Abstract
Human topoisomerase I plays an important role in removing positive DNA supercoils that accumulate ahead of replication forks. It also is the target for camptothecin-based anticancer drugs that act by increasing levels of topoisomerase I-mediated DNA scission. Evidence suggests that cleavage events most likely to generate permanent genomic damage are those that occur ahead of DNA tracking systems. Therefore, it is important to characterize the ability of topoisomerase I to cleave positively supercoiled DNA. Results confirm that the human enzyme maintains higher levels of cleavage with positively as opposed to negatively supercoiled substrates in the absence or presence of anticancer drugs. Enhanced drug efficacy on positively supercoiled DNA is due primarily to an increase in baseline levels of cleavage. Sites of topoisomerase I-mediated DNA cleavage do not appear to be affected by supercoil geometry. However, rates of ligation are slower with positively supercoiled substrates. Finally, intercalators enhance topoisomerase I-mediated cleavage of negatively supercoiled substrates but not positively supercoiled or linear DNA. We suggest that these compounds act by altering the perceived topological state of the double helix, making underwound DNA appear to be overwound to the enzyme, and propose that these compounds be referred to as ‘topological poisons of topoisomerase I’.
INTRODUCTION
Globally, DNA in all living systems ranging from eubacteria to humans is under torsional stress (1–4). The double helix is ∼6% underwound (i.e. negatively supercoiled) as compared to the ideal Watson–Crick structure (5). This underwinding puts energy into DNA and enhances the ability to open the double helix so that the genetic information can be duplicated or expressed. In contrast, the movement of tracking enzymes through the genetic material causes the DNA ahead of replication forks or transcription complexes to become overwound (1,2,4,6,7). The resulting positive DNA supercoils impair the ability to separate the two strands of the double helix and eventually block these and other essential nucleic acid processes (1,6,8–10).
The enzymes that remove (i.e. relax) negative and positive superhelical twists from DNA are known as topoisomerases (1,6,11–16). All topoisomerases function by generating transient breaks in the DNA backbone. There are two classes of topoisomerases, categorized by the number of strands that they cut. Type I and II enzymes generate transient single- and double-stranded breaks, respectively (1,6,11–16). Humans encode five nuclear topoisomerases: topoisomerase I, IIIα and IIIβ (which are type I enzymes) and topoisomerase IIα and IIβ (which are type II enzymes) (1,6,11–16). In order to maintain genomic integrity during enzyme function, all topoisomerases form covalent bonds between active site tyrosyl residues and the newly generated DNA termini (1,6,11–16). These covalent enzyme-cleaved DNA complexes are known as ‘cleavage complexes’.
Human topoisomerase I relaxes negative and positive superhelical twists by a controlled rotation mechanism (11,14,17–19). In contrast, topoisomerase IIIα and IIIβ utilize a single-stranded DNA passage mechanism (11,20). Since these latter enzymes require their DNA substrates to contain considerable single-stranded character, they relax only underwound molecules (11,20,21). Topoisomerase IIα and IIβ act by passing an intact DNA duplex through a transient double-stranded DNA break (11,15,20,22,23). Hence, they can relax positively or negatively supercoiled molecules and can also remove knots and tangles from the genetic material (6,11,15,20,22–24).
As a result of its catalytic mechanism, topoisomerase I plays an important role in removing positive DNA supercoils that accumulate ahead of replication forks and transcription complexes (1,6,11,14,17,25). Topoisomerase IIα also is capable of removing positive DNA supercoils that form during DNA replication (24). In light of their physiological roles, it is not surprising that these two enzymes can distinguish the geometry of DNA supercoils. Indeed, recent studies indicate that topoisomerase I and topoisomerase IIα both remove positive superhelical twists ∼10-fold faster than they do negative superhelical twists (17,24).
Beyond their important cellular functions, human topoisomerase I and topoisomerase IIα are targets for a number of highly effective anticancer agents that act at the enzyme–DNA interface (13,22,26–30). Topoisomerase I is the target of an emerging class of drugs based on the parent compound camptothecin (27,28,30). Two derivatives, topotecan (a water soluble formulation) and irinotecan (a prodrug that is activated in vivo) are used for the treatment of colorectal, gynecological and other cancers (27,28,30). Topoisomerase IIα is the target for a number of established anticancer drugs, including etoposide and adriamycin, that are used as front line therapy for a wide variety of human malignancies (13,22,26,29).
All of these drugs are referred to as topoisomerase ‘poisons’ as opposed to ‘catalytic inhibitors’ and function by increasing levels of enzyme–DNA cleavage complexes. The accumulation of these complexes ahead of DNA tracking systems is believed to kill cells by several different mechanisms (13,22,25–30). First, the presence of cleavage complexes or positive supercoils ahead of the replication or transcription machinery impairs these essential cellular functions. Second, the presence of blocked replication forks induces replication re-start pathways that generate DNA strand breaks. Third, collisions between DNA tracking systems and covalent topoisomerase roadblocks convert transient cleavage complexes to permanent DNA strand breaks. Since the DNA ahead of DNA tracking systems should be overwound, cleavage complexes that are most likely to block essential nuclear processes or generate permanent strand breaks are formed on positively supercoiled DNA.
Topoisomerase IIα maintains lower levels (∼2- to 4-fold) of cleavage complexes with positively supercoiled as opposed to negatively supercoiled DNA (24,31,32). While this feature makes the enzyme safer to function ahead of replication and transcription complexes, it may make it less sensitive to the actions of anticancer drugs. In contrast, preliminary reports suggest that human topoisomerase I maintains higher levels of cleavage complexes with positively supercoiled substrates (17,31). This property makes the type I enzyme a potentially better target for therapeutic agents. However, it also suggests that topoisomerase I is intrinsically more dangerous to the cell than the type II enzyme.
