LST-4/SNX9, SNX-1, and SNX-6 together drive the degradation of apoptotic cells, as PtdIns(3)P effectors, during Caenorhabditis elegans development. By inducing regional membrane curvature and maintaining RAB-7 GTPase on phagosomes, these three sorting nexins stimulate the fusion of endocytic organelles with phagosomes.
Abstract
Apoptotic cells are swiftly engulfed by phagocytes and degraded inside phagosomes. Phagosome maturation requires phosphatidylinositol 3-phosphate [PtdIns(3)P], yet how PtdIns(3)P triggers phagosome maturation remains largely unknown. Through a genome-wide PtdIns(3)P effector screen in the nematode Caenorhabditis elegans, we identified LST-4/SNX9, SNX-1, and SNX-6, three BAR domain-containing sorting nexins, that act in two parallel pathways to drive PtdIns(3)P-mediated degradation of apoptotic cells. We found that these proteins were enriched on phagosomal surfaces through association with PtdIns(3)P and through specific protein–protein interaction, and they promoted the fusion of early endosomes and lysosomes to phagosomes, events essential for phagosome maturation. Specifically, LST-4 interacts with DYN-1 (dynamin), an essential phagosome maturation initiator, to strengthen DYN-1’s association to phagosomal surfaces, and facilitates the maintenance of the RAB-7 GTPase on phagosomal surfaces. Furthermore, both LST-4 and SNX-1 promote the extension of phagosomal tubules to facilitate the docking and fusion of intracellular vesicles. Our findings identify the critical and differential functions of two groups of sorting nexins in phagosome maturation and reveal a signaling cascade initiated by phagocytic receptor CED-1, mediated by PtdIns(3)P, and executed through these sorting nexins to degrade apoptotic cells.
INTRODUCTION
In metazoan organisms, the rapid removal of apoptotic cells through phagocytosis is an evolutionarily conserved process and is important for tissue remodeling, prevention of tissue injury, and suppression of inflammatory and auto-immune responses (Savill and Fadok, 2000). During the development of the nematode Caenorhabditis elegans hermaphrodites, 131 somatic cells and 30–50% of germ cells undergo programmed cell death (Metzstein et al., 1998). These cells are easily recognized within living animals under the Nomarski Differential Interference Contrast (DIC) optics as highly refractive, button-like objects referred to as “cell corpses” (Sulston and Horvitz, 1977; Sulston et al., 1983). In C. elegans, as in other metazoans, apoptotic cells are swiftly engulfed by their neighboring cells and are degraded inside membrane vacuoles called phagosomes (reviewed in Kinchen and Ravichandran, 2008; Zhou and Yu, 2008). Well-established genetic techniques and recently developed in vivo time-lapse microscopic approaches helped to establish C. elegans as an efficient system in understanding both the internalization and the degradation of apoptotic cells within animal bodies (Reddien and Horvitz, 2004; Yu et al., 2006, 2008; Kinchen et al., 2008).
Membrane fusion events play fundamental roles during both the engulfment and degradation of apoptotic cells. The extension of pseudopods relies on the incorporation of intracellular vesicles, which provide both lipid and protein materials for the expansion of pseudopod membranes (Touret et al., 2005). The subsequent degradation of apoptotic cells inside phagosomes, or phagosome maturation, further involves a series of fusion events between phagosomes and intracellular organelles, including endosomes and lysosomes. These fusion events ultimately result in the formation of a phagolysosome, a compartment that is highly acidified and enriched with various digestive enzymes for apoptotic cells degradation (reviewed in Vieira et al., 2002; Kinchen and Ravichandran, 2008; Zhou and Yu, 2008).
Immediately after an apoptotic cells is internalized, a high level of phosphatidylinositol 3-phosphate [PtdIns(3)P] appears on the surface of a nascent phagosome, where it remains until the apoptotic cell is degraded (Mangahas et al., 2008; Yu et al., 2008). The robust production of PtdIns(3)P is a prominent feature observed on the surface of nascent phagosomes that contain various kinds of cells (Ellson et al., 2001a; Kinchen et al., 2008; Yu et al., 2008). We previously found that the phagocytic receptor CED-1, which was present on the surface of nascent phagosomes, led a signaling pathway that induced the production of PtdIns(3)P, an essential event for triggering phagosome maturation, on phagosomal surfaces (Yu et al., 2008). The CED-1 pathway also leads to the recruitment of RAB-5 and RAB-7 guanosine triphosphatases (GTPases), two important membrane tethering factors, to phagosomes (Yu et al., 2008; He et al., 2010). The RAB-7 GTPase plays an essential role in the recruitment and fusion of lysosomes to phagosomes (Harrison et al., 2003; Kinchen et al., 2008; Yu et al., 2008). Through these activities, CED-1 promotes the recruitment and fusion of early endosomes and lysosomes to phagosomes and the consequential degradation of apoptotic cells (Yu et al., 2008).
PtdIns(3)P is known to play important roles in multiple membrane trafficking events, including endosome trafficking, retrograde trafficking, and autophagy (reviewed in Lindmo and Stenmark, 2006; Backer, 2008). As a common feature, PtdIns(3)P recruits proteins with PtdIns(3)P-binding modules, such as the FYVE (Fab1p, YOTB, Vac1p, and EEA1) or PX (Phox Homology) domains, to particular membrane compartments for further regulation of distinct cellular events (Birkeland and Stenmark, 2004). A number of PtdIns(3)P effectors have been reported (Lindmo and Stenmark, 2006). EEA1 and Hrs1 were reported to participate in the maturation of phagosomes containing pathogens (Fratti et al., 2001; Vieira et al., 2004). p40phox is a core component of the phagocyte oxidase (Phox) complex that actively kills pathogens inside phagosomes (Ellson et al., 2001b, 2006; Kanai et al., 2001). However, PtdIns(3)P effector(s) specific for the removal of apoptotic cells have not been identified, nor is it known what specific molecular events they regulate. Whether EEA1 and Hrs1 play roles in phagosome maturation during animal development is also unknown. As a result, how PtdIns(3)P triggers the maturation of phagosomes and the degradation of apoptotic cells remains largely elusive.
C. elegans phagosomes that contain apoptotic cells, which are at 2.0–2.5-μm diameters, are much larger than the various kinds of small vesicles that fuse to them (Yu et al., 2006, 2008). The relatively flat surfaces of these phagosomes create a challenge to the membrane fusion events, which favor bilayers with high curvature (Chanturiya et al., 2002; Chernomordik and Kozlov, 2003; Marrink and Mark, 2003). Evidence indicates that fusions between relatively big vesicles (diameter >1 μm) need the assistance of specific protein factors capable of bending membranes in addition to the basic fusion machinery (Hui et al., 2009). So far, factors that facilitate the fusions between phagosomes and intracellular vesicles through generating higher membrane curvature have not been identified.
Reported here, by screening through all C. elegans proteins predicted to possess FYVE or PX domains for functions in the removal of cell corpses, we identified three PX domain proteins that act as novel PtdIns(3)P effectors that specifically control the degradation of apoptotic cells. These proteins belong to the SNX-BAR subfamily of sorting nexins, whose functions in the degradation of apoptotic cells, or, more generally, in phagosome maturation, were previously unknown. We revealed the molecular mechanisms used by these proteins to promote fusions of intracellular vesicles to phagosomes, including the regulation of local curvature of phagosomal membranes. Furthermore, we discovered how PtdIns(3)P regulates the activities of its effectors in response to an upstream signal initiated by CED-1 in C. elegans engulfing cells.
RESULTS
Identification of four PX domain proteins that actively promote the removal of apoptotic cells
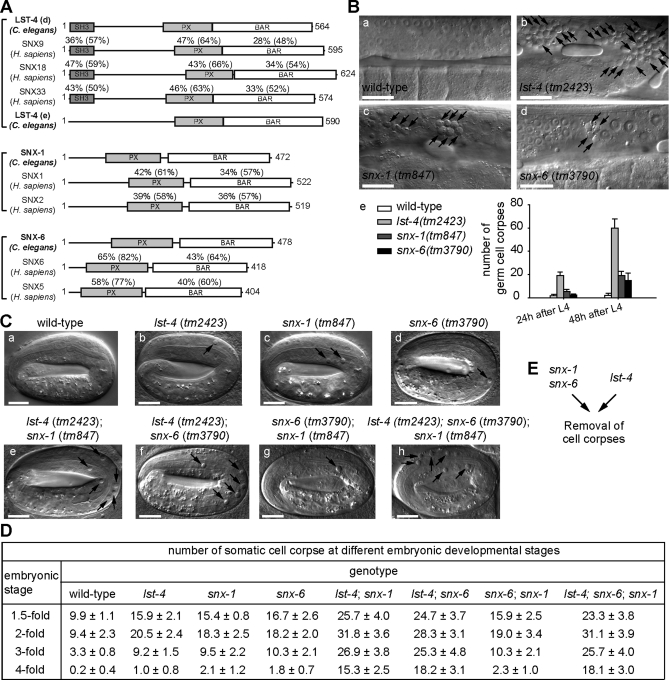
Using the Simple Modular Architecture Research Tool (SMART) program (Schultz et al., 2000), we identified genes encoding 16 FYVE domain-containing and 12 PX domain-containing proteins from the C. elegans genome (Table 1). To identify PtdIns(3)P effectors that specifically function in the removal of apoptotic cells, we analyzed the loss-of-function phenotypes of each of the 28 genes by scoring the number of persistent germ cell corpses, a readout of the cell-corpse removal defect (Ced phenotype), in the gonad of adult hermaphrodites bearing homozygous gene deletions (Table 1). When deletion alleles were not available or caused lethality, we scored animals in which the target genes were individually inactivated by RNA interference (RNAi) treatment (Materials and Methods) (Table 1). Inactivation of genes encoding FYVE domain-containing proteins did not result in an obvious Ced phenotype, except for a mild increase in the number of germ cell corpses observed in deletion mutants of C26H9A.2 (Table 1). In contrast, a large number of persistent germ cell corpses were observed from deletion mutants of F39B1.1, lst-4, snx-1, or snx-6, which encode PX domain-bearing proteins (Table 1; Figures 1B and S1, A–D). Moreover, each of these deletions also resulted in an excessive amount of somatic cell corpses in homozygous embryos throughout the mid- and late embryonic stages (Figure 1, C and D) (N. Lu and Z. Zhou, unpublished data). These results indicate that these four genes are required for the efficient removal of both somatic and germ cell corpses in C. elegans.
TABLE 1:
A candidate genetic screen for PtdIns(3)P effectors acting in the removal of apoptotic cells. The numbers of germ cell corpses were scored in the gonads of adult hermaphrodites at 48 h post-L4 stage. Data are presented as mean ± SD. n, number of animals scored. The names of C. elegans genes and deletion alleles were used according to the wormbase (http://www.wormbase.org). The name of Homo sapiens was used according to the unigene database of NCBI (http://www.ncbi.nlm.nih.gov).
| Genotype | Number of germ cell corpses | Mammalian homologue |
|---|---|---|
| wild-type | 2.8 ± 1.1 (n = 15) | N/A |
| FYVE domain-containing proteins | ||
| C26H9A.2(ok912) | 8.3 ± 3.5 (n = 15) | WDFY3 |
| mtm-6(ok330) | 5.5 ± 2.3 (n = 13) | MTMR6 |
| tag-77(tm702) | 1.9 ± 1.6 (n = 10) | FGD6 |
| aka-1(tm389) | 4.7 ± 1.6 (n = 15) | ZFYVE9 |
| eea-1(tm933) | 1.5 ± 2.0 (n = 15) | EEA1 |
| exc-5(rh232) | 1.2 ± 1.7 (n = 10) | FGD2 |
| rabs-5(ok1513) | 1.1 ± 1.2 (n = 10) | Rabenosyn-5 |
| rabn-5(tm1555) | 1.8 ± 1.2 (n = 10) | Rabaptin-5 |
| F22G12.4(tm3221) | 2.4 ± 1.2 (n = 15) | ANKFY1 |
| wdfy-2(tm3806) | 2.6 ± 2.4 (n = 15) | WDFY2 |
| lst-2(RNAi) | 4.3 ± 2.2 (n = 17) | ZFYVE28 |
| ZK632.12(RNAi) | 3.2 ± 1.6 (n = 10) | Phafin 2 |
| hgrs-1(RNAi) | 4.4 ± 1.4 (n = 11) | Hrs |
| mtm-3(RNAi) | 4.1 ± 2.7 (n = 20) | MTMR3 |
| R11D1.10(RNAi) | 2.8 ± 2.4 (n = 15) | ZFYVE19 |
| ppk-3(RNAi) | 3.7 ± 2.5 (n = 15) | PIKfyve |
| PX domain-containing proteins | ||
| pld-1(ok2222) | 1.9 ± 1.8 (n = 15) | PLD 1 |
| snx-1(tm847) | 19.1 ± 3.5 (n = 15) | SNX 1/2 |
| F13E9.1(tm1886) | 1.8 ± 1.7 (n = 10) | Nischarin |
| Y116A8C.26(tm2404) | 1.6 ± 1.3 (n = 10) | SNX 13 |
| snx-3(tm1595) | 2.7 ± 1.8 (n = 17) | SNX 3/12 |
| lst-4(tm2423) | 60.7 ± 7.8 (n = 15) | SNX 9/18/33 |
| F39B1.1(tm3171) | 38.7 ± 6.0 (n = 15) | PI3KC2α |
| F17H10.3(tm3643) | 2.3 ± 1.3 (n = 15) | SNX 17 |
| rskd-1(tm4031) | 1.7 ± 0.7 (n = 15) | RPS6KL1 |
| snx-6(tm3790) | 13.3 ± 4.0 (n = 15) | SNX 5/6 |
| Y48E1B.14(RNAi) | 4.3 ± 2.5 (n = 15) | SNX 14 |
| F25H2.2(RNAi) | 3.9 ± 1.9 (n = 16) | SNX 27 |
FIGURE 1:
Inactivation of lst-4, snx-1, and snx-6 results in the accumulation of apoptotic cells. (A) Domain structures of LST-4, SNX-1, SNX-6 and their mammalian orthologues. The percentage of amino acid identity (similarity in parentheses) of each domain between the C. elegans protein and each of its mammalian homologues are indicated. SH3: Src homology 3; PX: phox homology; BAR: Bin-amphiphysin-Rvs. (B) (a–d) DIC images of gonads of adult hermaphrodites at 48 h post-L4 stages. Arrows indicate cell corpses. Dorsal is to the top. Scale bars, 20 μm. (e) The number of germ cell corpses per gonad arm in adult hermaphrodites at 24 and 48 h post-L4 stage. Fifteen animals were scored for each datum. (C) DIC images of fourfold stage embryos with indicated genotypes. Arrows indicate persistent somatic cell corpses. Scale bars, 10 μm. (D) The number of somatic cell corpses at different embryonic stages. At least 15 embryos were scored for each datum. Data are presented as mean ± SD. (E) Diagram illustrating that snx-1 and snx-6 act in one genetic pathway and lst-4 acts in another, as indicated by the results in (D).
