Abstract
The site-selective encoding of noncanonical amino acids (NAAs) is a powerful technique for the installation of novel chemical functional groups in proteins. This is often achieved by recoding a stop codon and requires two additional components: an evolved aminoacyl tRNA synthetase (AARS) and a cognate tRNA. Analysis of the most successful AARSs reveals common characteristics. The highest fidelity NAA systems derived from the Methanocaldococcus jannaschii tyrosyl AARS feature specific mutations to two residues reported to interact with the hydroxyl group of the substrate tyrosine. We demonstrate that the restoration of just one of these determinants for amino acid specificity results in the loss of fidelity as the evolved AARSs become noticeably promiscuous. These results offer a partial explanation of a recently retracted strategy for the synthesis of glycoproteins. Similarly, we reinvestigated a tryptophanyl AARS reported to allow the site-selective incorporation of 5-hydroxy tryptophan within mammalian cells. In multiple experiments, the enzyme displayed elements of promiscuity despite its previous characterization as a high fidelity enzyme. Given the many similarities of the TyrRSs and TrpRSs reevaluated here, our findings can be largely combined, and in doing so they reinforce the long-established central dogma regarding the molecular basis by which these enzymes contribute to the fidelity of translation. Thus, our view is that the central claims of fidelity reported in several NAA systems remain unproven and unprecedented.
Keywords: directed evolution, protein engineering
Aminoacyl tRNA synthetases (AARSs) are a central feature of any effort to recode or reassign the genetic code (1). During translation AARSs enforce fidelity through a number of well-determined mechanisms (2–4). AARSs discriminate between amino acids, but also all endogenous tRNAs, thereby conferring specificity. A cognate tRNA molecule is aminoacylated based on sequence identity elements, and a given amino acid comes to be associated with the correct anticodon. Misacylation occurs, but for the AARSs where this is problematic, for example an IleRS (5) or LeuRS (6), additional editing domains result in the hydrolysis of the misacylated amino acid. However, TyrRSs and TrpRSs, the focus of this work, are not known to have additional editing mechanisms despite several decades of intensive molecular and biochemical investigation (7–9).
TyrRSs and TrpRSs, evolved or engineered, are responsible for a significant proportion of all noncanonical amino acids (NAAs) now routinely incorporated into proteins (10–14). These two AARSs probably share a common ancient origin given the similarities in protein sequence and overall structure (15–18). Thus, we believe that concepts about the determinants of specificity of one AARS are often applicable to the other and vice versa.
With some exceptions (19–21), the site-selective incorporation of NAAs is commonly achieved by recoding a single stop codon. This process has been reported using mutant TyrRSs or TrpRSs in a range of host organisms (22–25). In these systems, suppression of a stop codon is in constant competition with the translation system’s inherent tendency to terminate translation in response to the stop signal. As a result of this competitive recoding/suppression strategy, it is relatively easy to interpret results when a mutant AARS exhibits exquisite fidelity in charging its cognate NAA to tRNAstop (26–28). If the tRNAstop goes uncharged, the stop codon inserted into the gene of interest reverts to its natural role as the molecular signal for termination of translation, and truncated products of translation ensue. Mutant AARSs may also be capable of charging amino acids besides the desired NAA onto tRNAstop. In this situation, translation termination will not occur, and low-resolution analysis techniques such as SDS-PAGE designed to report solely on termination will not be able to discriminate between proteins containing successfully installed NAAs and unsuccessfully synthesized alloproteins with a substituting canonical amino acid. Selection schemes to minimize mischarging have been developed (29) and refined (30).
In NAA incorporation systems, the amino acid most likely to be charged in place of the NAA is the amino acid that the evolved or engineered AARS originally recognized. Translationally, the ribosome exerts little if any editing power, and so for AARSs without editing domains, such as TyrRSs and TrpRSs, the fidelity of NAA incorporation with these AARSs will be directly related to the relative activity that these synthetases have with their original amino acid versus the NAA. We show that single residue elements in these AARS enzymes confer the enzyme’s specificity and, ultimately, the fidelity of incorporation. The applicability of this concept has been extended to TyrRS (31–33) and TrpRS (25) derived systems whose reported fidelity is in question, or appears at odds with other work in the field.
Results and Discussion
TyrRS.
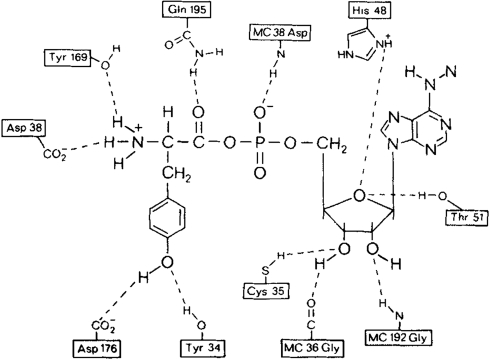
Fersht et al. identified two residues of the Bacillus stearothermophilus (Bst) TyrRS, Tyr 34, and Asp 176, as hydrogen bonding partners for the hydroxyl -OH group of substrate tyrosine (Fig. 1) (9, 34, 35). Their observations were based on crystallographic evidence (36), and their observations can be applied to the highly homologous Methanocaldococcus jannaschii (Mj) TyrRS (37). In the majority of Mj TyrRSs evolved to charge NAAs, these two residues (Mj numbering Tyr32 and Asp158) have been mutated to more hydrophobic residues as a requirement for achieving high fidelity incorporation of the NAA versus native tyrosine (Table S1).