Because of the fundamental role that topoisomerase I plays in a number of critical nuclear processes and in the treatment of human malignancies, it is important to more fully characterize the ability of the enzyme to cleave positively supercoiled DNA. Results confirm that topoisomerase I maintains higher levels of cleavage complexes with positively as opposed to negatively supercoiled substrates in the absence or presence of anticancer drugs. Furthermore, this effect correlates with a decreased rate of ligation with overwound DNA. Finally, intercalating agents that make covalently closed DNA appear to be positively supercoiled enhance topoisomerase I-mediated DNA cleavage in vitro and in cultured human cells. We propose that this latter class of compounds be referred to as ‘topological poisons of topoisomerase I’.
MATERIALS AND METHODS
Enzymes and materials
Human topoisomerase I was expressed in Saccharaomyces cerevisiae top1 null strain RS190 (a gift from R. Sternglanz, State University of New York at Stony Brook) and purified as described earlier (33).
Positively supercoiled pBR322 DNA was prepared by incubating negatively supercoiled plasmids with Archaeoglobus fulgidus reverse gyrase as described by McClendon et al. (24). The average number of superhelical twists present in DNA substrates and the resulting σ values were determined by electrophoretic band counting relative to relaxed molecules. Typical of plasmids isolated from Escherichia coli, negatively supercoiled plasmids contained ∼15–17 negative superhelical twists per molecule (σ ≈ −0.035 to −0.039). Positively supercoiled plasmids contained ∼15–17 positive superhelical twists per molecule (σ ≈ +0.035 to +0.039). Thus, the supercoiled substrates employed for this study contained equivalent numbers of superhelical twists but were of opposite handedness.
It should be noted that positively supercoiled plasmids bind less ethidium bromide than negatively supercoiled molecules (24). To ensure that equal amounts of plasmid were used in all experiments, the DNA concentration was assessed by spectrophotometric analysis and confirmed by ethidium bromide staining of linearized plasmid substrates.
[α-32P]dATP (6000 Ci/mmol) and [α-32P]TTP (3000 Ci/mmol) were obtained from New England Nuclear. Camptothecin, ethidium bromide and 9-aminoacridine were from Sigma, and topotecan was from Alexis Biochemicals. Tas-103 was a gift from Taiho Pharmaceuticals. Amsacrine was a gift from David Graves (University of Alabama at Birmingham). All other chemicals were analytical reagent grade. Camptothecin and topotecan were stored at −20°C as 10 mM stock solutions in 100% DMSO and water, respectively. Amsacrine and 9-aminoacridine were stored at 4°C as 20 mM stock solutions in 100% DMSO. TAS-103 and ethidium bromide were stored at 4°C as 10 mM and 2.5 mM stock solutions in water, respectively.
Cleavage of circular plasmid DNA by human topoisomerase I
Unless indicated otherwise, DNA cleavage reactions were carried out by incubating 5 nM positively or negatively supercoiled pBR322 plasmid DNA with 11 nM human topoisomerase I in 20 µl of cleavage buffer (10 mM Tris–HCl, pH 7.5, 15 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 5 mM CaCl2, 2.5% glycerol). Reactions were incubated for 2 min at 37°C, and enzyme–DNA cleavage complexes were trapped by the addition of 2 µl of 5% SDS followed by 1 µl of 375 mM EDTA, pH 8.0. Proteinase K (2 µl of a 0.8 mg/ml solution) was added, and protein samples were incubated for 30 min at 45°C to digest the topoisomerase I. Samples were mixed with 2 µl of agarose gel loading buffer (60% sucrose in 10 mM Tris–HCl, pH 7.9, 0.1% xylene cyanol and 0.1% bromophenol blue) heated for 5 min at 45°C, and subjected to electrophoresis in 1% agarose gels in 40 mM Tris–acetate, pH 8.3 and 2 mM EDTA containing 0.5 µg/ml ethidium bromide. DNA bands were visualized with ultraviolet light and quantified using an Alpha Innotech digital imaging system. DNA cleavage was monitored by the conversion of covalently closed circular supercoiled plasmid DNA to nicked molecules.
The effects of anticancer drugs on topoisomerase I-mediated cleavage of positively or negatively supercoiled plasmid DNA were assessed in the presence of 0–10 µM camptothecin or topotecan. The effects of intercalators were assessed in the presence of 0–20 µM ethidium bromide, 0–20 µM TAS-103, 0–100 µM 9-aminoacridine, or 0–400 µM amsacrine.
Site-specific cleavage of circular DNA by human topoisomerase I
DNA cleavage reactions contained 7 nM negatively or positively supercoiled pBR322 DNA and 14 nM human topoisomerase I (in the absence or presence of 5 µM topotecan) in a total of 160 µl of cleavage buffer. Samples were incubated for 2 min at 37°C and enzyme–DNA cleavage complexes were trapped by the addition of 16 µl of 1% SDS followed by 8 µl 375 mM EDTA. Proteinase K (3 µl of a 4 mg/ml solution) was added, and mixtures were incubated for 30 min at 45°C to digest the topoisomerase I. Reaction products were purified by passage through Qiaquick spin columns (Qiagen) as described by the manufacturer. Plasmids were linearized by treatment with EcoRI and labeled with Klenow (exo-) (New England Biolabs) in the presence of [α-32P]dATP and [α-32P]TTP. Samples were treated with HindIII, and the singly end-labeled 4330-bp DNA fragment was purified by passage through a CHROMA SPIN+ TE-10 column (Clontech). Reaction products were normalized for radioactivity. Equivalent counts were mixed with 5 µl of polyacrylamide gel loading buffer (80% formamide, 10 mM sodium hydroxide, 1 mM sodium EDTA, 0.1% xylene cyanol and 0.1% bromophenol blue), and subjected to electrophoresis in 6% polyacrylamide sequencing gels. Gels were dried in vacuo, and DNA cleavage products were visualized with a Bio-Rad Molecular Imager FX.
