphate, 0.1% Tween 20 and 5% dried milk). The filter was then rinsed twice in 25 ml PBS/T before being incubated for 1 h at room temperature in 25 ml PBS/T containing rabbit anti-Rep serum, raised against purified Rep (19) by Eurogentec, at a dilution of 1:6250. The blot was then rinsed briefly in 2 × 25 ml PBS/T, once in 25 ml PBS/T for 15 min and then 3 × 5 min in 25 ml PBS/T at room temperature. The blot was then incubated for 1 h at room temperature in 25 ml PBS/T containing goat anti-rabbit/peroxidase conjugate (Sigma) at a dilution of 1:50000. The blot was then rinsed as described above. Antibody was detected using the ECL Plus Western Blotting Detection System (GE Healthcare) according to the manufacturer’s recommendations. Amounts of Rep were quantified using a Bio-Rad Fluor-S MultiImager. The fluorescence intensity obtained with each known quantity of purified Rep was plotted against the amount of Rep to obtain a standard curve for the blot. Fluorescence intensity corresponding to the Rep signal in lanes containing 10 µg whole-cell extracts was then used in conjunction with this standard curve to estimate the number of Rep molecules within 10 µg of whole cell extract. The number of Rep molecules per cell was then calculated by dividing the number of Rep molecules per 10 µg total cell protein by the number of viable cells required to obtain 10 µg of total cell protein, as estimated above from the viable cell count. Rep concentrations in MKG08, MKG10, HB159, HB252 and HB254 were estimated in a similar manner using TB28 whole cell extracts as standards. Strains containing dnaA46 were grown at 30°C rather than 37°C.
Viability, cell length and growth rate assays
The ability of strains to form colonies in the absence of pRC7 derivatives was monitored as described (9). Growth rates of ΔuvrD strains bearing rep+<kan>, repΔC33<kan> or Δrep were monitored by initially plating pAM407-containing strains onto minimal agar containing Xgal and IPTG. After 4 days at 37°C, plasmid-free segregants were identified and inoculated into 10 ml of liquid minimal medium. Cultures were grown at 37°C to an A650 of 0.4 and then 125 µl of each culture was inoculated into 10 ml of LB. Incubation was continued at 37°C. Aliquots were removed at the indicated times after inoculation of the LB cultures, serial dilutions made in ice-cold 56/2 salts and the number of viable cells estimated after spotting of these dilutions onto minimal agar and incubation at 37°C for 3 days. The median length of cells was measured in minimal medium when an A650 of 0.4 was reached and then after 3 h in LB medium using a Zeiss Axioskop mot plus microscope and QCapture Pro v5.1 software.
RESULTS
Interaction of Rep with DnaB is required to minimize chromosome duplication time
The mean speed of replication fork movement in cells lacking Rep is half that in wild-type cells (12), implying an important role for Rep in underpinning genome duplication. However, the importance of localizing Rep at the replisome in maintaining replication fork speed is unknown. To measure the time required for chromosome duplication, synchronization of replication initiation was achieved using the temperature-sensitive dnaA46 allele (20). Synchronization was performed by shifting cells to the restrictive temperature for 2 h, generating cells with a single chromosome. Chromosome duplication was then initiated by shifting the cells to the permissive temperature for 10 min, before a return to 42°C to inhibit continued initiation. DNA content per cell was monitored by flow cytometry at 10-min intervals, with time zero being the point at which cells were shifted to the permissive temperature.
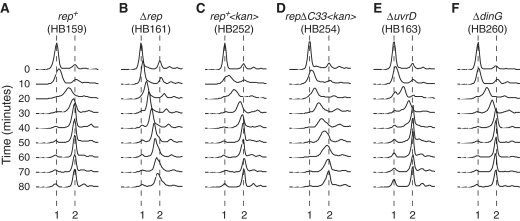
Wild-type cells took ∼40–50 min for the DNA content to increase from 1 to 2 chromosome equivalents (Figure 1A) consistent with a 40-min chromosome duplication time given the 10 min spent at the permissive temperature. Only a small fraction of cells underwent subsequent cell division, as indicated by the small increase between 40 and 80 min in the fraction of cells having 1 chromosome equivalent (Figure 1A). Absence of cell division after a single round of chromosome replication has been observed previously (20,21) and likely reflects the inability of chromosome duplication in itself to trigger cell division (22).
Figure 1.