Fig. 1.
Molecular interactions between hydrogen-bonding side chains from Bst tyrosyl tRNA synthetase with bound Tyr-AMP. Bst residues Tyr34 and Asp174 map onto Mj Tyr32 and Asp158, respectively. (Reproduced with permission from ref. 9 (Copyright 1985, Nature Publishing Group.)
Whereas most Mj TyrRS variants have changed Tyr32 to a smaller, more hydrophobic residue, the TyrRS evolved to charge 3-aminoTyrosine (3-aminoTyr) featured Gln, Glu, Lys, or Arg at this position (38). This can be rationalized from the overall structural similarity between 3-amino Tyr and Tyr and their relative ability to engage in salt bridging interactions. There are just three reports of an evolved TyrRS mutant preserving either of the Mj wild-type (WT) TyrRS residues Tyr32 or Asp158. The two TyrRSs that were reported to charge tRNACUA with GlcNAc-Ser or GalNAc-Thr retained Tyr32 or Asp158, respectively (39, 40). However, both reports have been corrected, (31, 32) and it seems these TyrRSs were likely mischarging tyrosine in place of the NAA (41). Similarly, the Mj TyrRS reported to incorporate 3-OH Tyr retained Asp158 (33). The latter report did not include SDS-PAGE analysis as is customary, but a mass spectrum was reported. Two masses were observed in the spectrum at 18,432.3 and 18,448.5 Da. It was previously established that a mass of ∼18,432 Da corresponds to incorporation of tyrosine into their common test protein myoglobin (42). These authors suggested that the signal at 18432.3 was due to fragmentation of the 3-hydroxy tyrosine side chain through, “….loss of oxygen, or oxygen and a proton.” However, this is not consistent with reported fragmentation patterns for the parent molecule (43, 44). We contend that it is more likely that the 18432.3 Da signal is due to competing incorporation of tyrosine, which is consistent with other mass spectra but also the importance of this particular TyrRS’s Asp158 binding the Tyr -OH group (vide infra).
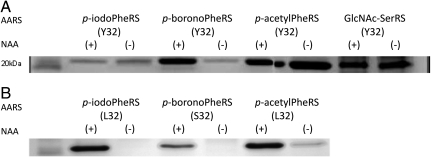
The importance of Tyr32 as a “gatekeeper” residue of TyrRS fidelity, especially in NAA systems, was investigated by reverting three evolved TyrRSs to code for Tyr at this position. The three TyrRSs chosen corresponded to the enzymes reported for p-iodoPhe (45), p-acetylPhe (28), and p-boronoPhe (46). The evolved enzymes were reported with either a Leu or Ser in place of Tyr at position 32. Each revertant and cognate tRNACUA were expressed along with a UAG mutant for T4 lysozyme in the presence (+) and absence (-) of each NAA. The results suggest high fidelity tRNACUA charging for the original enzymes with p-iodoPhe and p-boronoPhe, and marginal fidelity for charging with p-acetylPhe (Fig. 2). The results also show that the three TyrRS revertants would appear to charge tRNACUA in the absence of NAA. The presence of full-length protein in the absence of NAA (Fig. 2) is consistent with WT enzyme activity where tRNACUA is charged with a canonical amino acid, presumably tyrosine. It was also observed that the revertants displayed similar fidelity to the AARS reported to have high fidelity for GlcNAc-Ser. Again, this is consistent with the fact that the TyrRS variant reported to have high fidelity for GlcNAc-Ser possessed the same Tyr32 residue found in the WT enzyme (39).
Fig. 2.
Analysis of the revertants (A) and the original AARS mutants (B). Tyr32 was reintroduced into the three TyrRS variants indicated (the other mutations to these variants are listed in Table S1). The lanes are labeled (+) or (-) that designates whether the corresponding nonnatural amino acid was added to the culture. The SDS-PAGE analysis shown was performed after Ni2+-affinity purification of full-length T4 lysozyme purified by the C-terminal hexahistidine tag to remove the truncated material and simplify the analysis. The lane furthest left corresponds to a commercial protein ladder and a molecular weight band of 20 kDa.
Söll has suggested that an exogenous AARS produced in Escherichia coli requires as little as 1% of the activity of the WT enzyme to complement an auxotrophic phenotype (15). Our results suggest that the activity of the GlcNAc-SerRS and the revertants are well above this threshold. Other insight into why the GlcNAc-SerRS and revertant TyrRSs would charge tRNACUA with a canonical amino acid comes from Fersht et al.’s reports on the effects of mutating each of a Bst TyrRS’s molecular determinants. A Tyr34Phe mutation (Bst TyrRS numbering; equivalent to Mj Tyr32) resulted in a 15-fold decrease in the specificity of Tyr versus Phe (9). Attempts to mutate Asp176 (the equivalent of Mj Asp158) resulted in inactive enzymes, suggesting the latter residue is likely the more important molecular determinant of fidelity (34).