LST-4, SNX-1, and SNX-6 are members of the SNX-BAR subfamily of sorting nexins
C. elegans LST-4 (Lateral Signaling Target-4), SNX-1 (Sorting Nexin-1), and SNX-6 all display strong homology to proteins in the sorting nexin family, defined by the presence of an SNX-PX domain (Teasdale et al., 2001). LST-4, SNX-1, and SNX-6 also each have an additional BAR domain at their C termini and are further classified into the SNX-BAR subfamily of sorting nexins (Figure 1A) (van Weering et al., 2010). The BAR domain is a coiled-coil structure that homodimerizes and forms a rigidly curved surface that preferentially binds membranes with a certain degree of curvatures (Frost et al., 2009). Based on the domain structures and sequence homology, SNX-1 resembles mammalian SNX1 and SNX2, whereas SNX-6 resembles mammalian SNX5 and SNX6 (Figure 1A). LST-4, which possesses an additional SH3 domain at its N terminus, resembles mammalian SNX9, SNX18, and SNX33 (Figure 1A). Sorting nexins have diverse functions in endosomal trafficking events (Seet and Hong, 2006; van Weering et al., 2010). The PX and BAR domains together allow the SNX-BAR subfamily of sorting nexins to associate with membrane subdomains of high curvature and enriched for specific phosphoinositides, further stabilizing the local membrane curvature (McMahon and Gallop, 2005). We hypothesize that the three SNX-BAR proteins that we identified might act as effectors of PtdIns(3)P to assist the membrane fusion events involving phagosomes, the membranes of which display relatively low curvature (Yu et al., 2006). The characterization of F39B1.1, which encodes a Class II PI-3 kinase that we found to produce PtdIns(3)P on phagosomal membranes, will be reported elsewhere (N. Lu and Z. Zhou, unpublished data).
lst-4, snx-1, and snx-6 act in two parallel and partially redundant genetic pathways to drive the removal of apoptotic cells
To reveal the genetic interactions among lst-4, snx-1, and snx-6, we compared the severity of Ced phenotype in single-, double-, and triple-deletion mutant embryos during multiple stages throughout embryonic development (Figure 1, C and D). Each of the deletion alleles removes the majority of the protein coding sequence and introduces a stop codon or frame shift mutation, and is presumed to create a null mutation in the corresponding genes (Supplemental Figure S1, B–D; Supplemental text).
In embryos, the deletion of each gene resulted in a mild increase in the number of cell corpses. However, the numbers of persistent cell corpses increased synergistically when the deletion in either snx-1 or snx-6 was combined with the deletion in lst-4 (Figure 1, C[e, f] and D). The number of cell corpses in snx-6; snx-1 double mutants, on the other hand, is similar to that in snx-1 or snx-6 single mutants (Figure 1, C[g] and D). Moreover, the numbers of cell corpses in the lst-4; snx-6; snx-1 triple-mutant embryos is comparable to that of the lst-4; snx-6 or lst-4; snx-1 double mutants (Figure 1, C and D). Because all three are null mutations, the above results indicate that snx-6 and snx-1 are likely to act in a linear pathway, whereas lst-4 acts in another parallel pathway, and that the two pathways drive the removal of cell corpses in a partially redundant fashion (Figure 1E). We chose to further characterize mutant alleles of lst-4 and snx-1, genes that represent each of the two pathways.
Retromer complex components VPS-26 and VPS-35 are not involved in cell corpse removal
Both mammalian SNX-1 and SNX-6 are subunits of the retromer complex that mediates retrograde transport of transmembrane proteins from endosomes to the trans-Golgi network (Bonifacino and Hurley, 2008). To examine whether C. elegans SNX-1 and SNX-6 regulate the degradation of apoptotic cells as components of the retromer complex, we determined whether other known C. elegans retromer complex components were involved in the removal of cell corpses (Figure S2; Supplemental text). We found that the deletion alleles of the C. elegans vps-26 or vps-35, which encode subunits of the cargo-recognition subcomplex of the retromer, did not display any notable increase in the number of the somatic or germ cell corpse (Figure S2). These results suggest that SNX-1 and SNX-6 likely act in a protein complex distinct from the canonical retromer to regulate cell corpse removal. Similarly, SNX-3, a homologue of budding yeast Snx3/Grd19, a retromer-associating protein (Strochlic et al., 2007), does not appear to be involved in cell corpse removal (Table 1; Figure S2, Supplemental text).
LST-4 and SNX-1 pathways specifically control the degradation but not engulfment of somatic apoptotic cells
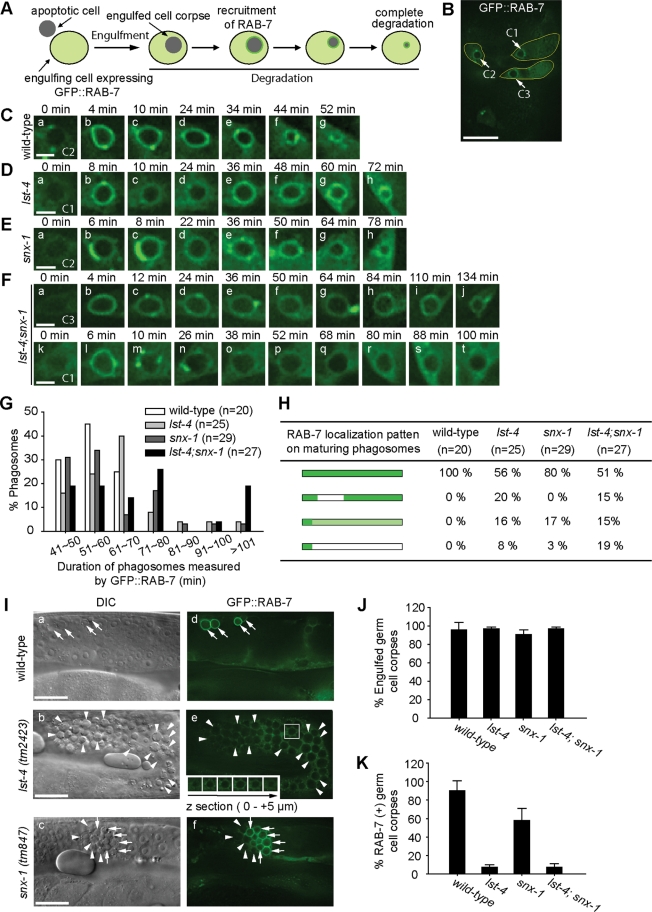
We first found that the lst-4 and snx-1 deletions resulted in the prolonged duration of cell corpses generated during embryogenesis but did not cause any increase in the number of cell death events (Supplemental text; Figure S1, F and G). The Ced phenotype thus resulted from defect(s) in either the engulfment or the degradation of cell corpses. To distinguish between these possibilities, we monitored the engulfment of embryonic cell corpse C3 using phagocytic receptor CED-1::GFP, which clustered on the extending pseudopods during engulfment and then quickly disappeared from nascent phagosomes (Materials and Methods; Figure S3B[a-i]) (Yu et al., 2006). Somatic apoptotic cells C1, C2, and C3 are located on the ventral surface, die almost simultaneously during mid-embryogenesis, and are quickly engulfed and degraded by three adjacent hypodermal cells that extend to the ventral midline (Figure 2B) (Yu et al., 2006; Lu et al., 2009). We found that both the timing of engulfment initiation and the period it took to complete engulfment are normal in lst-4; snx-1 double-mutant embryos (Figure S3, A–C). Normal pseudopod extension dynamics was also demonstrated by the dynamic localization pattern of GFP::moesin, a specific marker for filamentous actin polymerized under pseudopods (Figure S3D; Supplemental text).
FIGURE 2:
lst-4(tm2423) and snx-1(tm847) mutants are specifically defective in the degradation of cell corpses. (A) Diagram illustrating GFP::RAB-7 (Pced-1 gfp::rab-7) as a reporter for monitoring the degradation of cell corpses and the recruitment of RAB-7 onto phagosomes. (B) The epifluorecent image of a ∼330-min-old embryo. Arrows indicate the C1, C2, and C3 cell corpses. Engulfing cell of each cell corpse was outlined. Scale bar, 10 μm. (C–F) Time-lapse images of phagosome maturation in wild-type or mutant embryos expressing Pced-1 gfp::rab-7. “0 min” represents the time points when phagosomes were just sealed. Scale bars, 2 μm. (G) Histogram distribution of phagosome durations measured from the formation of a nascent phagosome until it was degraded. n, number of phagosomes C1, C2, and C3 scored. (H) The temporal enrichment pattern of GFP::RAB-7 on maturing phagosomes containing somatic cell corpses. Bars depict the duration of phagosomes, and dark and light green colors indicate the duration of the strong and weak signals of GFP::RAB-7 detected on phagosomes. n, number of phagosomes C1, C2, and C3 scored. (I) DIC (a–c) and GFP (d–f) images of adult hermaphrodite gonads expressing GFP::RAB-7 in gonadal sheath cells. Arrows and arrowheads indicated GFP::RAB-7(+) and (−) phagosomes, respectively. The inset in (e) is a serial z-section of the boxed area showing that the cell corpse in the images was fully internalized. Dorsal is to the top. Scale bars, 20 μm. (J) Percentage of engulfed germ cell corpses scored using GFP::RAB-7 as a phagosome marker in adult hermaphrodites at 48 h post-L4 stage. Data are presented as mean ± SD. Fifteen animals were scored for each sample. (K) Percentage of GFP::RAB-7(+) phagosomes in adult hermaphrodites at 48 h post-L4 stage. Data are presented as mean ± SD. Fifteen animals were scored for each sample.
We further characterized the degradation of somatic cell corpses in embryos by monitoring the gradual reduction of the volume of phagosomes containing cell corpses C1, C2, and C3 over time using GFP::RAB-7 as a marker (Figure 2B) (Yu et al., 2008). GFP::RAB-7, when expressed in engulfing cells from the Pced-1 promoter, displays bright and diffused GFP signal in the cytoplasm, allowing a nascent phagosome that contains an apoptotic cell to be first recognized as a dark, GFP(−) hole inside an engulfing cell (Figure 2A) (Yu et al., 2008). Once GFP::RAB-7 is enriched on the phagosomal surface a few minutes later, a phagosome could be tracked as a bright GFP(+) circle until it is fully degraded (Figure 2A) (Yu et al., 2008). The duration of a phagosome was measured from the moment when engulfment is complete (the “0 min” time point in Figure 2, C–F) to the moment when the phagosome disappeared. In wild-type embryos, the duration of a phagosome typically ranges between 40 and 60 min (Figure 2, C and G). In lst-4 or snx-1 single-mutant embryos, however, the duration varied within a broader range, with 20% or 28% phagosomes lasting longer than 70 min, respectively (Figure 2, D, E, and G). In lst-4; snx-1 double-mutant embryos, the delay in degradation was further enhanced, in which 48% of phagosomes lasted longer than 70 min and 19% even lasted longer than 100 min (Figure 2, F and G). These results indicate that, in the absence of lst-4 or snx-1, phagosome maturation is impaired, and further, when both lst-4 and snx-1 are inactivated, phagosome maturation is blocked. These defects result in the persistent cell corpses observed under the DIC optics (Figure 1, C and D).
LST-4 and SNX-1 are essential for the degradation, not engulfment, of germ cell corpses
We again used GFP::RAB-7 as a phagosomal marker to determine whether the many persistent germ cell corpses observed in the gonad of lst-4 or snx-1 single-mutant adult hermaphrodites were engulfed or not (Figures 2, I–J, and S4; Supplemental text). Although engulfed and unengulfed cell corpses display similar morphology under the DIC optics, only cell corpses inside phagosomes are surrounded by a bright ring of GFP::RAB-7 that was expressed in gonadal sheath cells, the engulfing cells for germ apoptotic cells (Figures 2I[d] and S4; Supplemental text). GFP::RAB-7 also allowed us to distinguish phagosomes as dark holes inside GFP(+) sheath cell cytoplasm even in cases in which the recruitment of RAB-7 to phagosomes was blocked (Figure 2I[e]). In lst-4(tm2423) mutants, all of the large number of DIC(+) germ cell corpses (mean = 58.9) were observed as dark holes imbedded inside the gonadal sheath cells (Figure 2, I and J). Due to the large number of phagosomes accumulating in the same sheath cells, a honeycomb pattern was generated inside the GFP(+) sheath cell cytoplasm (Figure 2I[e]). In snx-1(tm847) mutants, ∼95% of DIC(+) germ cell corpses (mean = 19.1) were inside phagosomes (Figure 2, I[f] and J). These results indicate that lst-4(tm2423) and snx-1(tm847) single mutants are exclusively defective in the degradation, not the internalization of germ cell corpses. We further performed transmission electron microscopy (TEM) to determine the engulfment status of germ cell corpses in the lst-4(tm2423); snx-1(tm847) double-mutant animals, the results of which indicated that 97% of the germ cell corpses analyzed were engulfed inside gonadal sheath cells and remained undegraded (Figure S5; Supplemental text). Together, these results demonstrated that lst-4 and snx-1 mutations abolished the cell corpse degradation activity, not the engulfment activity.