The impact of Rep, UvrD and DinG on chromosome duplication time. (A–F) Flow cytometry profiles of the indicated strains, all bearing the temperature sensitive dnaA46 allele. Cell samples were removed for analysis immediately prior to shifting the cultures from 42°C to 30°C (designated 0 min). After 10 min at 30°C the cultures were returned to 42°C. Samples were then removed at 10-min intervals after the temperature downshift. The positions of 1 and 2 chromosome equivalents are indicated.
In contrast to wild-type cells, chromosome duplication in Δrep cells was incomplete after 40 min (Figure 1B). Even after 80 min, a significant fraction of Δrep cells had not completed chromosome duplication (Figure 1B). Thus the time taken to replicate chromosomes in Δrep cells is at least twice that in rep+ cells. These data are consistent with absence of Rep causing a 2-fold reduction in mean replication fork speed (12), demonstrating the validity of this assay in monitoring chromosome duplication time.
To establish the importance of the Rep–DnaB interaction in maintaining chromosome duplication time, we exploited a mutant version of Rep lacking the C-terminal 33 amino acids that fails to interact with DnaB but retains helicase activity (9). The wild-type rep allele was replaced with a rep gene lacking the final 33 codons together with a downstream antibiotic resistance cassette. An otherwise isogenic strain was also constructed in which the antibiotic marker was placed in an identical position downstream of the wild-type rep gene. This rep+ strain again took ∼40–50 min for the number of chromosome equivalents to increase from 1 to 2 (Figure 1C). However, as with Δrep cells, chromosome duplication in the repΔC33 strain was incomplete after 40 min (Figure 1, compare parts B with D). This delay in completion of chromosome duplication in repΔC33 was not due to either decreased expression of repΔC33 or to instability of mutant as compared with wild-type Rep, as the number of molecules of RepΔC33 per cell were comparable with those of the wild-type protein (Table 1). We conclude that interaction of Rep with DnaB is required for minimization of chromosome duplication time.
Table 1.
Intracellular concentrations of Rep and RepΔC33
| Strain | Relevant genotype | Rep molecules per cell |
|---|---|---|
| TB28 | rep+ dnaA+ | 128 ± 16 |
| MKG08 | rep+<kan> dnaA+ | 120± 21 |
| MKG10 | repΔC33<kan> dnaA+ | 123 ± 16 |
| HB159 | rep+ dnaA46 | 122 ± 11 |
| HB252 | rep+<kan> dnaA46 | 117 ± 10 |
| HB254 | repΔC33<kan> dnaA46 | 145 ± 8 |
Maintenance of chromosome duplication time is specific to Rep
UvrD, like Rep, promotes replisome movement along protein-bound DNA in vitro and in vivo whilst DinG is also needed to facilitate fork movement through highly transcribed sequences (9,10). We tested therefore whether UvrD or DinG were necessary for maintaining wild-type rates of chromosome duplication. However, as with rep+ uvrD+ dinG+ cells, chromosome duplication in ΔuvrD and ΔdinG cells took approximately 40 min (Figure 1, compare parts E and F with A). Rep but not UvrD or DinG is critical therefore in maintaining wild-type rates of chromosome duplication in vivo.
Minimization of chromosome duplication time by Rep is not effected by PriC-directed replisome reassembly
The increased chromosome duplication time in Δrep cells could be explained by absence of Rep-catalysed promotion of replisome movement along protein-bound DNA. However, the possible involvement of Rep in PriC-directed replisome reassembly away from oriC could also explain reduced mean fork speed in Δrep cells (13). If forks stall and become inactive, creating a need for replisome reloading, then a reduced capacity for such reloading might increase the time needed to replicate the chromosome.
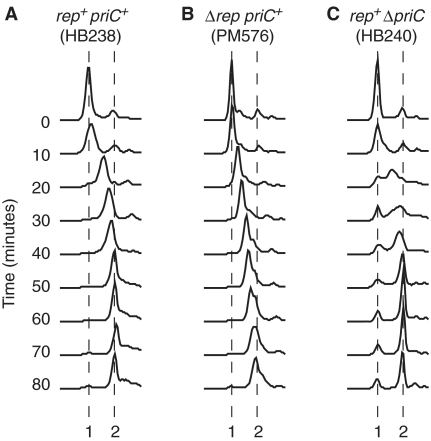
dnaA46 priC cells are inviable (23), preventing replication synchronization using this dnaA allele. However, we found that dnaA5, a priC-compatible allele (23), could also be used to synchronize replication initiation. rep+ priC+ dnaA5 cells took 40–50 min to complete chromosome duplication whereas Δrep priC+ dnaA5 cells took at least 80 min (Figure 2A and B), the same pattern as seen in dnaA46 cells (Figure 1A and B). These data confirm the duplication times seen in dnaA46-containing cells and demonstrate that the nature of the dnaA allele does not impact on the time required for genome duplication.