Crystallographic evidence suggests that the mutations to a TyrRS, such as those found in the GlcNAc-Ser TyrRS (total mutations: Glu107Pro, Asp158Cys, Ile159 Tyr, and Leu162Arg) would be expected to reconstitute some of the hydrogen bonding network lost by the Asp158Cys mutation (again, Tyr32 was retained). The marginal fidelity that the p-acetylPheRS demonstrated in the absence of p-acetylPhe (Fig. 2) may be similarly rationalized from this enzyme’s Ile159Cys and Leu162Arg mutations forming such a secondary hydrogen bonding network for the substrate Tyr -OH group. Previously, direct characterization of these TyrRSs was performed using isothermal calorimetry (41). Kd values of 330–522 μM were reported for the p-acetylRS with p-acetylPhe; the same experiment with the p-acetylRS and Tyr was below the limit of detection of the ITC method. Conversely, a Kd of 662–2659 μM was reported for the GlcNAc-Ser TyrRS with Tyr, but attempts to measure the binding affinity of this TyrRS mutant with GlcNAc-Ser were also below the limit of detection. Such results, along with Fersht’s analysis and available crystallographic evidence, can be used to rationalize why one amino acid (i.e., p-boronoPhe) but not others (i.e., GlcNAc-Ser or 3-OH Tyr) would be incorporated by high fidelity enzymes.
TrpRSs: Analysis of TrpRS Sequence and Structure.
TrpRS sequences from E. coli, Bacillus subtilis (Bs), and Bst are highly homologous and these enzymes are known to cross react with the others’ tRNATrp because of shared identity elements (47, 48). These bacterial TrpRSs also do not require mutations to charge tRNATrp with a variety of Trp analogs including: 4-nitroTrp, 4-flouroTrp, 5-fluoroTrp, 5-hydroxyTrp (5-OH Trp), and other azaTrp isosteres (13, 47, 49–52). The reported high fidelity incorporation of 5-OH Trp by a mutated Bs TrpRS thus caught our attention (25). In the report, the TrpRS was subjected to a single mutagenesis (V144P). The mutant enzyme’s amino acid binding pocket retained residue Asp133. This residue is homologous to residue Asp132 of Bst TrpRS and Asp 176 of the Bst TyrRS, and like in the TyrRSs, structurally forms a hydrogen bond with the original substrate; and with this TrpRS, the bond is to the indole N1 nitrogen (3, 16, 53) We suggest that because this residue serves as the primary molecular determinant of amino acid specificity for this enzyme, that any NAA-incorporating system based on this, or similar TrpRS should target this residue to achieve high fidelity incorporation over the canonical substrate.
In the original report V144 was targeted for saturation mutagenesis to create a library of 19 variants. Because the structure of the Bs TrpRS was unavailable, this residue was chosen based on a Bst TrpRS structure. A screen of 19 total variants identified a V144P variant as having high fidelity for 5-OH Trp. This result is unprecedented; all other evolved TyrRSs exhibiting high fidelity (10) have been selected from TyrRS libraries featuring randomization of 4–8 active site residues†. Furthermore, their target of incorporation, 5-OH Trp, is a well-documented substrate for the WT Bs TrpRSs (54). The burden on the V144P mutant is therefore twofold: it must ablate the enzyme’s inherent affinity for both Trp and 5-OH Trp and yet, switch the enzyme’s fidelity solely to 5-OH Trp. This is a different starting point from which each evolved Mj TyrRS mutant is selected; for example, the NAAs listed in Table S1 are not appreciable substrates for the Mj WT TyrRS.
There are reports from Tirrell and Yokoyama of AARSs requiring only one mutation to incorporate Tyr, Phe and Trp analogues with high fidelity (12, 55, 56). Yokoyama and coworkers demonstrated preferential incorporation of 3-iodo-L-tyrosine into a protein. Their SDS-PAGE analysis (figure 2 in ref. 12), however, shows that full-length protein is produced in the absence of 3-iodo-L-tyrosine. This result suggests their AARS exhibits only moderate fidelity in the absence of NAA. This is explainable because they only mutated one of the Bst TyrRS’s molecular determinants (Y34, equal to Y32 in Mj). Tirrell and coworkers (55, 56) reported incorporation of several Phe and Trp analogs with high fidelity. This required the use of auxotrophic (Phe) strains of E. coli, presumably to keep the pool of competing endogenous amino acids at a minimum. Furthermore, the kinetic parameters of their single-point mutant AARSs suggest that incorporation of Trp and/or Phe would occur in media with normal levels of these canonical amino acids. Whereas one AARS mutation may be sufficient to achieve incorporation of a NAA, either auxotrophic or depleted media conditions appear to be necessary for achieving high fidelity incorporation of an NAA, and neither of these conditions were reported for the incorporation of 5-OH Trp.
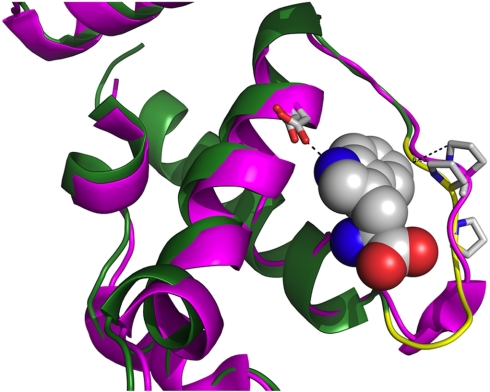
The Bs V144P TrpRS mutant crystallized in the presence of both Trp and 5-OH Trp. Crystals diffracted to 2.8 Å (Table S2), but electron density corresponding to either amino acid was not observed. A structural alignment of our V144P TrpRS structure with a Bst TrpRS structure with bound Trp shows that the alpha helix containing Asp133, again the major determinant for Trp specificity, is not appreciably displaced by the mutation at residue 144 (Fig. 3). Although the mutation does occur in the vicinity (∼6 Å) of the 5-position of Trp, the effect on the secondary structure appears rather modest. The most dramatic structural changes appear to occur where Asp146 binds ribose of ATP (57).