LST-4 and SNX-1 are both required for the incorporation of lysosomes and early endosomes into phagosomes
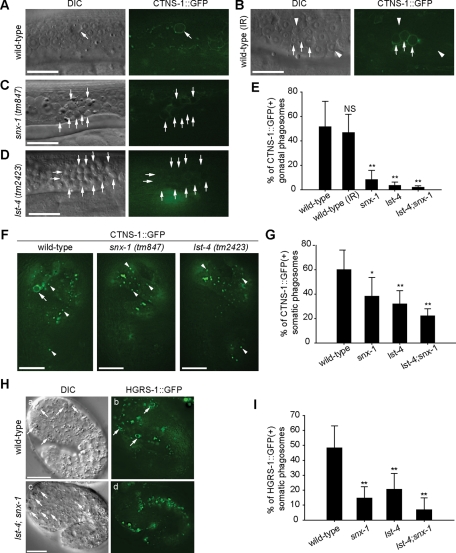
To determine the specific phagosome maturation defects caused by the deletions of lst-4 and snx-1, we examined the delivery of lysosomes and early endosomes to phagosomes, which are critical events for the degradation of phagosomal contents. We monitored the incorporation of lysosomes to phagosomes using CTNS-1, an integral lysosomal membrane protein and an established lysosomal marker (Mangahas et al., 2008; Yu et al., 2008). One group of wild-type animals was treated with γ-ray irradiation, which induced extra apoptosis events for the production of a larger number of phagosomes for characterization. Irradiation increased the average numbers of germ cell corpses from 3.5 to 13.9. In wild-type animals untreated or treated with γ-ray irradiation, continuous CTNS-1::GFP(+) signal was observed on the surfaces of ∼50% of phagosomes containing germ cell corpses, indicating that lysosomes were successfully recruited to and fused to phagosomal membranes (Figure 3, A, B, and E). In contrast, the CTNS-1::GFP signal was almost absent from the surfaces of phagosomes in the gonad of lst-4 (tm2423) or snx-1 (tm847) mutants and completely lost in lst-4; snx-1 double mutants (Figure 3, C–E). A similar defect in lysosomal incorporation was observed in 1.5-fold stage embryos of these mutants (Figure 3, F and G). Meanwhile, in lst-4 and snx-1 mutant embryos, relatively normal numbers of CTNS-1::GFP-labeled lysosomal particles were present in the cytoplasm (Figure 3F), indicating that in these mutants, lysosomal biogenesis and distribution in the cytoplasm were normal.
FIGURE 3:
lst-4 and snx-1 are both required for the efficient incorporation of lysosomes and early endosomes to phagosomes. (A–D) DIC and epifluorecent images of adult hermaphrodite gonads expressing Pced-1ctns-1::gfp. Arrows indicate phagosomes containing germ cell corpses. Arrowheads in (B) indicate nascent phagosomes not yet labeled with CTNS-1::GFP. γ-Irradiation (IR) was used in (B) to increase the number of apoptotic cells in wild-type animals. Scale bars, 20 μm. (E, G, and I) Data are presented as mean ± SD. Fifteen animals were scored for each sample. Independent Student’s t-test was used to calculate P values between wild-type and mutant animals. NS: not significant. *, P < 0.005; **, P < 0.001. (E) The percentage of CTNS-1::GFP(+) phagosomes in adult hermaphrodite gonads at 48 h post-L4 stage. (F) Epifluorecent images of 1.5-fold stage embryos expressing Pced-1ctns-1::gfp. Arrowheads indicate CTNS-1::GFP particles in hypodermal cells. An arrow indicates CTNS-1::GFP(+) phagosome. Scale bars: 10 μm. (G) The percentage of CTNS-1::GFP(+) phagosomes in 1.5-fold stage embryos. (H) DIC (a and c) and epifluorecent (b and d) images of 1.5-fold stage embryos expressing Pced-1hgrs-1::gfp. Arrows indicate phagosomes. Scale bars: 10 μm. (I) The percentage of HGRS-1::GFP(+) phagosomes in 1.5-fold stage embryos.
In addition, we monitored the incorporation of early endosomes to phagosomes using the HGRS-1::GFP reporter, a specific early endosomal marker expressed in engulfing cells (Yu et al., 2006). In 1.5-fold stage wild-type embryos, on average 47.7% of cell corpses distinguishable under DIC optics are labeled with HGRS-1::GFP(+) rings, indicating that the phagosomes enveloping these cell corpses received early endosomes from the host cells (Figure 3, H[a, b] and I). In lst-4 (tm2423); snx-1 (tm847) double-mutant embryos at the same stage, the fractions of phagosomes labeled with HGRS-1::GFP dropped to 6.9% (Figure 3, H[c, d] and I). In lst-4 and snx-1 single-mutant embryos, intermediate defects of endosome incorporation were observed (Figure 3I).
The above results indicate that LST-4 and SNX-1 act together to promote the efficient recruitment and fusion of early endosomes and lysosomes to phagosomes. We propose that the defect in incorporating intracellular vesicles to phagosomes is a primary cause for the phagosome maturation defect of the lst-4; snx-1 double mutants.
LST-4 and SNX-1 actively promote the extension of membrane tubules from phagosomes
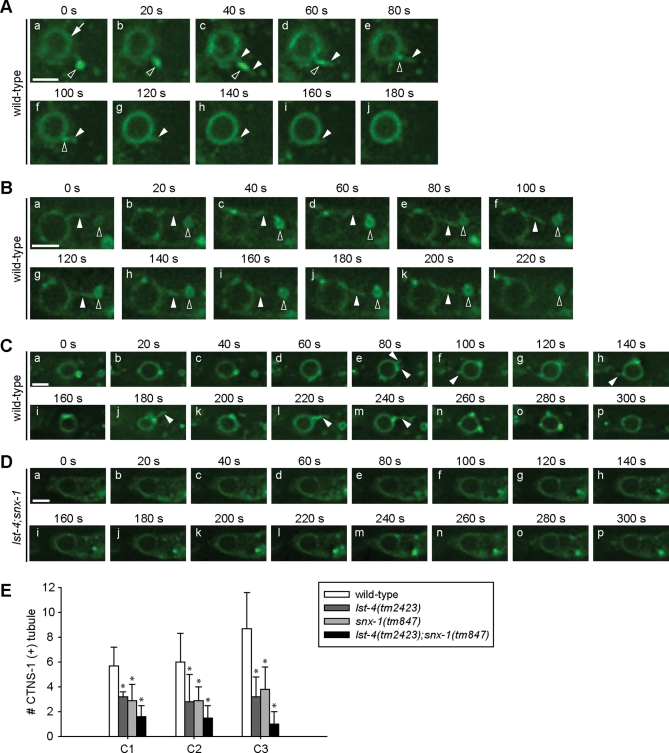
Previously, we observed that during the degradation of apoptotic cells, transient membrane tubules were frequently extended from phagosomal surfaces into the engulfing cell cytoplasm (Yu et al., 2008). Furthermore, we have established that lysosomal particles are recruited to phagosomes through two alternative routes, direct encounter with phagosomal surfaces or being captured by these phagosomal tubules, which subsequently retract and bring lysosomal particles to phagosomal surfaces (Yu et al., 2008). Because LST-4, SNX-1, and SNX-6 possess BAR domains known as sensors and inducers of membrane curvature, we analyzed the roles of LST-4 and SNX-1 in the formation of phagosomal tubules. We monitored tubule extension from phagosomes containing C1, C2, or C3 using the CTNS-1::GFP reporter, which labeled originally lysosomal membranes and subsequently phagosomal membranes once a detectable amount of lysosomes were incorporated into phagosomes (Figure 3F) (Yu et al., 2008). In wild-type embryos, phagosomal tubules were of lengths varying from ∼300 nm to ∼2.2 μm, and their durations varied from within 20 to ∼200 s (Figure 4, A–C; Supplemental Movies S1 and S2). We observed examples in which a tubule captured a nearby lysosomal particle and brought it to phagosomal surface through retraction (Figure 4A). In some of those examples, the volume and fluorescent intensity of a lysosomal particle rapidly reduced once it was attached to a membrane tubule, indicating that membrane fusion between the lysosome and tubule might have occurred before the lysosomal particle was brought to phagosomal surfaces (Figure 4A). Consistently, the successful capture of a lysosome particle by a membrane tubule was followed by a significant enrichment of the lysosomal marker CTNS-1::GFP on the phagosome (Figure 4A). Examples in which a phagosomal tubule repeatedly attempted to attach to a lysosomal particle yet eventually failed to bring the particle to phagosomal surface were also observed (Figure 4B), suggesting that first, the attachment between a tubule and a particle is unstable and transit, and secondly, attachment does not guarantee fusion. In lst-4; snx-1 double-mutant embryos, unlike in wild-type embryos (Figure 4, C and E; Movie S2), the frequency of observing phagosomal tubules was significantly reduced (Figure 4, D and E; Movie S3). In lst-4 or snx-1 single-mutant embryos, intermediate defects in the formation of phagosomal tubules were observed (Figure 4E). These results indicate that the functions of LST-4 and SNX-1 are both required for the formation and/or stabilization of phagosomal tubules. Furthermore, the lack of phagosomal tubules in lst-4; snx-1 double-mutant background was typically correlated with a slower enrichment and reduced signal intensity of CTNS-1::GFP on nascent C1, C2, and C3 phagosomes (Figure 4D), indicating that the defect in tubule formation is closely related to the inefficient targeting of lysosomes to phagosomes.
FIGURE 4:
LST-4 and SNX-1 are important for the extension of membrane tubules from the surface of phagosomes. (A–D) Time-lapse images of membrane tubules extended from phagosomes in embryos expressing Pced-1ctns-1::gfp. Arrows, arrowheads, and open arrowheads indicate phagosomes, membrane tubules, and lysosomal particles, respectively. Scare bar, 2 μm. (A) An extended membrane tubule from a C2 phagosome quickly captured and fused with a close-by lysosome in a wild-type embryo. (B) An attempted but failed capture of a lysosome by a membrane tubule extended from phagosome C3. (C and D) The time-lapse recording of the dynamic extension of membrane tubules from C3 phagosomes in a wild-type embryo (C) and in a lst-4(tm2423);snx-1(tm847) mutant embryo (D). (E) The average number of independent tubules detected on C1, C2, and C3 phagosomes in 15-min recording periods with 20-s intervals, starting immediately after the completion of engulfment. At least five C1, C2, or C3 phagosomes were scored for each datum point. Independent Student’s t-test was used to calculate the P values of comparisons between wild-type and mutant animals. *, P < 0.05.
LST-4 and SNX-1 differentially control the recruitment of RAB-7 to phagosomes
In lst-4(tm2423) mutant hermaphrodite gonad, GFP::RAB-7 was absent from all phagosomes observed inside gonadal sheath cells, indicating a complete failure in recruiting RAB-7 onto phagosomal surfaces (Figure 2, I and K). In contrast, ∼63% phagosomes were labeled by GFP::RAB-7 in snx-1(tm847) mutant gonadal sheath cells (Figure 2, I and K).
In lst-4 mutant embryos, the recruitment and maintenance of GFP::RAB-7 onto phagosomes was also defective, albeit less severe (Figure 2, D and H). Several types of RAB-7 maintenance defects have been observed, of which the premature dissociation of RAB-7 appears to be the common feature (Figure 2, D[c–d] and H). Defects in RAB-7 maintenance occurred less frequently in snx-1 single-mutant embryos; in addition, the reduction of phagosomal GFP::RAB-7 level was less severe (Figure 2, E and H). The types of defects and the distribution of each type observed from the lst-4; snx-1 double-mutant embryos are similar to that observed from lst-4 single-mutant embryos. These results indicate that, between LST-4 and SNX-1, LST-4 plays a major role in recruiting and maintaining RAB-7 on phagosomes.
LST-4, but not SNX-1, helps to maintain DYN-1’s association with phagosomal surfaces
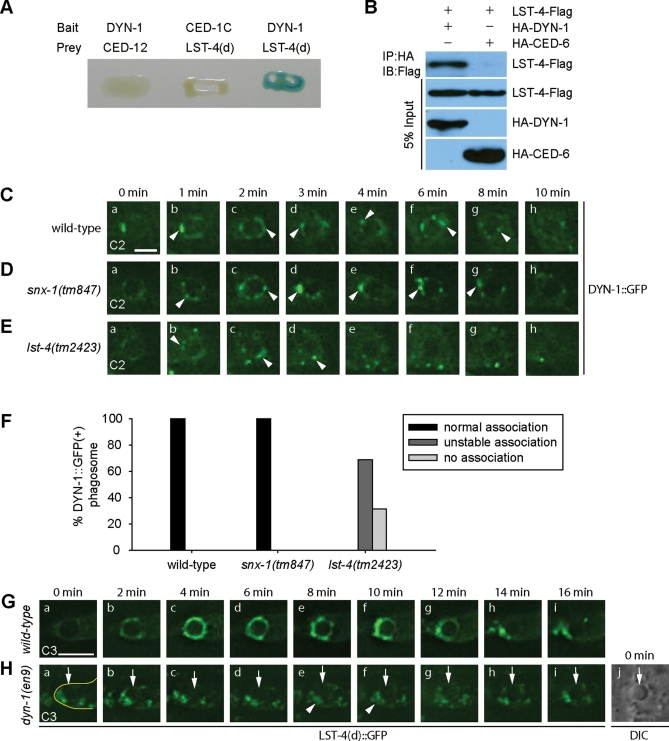
SNX-9, SNX-18, and SNX-30, the mammalian homologues of LST-4, are known to interact with mammalian dynamins (Lundmark and Carlsson, 2003; Soulet et al., 2005; Shin et al., 2007). In a yeast two-hybrid assay (Materials and Methods), we observed the physical interaction between LST-4 and C. elegans DYN-1 (Figure 5A). This specific interaction was further confirmed by coimmunoprecipitation assays of DYN-1 and LST-4 coexpressed in mammalian cell culture (Figure 5B) (Materials and Methods). During phagosome maturation, DYN-1 is transiently recruited to the surface of nascent phagosome (Yu et al., 2006). In lst-4 mutants, we found that the recruitment of DYN-1 was partially defective; on the surface of 69% of phagosomes, we detected fewer number of DYN-1::GFP puncta, almost all of which dissociated earlier from phagosomes than those observed in wild-type embryos (Figure 5, C, E, and F). On the other hand, the dynamic association pattern of DYN-1 with phagosomes was not affected by the snx-1(tm847) mutation (Figure 5, D and F). Thus, the inactivation of LST-4 but not SNX-1 appeared to impair the maintenance of DYN-1 on phagosomal surfaces. We propose that LST-4 might use multiple mechanisms to regulate phagosome maturation, one of which is to facilitate DYN-1 function.