Site-specific cleavage of linear DNA by human topoisomerase I
pBR322 was linearized, labeled, and the 4330-bp EcoRI-HindIII fragment was isolated as described in the preceding section. DNA cleavage reactions (20 µl) contained 4.4 nM labeled linear pBR322 DNA and 20 nM human topoisomerase I, in the absence of drugs or in the presence of topoisomerase I-targeted anticancer drugs (10 µM camptothecin or topotecan) or DNA intercalators (20 µM ethidium bromide, 20 µM TAS-103, 100 µM 9-aminoacridine or 200 µM amsacrine). Reactions were incubated for 2 min at 37°C, and enzyme–DNA cleavage complexes were trapped by the addition of 2 µl of 5% SDS followed by 1 µl of 375 mM EDTA, pH 8.0. Proteinase K (2 µl of a 0.8 mg/ml solution) was added, and protein samples were incubated for 30 min at 45°C to digest the topoisomerase I. Reaction products were ethanol precipitated and resuspended in 6 µl of polyacrylamide gel loading buffer. Samples were subjected to polyacrylamide gel electrophoresis and analyzed as described in the preceding section.
Ligation of cleaved DNA by human topoisomerase I
DNA cleavage-ligation equilibria were established for 2 min at 37°C in cleavage buffer that contained 10 µM camptothecin or 5 µM topotecan, as described in the section on cleavage of circular DNA. Ligation was initiated by the addition of NaCl to a final concentration of 300 mM and terminated from 5 to 45 s by the addition of 2 µl 5% SDS. Samples were processed and analyzed as described for circular DNA cleavage. The percent DNA cleavage at Time 0 was set to 100%, and ligation was monitored by quantifying the loss of nicked DNA over time.
DNA intercalation
Intercalation reaction mixtures contained 20 nM topoisomerase I and 5 nM pBR322 DNA in a total of 20 µl of 50 mM Tris–HCl (pH 7.5), 0.1 mM EDTA, 50 mM KCl, 10 mM MgCl2 and 0.5 mM DTT. Reactions contained 0–10 µM ethidium bromide or TAS–103, 0–50 µM 9-aminoacridine or 0–100 µM amsacrine. Mixtures were incubated at 37°C for 10 min, extracted with a phenol:chloroform:isoamyl alcohol mixture (25:24:1), and added to 3 µl of 0.77% SDS and 77 mM EDTA (pH 8.0). Samples were mixed with 2 µl of agarose gel loading buffer, heated at 45°C for 5 min, and subject to electrophoresis in a 1% agarose gel in 100 mM Tris–borate (pH 8.3) and 2 mM EDTA. Gels were stained with 1 µg/ml ethidium bromide, and DNA bands were visualized as described for plasmid DNA cleavage.
The DNA intercalation assay is based on the fact that intercalative agents induce constrained negative supercoils and compensatory unconstrained positive superhelical twists in covalently closed circular DNA (31,34). Therefore, as the concentration of an intercalative compound increases, a plasmid that is negatively supercoiled or relaxed (i.e. contains no superhelical twists) appears to become positively supercoiled. Treatment of an intercalated plasmid with topoisomerase I removes the unconstrained positive DNA superhelical twists. Subsequent extraction of the compound allows the local drug-induced unwinding to redistribute in a global manner and manifest itself as a net negative supercoiling of the plasmid. Thus, in the presence of an intercalative agent, topoisomerase treatment converts relaxed plasmids to negatively supercoiled molecules (see inset, Figure 5).
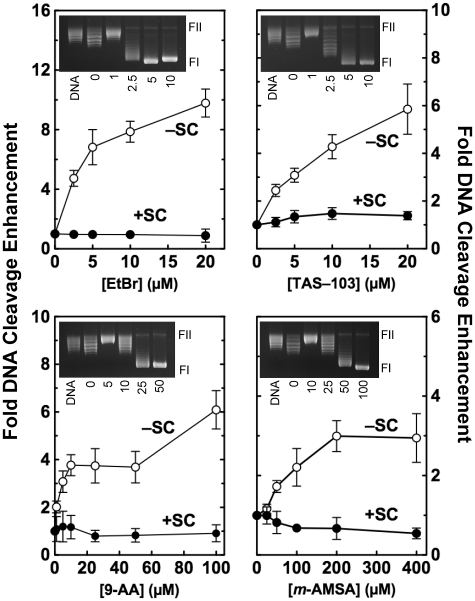
Figure 5.