Fig. 3.
Alignment of TrpRSs from B. stereothermophilus (magenta) and a V144P TrpRS from B. subtilis (green and yellow; PDB ID 3PRH). Bound substrate Trp (shown as space-filling model) is from Carter’s B. stereothermophilus structure (PDB ID 3FHJ1). Two Asp residues, one from each RS, are also shown near the indole nitrogen. These Asp residues are the determinants for amino acid specificity. The effect of the ProPro sequence on the B. subtilis secondary structure is shown in yellow and occurs where the nucleoside would normally reside.
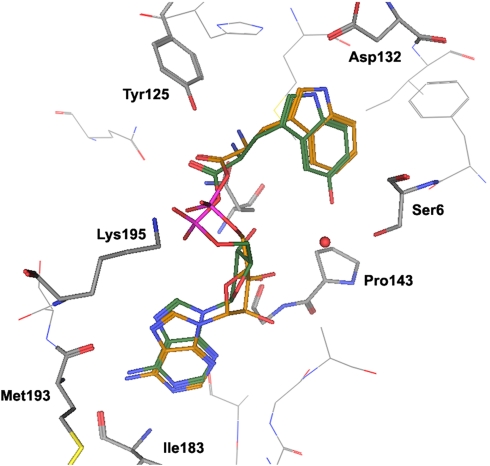
To rationalize the high fidelity of their V144P variant, Schultz and coworkers modeled the active site of the Bst TrpRS V144P variant with 5-OH Trp (25). They found that 5-OH Trp preferred to reorient its indole ring ∼180° so the indole nitrogen participates in a hydrogen bond with Ser7 and the 5-OH interacts with Asp132. We have conducted modelling experiments and found that both the Bst WT and Bs V144P TrpRSs prefer Trp and 5-OH Trp in the same orientation found in Schimmel, Crane, and Carter’s crystallographic work of homologous enzymes (Fig. 4) (17, 57, 58). Both models placed the indole nitrogen in close proximity to the key Asp residue, consistent with the residue’s role in determining fidelity. The docking results only became more variable when the critical Asp residue was modified with an Asp → Ala mutation (Fig. S1).
Fig. 4.
B. stereothermophilus active site with Val143Pro mutation to model the effects of the Bs Val144Pro (5-OH indole ring in green; L-Trp indole ring shown in orange). Highly similar results were observed for the WT Bst TrpRS.
Bs WT and V144P TrpRS Complementation of an Auxotrophic Phenotype.
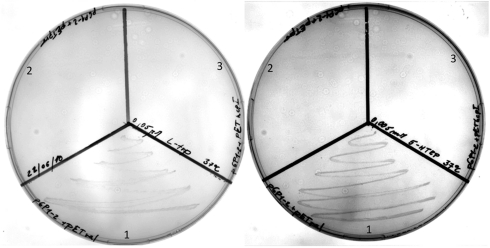
The E. coli strain trpS4040 is a Trp auxotroph that has been used to assess the activity of exogenous TrpRSs (15, 59). Briefly, the strain has a debilitated TrpRS gene that makes it difficult to culture in the absence of Trp. Growth on media enriched with Trp elevates the intracellular concentration to the point where tRNATrp become sufficiently charged. Alternatively, expressing an AARS gene that is able to efficiently charge E. coli tRNATrp complements the auxotrophic phenotype under Trp-starved conditions. Our results show complementation after 48 h on either 0.05 μM Trp or 5 μM 5-OH Trp plates in the presence of the Bs WT TrpRS, but not with the V144P mutant (Fig. 5). Even in liquid media supplemented at 50 μM 5-OH Trp, the Bs WT TrpRS allowed cultures to readily grow, whereas the cultures expressing the V144P mutant were still no different from the control experiments (Fig. S2).
Fig. 5.
Complementation of E. coli trpS4040 by (Left) 0.05 μM Trp, and (Right) 5 μM 5-OH Trp. Figure shows growth after 48 h at 37 °C. (1 = pET151 Bs TrpRS (wild type)/pGP1-2; 2 = pET151 Bs TrpRS (V144P)/pGP1-2; 3 = pET151ϵ (control)/pGP1-2; M9 minimal media + 50 μg/mL ampicillin and 25 μg/mL kanamycin). The results on plates were repeated three times.
Isothermal Calorimetry Suggests Lack of Fidelity of 5-OH TrpRS in Vitro.