FIGURE 5:
LST-4 and DYN-1 depend on each other for their association to phagosomal surfaces. (A and B) The interaction between DYN-1 and LST-4 was detected in the yeast two-hybrid assay (A) and the coimmunoprecipitation assay (B). Protein interactions were detected in (A) by X-Gal assays and in (B) by immunoblotting Flag-tagged LST-4 that was coimmuniprecipitated with HA-tagged proteins. CED-1C (the cytoplasmic domain of CED-1), CED-12, and CED-6 are negative controls to demonstrate the specificity of LST-4/DYN-1 interaction. (C–E) Time-lapse recording of the recruitment of DYN-1::GFP onto C2 phagosomes in a wild-type embryo (C), a snx-1(tm847) mutant embryo (D), and a lst-4(tm2423) mutant embryo (E). “0 min”: the time point when the engulfment was just completed. Arrowheads mark DYN-1::GFP puncta associating with phagosomes. Scale bars: 2 μm. (F) Quantification of different dynamic DYN-1::GFP localization patterns on phagosomes by monitoring multiple C1, C2, and C3 phagosomes over time. At least 16 phagosomes were monitored for each genotype. (G and H) Time-lapse images showing the dynamic recruitment of LST-4(d)::GFP onto a C3 phagosome in a wild-type embryo (G) or the lack of it in a dyn-1(en9) mutant embryo (H). “0 min”: the time point when the engulfment was just completed. Arrows in (H) indicate the C3 phagosome. Arrowheads in (H) indicate cytoplasmic LST(d)::GFP(+) puncta, which are surrounding but not associating with the C3 phagosome. The cell boundary of the engulfing cell for the C3 cell corpse is outlined in (H[a]). (H[j]) The DIC images corresponding to (H[a]) that indicates the position of the C3 phagosome. Scale bars: 4 μm.
LST-4 and SNX-1 are transiently recruited to the surfaces of nascent phagosomes and phagosomal tubules
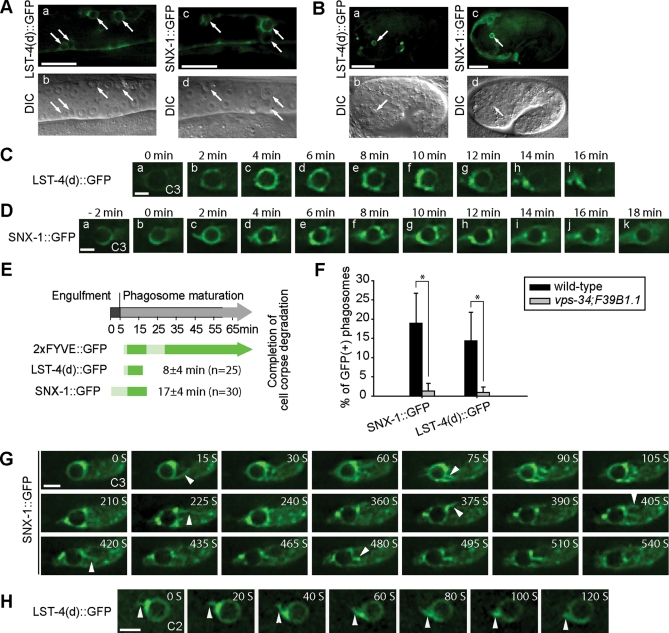
To determine whether LST-4 and SNX-1 are under the regulation of phagosomal PtdIns(3)P, we examined the phagosomal localization of GFP-tagged LST-4 and SNX-1. LST-4(d)::GFP and SNX-1::GFP, when expressed in engulfing cells, were fully functional for promoting cell corpses degradation (Supplemental Table S1; Supplemental text), indicating that the functions of LST-4 and SNX-1 in engulfing cells are sufficient for driving phagosome maturation. In wild-type animals expressing LST-4(d)::GFP or SNX-1::GFP, each reporter was primarily localized to the cytoplasm, with a portion enriched on cytoplasmic puncta (Supplemental Figure S6, A[a] and D[a]). Importantly, in these transgenic animals, we observed the association of GFP fusion proteins to phagosomes in both adult gonad and embryos (Figure 6, A and B), suggesting that LST-4 and SNX-1 act on phagosomal surfaces to promote phagosome maturation. In wild-type embryos, we observed rapid enrichment of both LST-4(d)::GFP and SNX-1::GFP on the surface of nascent phagosomes 2–4 min after phagosome formation (Figure 6, C–E). Later, the signal intensity of LST-4(d)::GFP on phagosomes gradually reduced (Figure 6, C[e–g] and E), and that of SNX-1::GFP reduced later and more gradually (Figure 6, D and E).
FIGURE 6:
LST-4::GFP and SNX-1::GFP are transiently localized on maturing phagosomes. All GFP reporters are expressed under the control of Pced-1 promotor. (A and B) GFP and DIC images of wild-type gonad arms (A) or embryos (B) expressing indicated reporters. Arrows indicate phagosomes. Scale bars in (A) and (B), 20 and 10 μm, respectively. (C and D) Time-lapse images displaying LST-4(d)::GFP (C) or SNX-1::GFP (D) on maturing phagosomes in wild-type embryos. “0 min” represents the time point when engulfment is just complete. Scale bars, 2 μm. (E) The temporal order and duration of each fluorescent reporter on extending pseudopods and maturing phagosomes. Data represent means obtained from time-lapse recording of multiple C1, C2, and C3 cell corpses (n, number of phagosomes measured). The light and dark green colors reflect weak and strong signal intensities, respectively. The average phagosomal durations of LST-4(d) and SNX-1 are indicated as mean ± SD. “0 min” represents the time when pseudopod extension is just initiated. (F) Percentage of phagosomes labeled with indicated GFP reporters in 1.5-fold stage wild-type or vps-34(h510); F39B1.1(tm3171) mutant embryos. At least 15 animals were scored for each sample. *, P < 0.001 by independent Student’s t-test. (G and H) Time-lapse images showing the localization of SNX-1::GFP (G) or LST-4(d)::GFP (H) on the tubules extended from phagosomes. Arrowheads indicate GFP-labeled phagosomal tubules. Scale bar: 2 μm.
In addition to phagosomal surfaces, SNX-1::GFP and LST-4::GFP were also observed on the highly dynamic membrane tubules extended from phagosomes (Figure 6, G and H; Supplemental Movie S4). Often particularly bright SNX-1::GFP and LST-4::GFP signals were observed on a few spots on phagosome surface, where the phagosomal tubules frequently emerged (Figure 6, G and H). This localization pattern is consistent with direct roles of SNX-1 and LST-4 in bending phagosome membrane and promoting phagosomal tubule formation.
PtdIns(3)P recruits LST-4 and SNX-1 to phagosomal surfaces via their PX domains
The temporal distribution patterns of SNX-1 and LST-4 on phagosomal surfaces are similar to that of PtdIns(3)P during its first of two signal intensity peaks on phagosomes (Figure 6E), suggesting that PtdIns(3)P on nascent phagosomes might act as a specific factor that recruits SNX-1 and LST-4. In in vitro protein–lipid overlap assays, both SNX-1 and LST-4 associated with a few phosphoinositide (PI) species, including PtdIns(3)P (Supplemental Figure S7; Supplemental text).
To directly test whether LST-4 and SNX-1 are PtdIns(3)P effectors in vivo, we compared the localization patterns of LST-4 and SNX-1 on phagosomes in wild-type embryos versus vps-34(h510); F39B1.1(tm3171) double-mutant embryos, in which the phagosomal PtdIns(3)P was depleted due to the inactivation of two classes of PI-3 kinases responsible for PtdIns(3)P production (N. Lu and Z. Zhou, unpublished data). Due to their transient enrichment nature, SNX-1::GFP and LST-4::GFP were visible on the surfaces of ∼19% and ∼15% of phagosomes in wild-type 1.5-fold stage embryos, respectively (Figure 6F). In contrast, in vsp-34; F39B1.1 double-mutant embryos, SNX-1::GFP and LST-4::GFP signals were completely absent from phagosomal surfaces (Figure 6F), demonstrating that PtdIns(3)P is essential for the recruitment of LST-4 and SNX-1 onto phagosomal surfaces.
We next investigated whether the PX domains in LST-4 and SNX-1 were the targets of phagosomal PtdIns(3)P. The three-dimensional structures of different PX domains display remarkable similarity, with a few highly conserved positively charged residues precisely positioned inside the binding pocket for phosphoinositides (Supplemental Figure S8A) (Ellson et al., 2002; Seet and Hong, 2006). Arginine 58 in the PX domain of p40phox is such a residue that plays a critical role in binding PtdIns(3)P (Bravo et al., 2001). We mutated R263 and R132, the corresponding arginine residues in LST-4 and SNX-1 (Figure S8A), respectively, to glutamine, and examined the phagosomal localization as well as the in vivo functions of these mutant proteins in transgenic animals [Pced-1lst-4(d)(PXm)::gfp or Pced-1snx-1(PXm)::gfp]. LST-4(d)(PXm)::GFP and SNX-1(PXm)::GFP displayed diffused cytoplasmic localization pattern (Figure S6, B and E) instead of the punctate localization pattern observed from the wild-type reporters (Figure S6, A and D). In time-lapse recording experiments, no enrichment of LST-4(PXm)::GFP or SNX-1(PXm)::GFP to the surface of maturing phagosomes was detected over time (Figure S6, H and I; Supplemental Movies S5 and S6). These results indicate that the PX domains are essential for the association of LST-4 and SNX-1 to cellular organelles, in particular to phagosomes. Furthermore, the R263Q mutation completely abolished the activity of Pced-1lst-4(d)(PXm)::gfp in rescuing the lst-4 mutant phenotype (Table S1). Similarly, Pced-1snx-1(PXm)::gfp in snx-1 mutant background displayed reduced rescuing activity (Supplemental Table S1). These results demonstrate that the binding to PtdIns(3)P and the resulting association to phagosomes are critical for the functions of LST-4 and SNX-1 in phagosome maturation.
The BAR domain is critical for the phagosomal localization and functions of SNX-1 and LST-4
A BAR domain is composed of three long coiled-coil α helices and is capable of forming a crescent-shaped homodimer (Peter et al., 2004). The concave surface of the BAR-domain dimer, which mainly consists of the C-terminal part of helix 2 and the loop between helices 2 and 3, is enriched with basic residues and proposed to be the interface that interacts with the negatively charged membranes of particular curvatures (Supplemental Figure S8, B and C) (Peter et al., 2004). The PX and BAR domains of a number of mammalian SNX-BAR sorting nexins act together to facilitate the protein–membrane association, in a manner referred to as “coincidence detection” (Carlton et al., 2004; Pylypenko et al., 2007; Traer et al., 2007; Haberg et al., 2008; Yarar et al., 2008; van Weering et al., 2010). In LST-4 and SNX-1, we simultaneously mutated a few conserved basic residues known to be essential for the functions of their mammalian orthologues (K492E and R499E in LST-4, and R376E, K377E, and R378E in SNX-1) (Figure S8, B and C) (Carlton et al., 2004; Peter et al., 2004; Pylypenko et al., 2007) and examined the effects on the phagosomal localization of the host proteins. Similar to the mutations in the PX domains, the mutations in these BAR domains resulted in a diffused localization pattern of the GFP-tagged reporters in the cytoplasm, and further greatly reduced the GFP signals on the surfaces of nascent phagosomes (Figure S6, C, F, H, and I), indicating that BAR domains are indeed necessary for the attachment of these proteins to cellular organelles and to phagosomes. Moreover, transgenes carrying these mutations lost part of the activities toward rescuing the Ced phenotypes of the corresponding mutants (Supplemental Table S1). Together, our data indicate that both the PX and the BAR domains play important roles in promoting the association of SNX-1 and LST-4 to phagosomal surfaces and in driving phagosome maturation. The combined point mutations in both PX domain and BAR domain of SNX-1 completely abolished its rescue activity (Table S1), indicating complementary roles of both domains in phagosome maturation.
DYN-1 is another essential factor for recruiting LST-4 to phagosomal surfaces
Consistent with the physical interaction observed between LST-4 and DYN-1 in vitro, the recruitment of LST-4 to phagosomal relies on DYN-1. Unlike in wild-type embryos, in dyn-1(en9) mutants, in which DYN-1’s self-assembly and its association with phagosomes were abolished (He et al., 2010), LST-4(d)::GFP signal was not detected on the surface of maturing phagosomes (Figure 5, G and H); rather, LST-4(d)::GFP were detected on puncta that are distributed inside the host cell cytoplasm, allowing a phagosome to be visualized as a dark hole (Figure 5H). Given that the transient enrichment pattern of LST-4 overlaps with the dynamic enrichment of DYN-1 and the initial production of PtdIns(3)P on phagosomal surfaces, that LST-4 interacts with both DYN-1 and PtdIns(3)P, and that LST-4 depends on both PtdIns(3)P and DYN-1 for enrichment on phagosomal surfaces, we propose that DYN-1 and PtdIns(3)P are two essential factors that together attract LST-4 to maturing phagosomes. The accumulation of DYN-1 and LST-4 on phagosomes thus appears to be mutually dependent.