Effects of DNA intercalators on topoisomerase I-mediated cleavage of plasmid DNA. Cleavage of positively [(+)SC, closed circles] and negatively [(−)SC, open circles] supercoiled pBR322 plasmid DNA was monitored in the presence of ethidium bromide (EtBr), TAS-103, 9-aminoacridine (9-AA) or amsacrine (m-AMSA). Data were plotted as relative (i.e. fold) DNA cleavage enhancement for simplicity and to aid in visualizing the effects of intercalators on topoisomerase I-mediated cleavage of positively and negatively supercoiled substrates. Fold DNA cleavage enhancement was calculated by normalizing levels of scission in the absence of intercalator to a relative value of 1.0. Error bars represent the standard deviation of at least three independent experiments. Insets show representative gels of DNA intercalation assays using relaxed plasmids in the absence of enzyme (DNA) or in the presence of the indicated concentration (µM) of compound (see ‘Materials and Methods’ section for the interpretation of intercalation assays). Note that intercalation assays are designed to monitor the DNA relaxation activity of topoisomerase I. Consequently, reactions are terminated under conditions that do not trap enzyme–DNA cleavage complexes. The positions of supercoiled (FI) and nicked circular (FII) molecules are indicated.
Formation of topoisomerase I-DNA cleavage complexes in cultured human cells
Human CEM leukemia cells were cultured in <5% CO2 at 37°C in RPMI 1640 medium (Cellgro by Mediatech, Inc.) containing 10% heat-inactivated fetal calf serum (Hyclone). The in vivo complex of enzyme (ICE) bioassay (as modified on the TopoGen, Inc., website) was utilized to determine levels of topoisomerase I-DNA cleavage complexes formed in the presence of anticancer drugs and/or intercalative compounds. Exponentially growing cultures were treated with no drug, 10 µM ethidium bromide, 5 µM topotecan or 10 µM ethidium bromide + 5 µM topotecan for 1 h. Cells (∼5 × 106) were harvested by centrifugation and lysed by the immediate addition of 3 ml of 1% sarkosyl. Following gentle homogenization in a Dounce homogenizer, cell lysates were layered onto a 2 ml cushion of CsCl (1.5 g/ml) and centrifuged at 45 000 rpm for 15 h at 20°C. DNA pellets were isolated, resuspended in 5 mM Tris–HCl, pH 8.0 and 0.5 mM EDTA, normalized for the amount of DNA present, and blotted onto nitrocellulose membranes using a Schleicher and Schuell slot blot apparatus. Covalent complexes formed between human topoisomerase I and DNA were detected using a polyclonal antibody directed against human topoisomerase I (Topogen) at a 1:3000 dilution.
RESULTS
Cleavage of positively supercoiled DNA by human topoisomerase I
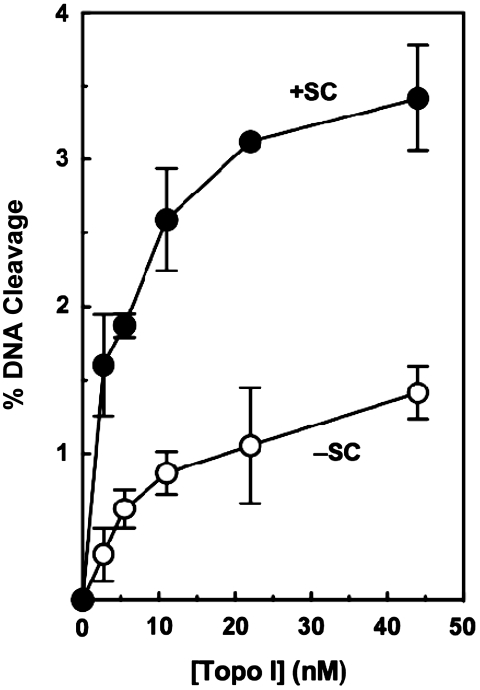
As discussed earlier, topoisomerase I cleavage complexes formed ahead of DNA tracking systems (i.e. on positively supercoiled portions of the genome) are most likely to be converted to permanent strand breaks (4,6,7,25,28,30,35). Previous data generated using a single concentration of human topoisomerase I suggested that the enzyme maintained higher levels of cleavage complexes with positively supercoiled DNA as compared to negatively supercoiled molecules (17,31). Given the importance of the topoisomerase I-DNA cleavage reaction to the physiological and pharmacological functions of the enzyme, we explored the preference for overwound substrates in greater detail. To begin this characterization, the ability of human topoisomerase I to cleave overwound and underwound DNA was assessed over a broad enzyme concentration range (∼3–45 nM). As seen in Figure 1, topoisomerase I retained its ability to discern the geometry of DNA over the entire concentration range, maintaining a concentration of cleavage complexes that was approximately three times greater with substrates that contained positive as compared to negative supercoils. Taken together with the preferential relaxation of positive DNA supercoils (17), these findings strongly suggest that human topoisomerase I is an enzyme that is designed to act primarily on overwound substrates.
Figure 1.
Topoisomerase I maintains higher levels of cleavage complexes with positively supercoiled DNA. The ability of increasing concentrations of human topoisomerase I to cleave positively [(+)SC, closed circles] and negatively [(−)SC, open circles] supercoiled pBR322 plasmid DNA is shown. Error bars represent the standard deviation of at least three independent experiments.
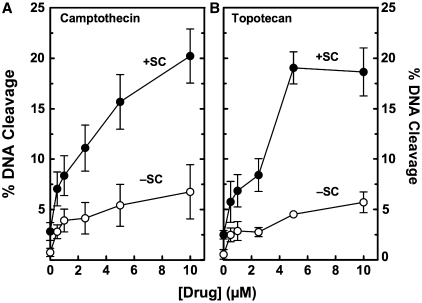
The effects of DNA handedness on topoisomerase I-mediated cleavage in the presence of anticancer drugs are shown in Figure 2. As shown earlier (17,31), higher levels of cleavage were observed with camptothecin and positively supercoiled DNA. Topotecan also induced higher levels of DNA scission with overwound plasmids. In both cases, ∼3-fold more drug-induced scission was observed with positively supercoiled (as compared to negatively supercoiled) substrates over the range of camptothecin and topotecan examined. Since this level of enhancement is similar to that seen in the absence of topoisomerase I poisons, it is proposed that increased drug efficacy on overwound DNA is due primarily to an increase in baseline levels of cleavage rather than an altered drug interaction in the enzyme–DNA complex.