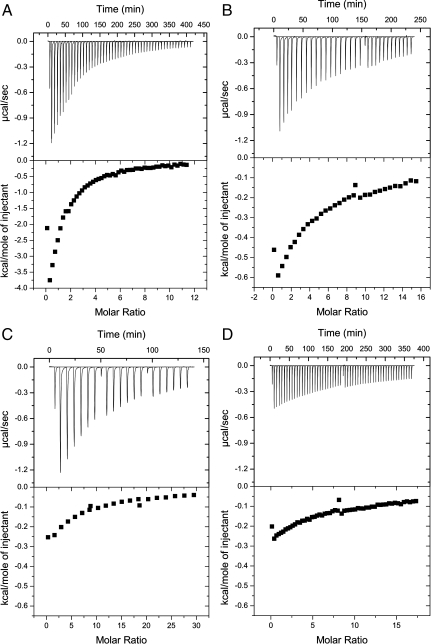
We used isothermal calorimetry (ITC) to directly characterize the fidelity of the Bs TrpRSs (Fig. 6 and Fig. S3). ITC can be used to measure binding constants of TrpRSs, and when two substrates are directly compared, a greater relative binding affinity (lower dissociation constant) gives some insight into the enzyme’s activity. By ITC, the Bs WT TrpRS exhibited characteristics similar to other bacterial (60) and human TrpRSs (61). The WT enzyme favors Trp (Kd = 149 μM) over 5-OH Trp (Kd = 1,079 μM) as would be expected as a result of evolutionary optimization. The V144P mutant showed decreased binding to Trp (Kd≥2,000 μM) relative to WT and, due to experimental limitations in fitting, we place a lower bound on the Kd at greater than 2,500 μM for the mutant with 5-OH Trp. The observation that the V144P TrpRS consistently exhibited greater binding affinity for Trp vs. 5-OH Trp conflicts with the premise of the prior study. It was reported that the Bs TrpRS V144P variant failed to charge Bs tRNATrp in the absence of 5-OH Trp (see ref. 25 and Fig. 4A, lane 2). However, the previous experiments were performed in 293T cells, so contextual issues within the mammalian expression system cannot be ruled out. But given the importance of intracellular expression levels of the orthogonal pair on the efficiency of incorporation, (62–64) it is unclear how the V144P mutant, an enzyme with a fraction of the activity of the WT enzyme, achieved efficient charging of Bs tRNATrp with 5-OH Trp (to the complete exclusion of Trp) in a more challenging mammalian expression system.
Fig. 6.
Isothermal calorimetry results with tryptophan and 5-OH Trp ligands. (A) Bs WT TrpRS with L-Trp; (B) Bs WT TrpRS with 5-OH Trp; (C) Bs TrpRS V144P variant with L-Trp; (D) Bs TrpRS V144P variant with 5-OH Trp.
Söll and coworkers reported the aminoacylation kinetics for Bst TrpRS single and double mutants (15), including a V141Q mutant. This one mutation, in a similar region as the Bs V144P mutation, caused reductions of ∼300 and 1,000-fold in catalytic efficiency for Trp and ATP activation, respectively. Schimmel and coworkers, however, were able to completely disrupt a human TrpRS’s affinity for Trp by introducing targeted mutations to the enzyme’s determinants of amino acid specificity (61). Consistent with the ideas exposed here, such targeted mutations would be a requirement to break the enzyme’s capacity to bind its canonical substrate and any closely related analog; and with these bacterial TrpRSs, this involves a critical Asp interaction to the indole nitrogen of Trp.
Schultz and coworkers reported the utility of incorporating a single 5-OH Trp by performing fluorescence and electrochemical crosslinking experiments on their purified 5-OH Trp labeled protein (25). However, because their targeted recombinant protein contained a single Trp residue (25), the same single substitution experiment was already permissive using standard mammalian (6965) and bacterial hosts; a bacterial host would have been easier to culture, have produced significantly more labeled protein and was already well-precedented to permit such substitutions due to the inherent promiscuity of E. coli and Bs TrpRSs for Trp analogs, including 5-OH Trp (14, 51, 54, 66).
Conclusion
The reinvestigation of unprecedented results is an important component of the scientific endeavour (67). As such, we are not able to confirm the conclusions offered in multiple studies in the field of expanded genetic codes. In some instances we are able to suggest explanations for observations that are, in our opinion, more consistent of a wild type, or wild type-like enzyme. Other experimental features were found to be more anomalous and difficult to rationalize. Our analysis is important because there are many functional groups that would be useful if incorporated into a given alloprotein. Indeed, strides have been taken to hasten the introduction of NAAs without extensive directed evolution (68). However, understanding the role of fidelity, and what is required of an enzyme to achieve it, will always be key in successful NAA systems. To this end, we have shown that with some TyrRSs and TrpRSs, that there is a clear molecular basis for achieving high fidelity and it involves a balance between the chemical identity of the amino acid and specific mutations to the enzyme.
Materials and Methods
Custom gene synthesis (Epoch Biolabs) was used to construct the Mj and Bs expression vectors. Synthetic genes were provided as either PCR products or cloned products. ITC data were fit according to reported methods (69). E. coli strain 4040 was provided by the Yale E. coli Genetic Stock Center. Further methods are given in the SI Appendix.
Supplementary Material
Acknowledgments.
The authors thank Drs. Niek Buurma and Livinia Onel for useful discussions regarding ITC. This work was in part supported by EC FP7 Grant (HEALTH-PROT, GA No 229676) and the Engineering and Physical Sciences Research Council EPSRC/EP/H045848/1.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3PRH and 3FHJ-1).