SNX-1 mediates the association of SNX-6 to phagosomal surfaces
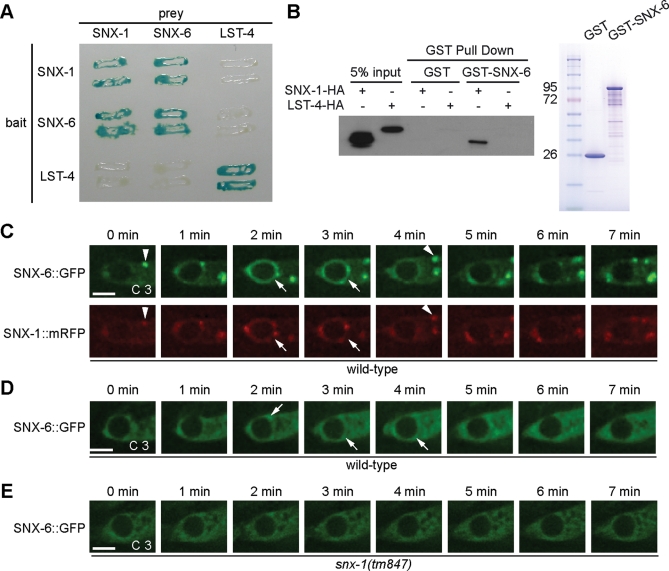
Both mammalian SNX-1 and SNX-6 and their budding yeast counterpart (Vps5 and Vps17) interact with each other (reviewed in Bonifacino and Hurley, 2008). In yeast two-hybrid assays, we observed that C. elegans SNX-1 and SNX-6 displayed strong and specific interaction with each other but not with LST-4 (Figure 7A). In addition, all three proteins displayed self-association activities, suggesting that they each might exist in multimeric forms in vivo (Figure 7A). In in vitro pull-down assays (Materials and Methods), we further found that GST-SNX-6 expressed and affinity-purified from Escherichia coli specifically pulled down hemagglutinin (HA)-tagged SNX-1 but not LST-4 from extracts of mammalian cells ectopically expressing each of the above proteins (Figure 7B). The above results have demonstrated the specific physical interaction between SNX-1 and SNX-6.
FIGURE 7:
SNX-1 physically interacts with SNX-6 and recruits SNX-6 onto phagosomes. (A) A yeast two-hybrid assay testing the binary interaction among SNX-1, SNX-6, and LST-4. Protein interactions were visualized by X-Gal assays. (B) GST-pull down assay showing that purified recombinant GST-SNX-6 specifically interacts with SNX-1-HA expressed in 293T cells. The SDS–PAGE gel image (right) shows purified proteins used for GST-pull down assay. (C–E) Time-lapse images monitoring GFP and mRFP signals on maturing phagosomes in wild-type (C and D) or snx-1(tm847) mutant (E) embryos expressing Pced-1 snx-6::gfp alone (D and E) or coexpressing Pced-1 snx-6::gfp and Pced-1 snx-1::mrfp (C). SNX-6::GFP and SNX-1::mRFP signals on phagosomes are indicated by arrows. Colocalization of SNX-6::GFP and SNX-1::mRFP on cytoplasmic puncta are indicated by arrowheads. “0 min” represents the time point when phagosomes are just sealed. Scale bars, 2 μm.
We next investigated the functional significance of the SNX-1/SNX-6 interaction. We constructed a Pced-1 snx-6::gfp transgene that fully rescued the Ced phenotype of snx-6 mutants (Supplemental Table S1). In wild-type embryos co-overexpressing both SNX-6::GFP and SNX-1::mRFP, a prominent level of SNX-6::GFP was detected on the surfaces of nascent phagosomes (Figure 7C). Moreover, the kinetics of SNX-6::GFP’s enrichment onto and dissociation from phagosomal surfaces was highly similar to that of SNX-1::mRFP (Figure 7C). Interestingly, in wild-type embryos expressing Pced-1 snx-6::gfp alone, the enrichment of SNX-6::GFP on phagosomal surfaces is much reduced (Figure 7D). Furthermore, in snx-1(tm847) mutant embryos, SNX-6::GFP was not detected on nascent phagosomes (Figure 7E). These results indicate that SNX-6 is recruited to the surfaces of nascent phagosomes in an SNX-1-dependent manner, a mechanism supported by their physical interaction detected in vitro.
The ced-1 pathway controls the localization of LST-4 and SNX-1 on phagosomes
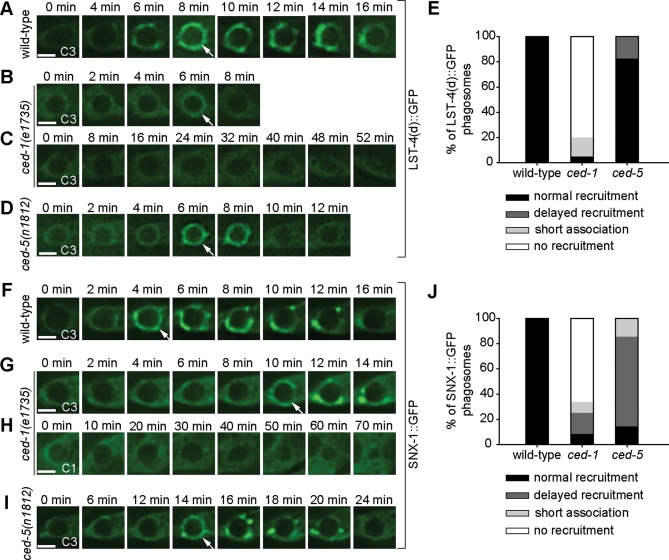
In C. elegans, the engulfment and degradation of apoptotic cells are controlled by two parallel genetic pathways (Yu et al., 2008; Zhou and Yu, 2008). The production of PtdIns(3)P on phagosomal surfaces is primarily controlled by the pathway led by phagocytic receptor CED-1 and, to a much less extent, by the second pathway composed of ced-2, ced-5, ced-10, and ced-12 (Yu et al., 2008). If LST-4 and SNX-1 act directly downstream of PtdIns(3)P, we anticipated that the phagosomal recruitment of LST-4 and SNX-1 was also under the control of the ced-1 pathway. To examine this model, we monitored the recruitment of LST-4 and SNX-1 to phagosomes in ced-1 and ced-5 null mutant embryos. In wild-type embryos, 100% of phagosomes acquired LST-4 and SNX-1 within 8 min after enclosure (Figure 8, A, E, F, and J). In contrast, in ced-1 mutant embryos, 80% of phagosomes never acquired LST-4::GFP (Figure 8, C and E) and 66.7% of phagosomes never acquired SNX-1::GFP during a 50-min recording period starting at the enclosure of a phagosome (Figure 8, H and J). In some cases, GFP signals disappeared from phagosomal surfaces much sooner than in wild-type background (Figure 8, B, E, and J), indicating an additional defect in signal maintenance. Furthermore, the LST-4(d)::GFP or SNX-1::GFP signal intensity on phagosomal surfaces was in general lower than normal, indicating another aspect of recruitment or maintenance defects (Figure 8, B and G). On the other hand, the localization patterns of LST-4 and SNX-1 on cytoplasmic puncta were not affected in ced-1 mutant embryos (Supplemental Figure S6, A[b] and D[b]), indicating that CED-1 specifically regulates the phagosomal recruitment of LST-4 and SNX-1.
FIGURE 8:
The localization patterns of LST-4 and SNX-1 on phagosomes are primarily controlled by the ced-1 pathway. (A–D) and (F–I) Time-lapse images of indicated GFP reporters on maturing phagosomes in wild-type (A and F) or ced mutant embryos (B–D and G–I) expressing Pced-1lst-4(d)::gfp or Pced-1 snx-1::gfp, respectively. Arrows indicated GFP signals on phagosomes. “0 min” represents the time point when the phagosome is just sealed. Scale bars, 2 μm. (E and J) Quantification of the different localization patterns of LST-4(d)::GFP (E) and SNX-1::GFP (J) on phagosomes in wild-type and ced-1 or ced-5 mutant embryos. Types of localization patterns are defined as follows: “normal recruitment,” GFP(+) circles were detected on phagosomes within 8 min after the engulfment was completed; “delayed recruitment,” GFP(+) circles were detected on the phagosomes at least 10 min after the engulfment was completed; “no recruitment,” GFP signals were not detected on the phagosomes for a period > 50 min starting from the completion of engulfment; “short association,” the GFP signal was recruited to phagosomes at the normal time point, yet quickly disappeared from phagosomal surfaces.
In ced-5 null mutant embryos, phagosomal recruitment of LST-4(d)::GFP followed a relatively normal kinetics (Figure 8, D and E). However, 71% of phagosomes acquired SNX-1::GFP with significantly delayed kinetics but obtained nevertheless strong GFP signals (Figure 8, I and J). Consistent with these results, the defects in the maturation of C1, C2, and C3 phagosomes were much weaker in ced-5 mutants than in ced-1 mutants (Yu et al., 2006). Together, these results indicate that, like the production of phagosomal PtdIns(3)P, the recruitment of LST-4 and SNX-1 to phagosomes is primarily controlled by the ced-1 pathway, whereas the ced-5 pathway also contributes to these events.
Deletion of snx-1 does not affect the presentation of CED-1 to engulfing cell surfaces or the activity of CED-1 in promoting phagosome maturation
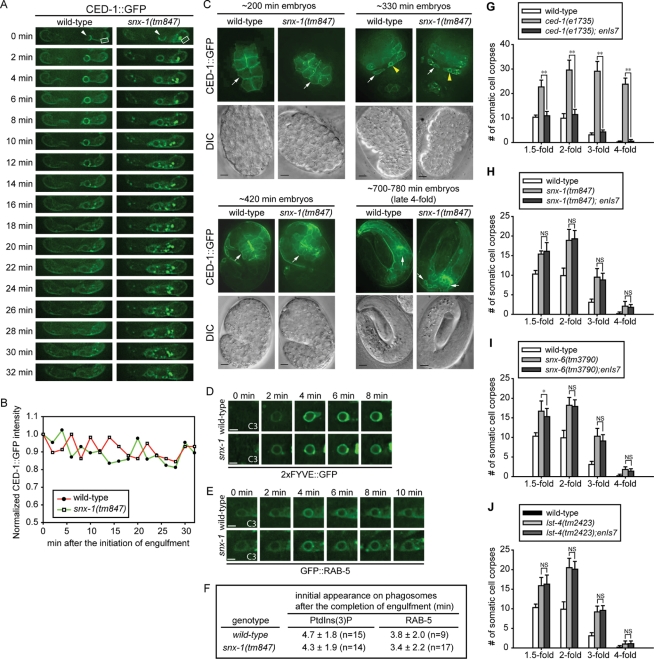
The disappearance of CED-1 from the surfaces of nascent phagosomes is slower in snx-1 single-mutant than in wild-type or lst-4 single-mutant embryos (Figures 9A and S3, B and C). This phenotype suggests that, in snx-1 deletion mutant embryos, phagosome maturation might be arrested at an early stage before the dissociation of CED-1 from phagosomes. Alternatively, snx-1 may directly regulate the dissociation of CED-1 from phagosomal membranes. A recent report indicated that SNX-1 and SNX-6 act in the retromer complex to recycle CED-1 from phagosomal surfaces to the plasma membrane, and in this manner regulate CED-1-mediated engulfment of apoptotic cells (Chen et al., 2010). To determine whether the prolonged presence of CED-1 on phagosomes affect the recycling of CED-1 to the plasma membrane of the host cell, we measured the level of CED-1::GFP on the plasma membrane of ABplaapppp, the engulfing cell for C3, during and after the internalization of C3, in embryos carrying enIs7 [Pced-1ced-1::gfp], an integrated array (Venegas and Zhou, 2007). Over a 32-min time course starting from the initiation of engulfment, in both wild-type and snx-1 mutant embryos, the levels of GFP signal on the plasma membrane merely fluctuated within 15% of that at the “0 min” time point (Figure 9, A and B). Moreover, no significant differences in signal intensity levels were observed between wild-type and snx-1(tm847) backgrounds (Figure 9, A and B). This set of results indicates that, in snx-1 mutants, the prolonged existence of CED-1 on phagosomal surfaces does not significantly affect the level of CED-1 on the surface of the host cell. In addition, we observed that, throughout the entire embryogenesis, CED-1::GFP is indeed presented on the surfaces of cells expressing Pced-1 in snx-1(tm847), snx-6(tm3790), and lst-4(n2423) single-mutant embryos as in wild-type embryos (Figure 9C and data not shown), indicating that, unlike that reported by Chen et al. (2010), CED-1’s plasma membrane presentation in embryos was not affected by the deletion of any of these three genes. Furthermore, we quantified the CED-1::GFP signal intensity in entire individual embryos at four embryonic stages, including a period spanning from 200 to 420 min after the first embryonic cell division, during which most (88 of 118) embryonic cell death events occur and cell corpses are removed (Materials and Methods) (Sulston et al., 1983). We found that, at most stages, the mean GFP intensity levels were comparable in wild-type, snx-1(tm847), snx-6(tm3790), and lst-4(tm2423) single-mutant embryos (Supplemental Table S2). Only at the late fourfold stage, which is at least 100 min after the last cell death event occurs (Sulston et al., 1983), were relatively weaker CED-1::GFP signal intensity levels observed from snx-1(tm847) and snx-6(tm3790) mutant embryos (77% and 50% of that of wild-type level, respectively) (Table S2). These results are different from the nearly complete loss of CED-1::GFP protein in snx-1(tm847) embryos reported by Chen et al. (2010), and indicate that, during the period when cell-corpse engulfment and degradation events are most active, CED-1::GFP level is not reduced by mutations in snx-1 or snx-6.
FIGURE 9:
The membrane localization of CED-1 in engulfing cells and its phagosome maturation-promoting activity are not affected by the snx mutations. (A) Time-lapse images monitoring the level of CED-1::GFP on the plasma membrane of engulfing cell ABplaapppp during and after the engulfment of cell corpse C3 (arrowhead) in wild-type and snx-1(tm847) mutant embryos. “0 min” indicates the time point when pseudopods start to extend around C3. The rectangle frames label the region from which the plasma membrane-associated GFP levels were measured and plotted in (B). (B) The fluorescence intensity of plasma membrane-associated CED-1::GFP within the framed region in (A) were measured at different time points, normalized to the GFP intensity at “0 min,” and plotted overtime. (C) CED-1::GFP is presented on the plasma membranes in snx-1(tm847) mutant embryos as in wild-type embryos at four different embryonic stages. CED-1::GFP is expressed from enIs7, an integrated transgenic array of the Pced-1ced-1::gfp reporter. White arrows indicate plasma membrane-localized CED-1::GFP. Yellow arrowheads indicate CED-1::GFP on phagosomal surfaces. Scale bar: 5 μm. (D–F) Time-lapse images monitoring the dynamic pattern of PtdIns(3)P production (D) and GFP::RAB-5 recruitment (E) on phagosomal surfaces. “0 min” represents the time point when engulfment is just completed. Scar bar: 2 μm. The quantification data of (D) and (E), obtained by monitoring multiple C1, C2, and C3 phagosomes over time, are shown in (F). n, number of phagosomes analyzed. (G–J) Overexpression of CED-1::GFP does not significantly rescue the Ced defect of snx mutant embryos. Bars represent average numbers of somatic cell corpses scored at different embryonic stages. Error bars represent standard deviation. Fifteen embryos were scored for each data point. enIs7 is an integrated transgenic array that expressed multiple copies of Pced-1 ced-1::gfp. *, **, and NS indicate p < 0.05, p < 0.01, and not significant (p > 0.05), respectively (independent Student’s t-test).