Figure 2.
Effects of DNA supercoil handedness on topoisomerase I-mediated DNA cleavage in the presence of anticancer drugs. The ability of topoisomerase I to cleave positively [(+)SC, closed circles] and negatively [(−)SC, open circles] supercoiled pBR322 plasmid DNA in the presence of 0–10 µM camptothecin (A) or topotecan (B) is shown. Error bars represent the standard deviation of four independent experiments.
Mechanistic basis for increased topoisomerase I-mediated DNA cleavage of positively supercoiled substrates
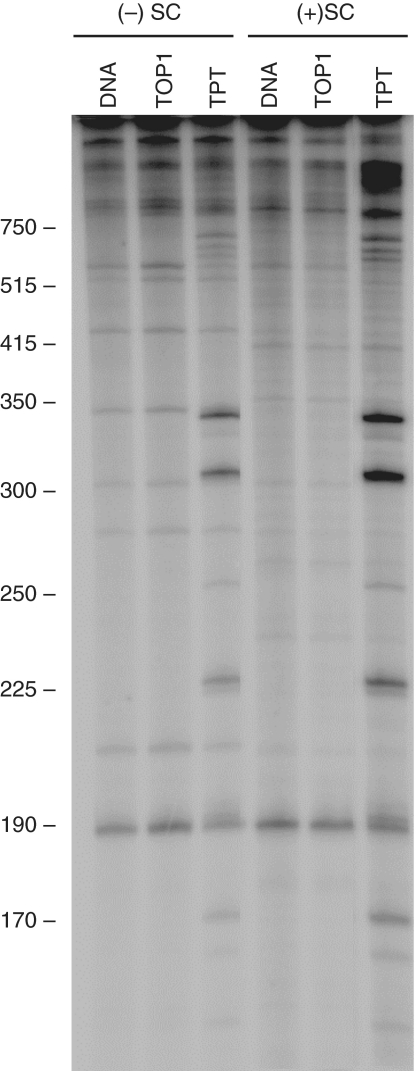
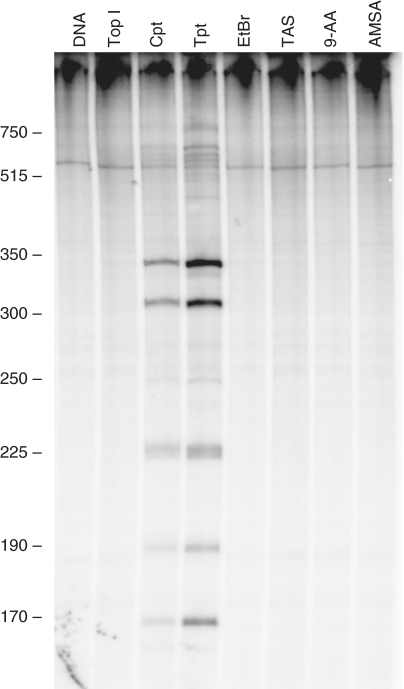
Although topoisomerase I preferentially relaxes and cleaves overwound molecules, it binds positively and negatively supercoiled DNA with similar affinities (17). Therefore, the enhanced cleavage of overwound molecules must result from a different aspect of the enzyme–DNA interaction. One possibility is that topoisomerase I cleaves a broader selection of sites in overwound DNA. Therefore, sites of enzyme-mediated scission were mapped in positively and negatively supercoiled substrates. Mapping in the absence of drugs is difficult due to the low level of cleavage. Consequently, topotecan was included in experiments to increase the overall level of scission. Four to five major and several minor sites of cleavage were observed in the presence of the anticancer drug (Figure 3). In general, corresponding sites were observed in both substrates, but levels of scission were higher when positively supercoiled plasmid was used. Thus, differences in site specificity probably are not the major cause for the enhanced cleavage with overwound substrates.
Figure 3.
Effects of DNA supercoil handedness on sites of topoisomerase I-mediated DNA cleavage. DNA sites cleaved by topoisomerase I were mapped in negatively [(−)SC] and positively [(+)SC] supercoiled pBR322 plasmid DNA in the absence (TOP1) or presence (TPT) of 5 µM topotecan. Untreated DNA is shown as a control (DNA). Following cleavage assays, plasmids were linearized and singly-end-labeled with [32P]-phosphate as described under ‘Materials and Methods’ section. The autoradiogram is representative of three independent experiments. Size markers that were 300 bp and smaller were derived from DNA sequence ladders and those that were 350 bp and larger were derived from restriction digests.