Figure 2.
Increased chromosome duplication time in the absence of Rep is not due to the absence of PriC-directed replisome reassembly. (A–C) Flow cytometry profiles of the indicated dnaA5-containing strains. Samples were removed just before shifting the cells to 30°C (time zero) and every 10 min thereafter.
Chromosome duplication in rep+ ΔpriC dnaA5 took 40–50 min, a similar time as compared with rep+ priC+ dnaA5 (Figure 2, compare parts C with A). Thus PriC-directed replisome reloading does not have a critical role in maintaining chromosome duplication time. The corollary of this observation is that the increased chromosome duplication time in Δrep and repΔC33 cells (Figure 1) is not due to a defect in PriC-directed replisome reloading.
The Rep–DnaB interaction facilitates resolution of conflicts between replication and transcription
Promotion of fork movement along protein-bound DNA by an accessory helicase is essential for viability under rapid but not restricted growth conditions (9,10). However, Δrep cells are viable on rich medium since UvrD, like Rep, can promote fork movement through nucleoprotein complexes (9). Inviability is only apparent therefore when both Rep and UvrD are absent.
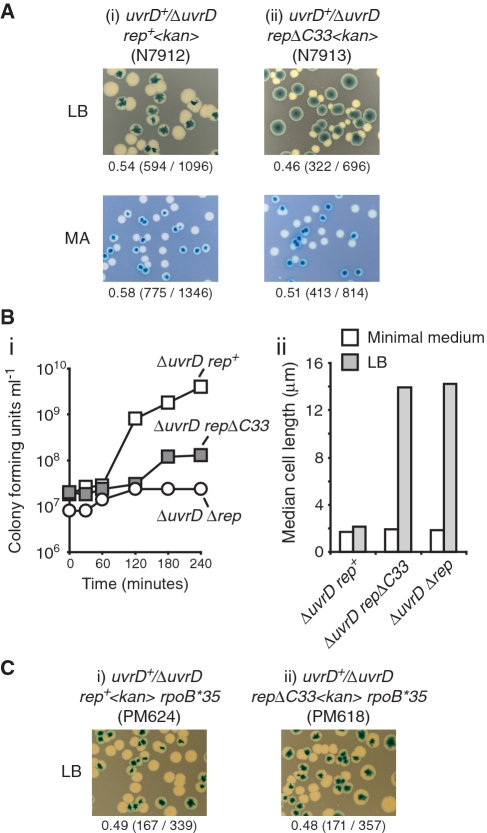
To analyse the effect of absence of the Rep–DnaB interaction on the viability of ΔuvrD cells, repΔC33 and rep+ alleles were introduced into a ΔuvrD strain in which the uvrD mutation was complemented by a very low copy, highly unstable plasmid (pRC7) bearing wild-type uvrD (9,24). This plasmid also encodes lacZYA and so retention or loss of the plasmid can be monitored by blue/white colony colour in strains bearing a chromosomal deletion of the lac operon. As expected, colonies of plasmid-free segregants of the rep+ ΔuvrD strain formed readily on rich medium, with plasmid-free colonies being of similar size to those retaining the plasmid (Figure 3Ai). The similar viability of cells with and without the uvrD+ plasmid was also reflected in the formation of mosaic blue and white colonies, formed only when plasmid-containing cells do not rapidly outgrow plasmid-free cells (18). The repΔC33 ΔuvrD strain also formed plasmid-free colonies on LB but these colonies were reduced in size as compared with those retaining the uvrD+ plasmid (Figure 3A, compare ii and i). The growth defect of repΔC33 ΔuvrD cells was reflected in the absence of mosaic colonies, reflecting the inability of plasmid-free cells to compete with uvrD+ plasmid-containing cells. Thus repΔC33 ΔuvrD cells displayed a viability defect under rapid growth conditions although this defect was not as extreme as that seen with Δrep ΔuvrD cells which fail to form any visible plasmid-less colonies on rich medium (9). In contrast, analysis of plasmid segregation under reduced growth conditions on minimal agar demonstrated that plasmid-free segregants of repΔC33 ΔuvrD formed with a colony size similar to that of rep+ ΔuvrD (Figure 3A). Limiting the growth rate thus restores the viability of repΔC33 ΔuvrD, as seen with Δrep ΔuvrD (9). The rich medium-dependent viability defect of repΔC33 ΔuvrD cells was confirmed by isolating plasmid-free segregants on minimal agar, subsequent growth in liquid minimal medium and then monitoring growth upon subculturing into LB. repΔC33 ΔuvrD cells did continue to produce viable progeny in LB, unlike Δrep ΔuvrD cells, but the growth rate was substantially lower than with rep+ ΔuvrD (Figure 3Bi). Indeed, although repΔC33 ΔuvrD cells could continue to divide in rich medium they were highly filamented as compared with rep+ ΔuvrD (Figure 3Bii).