Chen et al. (2010) reported that overexpressing CED-1::GFP under the control of Pced-1 from an integrated array (smIs34) resulted in a complete rescue of the Ced phenotype displayed by snx-1(tm847) and snx-6(tm3790) mutant embryos. In contrast, we found that the overexpression of CED-1::GFP from a reporter plasmid constructed identically and was also integrated into the genome (enIs7[Pced-1ced-1::gfp]), did not result in any significant rescuing activity of the Ced phenotype in the snx-1(tm847), snx-6(tm3790), or lst-4(n2423) single-mutant embryos at multiple embryonic stages identical to that examined by Chen et al. (2010) (Figure 9, G–J; Supplemental text). Together, the above results strongly suggest that SNX-1, SNX-6, and LST-4 must control phagosome maturation through a mechanism other than regulating the cell-surface presentation or overall level of CED-1.
We thus directly measured whether the activity of CED-1 in promoting phagosome maturation was affected by the deletion of snx-1. As established previously, CED-1 triggers phagosome maturation through recruiting the large GTPase DYN-1 to the surface of nascent phagosomes. DYN-1 subsequently induces the robust production of PtdIns(3)P on phagosomal membranes and also recruits membrane tethering factors RAB-5 and RAB-7 to phagosomal surfaces (Yu et al., 2008; He et al., 2010). The enrichment of all of these molecules on phagosomal surfaces thus is dependent on CED-1 and could serve as readouts for CED-1 activity. We already observed that in snx-1(tm847) mutant embryos, the enrichment of DYN-1 onto the surface of nascent phagosomes was normal (Figure 5, D and F). We further monitored the presentation of PtdIns(3)P and RAB-5 on phagosomal surfaces and found no difference in the level or timing of the dynamic presence of these molecules in either wild-type or snx-1(tm847) mutant embryos (Figure 9, D–F). These results demonstrate that SNX-1 does not influence CED-1’s activity in triggering phagosome maturation, a conclusion consistent with our results that indicate that SNX-1 and LST-4 both act downstream of CED-1 for promoting phagosome maturation.
DISCUSSION
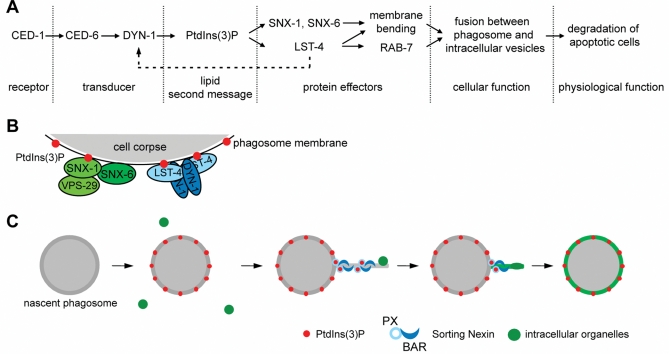
Through a screen for candidate PtdIns(3)P-binding proteins that specifically participate in the removal of apoptotic cells, we have identified LST-4/SNX9, SNX-1, and SNX-6, three sorting nexins that promote phagosome maturation in two parallel pathways in response to PtdIns(3)P signaling. Our findings have revealed how PtdIns(3)P triggers the degradation of apoptotic cells during C. elegans development (Figure 10). Furthermore, we discovered that these SNX-BAR sorting nexins specifically control the fusion of intracellular organelles with phagosomes through two distinct mechanisms: induction and stabilization of membrane curvature and maintenance of membrane-tethering factor RAB-7 on phagosomal surfaces (Figure 10, A and C). Our work further indicates that SNX-1 and SNX-6 directly regulate membrane fusion in a manner independent of the canonical retromer complex, whereas LST-4 helps retain RAB-7 on phagosomes through a DYN-1-mediated mechanism (Figure 10B). Sorting nexins are involved in multiple cellular events, such as endocytosis, endosomal trafficking, and cytoskeleton reorganization (reviewed in van Weering et al., 2010). To our knowledge, this is the first time that the critical physiological functions of sorting nexins in promoting phagosome maturation are demonstrated during animal development. LST-4, SNX-1, and SNX-6 are the only SNX-BAR subfamily sorting nexins encoded by the C. elegans genome. Their mammalian homologues are likely to play conserved roles in the degradation of apoptotic cells, an important event that prevents autoimmune and inflammatory responses and ensures homeostasis. Moreover, in other cellular and developmental context, SNX-BAR sorting nexins might promote the degradation of other kinds of phagosomal cargos, such as invading pathogens or necrotic cells.
FIGURE 10:
A PtdIns(3)P-mediated signaling pathway that drives the degradation of apoptotic cells. (A) Diagram of the pathway. The phagocytic receptor CED-1 and its adaptor CED-6 recruit the large GTPase DYN-1 to phagosomal surfaces, an event resulting in the robust production of phagosomal PtdIns(3)P, which in turn recruits LST-4, SNX-1, and SNX-6 from cytosol to phagosomal surfaces. These PtdIns(3)P effectors act in two parallel and partially redundant pathways to induce the extension of membrane tubules from phagosomes. In addition, LST-4 interacts with DYN-1 (Dynamin) and stabilizes its association with phagosome, which recruits and stabilizes the association of RAB-7 with phagosomes. Both RAB-7 and phagosome tubules facilitate efficient endosomes/phagosome and lysosomes/phagosome fusions and ultimately result in the degradation of apoptotic cells. (B) A model indicating the two distinct PtdIns(3)P effector complexes on phagosomal surfaces. One complex include SNX-1 and SNX-6 (may also contain VPS-29), in which SNX-1 interacts with PtdIns(3)P and brings SNX-6 onto phagosomes. In the second complex, LST-4, perhaps as homodimers, interacts with both PtdIns(3)P and DYN-1, which are enriched on phagosomes, and stabilize the association of the whole complex with phagosomes. (C) Model proposing that the PtdIns(3)P effectors use their BAR domains to induce and/or stabilize the formation of phagosomal tubules, which facilitates the recruitment and fusion of intracellular organelles to phagosomes.
LST-4, SNX-1, and SNX-6 control the degradation but not the engulfment of apoptotic cells
The engulfment and degradation of phagocytic targets are two different cellular events that are executed through distinct mechanisms (Caron and Hall, 2001; Vieira et al., 2002). Determining which of these two events these PtdIns(3)P effectors are involved in is pivotal for further understanding their molecular functions. Two lines of experimental evidence demonstrate that LST-4 and SNX-1 specifically control the degradation, not the engulfment of apoptotic cells. First, analyses using TEM technique and fluorescence microscopy both strongly indicate that, in each of the snx-1(tm847) and lst-4(tm2423) deletion alleles as well as the snx-1; lst-4 double mutants, germ cell corpses were normally internalized yet remained in phagosomes without being promptly degraded. Second, time-lapse recording monitoring somatic cell corpses revealed that the kinetics of engulfment were normal in lst-4; snx-1 double mutant embryos; rather, the degradation of cell corpses was blocked. This conclusion is consistent with the observations that PtdIns(3)P, the upstream regulating molecule of SNX-1 and LST-4, is not detected on phagosomal surfaces until engulfment is complete and phagosomes are sealed (Yu et al., 2008). This conclusion is different from a recent report that concludes that SNX-1 and the retromer complex in which SNX-1 acts primarily control the engulfment of apoptotic cells (Chen et al., 2010).
SNX-1 and SNX-6 promote phagosome maturation as downstream effectors of CED-1 and in a manner independent of the canonical retromer complex
Mammalian SNX1 and SNX6 are the known members of the membrane deformation subcomplex of the evolutionarily conserved retromer complex that mediates endosome-to-Golgi retrieval of transmembrane receptors and other trafficking-related proteins (reviewed in Bonifacino and Hurley, 2008; van Weering et al., 2010). Retromer’s cargo recognition subcomplex, a heterotrimer consisting of VPS26, VPS29, and VPS35, targets specific transmembrane receptors for retrograde transport (Bonifacino and Hurley, 2008; van Weering et al., 2010). C. elegans VPS-26 and VPS-35 were known to act in the retromer-dependent recycling of transmembrane protein MIG-14/Wntless (Pan et al., 2008; Yang et al., 2008). Our observation that the inactivation of VPS-26 and VPS-35 do not result in the persistence of cell corpses indicates that the canonical retromer complex is not involved in the degradation of apoptotic cells and further suggests that the phagosome-maturation functions of SNX-1 and SNX-6 are likely executed through a distinct protein complex and a novel mechanism. We found that the deletion of vps-29 caused a mild defect in the removal of germ cell but not somatic cell corpses. Because VPS29 possesses an SNX1-interacting motif (Collins et al., 2005; Wang et al., 2005), in the gonad, this distinct protein complex might also include VPS-29 (Figure 10B).
C. elegans SNX-1 was implicated in the regulation of endosomal clathrin dynamics (Shi et al., 2009). Chen et al. (2010) proposed that SNX-1 and SNX-6 act in the retromer complex to recycle CED-1 from phagosomal surfaces back to the plasma membrane, and in this manner regulate apoptotic cell engulfment. Our results, many obtained under highly comparable experimental conditions, are different from that reported by Chen et al. (2010) and do not support this model. In particular, we observed that, in snx-1(tm847) null mutant embryos, CED-1 was presented on engulfing cell surfaces at normal levels. Furthermore, the internalization of an apoptotic cell and the prolonged presence of CED-1 on the phagosome did not appear to affect the level of CED-1 on the surface of the host cell. In addition, in snx-1(tm847), snx-6(tm3790), or lst-4(tm2423) mutant backgrounds, the overall levels of CED-1::GFP in entire embryos are similar to that in wild-type embryos at the same developmental stages during early to mid-embryogenesis, when most cell death events occur (Supplemental Table S2). More importantly, we found that, in contrast to the full rescuing activity reported by Chen et al. (2010), overexpression of CED-1 did not rescue the Ced phenotype of snx-1 or snx-6 mutant embryos. These results, together with our finding that snx-1 mutants are not defective in cell corpse engulfment, indicate that the level and, more importantly, the function of CED-1 on engulfing cell surfaces are not primarily controlled by SNX-1 or SNX-6.
The direct evidence that the snx-1 deletion does not affect CED-1’s phagosome maturation activity comes from the observation that, in snx-1 mutant embryos, the dynamic phagosomal enrichment patterns of DYN-1, PtdIns(3)P, and RAB-5, three phagosome maturation factors that are dependent on CED-1 for their enrichment on phagosomal surfaces, occur in the normal manner. On the other hand, SNX-1 is recruited to phagosomes in a PtdIns(3)P- and CED-1-dependent manner. Together, these results have established that SNX-1 acts downstream of CED-1 to control phagosome maturation (Figure 10A). Recently, several reports revealed cases in which mammalian SNX1 regulates membrane trafficking and membrane remodeling in retromer-independent mechanisms (Gullapalli et al., 2006; Nisar et al., 2010; Prosser et al., 2010). Our finding reveals a new retromer-independent function of the SNX-1/SNX-6 complex.
LST-4, SNX-1, and SNX-6 promote the fusion of lysosomes to phagosomes through two distinct molecular mechanisms
In lst-4 and snx-1 single-mutant and lst-4; snx-1 double-mutant animals, a striking set of defects reside in the incorporation of early endosomes and lysosomes to phagosomes. These defects might be the primary cause for the defects in phagosome maturation, since the recruitment and fusion of these organelles are essential for the delivery of digestive enzymes to phagosomes and the acidification of phagosomal lumen. Our further analyses indicate the presence of two distinct molecular mechanisms used by these SNX-BAR proteins in the recruitment and fusion of intracellular vesicles to phagosomes (Figure 10).
The first mechanism involves the functions of BAR domains in inducing and stabilizing membrane curvature. High membrane curvature has been proposed to facilitate fusion (Chanturiya et al., 2002; Chernomordik and Kozlov, 2003; Marrink and Mark, 2003). The C. elegans phagosomes that contain apoptotic cells, which are of 2.0–2.5-μm diameters, are relatively flat compared with various intracellular vesicles that fuse to them (Yu et al., 2006, 2008). Regional membrane bending may thus be necessary for the extensive fusion events to occur between maturing phagosomes and the smaller endosomal and lysosomal particles. BAR domains are capable of using their rigid concave surfaces to induce and/or stabilize membrane curvature (Frost et al., 2009). Both mammalian SNX1 and SNX9 are able to tubulate liposomes in vitro and induce tubular structure formation when overexpressed in cultured cells (Carlton et al., 2004; Pylypenko et al., 2007; Haberg et al., 2008). Recruitment of SNX1 to Salmonella-containing vacuoles (SCV) induces the formation of tubules from SCV surfaces (Bujny et al., 2008). Previously, it has been established that phagosomes containing immunoglobulin G–opsonized particles or apoptotic cells extend membrane tubules, which facilitate the capture and incorporation of lysosomal particles (Figure 10B) (Harrison et al., 2003; Yu et al., 2008). These tubules are an extreme form of regional high membrane curvature. Here we report that the extension of phagosomal tubules requires the additive functions of both LST-4 and SNX-1, which are enriched on tubules. The BAR domains of LST-4, SNX-1, and SNX-6 may thus directly modulate membrane curvature by initiating and/or stabilizing phagosomal tubules and smaller protrusions. We further propose that the putative LST-4 homodimer and the SNX-1/SNX-6 heterodimer might perform such functions in a cooperative manner that has not been described previously, since lst-4; snx-1 double mutants displayed a much enhanced tubule extension defect than each of the single mutants (Figure 10, B and C).