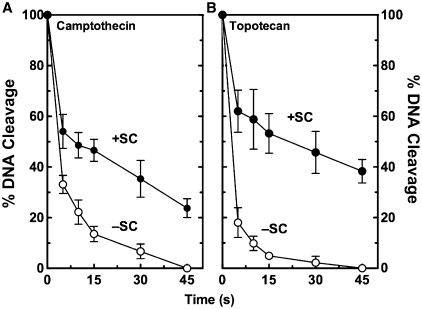
A second possibility is that topoisomerase I maintains higher concentrations of cleavage complexes with overwound molecules because the enzyme ligates these substrates more slowly. In the absence of drugs, rates of enzyme-mediated ligation are too quick to monitor at the bench. Therefore, camptothecin or topotecan (both of which reduce the rate of ligation) were included in assays. In the presence of either drug, topoisomerase I ligated positively supercoiled plasmids more slowly than it did negatively supercoiled DNA (Figure 4). Relative rates of ligation (based on calculated apparent first order constants) for positively versus negatively supercoiled DNA were ∼2.6- and 7.0-fold slower in reactions that contained camptothecin and topotecan, respectively. On the basis of this finding, it is proposed that topoisomerase I maintains higher levels of cleavage complexes with overwound substrates (at least in part) because it ligates them more slowly than it does underwound DNA.
Figure 4.
Effects of DNA supercoil handedness on topoisomerase I-mediated DNA ligation in the presence of anticancer drugs. The ability of topoisomerase I to ligate positively [(+)SC, closed circles] and negatively [(−)SC, open circles] supercoiled pBR322 plasmid DNA was monitored in the presence of 10 µM camptothecin (A) or 5 µM topotecan (B). DNA ligation was initiated by the addition of 300 mM NaCl. Levels of cleavage at time zero were set to 100%. Error bars represent the standard deviation of three independent experiments.
DNA intercalators as topological poisons of topoisomerase I
The binding of intercalators to DNA locally opens (i.e. underwinds) the double helix (31,36,37). In a covalently closed plasmid, this local underwinding is balanced by a compensatory global overwinding of the unconstrained (i.e. unbound) DNA. Thus, even though the overall topological state of the plasmid has not changed, the presence of intercalative agents make the DNA available to the enzyme appear to be positively supercoiled.
Since topoisomerase I maintains higher levels of cleavage complexes with overwound substrates, we investigated the effects of intercalators on enzyme-mediated DNA scission. Four different intercalators, ethidium bromide, 9-aminoacridine, TAS-103 and amsacrine, were employed. The latter two compounds are topoisomerase II poisons (38,39). Significant intercalation was observed over the concentration ranges employed (Figure 5, insets).
As seen in Figure 5, all of the intercalators examined enhanced topoisomerase I-mediated DNA cleavage when added to negatively supercoiled plasmids. Ethidium bromide was the most potent and efficacious of the compounds tested. Approximately 10-fold enhancement of DNA cleavage was observed at 20 µM ethidium bromide. The other three compounds enhanced cleavage ∼3- to 6-fold.
Two possible conclusions can be drawn from the above results. The intercalators may be ‘topological poisons’ of topoisomerase I, enhancing DNA scission by making the negatively supercoiled substrate appear to be positively supercoiled. Alternatively, they may be previously undescribed ‘interfacial’ topoisomerase I poisons (i.e. poisons such as camptothecin that function at the enzyme–DNA interface) (30). Three experiments were carried out to distinguish between these possibilities. In the first, the effects of intercalators on topoisomerase I-mediated DNA cleavage were determined using positively supercoiled substrates. Since these plasmids are already overwound, the addition of intercalative compounds should have very little effect on the apparent topology of the DNA. In all cases, virtually no enhancement of cleavage was observed when intercalators were present in assays that examined overwound substrates (Figure 5).
In the second experiment, levels and sites of topoisomerase I-mediated cleavage in the presence of ethidium bromide, 9-aminoacridine, TAS-103 or amsacrine were monitored using radioactively end-labeled linear DNA (Figure 6). Since linear DNA is a topologically open system, the opening of the double helix by intercalators does not result in the accumulation of positive superhelical twists. Intercalator concentrations corresponded to those that generated maximal cleavage with negatively supercoiled substrates (Figure 5). No enhancement of cleavage was seen with any of the compounds. This is in marked contrast to camptothecin or topotecan, both of which greatly increased levels of scission.
Figure 6.
Effects of anticancer drugs and DNA intercalators on topoisomerase I-mediated cleavage of linear DNA. The ability of anticancer drugs [5 µM camptothecin (Cpt) or 10 µM topotecan (Tpt)] and DNA intercalating agents [20 µM ethidium bromide (EtBr), 20 µM TAS-103 (TAS), 100 µM 9-aminoacridine (9-AA) or 200 µM amsacrine (AMSA)] to enhance cleavage of a 3′-end labeled DNA substrate was determined. The autoradiograph is representative of three independent experiments. Size markers are described in Figure 3.
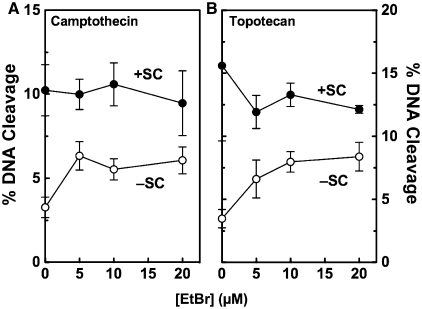
In the third experiment, the effects of ethidium bromide on topoisomerase I-mediated DNA cleavage were assessed in the presence of camptothecin or topotecan (Figure 7). Once again, the intercalator increased scission only when negatively supercoiled plasmid was used. These results suggest that ethidium bromide affects topoisomerase I cleavage by a mechanism that is distinct from that of interfacial poisons such as camptothecin and topotecan.
Figure 7.
Effects of DNA intercalators on topoisomerase I-mediated cleavage of plasmid DNA in the presence of anticancer drugs. Cleavage of positively [(+)SC, closed circles] and negatively [(−)SC, open circles] supercoiled pBR322 plasmid DNA was monitored in the presence of 0–20 µM ethidium bromide and either 2.5 µM camptothecin (A) or 5 µM topotecan (B). Error bars represent the standard deviation of at least three independent experiments.