†By design, these libraries targeted and changed the determinants for amino acid specificity (Table S1), which along with the fact the amino acids are mostly phenylalanine derivatives substituted at the 4-position with nonpolar functional groups, contributes to the overall success of the method.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012276108/-/DCSupplemental.
References
- 1.Budisa N. Engineering the Genetic Code. Weinheim, Germany: Wiley; 2006. [Google Scholar]
- 2.Delarue M, Moras D. The aminoacyl-tRNA synthetase family: Modules at work. Bioessays. 1993;15:675–687. doi: 10.1002/bies.950151007. [DOI] [PubMed] [Google Scholar]
- 3.Carter CW., Jr Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu Rev Biochem. 1993;62:715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 4.Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 5.Silvian LF, Wang J, Steitz TA. Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin. Science. 1999;285:1074–1077. [PubMed] [Google Scholar]
- 6.Mursinna RS, Lincecum TL, Jr, Martinis SA. A conserved threonine within Escherichia coli leucyl-tRNA synthetase prevents hydrolytic editing of leucyl-tRNALeu. Biochemistry. 2001;40:5376–5381. doi: 10.1021/bi002915w. [DOI] [PubMed] [Google Scholar]
- 7.Yarus M, Berg P. Recognition of tRNA by aminoacyl tRNA synthetases. J Mol Biol. 1967;28:479–490. doi: 10.1016/s0022-2836(67)80098-6. [DOI] [PubMed] [Google Scholar]
- 8.Fersht AR, Mulvey RS, Koch GLE. Ligand binding and enzymic catalysis coupled through subunits in tyrosyl-tRNA synthetase. Biochemistry. 1975;14:13–18. doi: 10.1021/bi00672a003. [DOI] [PubMed] [Google Scholar]
- 9.Fersht AR, et al. Hydrogen bonding and biological specificity analysed by protein engineering. Nature. 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- 10.Xie J, Schultz PG. A chemical toolkit for proteins—An expanded genetic code. Nat Rev Mol Cell Biol. 2006;7:775–782. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- 11.Hino N, et al. Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nat Methods. 2005;2:201–206. doi: 10.1038/nmeth739. [DOI] [PubMed] [Google Scholar]
- 12.Kiga D, et al. An engineered Escherichia coli tyrosyl-tRNA synthetase for site-specific incorporation of an unnatural amino acid into proteins in eukaryotic translation and its application in a wheat germ cell-free system. Proc Natl Acad Sci USA. 2002;99:9715–9720. doi: 10.1073/pnas.142220099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lepthien S, Hoesl MG, Merkel L, Budisa N. Azatryptophans endow proteins with intrinsic blue fluorescence. Proc Natl Acad Sci USA. 2008;105:16095–16100. doi: 10.1073/pnas.0802804105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bae JH, et al. Expansion of the genetic code enables design of a novel “gold” class of green fluorescent proteins. J Mol Biol. 2003;328:1071–1081. doi: 10.1016/s0022-2836(03)00364-4. [DOI] [PubMed] [Google Scholar]
- 15.Praetorius-Ibba M, et al. Ancient adaptation of the active site of tryptophanyl-tRNA synthetase for tryptophan binding. Biochemistry. 2000;39:13136–13143. doi: 10.1021/bi001512t. [DOI] [PubMed] [Google Scholar]
- 16.Doublie S, Bricogne G, Gilmore C, Carter CW., Jr Tryptophanyl-tRNA synthetase crystal structure reveals an unexpected homology to tyrosyl-tRNA synthetase. Structure. 1995;3:17–31. doi: 10.1016/s0969-2126(01)00132-0. [DOI] [PubMed] [Google Scholar]
- 17.Yang XL, et al. Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains. Proc Natl Acad Sci USA. 2003;100:15376–15380. doi: 10.1073/pnas.2136794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landes C, et al. A structure-based multiple sequence alignment of all class I aminoacyl-tRNA synthetases. Biochimie. 1995;77:194–203. doi: 10.1016/0300-9084(96)88125-9. [DOI] [PubMed] [Google Scholar]
- 19.Kwon I, Kirshenbaum K, Tirrell DA. Breaking the degeneracy of the genetic code. J Am Chem Soc. 2003;125:7512–7513. doi: 10.1021/ja0350076. [DOI] [PubMed] [Google Scholar]
- 20.Magliery TJ, Anderson JC, Schultz PG. Expanding the genetic code: Selection of efficient suppressors of four-base codons and identification of “shifty” four-base codons with a library approach in Escherichia coli. J Mol Biol. 2001;307:755–769. doi: 10.1006/jmbi.2001.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan W, et al. A facile system for genetic incorporation of two different noncanonical amino acids into one protein in Escherichia coli. Angew Chem Int Ed Engl. 2010;49:3211–3214. doi: 10.1002/anie.201000465. [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto K, et al. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young TS, Ahmad I, Brock A, Schultz PG. Expanding the genetic repertoire of the methylotrophic yeast Pichia pastoris. Biochemistry. 2009;48:2643–2653. doi: 10.1021/bi802178k. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 25.Zhang ZW, et al. Selective incorporation of 5-hydroxytryptophan into proteins in mammalian cells. Proc Natl Acad Sci USA. 2004;101:8882–8887. doi: 10.1073/pnas.0307029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HS, Spraggon G, Schultz PG, Wang F. Genetic incorporation of a metal-ion chelating amino acid into proteins as a biophysical probe. J Am Chem Soc. 2009;131:2481–2483. doi: 10.1021/ja808340b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tippmann EM, Liu W, Surnmerer D, Mack AV, Schultz PG. A genetically encoded diazirine photocrosslinker in Escherichia coli. Chembiochem. 2007;8:2210–2214. doi: 10.1002/cbic.200700460. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, Zhang Z, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc Natl Acad Sci USA. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santoro SW, Wang L, Herberich B, King DS, Schultz PG. An efficient system for the evolution of aminoacyl-tRNA synthetase specificity. Nat Biotechnol. 2002;20:1044–1048. doi: 10.1038/nbt742. [DOI] [PubMed] [Google Scholar]
- 30.Melançon Cr, Schultz P. One plasmid selection system for the rapid evolution of aminoacyl-tRNA synthetases. Bioorg Med Chem Lett. 2009;19:3845–3847. doi: 10.1016/j.bmcl.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, et al. A new strategy for the synthesis of glycoproteins. Science. 2009;326:1187. doi: 10.1126/science.1089509. [DOI] [PubMed] [Google Scholar]
- 32.Xu R, et al. Site-specific incorporation of the mucin-type N-acetylgalactosamine-alpha-O-threonine into protein in Escherichia coli. J Am Chem Soc. 2009;131:13883. doi: 10.1021/ja906705a. [DOI] [PubMed] [Google Scholar]
- 33.Alfonta L, Zhang Z, Uryu S, Loo J, Schultz P. Site-specific incorporation of a redox-active amino acid into proteins. J Am Chem Soc. 2003;125:14662–14663. doi: 10.1021/ja038242x. [DOI] [PubMed] [Google Scholar]
- 34.de Prat Gay G, Duckworth HW, Fersht AR. Modification of the amino acid specificity of tyrosyl-tRNA synthetase by protein engineering. FEBS Lett. 1993;318:167–171. doi: 10.1016/0014-5793(93)80014-l. [DOI] [PubMed] [Google Scholar]
- 35.Fersht AR. Dissection of the structure and activity of the tyrosyl-tRNA synthetase by site-directed mutagenesis. Biochemistry. 1987;26:8031–8037. doi: 10.1021/bi00399a001. [DOI] [PubMed] [Google Scholar]
- 36.Bhat TN, Blow DM, Brick P, Nyborg J. Tyrosyl-tRNA synthetase forms a mononucleotide-binding fold. J Mol Biol. 1982;158:699–709. doi: 10.1016/0022-2836(82)90255-8. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi T, et al. Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion. Nat Struct Biol. 2003;10:425–432. doi: 10.1038/nsb934. [DOI] [PubMed] [Google Scholar]
- 38.Seyedsayamdost MR, Xie J, Chan CTY, Schultz PG, Stubbe J. Site-specific insertion of 3-aminotyrosine into subunit α2 of E. coli ribonucleotide reductase: Direct evidence for involvement of Y730 and Y731 in radical propagation. J Am Chem Soc. 2007;129:15060–15071. doi: 10.1021/ja076043y. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z, et al. A new strategy for the synthesis of glycoproteins. Science. 2004;303:371–373. doi: 10.1126/science.1089509. [DOI] [PubMed] [Google Scholar]
- 40.Xu R, et al. Site-specific incorporation of the mucin-type N-acetylgalactosamine-?-O-threonine into protein in Escherichia coli. J Am Chem Soc. 2004;126:15654–15655. doi: 10.1021/ja044711z. [DOI] [PubMed] [Google Scholar]
- 41.Antonczak AK, Simova Z, Tippmann EM. A critical examination of Escherichia coli esterase activity. J Biol Chem. 2009;284:28795–28800. doi: 10.1074/jbc.M109.027409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yonemoto IT, Tippmann EM. The juggernauts of biology. Bioessays. 2010;32:314–321. doi: 10.1002/bies.200900142. [DOI] [PubMed] [Google Scholar]
- 43.Igarashi K, Hotta K, Kasuya F, Abe K, Sakoda S. Determination of cabergoline and L-dopa in human plasma using liquid chromatography-tandem mass spectrometry. J Chromatogr B. 2003;792:55–61. doi: 10.1016/s1570-0232(03)00279-4. [DOI] [PubMed] [Google Scholar]
- 44.Bourcier S, et al. Detection of 28 neurotransmitters and related compounds in biological fluids by liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2006;20:1405–1421. doi: 10.1002/rcm.2459. [DOI] [PubMed] [Google Scholar]
- 45.Xie J, et al. The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination. Nat Biotechnol. 2004;22:1297–1301. doi: 10.1038/nbt1013. [DOI] [PubMed] [Google Scholar]
- 46.Brustad E, et al. A genetically encoded boronate-containing amino acid. Angew Chem Int Edit. 2008;47:8220–8223. doi: 10.1002/anie.200803240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jia J, et al. Two essential regions for tRNA recognition in Bacillus subtilis tryptophanyl-tRNA synthetase. Biochem J. 2002;365:749–756. doi: 10.1042/BJ20020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xue H, Shen W, Giegé R, Wong J. Identity elements of tRNA(Trp). Identification and evolutionary conservation. J Biol Chem. 1993;268:9316–9322. [PubMed] [Google Scholar]