The second mechanism, which is LST-4-specific, involves the recruitment and maintenance of the membrane-tethering factor RAB-7 to phagosomal surfaces. The association of RAB-7 to phagosomes is essential for the extension of phagosomal tubules as well as the fusion between lysosomal particles and phagosomal membranes (Harrison et al., 2003; Yu et al., 2008). In the gonad of lst-4 but not snx-1 null mutant hermaphrodites, no RAB-7 signal is observed on phagosomal surfaces, indicating an essential role of LST-4 in this event. In lst-4 mutant embryos, time-lapse recording revealed frequent premature dissociation of RAB-7 from phagosomal surfaces. We thus propose that, in addition to directly promoting tubule formation, LST-4 also indirectly facilitates the incorporation of lysosomes to phagosomes through retaining RAB-7 on phagosomal surfaces.
The interaction between mammalian SNX9 and dynamin stimulates dynamin’s GTPase activity in vitro (Yarar et al., 2008) and facilitates dynamin’s membrane association in cells (Lundmark and Carlsson, 2004). In lst-4 mutant embryos, the defect in maintaining RAB-7 on phagosomes correlates well with that in maintaining DYN-1. As DYN-1 and LST-4 rely on each other for stable association with phagosomes (Figure 10B), and as the association of RAB-7 with phagosome is DYN-1-dependent (Yu et al., 2008; He et al., 2010), we propose that LST-4 acts to ensure the retention of RAB-7 on phagosomes at least partially through stabilizing the DYN-1-phagosome association.
Mammalian PtdIns(3)P-binding proteins EEA1 and Hrs1 were both implicated in the maturation of phagosomes containing latex beads (Fratti et al., 2001; Vieira et al., 2004). To our surprise, eea-1 null mutants or hgrs-1 (RNAi) are not defective in the removal of apoptotic cells (Table 1). This discrepancy may reflect the difference in the regulation of phagosome maturation in cultured mammalian cells versus C. elegans or, alternatively, in the nature of engulfed particles. Differential dynamic enrichment patterns of PtdIns(3)P on phagosomes containing different objects have been observed (Vieira et al., 2001; Yu et al., 2008), which might differentially influence the involvement of distinct PtdIns(3)P effectors.
The dynamic phagosomal association patterns of LST-4, SNX-1, and SNX-6 are regulated by complex PtdIns(3)P-mediated mechanisms
We have demonstrated that the phagosomal association of LST-4, SNX-1, and SNX-6, which is essential for their in vivo functions in promoting phagosome maturation, is dependent on the PX domain–PtdIns(3)P interaction. PX domains alone, however, often are not sufficient for targeting their host proteins to membranes (Lemmon, 2008). This seems to be the case for LST-4 and SNX-1 because the phagosomal association of both proteins also depends on their C-terminal BAR domains. BAR domains are likely to facilitate the protein-membrane association through two different mechanisms. The intrinsic dimerization of BAR domain could drive the bivalent binding of PX domain to PtdIns(3)P-containing phagosome membranes (Frost et al., 2009). In addition, the basic patches on the concave surface of the BAR domains might directly interact with negatively charged membranes (Peter et al., 2004). A number of mammalian SNX-BAR sorting nexins, including SNX-1, SNX-5, SNX9, and SNX-18, rely on the “coincidence detection” mechanism, through which the PX domain recognizes specific membrane phosphoinositides and the BAR domain senses membrane curvature, for targeting to restricted regions of membranes (Carlton et al., 2004; Pylypenko et al., 2007; Traer et al., 2007; Haberg et al., 2008; Yarar et al., 2008; van Weering et al., 2010). Our observations support the model that the PX and BAR domains act in distinct manners yet cooperatively to target LST-4 and SNX-1 to phagosomal surfaces.
We found that SNX-6 relied on the physical interaction with SNX-1 for targeting to phagosomes. The physical and genetic interactions between SNX-1 and SNX-6 indicate that SNX-1 and SNX-6 form a protein complex on phagosomal surfaces to control phagosome maturation in one of the two parallel pathways (Figure 10B). The SNX-1/SNX-6 interaction is likely to enable the formation of a BAR domain dimer that could strengthen the association of both proteins with phagosomal membranes and PtdIns(3)P.
Interestingly, LST-4 and SNX-1 associate with phagosomes transiently, concurrent only with the first of the two waves of PtdIns(3)P on phagosomes. This phenomenon suggests that PtdIns(3)P alone is not sufficient to retain the SNX-BAR proteins on phagosomal surfaces efficiently. We found that C. elegans LST-4 interacted with DYN-1 (dynamin) in vitro like its mammalian orthologue and is dependent on DYN-1 for association with phagosomal surfaces. DYN-1 is transiently recruited to the surface of nascent phagosomes prior to the first wave of PtdIns(3)P (Yu et al., 2006, 2008). We propose that DYN-1 and PtdIns(3)P together determine the transient enrichment pattern of LST-4 on phagosomal surfaces. On the other hand, phagosomal-localized LST-4 further stabilizes DYN-1’s association with phagosomes via interaction with DYN-1. Such mutual dependency strengthens the membrane association of both DYN-1 and LST-4.
MATERIALS AND METHODS
Mutations and strains
C. elegans strains were grown at 20°C as previously described (Brenner, 1974). The N2 Bristol strain was used as the reference wild-type strain. Mutations and integrated arrays are described by Riddle et al. (1997) and the Worm Base (http://www.wormbase.org), except when noted otherwise: LGI, ced-1(n1735), F22G12.4(tm3221), snx-3(tm1595); LGII, rab-7(ok511), aka-1(tm389), wdfy-2(tm3806), pld-1(ok2222); LGIII, mtm-6(ok330), rabn-5(tm1555); LGIV, ced-5(n1812), lst-4(tm2423), tag-77(tm702), exc-5(rh232), rabs-5(ok1513), F13E9.1(tm1886), Y116A8C.26(tm2404); LGV, unc-76(e911), eea-1(tm933), rskd-1(tm4031), snx-6(tm3790); LGX, snx-1(tm847), F39B1.1(tm3171), F17H10.3(tm3643), enIs7[Pced-1ced-1::gfp] (Venegas and Zhou, 2007). All ok alleles are generated by the C. elegans Gene Knockout Consortium and distributed by Caenorhabditis Genetics Center (CGC). All tm alleles were generated and provided by the National Bioresource Project of Japan. Strains carrying lst-4(tm2423), snx-1(tm847), and snx-6(tm3790) used here were outcrossed four times. Transgenic worms were generated by microinjection as previously described (Jin, 1999). The plasmids were injected with coinjection marker pUNC76 [unc-76(+)] (Bloom and Horvitz, 1997) into unc-76 (e911) mutant adult hermaphrodites and transgenic animals were identified as non-Unc animals.
Plasmid construction
The snx-1, snx-6, lst-4(d), and lst-4(e) cDNAs were amplified from a mixed-stage C. elegans cDNA library (Z. Zhou and H.R. Horvitz, unpublished data) using the PCR. The engulfing cell-specific expressing plasmids Pced-1snx-1::gfp, Pced-1snx-6::gfp, Pced-1lst-4(d)::gfp, and Pced-1lst-4(e)::gfp were constructed by cloning the snx-1, snx-6, lst-4(d), and lst-4(e) cDNAs into the multicloning sites of pZZ829, a plasmid carrying Pced-1 (the ced-1 promotor), a 3′ gfp tag, and the unc-54 3′ UTR (Yu et al., 2006). The (R132Q), (R376E K377E R378E), and (R132Q R376E K377E R378E) mutations were introduced into Pced-1snx-1::gfp using the QuickChange Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA) to generate Pced-1snx-1(PXm)::gfp, Pced-1snx-1(BARm)::gfp, and Pced-1snx-1(PXm BARm)::gfp, respectively. Using the same strategy, the (R263Q) and (K492E R499E) mutations were introduced into Pced-1lst-4(d)::gfp to generate Pced-1lst-4(d)(PXm)::gfp and Pced-1lst-4(d)(BARm)::gfp, respectively. snx-1, snx-6, and lst-4(d) cDNAs were cloned into vector pGEX4T-1 (GE Healthcare, Piscataway, NJ) to generate pGST-snx-1, pGST-snx-6, and pGST-lst-4(d) plasmids, respectively. pGST-PXSNX-1 (GST-PX domain of SNX-1) was constructed by cloning the cDNA encoding amino acid 75–229 of SNX-1 into vector pGEX4T-1. To generate yeast two-hybrid assay constructs, snx-1, snx-6, and lst-4(d) cDNA were first cloned into the pENTR/D-TOPO (Invitrogen, Carlsbad, CA) vector and then transferred into Gateway destination vectors pLexAgtwy or pACT2.2gtwy (Caldwell et al., 2006) by LR recombination (Invitrogen), fused with the DNA binding domain of LexA or the transcriptional activation domain of GAL4, respectively.
Nomarski DIC microscopy
DIC microscopy was performed with an Axionplan 2 compound microscope (Carl Zeiss, Thornwood, NY) equipped with Nomarski DIC optics, a digital camera (AxioCam MRm; Carl Zeiss), and imaging software (AxioVision; Carl Zeiss). Previously established protocols were used to score cell corpses under DIC microscopy (Yu et al., 2006). Somatic embryonic cell corpses were scored in the head region of embryos at different developmental stages. Germ cell corpses were scored in one of the two gonadal arms of adult hermaphrodites 48 h post-L4 stage unless otherwise stated. Time-lapse DIC microscopy was used to count the total number of apoptotic events and record the duration of each cell corpse for a 300-min time window during embryogenesis as described (Yu et al., 2006). Embryos undergoing first cell cleavage were identified and defined as “0 min” embryo. The same embryos were recorded from “160 min” to “460 min” at 3-min intervals. For each time point, DIC images of 40 z-sections were captured at 0.5-μm intervals.
Fluorescence microscopy and time-lapse recording
An Olympus IX70-Applied Precision DeltaVision microscope equipped with a Photometris Coolsnap digital camera was used to capture fluorescence images, and the Applied Precision SoftWoRx software was used for image deconvolution and processing (Yu et al., 2008). To monitor the kinetics of the subcellular localization of various GFP reporters during the engulfment and degradation of cell corpses C1, C2, and C3, the procedure followed an established protocol (Yu et al., 2008; Lu et al., 2009). Recording of the ventral surface of embryos started at 310–320 min post–first cleavage and lasted for 60–120 min in 1- or 2-min intervals, except when the extension of phagosomal tubules were monitored, when 20-s intervals were chosen and the recording lasted 20–30 min. At each time point, 10–15 serial z-sections at a 0.5-μm interval were recorded. Signs such as embryo elongation and movement were closely monitored to ensure that the embryo being recorded developed normally. Images were analyzed using the ImageJ software and fluorescence signal intensity was measured (Lu et al., 2009). The enrichment of fluorescence signal on phagosomal surfaces was considered significant when the ratio of the signal intensity on phagosomal surfaces versus the cytoplasm was ≥1.2 (Lu et al., 2009).
RNAi
RNAi experiments were performed using feeding protocol as previously described (Kamath et al., 2001). In brief, mid–L4-stage hermaphrodites were transferred to RNAi plates. After 48 h, the numbers of germ cell corpses per gonad arm were scored under DIC optics. The RNAi feeding constructs for lst-2, ZK632.12, hgrs-1, mtm-3, R11D1.10, ppk-3, and F25H2.2 were from a C. elegans RNAi library (Kamath et al., 2003). The RNAi feeding construct for Y48E1B.14 was constructed by cloning PCR amplified cDNA fragment into vector pPD129.36 using the primer pairs: 5′-atgatgaacatgtccgatcc-3′ and 5′-acgcgtcggatgcaccggatg-3′. The vector pPD129.36 was used as a control RNAi construct.
Yeast two-hybrid assay
The yeast two-hybrid assay to detect possible interaction among SNX-1, SNX-6, and LST-4 was performed as described (Vojtek and Hollenberg, 1995; Bai and Elledge, 1997). Different combinations of bait and prey constructs were transformed into yeast strain CTYLH, and transformants were selected on leucine and tryptophan drop-out synthetic plates. Interactions between baits and preys were examined at 30°C using the β-galactosidase reporter assay.
GST pull-down and coimmunoprecipitation
For GST pull-down assays, GST and GST-tagged SNX-6 were purified from E. coli lysates using Glutathione Sepharose 4B (GE Heathcare) according to the manufacturer’s instructions. snx-1 and lst-4 cDNA were cloned into pcDNA3.1/C-HA and transiently transfected into HEK 293T cell individually using Lipofectamine 2000 (Invitrogen). Transfected cells were lysed with NETN buffer (50 mM Tris-HCl, pH 8.0, 1 mM EDTA, 150 mM NaCl, 0.5% NP-40 supplemented with protease inhibitor cocktail [Roche Applied Science, Mannheim, Germany and phenylmethylsulfonyl fluoride [PMSF]). Equal amounts of cell lysates expressing SNX-1-HA or LST-4-HA were incubated with immobilized GST or GST-tagged SNX-6 at 4°C for 3 h. After three washes with NETN, proteins bound on resins were separated by SDS–PAGE and detected by Western blot with anti-HA antibody (Sigma Chemical, St. Louis, MO).
For coimmunoprecipitation assays, dyn-1 and ced-6 cDNAs were cloned into pcDNA3.1/N-HA, and the lst-4(d) cDNA into pcDNA3.1/C-Flag. Pairs of constructs expressing HA- and FLAG-tagged proteins were transiently transfected into HEK 293T cells. Approximately 500 μg transfected cell lysates in NETN buffer was incubated with 5 μg anti-HA antibody (Sigma Chemical) at 4°C for 1 h and then with Protein A agarose beads (GE Healthcare) overnight at 4°C. After three washes with NETN, the precipitated proteins were analyzed by SDS–PAGE and Western blots.
Acknowledgments
We thank X. He for the DeltaVision and X. He, A. Kuspa, and H. Bellen for helpful comments. We thank the C. elegans Genetics Center (CGC) and the National BioResource Project in Japan (Shohei Mitani) for deletion alleles. Z.Z. is supported by the NIH (GM067848). T.R.M was funded by NIH Training Grant T32-NS043124 and NIH IRACDA Training Grant K12-GM084897–03.