Taken together, these findings provide strong evidence that the intercalative compounds examined have no intrinsic activity against topoisomerase I and are not classical interfacial poisons. Since intercalators only affected enzyme-mediated DNA cleavage when underwound covalently closed substrates were employed, it is concluded that these compounds are topological poisons of topoisomerase I and enhance enzyme-mediated scission by altering the apparent superhelical state of the double helix.
Effects of intercalators on topoisomerase I-mediated DNA cleavage in cultured human cells
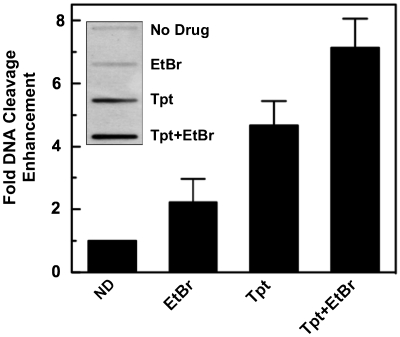
Since the genetic material in human cells is globally underwound, DNA intercalators might influence the ability of topoisomerase I to cleave the double helix in vivo. To assess this possibility, the effects of ethidium bromide on topoisomerase I-mediated DNA scission were determined in human CEM cells (Figure 8). Levels of topoisomerase I-DNA cleavage complexes rose ∼2.0- to 2.5-fold when cells were treated with 10 µM ethidium bromide. Similar to the in vitro results seen in Figure 7, the intercalator also enhanced scission in cells that were treated with 5 µM topotecan. These results indicate that ethidium bromide can poison topoisomerase I in human cells.
Figure 8.
Effects of ethidium bromide on topoisomerase I-mediated DNA cleavage in cultured human CEM cells. Cells were treated for 1 h with no drug (ND), 10 µM ethidium bromide (EtBr), 5 µM topotecan (Tpt) or both ethidium bromide and topotecan (Tpt + EtBr). Topoisomerase I-DNA complexes were monitored using the ICE bioassay (see inset for a representative blot). Levels of topoisomerase I-mediated DNA cleavage in the absence of drug were set to a relative value of 1. Error bars represent the standard deviation of six independent experiments.
DISCUSSION
Topoisomerase I is an important enzyme that functions in a number of essential nuclear processes (1,6,11,12,14,15). Previous work demonstrated that the enzyme can distinguish the handedness of DNA supercoils and relaxes positively supercoiled substrates ∼10-fold faster than negatively supercoiled molecules (17). The present study provides further evidence that the enzyme also maintains ∼3-fold higher levels of cleavage complexes with overwound substrates in the absence or presence of anticancer drugs. The above findings suggest that topoisomerase I is designed to function primarily on positively supercoiled DNA. While the high levels of cleavage that topoisomerase I potentially generates ahead of DNA tracking systems makes the enzyme a potent target for anticancer drugs, it also makes it an intrinsic danger to human cells. This may explain why eukaryotic cells encode an enzyme, tyrosyl–DNA phosphodiesterase 1, that specifically removes processed topoisomerase I from the 3′-terminus of cleaved nucleic acids (40,41).
The increased concentration of cleavage complexes generated with positively supercoiled substrates appears to correlate with decreased rates of enzyme-mediated DNA ligation. This seems counterintuitive given the fact that topoisomerase I preferentially relaxes positive supercoils (17). However, since the enzyme removes multiple superhelical twists per event, ligation may not be the limiting step of the relaxation reaction. Alternatively, if the enzyme is less likely to ligate cleaved DNA, it may actually remove superhelical twists more rapidly.
Finally, intercalators that have little or no intrinsic effect on topoisomerase I function enhance the ability of the enzyme to cleave covalently closed negatively supercoiled substrates. We suggest that these compounds act by altering the perceived topological state of the double helix, making underwound DNA appear to be overwound. Due to the novel mechanism of action of intercalators on the type I enzyme, it is proposed that these compounds be referred to as topological poisons of topoisomerase I.
FUNDING
National Institutes of Health grant GM033944 (to N.O.); Danish Research Councils; the Danish Cancer Society; Aase and Ejnar Danielsen Foundation; Civilingeniør Frode V. Nyegaard og hustrus Foundation; Karen Elise Jensens Foundation; Købmand Sven Hansen og hustra Ina Hansen Foundation (to B.R.K.). A.C.G. was a trainee under grant T32 CA09582 from the National Institutes of Health. Funding for open access charge: National Institutes of Health grant GM033944; Laboratory Development Funds (to N.O.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Qasim Khan, Steven L. Pitts and Adam C. Ketron for critical reading of the article.