- 49.Budisa N, et al. Global replacement of tryptophan with aminotryptophans generates non-invasive protein-based optical pH sensors. Angew Chem Int Edit. 2002;41:4066–4069. doi: 10.1002/1521-3773(20021104)41:21<4066::AID-ANIE4066>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 50.Lepthien S, Wiltschi B, Bolic B, Budisa N. In vivo engineering of proteins with nitrogen-containing tryptophan analogs. Appl Microbiol Biotechnol. 2006;73:740–754. doi: 10.1007/s00253-006-0665-2. [DOI] [PubMed] [Google Scholar]
- 51.Wong CY, Eftink MR. Biosynthetic incorporation of tryptophan analogues into staphylococcal nuclease: Effect of 5-hydroxytryptophan and 7-azatryptophan on structure and stability. Protein Sci. 1997;6:689–697. doi: 10.1002/pro.5560060318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Azim MK, Budisa N. Docking of tryptophan analogs to trytophanyl-tRNA synthetase: implications for non-canonical amino acid incorporations. Biol Chem. 2008;389:1173–1182. doi: 10.1515/BC.2008.133. [DOI] [PubMed] [Google Scholar]
- 53.Retailleau P, Weinreb V, Hu M, Carter CW., Jr Crystal structure of tryptophanyl-tRNA synthetase complexed with adenosine-5′ tetraphosphate: Evidence for distributed use of catalytic binding energy in amino acid activation by class I aminoacyl-tRNA synthetases. J Mol Biol. 2007;369:108–128. doi: 10.1016/j.jmb.2007.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barlati S, Ciferri O. Incorporation of 5-methyl- and 5-hydroxy-tryptophan into the protein of Bacillus subtilis. J Bacteriol. 1970;101:166–172. doi: 10.1128/jb.101.1.166-172.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwon I, Tirrell DA. Site-specific incorporation of tryptophan analogues into recombinant proteins in bacterial cells. J Am Chem Soc. 2007;129:10431–10437. doi: 10.1021/ja071773r. [DOI] [PubMed] [Google Scholar]
- 56.Kwon I, Wang P, Tirrell DA. Design of a bacterial host for site-specific incorporation of p-bromophenylalanine into recombinant proteins. J Am Chem Soc. 2006;128:11778–11783. doi: 10.1021/ja0626281. [DOI] [PubMed] [Google Scholar]
- 57.Laowanapiban P, et al. Independent saturation of three TrpRS subsites generates a partially assembled state similar to those observed in molecular simulations. Proc Natl Acad Sci USA. 2009;106:1790–1795. doi: 10.1073/pnas.0812752106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buddha MR, Crane BR. Structure and activity of an aminoacyl-tRNA synthetase that charges tRNA with nitro-tryptophan. Nat Struct Mol Biol. 2005;12:274–275. doi: 10.1038/nsmb907. [DOI] [PubMed] [Google Scholar]
- 59.Sever S, Rogers K, Rogers MJ, Carter C, Soll D. Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer interface in optimizing amino acid binding. Biochemistry. 1996;35:32–40. doi: 10.1021/bi952103d. [DOI] [PubMed] [Google Scholar]
- 60.Buddha MR, Tao T, Parry RJ, Crane BR. Regioselective nitration of tryptophan by a complex between bacterial nitric-oxide synthase and tryptophanyl-tRNA synthetase. J Biol Chem. 2004;279:49567–49570. doi: 10.1074/jbc.C400418200. [DOI] [PubMed] [Google Scholar]
- 61.Zhou Q, et al. Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality. Nat Struct Mol Biol. 2010;17:57–61. doi: 10.1038/nsmb.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E. coli. J Mol Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 63.Iraha F, et al. Functional replacement of the endogenous tyrosyl-tRNA synthetase-tRNATyr pair by the archaeal tyrosine pair in Escherichia coli for genetic code expansion. Nucleic Acids Res. 2010;38:3682–3691. doi: 10.1093/nar/gkq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu W, Brock A, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 65.James NG, Byrne SL, Mason AB. Incorporation of 5-hydroxytryptophan into transferrin and its receptor allows assignment of the pH induced changes in intrinsic fluorescence when iron is released. Biochem Biophys Acta. 2009;1794:532–540. doi: 10.1016/j.bbapap.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Majerfeld I, Barlati S, Ciferri O. Tryptophanless death in Bacillus subtilis. J Bacteriol. 1970;101:350–354. doi: 10.1128/jb.101.2.350-354.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wells JA, Fairbrother WJ, Otlewski J, Laskowski M, Jr, Burnier J. A reinvestigation of a synthetic peptide (TrPepz) designed to mimic trypsin. Proc Natl Acad Sci USA. 1994;91:4110–4114. doi: 10.1073/pnas.91.10.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miyake-Stoner SJ, et al. Generating permissive site-specific unnatural aminoacyl-tRNA synthetases. Biochemistry. 2010;49:1667–1677. doi: 10.1021/bi901947r. [DOI] [PubMed] [Google Scholar]
- 69.Buurma NJ, Haq I. Advances in the analysis of isothermal titration calorimetry data for ligand-DNA interactions. Methods. 2007;42:162–172. doi: 10.1016/j.ymeth.2007.01.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.