Figure 3.
The Rep–DnaB interaction promotes viability of ΔuvrD cells in a transcription-dependent manner. (A) The ability of the indicated strains to form colonies in the absence of plasmid-encoded UvrD, as indicated by the formation of white colonies in the presence of Xgal and IPTG, was monitored under rapid growth conditions (LB) and under restricted growth conditions (MA, minimal agar). Numbers under each panel refer to the fraction of white colonies obtained, whilst the numbers in parentheses indicate the actual number of white colonies and total number of colonies. (B) (i) Growth rates of ΔuvrD rep+ (N7912), ΔuvrD repΔC33 (N7913) and ΔuvrD Δrep (N6644), all lacking pRC7 encoding UvrD, upon transfer of cells from minimal medium into LB. (ii) Median lengths of cells grown in minimal medium immediately prior to transfer into LB, and after 3 h in LB. (C) Retention or loss of pRC7-encoded UvrD in (i) ΔuvrD rep+ and (ii) ΔuvrD repΔC33 strains bearing rpoB*35 was monitored on LB plates containing Xgal and IPTG.
The inviability of Δrep ΔuvrD cells on rich medium can be suppressed by mutations in RNA polymerase, reflecting the importance of transcribing or stalled RNA polymerases as nucleoprotein blocks to replication (9–11). The ability of a mutation in rpoB, known to destabilize transcription complexes (25) and to suppress Δrep ΔuvrD inviability (9), was tested for suppression of the repΔC33 ΔuvrD viability defect. Loss of plasmid-encoded UvrD in repΔC33 ΔuvrD rpoB*35 resulted in plasmid-less colonies of a size similar to those seen with strains bearing rep+ (Figure 3C), in marked contrast to repΔC33 ΔuvrD rpo+ (Figure 3A). Destabilization of transcription complexes can suppress therefore the viability defect observed in repΔC33 ΔuvrD cells.
For Δrep and repΔC33 the correlation between increased chromosome duplication time in uvrD+ cells (Figure 1), growth defects in ΔuvrD cells (Figure 3) and suppression of these viability defects by rpoB*35 (Figure 3C) indicates that inefficient fork movement along transcribed DNA is responsible for decreased mean fork speed. Moreover, since the repΔC33 ΔuvrD growth defect on rich medium was not as pronounced as that seen with Δrep ΔuvrD cells (Figure 3Bi) (9), we conclude that Rep helicase can promote fork movement along protein-bound DNA in the absence of interaction with DnaB but that this promotion is restricted.
Rep localization with DnaB minimizes the need for blocked fork processing
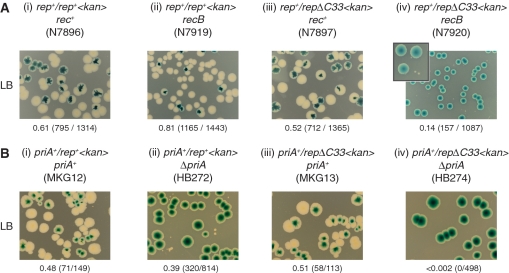
Cells lacking both Rep and the helicase/exonuclease RecBCD are inviable (26) indicating that increased fork blockage in the absence of Rep creates a requirement for blocked fork processing by RecBCD (27). We tested if repΔC33 also rendered recB cells inviable by establishing whether recB strains bearing either rep+ or repΔC33 could survive in the absence of pRC7 encoding Rep. Plasmid-free colonies were formed by repΔC33 rec+ with a frequency and size comparable to rep+ rec+ (Figure 4Aiii and i). However, repΔC33 recB formed large blue colonies but no white colonies of comparable size, in contrast to rep+ recB (Figure 4Aiv and ii). Micro-colonies did also form with repΔC33 recB (Figure 4Aiv, inset) but these could not be subcultured (data not shown). The absence of RecBCD caused therefore a major growth defect in repΔC33 cells, consistent with absence of the Rep–DnaB interaction increasing the need for blocked fork processing by RecBCD.