REFERENCES
- 1.Wang JC. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 2.Cozzarelli NR, Wang JC, editors. DNA Topology and its Biological Effects. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; 1990. [Google Scholar]
- 3.Kanaar R, Cozzarelli NR. Roles of supercoiled DNA structure in DNA transactions. Curr. Opin. Struct. Biol. 1992;2:369–379. [Google Scholar]
- 4.Schvartzman JB, Stasiak A. A topological view of the replicon. EMBO Rep. 2004;5:256–261. doi: 10.1038/sj.embor.7400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates AD, Maxwell A. DNA Topology. 2nd edn. Oxford: Oxford University Press; 2005. [Google Scholar]
- 6.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 7.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326:414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- 9.Kim RA, Wang JC. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J. Mol. Biol. 1989;208:257–267. doi: 10.1016/0022-2836(89)90387-2. [DOI] [PubMed] [Google Scholar]
- 10.Peter BJ, Ullsperger C, Hiasa H, Marians KJ, Cozzarelli NR. The structure of supercoiled intermediates in DNA replication. Cell. 1998;94:819–827. doi: 10.1016/s0092-8674(00)81740-7. [DOI] [PubMed] [Google Scholar]
- 11.Champoux JJ. DNA topisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 12.Nitiss JL. Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim. Biophys. Acta. 1998;1400:63–81. doi: 10.1016/s0167-4781(98)00128-6. [DOI] [PubMed] [Google Scholar]
- 13.Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res. 2009;37:738–748. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leppard JB, Champoux JJ. Human DNA topoisomerase I: relaxation, roles, and damage control. Chromosoma. 2005;114:75–85. doi: 10.1007/s00412-005-0345-5. [DOI] [PubMed] [Google Scholar]
- 15.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 16.Pommier Y, Pourquier P, Fan Y, Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta. 1998;1400:83–106. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 17.Frohlich RF, Veigaard C, Andersen FF, McClendon AK, Gentry AC, Andersen AH, Osheroff N, Stevnsner T, Knudsen BR. Tryptophane-205 of human topoisomerase I is essential for camptothecin inhibition of negative but not positive supercoil removal. Nucleic Acids Res. 2007;35:6170–6180. doi: 10.1093/nar/gkm669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 19.Sari L, Andricioaei I. Rotation of DNA around intact strand in human topoisomerase I implies distinct mechanisms for positive and negative supercoil relaxation. Nucleic Acids Res. 2005;33:6621–6634. doi: 10.1093/nar/gki935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 21.Goulaouic H, Roulon T, Flamand O, Grondard L, Lavelle F, Riou J-F. Purification and characterization of human DNA topoisomerase IIIα. Nucleic Acids Res. 1999;27:2443–2450. doi: 10.1093/nar/27.12.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog. Nucleic Acid Res. Mol. Biol. 2000;64:221–253. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 23.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 24.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIα rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J. Biol. Chem. 2005;280:39337–39345. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 25.Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 26.Hande KR. Clinical applications of anticancer drugs targeted to topoisomerase II. Biochim. Biophys. Acta. 1998;1400:173–184. doi: 10.1016/s0167-4781(98)00134-1. [DOI] [PubMed] [Google Scholar]
- 27.Takimoto CH, Wright J, Arbuck SG. Clinical applications of the camptothecins. Biochim. Biophys. Acta. 1998;1400:107–119. doi: 10.1016/s0167-4781(98)00130-4. [DOI] [PubMed] [Google Scholar]
- 28.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 29.McClendon AK, Osheroff N. DNA topoisomerase II, genotoxicity and cancer. Mutat. Res. 2007;623:83–97. doi: 10.1016/j.mrfmmm.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem. Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClendon AK, Osheroff N. The geometry of DNA supercoils modulates topoisomerase-mediated DNA cleavage and enzyme response to anticancer drugs. Biochemistry. 2006;45:3040–3050. doi: 10.1021/bi051987q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McClendon AK, Gentry AC, Dickey JS, Brinch M, Bendsen S, Andersen AH, Osheroff N. Bimodal recognition of DNA geometry by human topoisomerase IIα: preferential relaxation of positively supercoiled DNA requires elements in the C-terminal domain. Biochemistry. 2008;47:13169–13178. doi: 10.1021/bi800453h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lisby M, Olesen JR, Skouboe C, Krogh BO, Straub T, Boege F, Velmurugan S, Martensen PM, Andersen AH, Jayaram M, et al. Residues within the N-terminal domain of human topoisomerase I play a direct role in relaxation. J. Biol. Chem. 2001;276:20220–20227. doi: 10.1074/jbc.M010991200. [DOI] [PubMed] [Google Scholar]
- 34.Pommier Y, Covey JM, Kerrigan D, Markovits J, Pham R. DNA unwinding and inhibition of mouse leukemia L1210 DNA topoisomerase I by intercalators. Nucleic Acids Res. 1987;15:6713–6731. doi: 10.1093/nar/15.16.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: conformations at the fork. Proc. Natl Acad. Sci. USA. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waring M. Variation of the supercoils in closed circular DNA by binding of antibiotics and drugs: evidence for molecular models involving intercalation. J. Mol. Biol. 1970;54:247–279. doi: 10.1016/0022-2836(70)90429-8. [DOI] [PubMed] [Google Scholar]
- 37.Waring MJ. Drugs and DNA: uncoiling of the DNA double helix as evidence of intercalation. Humangenetik. 1970;9:234–236. doi: 10.1007/BF00279229. [DOI] [PubMed] [Google Scholar]
- 38.Byl JA, Fortune JM, Burden DA, Nitiss JL, Utsugi T, Yamada Y, Osheroff N. DNA topoisomerases as targets for the anticancer drug TAS-103: primary cellular target and DNA cleavage enhancement. Biochemistry. 1999;38:15573–15579. doi: 10.1021/bi991791o. [DOI] [PubMed] [Google Scholar]
- 39.Nelson EM, Tewey KM, Liu LF. Mechanism of antitumor drug action: poisoning of mammalian DNA topoisomerase II on DNA by 4′-(9-acridinylamino)-methanesulfon-m-anisidide. Proc. Natl Acad. Sci. USA. 1984;81:1361–1365. doi: 10.1073/pnas.81.5.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Interthal H, Pouliot JJ, Champoux JJ. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc. Natl Acad. Sci. USA. 2001;98:12009–12014. doi: 10.1073/pnas.211429198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]