Abbreviations used:
- BAR domain
(Bin–amphiphysin–Rvs) domain
- Ced
cell corpse removal defective
- DIC
differential interference contrast
- DYN-1
dynamin 1
- EEA1
early endosome antigen 1
- FYVE domain
(Fab1p, YOTB, Vac1p, and EEA1) domain
- Hrs1
hepatocyte growth factor–regulated tyrosine kinase substrate 1
- LST-4
lateral signaling target 4
- PtdIns(3)P
phosphatidylinositol 3-phosphate
- PX domain
(phox homology) domain
- SH3 domain
Src homology 3 domain
- SNX-1
sorting nexin 1
Footnotes
This article was published online ahead of print in MBoC in Press http://www.molbiolcell.org/cgi.doi/10.1091/mbc.E10-09-0756) on December 9, 2010.
REFERENCES
- Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- Bai C, Elledge SJ. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1997;283:141–156. doi: 10.1016/s0076-6879(97)83013-3. [DOI] [PubMed] [Google Scholar]
- Birkeland HC, Stenmark H. Protein targeting to endosomes and phagosomes via FYVE and PX domains. Curr Top Microbiol Immunol. 2004;282:89–115. doi: 10.1007/978-3-642-18805-3_4. [DOI] [PubMed] [Google Scholar]
- Bloom L, Horvitz HR. The Caenorhabditis elegans gene unc-76 and its human homologs define a new gene family involved in axonal outgrowth and fasciculation. Proc Natl Acad Sci USA. 1997;94:3414–3419. doi: 10.1073/pnas.94.7.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Hurley JH. Retromer. Curr Opin Cell Biol. 2008;20:427–436. doi: 10.1016/j.ceb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J, et al. The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol Cell. 2001;8:829–839. doi: 10.1016/s1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujny MV, Ewels PA, Humphrey S, Attar N, Jepson MA, Cullen PJ. Sorting nexin-1 defines an early phase of Salmonella-containing vacuole-remodeling during Salmonella infection. J Cell Sci. 2008;121:2027–2036. doi: 10.1242/jcs.018432. [DOI] [PubMed] [Google Scholar]
- Caldwell GA, Williams SN, Caldwell KA. Integrated Genomics: A Discovery-Based Laboratory Course. Chichester, UK: John Wiley & Sons; 2006. [Google Scholar]
- Carlton J, Bujny M, Peter BJ, Oorschot VM, Rutherford A, Mellor H, Klumperman J, McMahon HT, Cullen PJ. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr Biol. 2004;14:1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- Caron E, Hall A. Phagocytosis. In: Marsh M, editor. In: Endocytosis. Oxford, UK:: Oxford University Press,; 2001. pp. 58–77. [Google Scholar]
- Chanturiya A, Scaria P, Kuksenok O, Woodle MC. Probing the mechanism of fusion in a two-dimensional computer simulation. Biophys J. 2002;82:3072–3080. doi: 10.1016/S0006-3495(02)75648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Xiao H, Zhang K, Wang B, Gao Z, Jian Y, Qi X, Sun J, Miao L, Yang C. Retromer is required for apoptotic cell clearance by phagocytic receptor recycling. Science. 2010;327:1261–1264. doi: 10.1126/science.1184840. [DOI] [PubMed] [Google Scholar]
- Chernomordik LV, Kozlov MM. Protein-lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- Collins BM, Skinner CF, Watson PJ, Seaman MN, Owen DJ. Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly. Nat Struct Mol Biol. 2005;12:594–602. doi: 10.1038/nsmb954. [DOI] [PubMed] [Google Scholar]
- Ellson CD, Anderson KE, Morgan G, Chilvers ER, Lipp P, Stephens LR, Hawkins PT. Phosphatidylinositol 3-phosphate is generated in phagosomal membranes. Curr Biol. 2001a;11:1631–1635. doi: 10.1016/s0960-9822(01)00447-x. [DOI] [PubMed] [Google Scholar]
- Ellson CD, Andrews S, Stephens LR, Hawkins PT. The PX domain: a new phosphoinositide-binding module. J Cell Sci. 2002;115:1099–1105. doi: 10.1242/jcs.115.6.1099. [DOI] [PubMed] [Google Scholar]
- Ellson CD, et al. PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40(phox). Nat Cell Biol. 2001b;3:679–682. doi: 10.1038/35083076. [DOI] [PubMed] [Google Scholar]
- Ellson C, Davidson K, Anderson K, Stephens LR, Hawkins PT. PtdIns3P binding to the PX domain of p40phox is a physiological signal in NADPH oxidase activation. EMBO J. 2006;25:4468–4478. doi: 10.1038/sj.emboj.7601346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti RA, Backer JM, Gruenberg J, Corvera S, Deretic V. Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J Cell Biol. 2001;154:631–644. doi: 10.1083/jcb.200106049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullapalli A, Wolfe BL, Griffin CT, Magnuson T, Trejo J. An essential role for SNX1 in lysosomal sorting of protease-activated receptor-1: evidence for retromer-, Hrs-, and Tsg101-independent functions of sorting nexins. Mol Biol Cell. 2006;17:1228–1238. doi: 10.1091/mbc.E05-09-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberg K, Lundmark R, Carlsson SR. SNX18 is an SNX9 paralog that acts as a membrane tubulator in AP-1-positive endosomal trafficking. J Cell Sci. 2008;121:1495–1505. doi: 10.1242/jcs.028530. [DOI] [PubMed] [Google Scholar]
- Harrison RE, Bucci C, Vieira OV, Schroer TA, Grinstein S. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol Cell Biol. 2003;23:6494–6506. doi: 10.1128/MCB.23.18.6494-6506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, et al. Live-cell imaging in Caenorhabditis elegans reveals the distinct roles of dynamin self-assembly and guanosine triphosphate hydrolysis in the removal of apoptotic cells. Mol Biol Cell. 2010;21:610–629. doi: 10.1091/mbc.E09-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui E, Johnson CP, Yao J, Dunning FM, Chapman ER. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca(2+)-regulated fusion. Cell. 2009;138:709–721. doi: 10.1016/j.cell.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y. In: Transformation. In: C. elegans, A Practical Approach. Hope IA, editor. Oxford, UK: Oxford University Press,; 1999. pp. 69–96. [Google Scholar]
- Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai F, Liu H, Field SJ, Akbary H, Matsuo T, Brown GE, Cantley LC, Yaffe MB. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat Cell Biol. 2001;3:675–678. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Doukoumetzidis K, Almendinger J, Stergiou L, Tosello-Trampont A, Sifri CD, Hengartner MO, Ravichandran KS. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol. 2008;10:556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen JM, Ravichandran KS. Phagosome maturation: going through the acid test. Nat Rev Mol Cell Biol. 2008;9:781–795. doi: 10.1038/nrm2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- Lu N, Yu X, He X, Zhou Z. Detecting apoptotic cells and monitoring their clearance in the nematode Caenorhabditis elegans. Methods Mol Biol. 2009;559:357–370. doi: 10.1007/978-1-60327-017-5_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark R, Carlsson SR. Sorting nexin 9 participates in clathrin-mediated endocytosis through interactions with the core components. J Biol Chem. 2003;278:46772–46781. doi: 10.1074/jbc.M307334200. [DOI] [PubMed] [Google Scholar]
- Lundmark R, Carlsson SR. Regulated membrane recruitment of dynamin-2 mediated by sorting nexin 9. J Biol Chem. 2004;279:42694–42702. doi: 10.1074/jbc.M407430200. [DOI] [PubMed] [Google Scholar]
- Mangahas PM, Yu X, Miller KG, Zhou Z. The small GTPase Rab2 functions in the removal of apoptotic cells in Caenorhabditis elegans. J Cell Biol. 2008;180:357–373. doi: 10.1083/jcb.200708130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink SJ, Mark AE. Molecular dynamics simulation of the formation, structure, and dynamics of small phospholipid vesicles. J Am Chem Soc. 2003;125:15233–15242. doi: 10.1021/ja0352092. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Metzstein MM, Stanfield GM, Horvitz HR. Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet. 1998;14:410–416. doi: 10.1016/s0168-9525(98)01573-x. [DOI] [PubMed] [Google Scholar]
- Nisar S, Kelly E, Cullen PJ, Mundell SJ. Regulation of P2Y1 receptor traffic by sorting Nexin 1 is retromer independent. Traffic. 2010;11:508–519. doi: 10.1111/j.1600-0854.2010.01035.x. [DOI] [PubMed] [Google Scholar]
- Pan CL, Baum PD, Gu M, Jorgensen EM, Clark SG, Garriga G. C. elegans AP-2 and retromer control Wnt signaling by regulating mig-14/Wntless. Dev Cell. 2008;14:132–139. doi: 10.1016/j.devcel.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Prosser DC, Tran D, Schooley A, Wendland B, Ngsee JK, A novel, retromer-independent role for sorting nexins 1 and 2 in RhoG-dependent membrane remodeling. Traffic. 2010;11:1347–1362. doi: 10.1111/j.1600-0854.2010.01100.x. [DOI] [PubMed] [Google Scholar]
- Pylypenko O, Lundmark R, Rasmuson E, Carlsson SR, Rak A. The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J. 2007;26:4788–4800. doi: 10.1038/sj.emboj.7601889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR. The engulfment process of programmed cell death in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2004;20:193–221. doi: 10.1146/annurev.cellbio.20.022003.114619. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Blumenthal T, Meyer BJ, Priess JR (eds.), editors. C. elegans II. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- Savill J, Fadok V. Corpse clearance defines the meaning of cell death. Nature. 2000;407:784–788. doi: 10.1038/35037722. [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet LF, Hong W. The Phox (PX) domain proteins and membrane traffic. Biochim Biophys Acta. 2006;1761:878–896. doi: 10.1016/j.bbalip.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Shi A, Sun L, Banerjee R, Tobin M, Zhang Y, Grant BD. Regulation of endosomal clathrin and retromer-mediated endosome to Golgi retrograde transport by the J-domain protein RME-8. EMBO J. 2009;28:3290–3302. doi: 10.1038/emboj.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N, Lee S, Ahn N, Kim SA, Ahn SG, YongPark Z, Chang S. Sorting nexin 9 interacts with dynamin 1 and N-WASP and coordinates synaptic vesicle endocytosis. J Biol Chem. 2007;282:28939–28950. doi: 10.1074/jbc.M700283200. [DOI] [PubMed] [Google Scholar]
- Soulet F, Yarar D, Leonard M, Schmid SL. SNX9 regulates dynamin assembly and is required for efficient clathrin-mediated endocytosis. Mol Biol Cell. 2005;16:2058–2067. doi: 10.1091/mbc.E04-11-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic TI, Setty TG, Sitaram A, Burd CG. Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J Cell Biol. 2007;177:115–125. doi: 10.1083/jcb.200609161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Teasdale RD, Loci D, Houghton F, Karlsson L, Gleeson PA. A large family of endosome-localized proteins related to sorting nexin 1. Biochem J. 2001;358:7–16. doi: 10.1042/0264-6021:3580007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touret N, et al. Quantitative and dynamic assessment of the contribution of the ER to phagosome formation. Cell. 2005;123:157–170. doi: 10.1016/j.cell.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Traer CJ, Rutherford AC, Palmer KJ, Wassmer T, Oakley J, Attar N, Carlton JG, Kremerskothen J, Stephens DJ, Cullen PJ. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat Cell Biol. 2007;9:1370–1380. doi: 10.1038/ncb1656. [DOI] [PubMed] [Google Scholar]
- van Weering JR, Verkade P, Cullen PJ. SNX-BAR proteins in phosphoinositide-mediated, tubular-based endosomal sorting. Semin Cell Dev Biol. 2010;21:371–380. doi: 10.1016/j.semcdb.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venegas V, Zhou Z. Two alternative mechanisms that regulate the presentation of apoptotic cell engulfment signal in Caenorhabditis elegans. Mol Biol Cell. 2007;18:3180–3192. doi: 10.1091/mbc.E07-02-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira OV, Botelho RJ, Grinstein S. Phagosome maturation: aging gracefully. Biochem J. 2002;366:689–704. doi: 10.1042/BJ20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira OV, Botelho RJ, Rameh L, Brachmann SM, Matsuo T, Davidson HW, Schreiber A, Backer JM, Cantley LC, Grinstein S. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol. 2001;155:19–25. doi: 10.1083/jcb.200107069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira OV, Harrison RE, Scott CC, Stenmark H, Alexander D, Liu J, Gruenberg J, Schreiber AD, Grinstein S. Acquisition of Hrs, an essential component of phagosomal maturation, is impaired by mycobacteria. Mol Cell Biol. 2004;24:4593–4604. doi: 10.1128/MCB.24.10.4593-4604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM. Ras-Raf interaction: two-hybrid analysis. Methods Enzymol. 1995;255:331–342. doi: 10.1016/s0076-6879(95)55036-4. [DOI] [PubMed] [Google Scholar]
- Wang D, et al. Crystal structure of human vacuolar protein sorting protein 29 reveals a phosphodiesterase/nuclease-like fold and two protein-protein interaction sites. J Biol Chem. 2005;280:22962–22967. doi: 10.1074/jbc.M500464200. [DOI] [PubMed] [Google Scholar]
- Yang PT, Lorenowicz MJ, Silhankova M, Coudreuse DY, Betist MC, Korswagen HC. Wnt signaling requires retromer-dependent recycling of MIG-14/Wntless in Wnt-producing cells. Dev Cell. 2008;14:140–147. doi: 10.1016/j.devcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Yarar D, Surka MC, Leonard MC, Schmid SL. SNX9 activities are regulated by multiple phosphoinositides through both PX and BAR domains. Traffic. 2008;9:133–146. doi: 10.1111/j.1600-0854.2007.00675.x. [DOI] [PubMed] [Google Scholar]
- Yu X, Lu N, Zhou Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 2008;6:e61. doi: 10.1371/journal.pbio.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Odera S, Chuang CH, Lu N, Zhou Z. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev Cell. 2006;10:743–757. doi: 10.1016/j.devcel.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Yu X. Phagosome maturation during the removal of apoptotic cells: receptors lead the way. Trends Cell Biol. 2008;18:474–485. doi: 10.1016/j.tcb.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]