Figure 4.
The Rep–DnaB interaction minimizes the need for blocked fork processing and replication restart. (A) Retention or loss of pRC7-encoded Rep in rec+ and recB strains bearing chromosomal rep+ or repΔC33 alleles monitored on LB plates containing Xgal and IPTG. The inset panel in (iv) is an expanded view of blue and white colonies, illustrating the formation of micro-colonies alongside the large plasmid-containing blue colonies. (B) Retention or loss of pRC7-encoded PriA in priA+ and ΔpriA strains bearing chromosomal rep+ or repΔC33 alleles monitored on LB containing Xgal and IPTG.
Processing of blocked replication forks results ultimately in a need to reload the replication machinery back onto the chromosome, either by PriA or by PriC (28–30). Cells lacking either PriA or PriC are viable whereas cells lacking both are inviable, indicating the importance of replisome reloading away from oriC (29). Cells lacking both PriA and Rep are also inviable which may reflect a requirement for Rep in PriC-directed restart (29) but might also be caused by absence of Rep increasing the frequency of fork blockage and a consequent increased need for PriA-dependent fork reassembly.
The ability of ΔpriA cells to survive in combination with repΔC33 was tested. As shown previously (18), rep+ ΔpriA cells bearing pRC7 encoding PriA could form plasmid-free colonies (Figure 4Bii). These plasmid-free segregants were much smaller than those obtained in rep+ priA+ cells (Figure 4B, compare ii with i), reflecting the decreased viability of ΔpriA cells (31). In contrast, whilst large plasmid-free segregants were formed by repΔC33 priA+ cells, no segregants were formed from repΔC33 ΔpriA cells (Figure 4B, compare iii and iv). PriA was essential therefore in repΔC33 cells, indicating that absence of the Rep–DnaB interaction increased the requirement for PriA-directed replication restart. We cannot exclude the possibility that the growth defects noted in repΔC33 ΔpriA cells are associated with defects in Rep/PriC-dependent replisome reloading (29) rather than Rep-promoted fork movement. However, the viability defects of repΔC33 recB cells (Figure 4A) support a model in which there is an elevated requirement for RecBCD-dependent blocked fork processing and consequent PriA-directed replication restart due to increased fork blockage in the absence of a Rep–DnaB interaction.
Interaction of DnaB with Rep does not impact directly on replisome movement
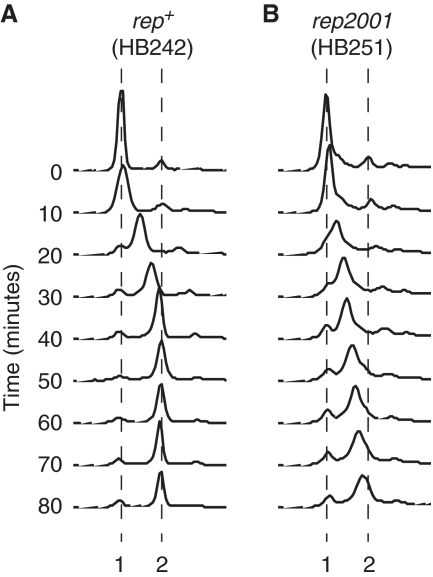
The above data demonstrate that the Rep–DnaB interaction is critical for Rep helicase function in vivo. We also tested whether the Rep–DnaB interaction has an allosteric effect on replisome function. In other words, does the interaction of DnaB with Rep contribute to the maintenance of wild-type rates of replication fork movement independent of Rep helicase activity? Chromosome duplication time was measured in a rep2001 strain encoding RepK28R in which the lysine residue within helicase motif I (32,33) was mutated to arginine. This lysine residue interacts with the phosphate tail of ATP in Rep and other Superfamily 1 helicases (34–36) and has been shown to be required for ATP hydrolysis and hence helicase activity in many helicases (37). Thus rep2001 is likely to encode a helicase-defective Rep but one that retains the C-terminal 33 amino acids known to interact with DnaB (9). As seen in Δrep cells (Figure 1B), chromosome duplication in rep2001 cells was incomplete after 40 min, with a significant fraction of cells having less than two chromosome equivalents even after 80 min (Figure 5, compare parts A and B). Thus the Rep–DnaB interaction per se has no detectable impact on mean replication fork speed, indicating the absence of any significant direct positive effect of this interaction on genome duplication time. The Rep–DnaB interaction is critical therefore only in terms of Rep helicase function, not in terms of the integral speed of the replisome.
Figure 5.
The Rep–DnaB interaction does not alter the mean integral speed of the replisome. Chromosome duplication was monitored by flow cytometry in dnaA46 strains bearing rep alleles encoding (A) wild-type Rep and (B) RepK28R. Time zero indicates samples removed after 2 h at 42°C immediately prior to shifting the cultures to 30°C. Positions of 1 and 2 chromosome equivalents are indicated.
DISCUSSION
We have shown that although absence of Rep increases the time required to replicate a chromosome, absence of either UvrD or DinG has no detectable effect (Figure 1). Moreover, this decrease in mean replication fork speed in Δrep cells is not due to involvement of Rep in replication restart (Figure 2). Rapid genome duplication in E. coli requires therefore only one type of accessory helicase, Rep, and this underpinning occurs via promotion of fork movement along protein-bound DNA rather than replication restart.
Our data also demonstrate that interaction of Rep with DnaB is critical for the ability of wild-type levels of Rep to maintain wild-type rates of chromosome duplication (Figure 1) and to promote viability in the absence of UvrD (Figure 3), a phenotype associated with problems replicating protein-bound DNA (9,10). Moreover, suppression of the repΔC33 ΔuvrD viability defect by an RNA polymerase mutation (Figure 3C) demonstrates that conflicts between replication and gene expression create the need for Rep colocalization with DnaB. Absence of the Rep–DnaB interaction also increases the requirement for RecBCD (Figure 4A) indicating an increased need for blocked fork processing by RecBCD (27). RepΔC33 also cannot support colony formation in cells lacking the replication restart protein PriA (Figure 4B). Although we cannot distinguish whether repΔC33 ΔpriA lethality is due to increased fork blockage or to defective PriC-catalysed replication restart (29), this lethality indicates that the Rep–DnaB interaction is important for all aspects of Rep function in vivo. In contrast, this interaction in itself did not have any major allosteric effect on replisome movement (Figure 5). The Rep–DnaB interaction appears therefore to modulate Rep function only.
Comparison of the viability of cells bearing the repΔC33 allele in combination with ΔuvrD, recB or ΔpriA (Figures 3 and 4) indicates that Rep helicase in the absence of interaction with DnaB can sustain cell division, albeit at a reduced efficiency, in the absence of UvrD but not of RecBCD or PriA. It is tempting to speculate that recombinational processing of blocked forks and replisome reassembly are more important therefore than clearance of protein blocks ahead of replication forks. However, the multiple blocked fork processing pathways existing in vivo, with the attendant possibility for off pathway reactions to occur in mutant strains that are never normally operative in wild-type cells, make such speculation meaningless. The severe viability defects seen with all the above mutant combinations suggest, though, that a combination of accessory helicase activity, blocked fork processing and replisome reloading is important in underpinning genome duplication.
Why might localization with DnaB be critical for Rep function? Functional cooperativity between helicases translocating along ssDNA may promote displacement of proteins from the DNA (38). Increasing the local concentration of Rep near the replisome may facilitate therefore the loading of multiple Rep monomers onto ssDNA at forks, aiding the clearance of proteins ahead of blocked forks and subsequent resumption of fork movement (9). In this view of the function of the Rep–DnaB interaction, this interaction does not impact directly on either DnaB or Rep catalysis, it merely increases the effective concentration of Rep at the required site within the cell. This model also implies that absence of a UvrD–DnaB interaction renders the 500–900 UvrD molecules per cell (39) (and data not shown) incapable of sustaining wild-type rates of chromosome duplication, given the 2-fold increase in chromosome duplication time in uvrD+ Δrep cells (Figure 1). However, we cannot exclude the possibility that other differences between Rep and UvrD, in addition to the interaction with DnaB, may contribute to the ability of Rep but not UvrD to sustain wild-type rates of chromosome duplication.
Regardless of whether interaction with DnaB facilitates functional cooperativity between multiple Rep molecules, it remains unknown whether Rep is always present at replication forks or whether Rep associates only with blocked replisomes. The high affinity association between Rep and DnaB (9) might suggest that Rep is continuously associated with moving forks. However, it remains possible that other components within a moving replisome inhibit the Rep–DnaB interaction, possibly by masking the interaction domain on DnaB, and that such inhibition is relieved only within the context of a blocked fork.
If a high concentration of an accessory helicase at the fork is all that is required to underpin replisome movement, is it possible that other organisms rely on accessory helicases that operate at replisomes by virtue of high intracellular concentration rather than colocalization with replisomes? Such a system would imply non-specific targeting of accessory helicases to DNA. However, an emerging theme in regulation of helicase catalysis in vivo is that other factors target helicases to specific substrates, in effect conferring DNA substrate specificity on helicases. Untargeted and hence unregulated helicase activity may therefore be deleterious with respect to maintenance of genome stability. Note that although the absence of any significant impact of high level plasmid-based expression of Rep on viability suggests that unrestricted Rep activity is not toxic (9,40), more subtle detrimental effects cannot be excluded. DnaB could be viewed therefore as a substrate specificity factor that allows low concentrations of Rep to underpin genome duplication effectively in vivo.
Localization at the replisome may be a conserved feature of accessory helicases. The only other known accessory replicative helicase, S. cerevisiae Rrm3p, is also associated with the replisome although this association may be via the sliding clamp and/or the leading strand polymerase rather than the replicative helicase (14,15). Our work with Rep demonstrates that physical association with the replisome has an important functional impact on accessory helicase activity. The work presented here also suggests that the identity of the interacting partner within the replisome may not be critical so long as the local concentration of the accessory helicase at the fork is high.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biotechnology and Biological Sciences Research Council (BB/G005915/1 and BB/E0020690 to P.M.); MRC (G0800970 to R.G.L.); Leverhulme Trust (to C.J.R.). Funding for open access charge: BBSRC.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Steve Sandler and Bénédicte Michel for supplying SS1076 and JJC478, respectively, and Carol Brown and Lynda Harris for superlative technical support.
REFERENCES
- 1.French S. Consequences of replication fork movement through transcription units in vivo. Science. 1992;258:1362–1365. doi: 10.1126/science.1455232. [DOI] [PubMed] [Google Scholar]
- 2.Yancey-Wrona JE, Matson SW. Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res. 1992;20:6713–6721. doi: 10.1093/nar/20.24.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 4.Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009;37:3475–3492. doi: 10.1093/nar/gkp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matson SW, Bean DW, George JW. DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. Bioessays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 6.Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol. Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keil RL, McWilliams AD. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae. Genetics. 1993;135:711–718. doi: 10.1093/genetics/135.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taucher-Scholtz G, Abdel-Monem M, Hoffmann-Berling H. In: Mechanisms of DNA Replication and Recombination. Cozzarelli NR, editor. New York: Alan R. Liss Inc.; 1983. pp. 65–76. [Google Scholar]
- 9.Guy CP, Atkinson J, Gupta MK, Mahdi AA, Gwynn EJ, Rudolph CJ, Moon PB, van Knippenberg IC, Cadman CJ, Dillingham MS, et al. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol. Cell. 2009;36:654–666. doi: 10.1016/j.molcel.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boubakri H, de Septenville AL, Viguera E, Michel B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 2010;29:145–157. doi: 10.1038/emboj.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baharoglu Z, Lestini R, Duigou S, Michel B. RNA polymerase mutations that facilitate replication progression in the rep uvrD recF mutant lacking two accessory replicative helicases. Mol. Microbiol. 2010;77:324–336. doi: 10.1111/j.1365-2958.2010.07208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lane HE, Denhardt DT. The rep mutation. IV. Slower movement of replication forks in Escherichia coli rep strains. J. Mol. Biol. 1975;97:99–112. doi: 10.1016/s0022-2836(75)80025-8. [DOI] [PubMed] [Google Scholar]
- 13.Heller RC, Marians KJ. Unwinding of the nascent lagging strand by Rep and PriA enables the direct restart of stalled replication forks. J. Biol. Chem. 2005;280:34143–34151. doi: 10.1074/jbc.M507224200. [DOI] [PubMed] [Google Scholar]
- 14.Azvolinsky A, Dunaway S, Torres JZ, Bessler JB, Zakian VA. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 2006;20:3104–3116. doi: 10.1101/gad.1478906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt KH, Derry KL, Kolodner RD. Saccharomyces cerevisiae RRM3, a 5′ to 3′ DNA helicase, physically interacts with proliferating cell nuclear antigen. J. Biol. Chem. 2002;277:45331–45337. doi: 10.1074/jbc.M207263200. [DOI] [PubMed] [Google Scholar]
- 16.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahdi AA, Buckman C, Harris L, Lloyd RG. Rep and PriA helicase activities prevent RecA from provoking unnecessary recombination during replication fork repair. Genes Dev. 2006;20:2135–2147. doi: 10.1101/gad.382306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atkinson J, Guy CP, Cadman CJ, Moolenaar GF, Goosen N, McGlynn P. Stimulation of UvrD helicase by UvrAB. J. Biol. Chem. 2009;284:9612–9623. doi: 10.1074/jbc.M808030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maisnier-Patin S, Nordstrom K, Dasgupta S. Replication arrests during a single round of replication of the Escherichia coli chromosome in the absence of DnaC activity. Mol. Microbiol. 2001;42:1371–1382. doi: 10.1046/j.1365-2958.2001.02718.x. [DOI] [PubMed] [Google Scholar]
- 21.Bernander R, Akerlund T, Nordstrom K. Inhibition and restart of initiation of chromosome replication: effects on exponentially growing Escherichia coli cells. J. Bacteriol. 1995;177:1670–1682. doi: 10.1128/jb.177.7.1670-1682.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernander R, Nordstrom K. Chromosome replication does not trigger cell division in E. coli. Cell. 1990;60:365–374. doi: 10.1016/0092-8674(90)90588-6. [DOI] [PubMed] [Google Scholar]
- 23.Hinds T, Sandler SJ. Allele specific synthetic lethality between priC and dnaAts alleles at the permissive temperature of 30°C in E. coli K-12. BMC Microbiol. 2004;4:47. doi: 10.1186/1471-2180-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernhardt TG, de Boer PA. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol. Microbiol. 2004;52:1255–1269. doi: 10.1111/j.1365-2958.2004.04063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol. Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Uzest M, Ehrlich SD, Michel B. Lethality of rep recB and rep recC double mutants of Escherichia coli. Mol. Microbiol. 1995;17:1177–1188. doi: 10.1111/j.1365-2958.1995.mmi_17061177.x. [DOI] [PubMed] [Google Scholar]
- 27.Seigneur M, Bidnenko V, Ehrlich SD, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Xu L, Sandler SJ, Marians KJ. Replication fork assembly at recombination intermediates is required for bacterial growth. Proc. Natl Acad. Sci. USA. 1999;96:3552–3555. doi: 10.1073/pnas.96.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandler SJ. Multiple genetic pathways for restarting DNA replication forks in Escherichia coli K-12. Genetics. 2000;155:487–497. doi: 10.1093/genetics/155.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heller RC, Marians KJ. The disposition of nascent strands at stalled replication forks dictates the pathway of replisome loading during restart. Mol. Cell. 2005;17:733–743. doi: 10.1016/j.molcel.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 31.Nurse P, Zavitz KH, Marians KJ. Inactivation of the Escherichia coli priA DNA replication protein induces the SOS response. J. Bacteriol. 1991;173:6686–6693. doi: 10.1128/jb.173.21.6686-6693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorbalenya AE, Koonin EV. Helicases – amino-acid-sequence comparisons and structure-function-relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 33.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 35.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 36.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall MC, Matson SW. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 1999;34:867–877. doi: 10.1046/j.1365-2958.1999.01659.x. [DOI] [PubMed] [Google Scholar]
- 38.Byrd AK, Raney KD. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- 39.George JW, Brosh RM, Jr, Matson SW. A dominant negative allele of the Escherichia coli uvrD gene encoding DNA helicase II. A biochemical and genetic characterization. J. Mol. Biol. 1994;235:424–435. doi: 10.1006/jmbi.1994.1003. [DOI] [PubMed] [Google Scholar]
- 40.Colasanti J, Denhardt DT. The Escherichia coli rep mutation. X. Consequences of increased and decreased Rep protein levels. Mol. Gen. Genet. 1987;209:382–390. doi: 10.1007/BF00329669. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.