Abstract
Background: Epidemiologic data have shown that obesity independently increases colorectal cancer (CRC) risk, but the mechanisms are poorly understood. Obesity is an inflammatory state, and chronic colonic inflammation induces CRC.
Objective: We conducted this proof-of-principle study to seek evidence of obesity-associated colorectal inflammation and to evaluate effects of diet-induced weight loss.
Design: We measured inflammatory cytokines, gene arrays, and macrophage infiltration in rectosigmoid mucosal biopsies of 10 obese premenopausal women [mean ± SD body mass index (in kg/m2): 35 ± 3.5] before and after weight loss induced by a very-low-calorie diet.
Results: Subjects lost a mean (±SD) of 10.1 ± 1% of their initial weight. Weight loss significantly reduced fasting blood glucose, total cholesterol, triglycerides, LDL, tumor necrosis factor-α (TNF-α), and interleukin (IL)-8 concentrations (P < 0.05). After weight loss, rectosigmoid biopsies showed a 25–57% reduction in TNF-α, IL-1β, IL-8, and monocyte chemotactic protein 1 concentrations (P < 0.05). T cell and macrophage counts decreased by 28% and 42%, respectively (P < 0.05). Gene arrays showed dramatic down-regulation of proinflammatory cytokine and chemokine pathways, prostaglandin metabolism, and the transcription factors STAT3 (signal transducer and activator of transcription 3) and nuclear transcription factor κB. Weight loss reduced expression of FOS and JUN genes and down-regulated oxidative stress pathways and the transcription factors ATF (activating transcription factor) and CREB (cyclic AMP response element-binding).
Conclusions: Our data show that diet-induced weight loss in obese individuals reduces colorectal inflammation and greatly modulates inflammatory and cancer-related gene pathways. These data imply that obesity is accompanied by inflammation in the colorectal mucosa and that diet-induced weight loss reduces this inflammatory state and may thereby lower CRC risk.
INTRODUCTION
Epidemiologic evidence suggests obesity as an independent risk factor for the development of several cancers, including colorectal cancer (CRC) (1). Every 5-unit increase in body mass index (BMI) increases risk of CRC by 30% in men and by 13% in women (2). The prevalence of obesity has increased dramatically in adult Americans from 14% in the 1970s to the current level of 34% (3–5). The increased prevalence of obesity indicates that the incidence of CRC and other forms of cancer will become a greater public health problem in the future.
In mouse models of genetic and chemically induced colitis, obesity enhanced the development of colorectal neoplasia (6, 7). Western-style diet consumption in mice caused obesity accompanied by increased oxidative stress, inflammation in the colon, and development of colon tumors without genetic or chemical manipulation (8, 9). Consistent are findings that caloric restriction protected mice from developing aberrant crypt foci and colonic neoplasia (10) and decreased expression of colonic cyclooxygenase-2 (COX-2) in the colon (11). In the Iowa Women's Health Study, an intentional weight loss of >20 pounds reduced colon cancer incidence by 9% and obesity-related cancer mortality by 40% (12).
Several factors have been proposed to mediate the relation between obesity and increased CRC risk. Obesity is associated with elevated concentrations of insulin-like growth factor (IGF)-1, which is a known stimulator of epithelial growth (13). Increased plasma concentrations of inflammatory cytokines including tumor necrosis factor-α (TNF-α) and interleukin (IL)-6 occur in obesity and are elevated in subjects with colorectal adenomas (14). Furthermore, antiinflammatory drugs, such as aspirin and celecoxib, reduce colorectal adenoma incidence (15, 16). Subgroup analyses in one study revealed aspirin to have a greater protective effect in obese subjects than in lean subjects (17).
Obesity is associated with chronic low-grade inflammation in the adipose tissue, liver, and coronary endothelium and is accompanied by increased oxidative stress (18), which through intermediary molecules, increase the expression of the proto-oncogenes FOS and JUN that leads to increased transcription of proinflammatory and cell-cycle regulatory genes that promote carcinogenesis (19). Chronic inflammation can enhance the initiation and progression of CRC (20).
Therefore, we hypothesized that obese humans would show signs of chronic inflammation in their colorectal mucosa. To test this hypothesis, obese women were studied before and after a 10% weight loss induced by a very-low-calorie diet (VLCD), which reduced rectosigmoid mucosal inflammatory cytokine concentrations and inflammatory cell infiltration accompanied by a decreased expression of gene pathways involved in inflammation and cancer, including gene sets regulated by TNF-α, IL-6 and IL-17, prostaglandins, and oxidative stress. Weight loss also decreased the expression of gene sets regulated by transcription factors involved in colorectal inflammation and CRC, such as signal transducer and activator of transcription 3 (STAT3), nuclear transcription factor κB (NF-κB), activating transcription factor (ATF), and cyclic AMP response element-binding (CREB).
SUBJECTS AND METHODS
Subjects
For this unblinded study, 10 healthy, obese [average (±SD) BMI (in kg/m2): 34.8 ± 3.5], premenopausal women (mean age: 43.2 ± 8.3 y) were recruited from the community through advertisements. Study volunteers included 6 whites, 1 Hispanic, 2 African Americans, and 1 subject with a mixed racial background (Table 1). All subjects underwent a complete medical examination, standard blood and urine tests, and an electrocardiogram. Subjects were weight stable (<2% change) for ≥6 mo before the study. Exclusion criteria included a history of cancer, current treatment of weight loss, previous intestinal surgery, a history suggestive of malabsorption, major medical problems, consumption of medications with antiinflammatory properties (eg, nonsteroidal antiinflammatory drugs, statins, glitazones, and COX-2 inhibitors) or medications that could increase risk of complications associated with consuming a VLCD (eg, lithium), an allergy to soy products or aspartame, or contraindications to undergoing a sigmoidoscopy and biopsy, including any history of excessive bleeding. The study was approved by the Institutional Review Board of The Rockefeller University (New York, NY). Subjects were recruited between September 2007 and May 2008. Written informed consent was obtained from each subject before participation in the study.
TABLE 1.
Baseline characteristics of the 10 obese volunteers1
| Subject | Age | Weight | Height | BMI | Race |
| y | kg | cm | kg/m2 | ||
| 1 | 49 | 74.8 | 158 | 30.2 | H |
| 2 | 43 | 104 | 159 | 41.3 | W |
| 3 | 39 | 92 | 171 | 31.4 | W |
| 4 | 49 | 95 | 158 | 38.3 | M |
| 5 | 52 | 97 | 172 | 32.8 | AA |
| 6 | 24 | 93 | 167.5 | 33.0 | W |
| 7 | 49 | 120 | 178 | 37.9 | AA |
| 8 | 37 | 98 | 170 | 34.0 | W |
| 9 | 47 | 83 | 158 | 33.4 | W |
| 10 | 43 | 103 | 170 | 35.5 | W |
| Combined | 43 ± 822 | 96 ± 12 | 166 ± 7 | 34.8 ± 4 | 6 W, 2 AA, 1 H, and 1 M |
H, Hispanic; W, white; M, mixed; AA, African American.
Mean ± SD (all such values).
Experimental design
Seven subjects chose to complete the weight-loss phase of the study in The Rockefeller University Hospital inpatient unit, and 3 subjects were closely monitored as outpatients. After screening, subjects underwent baseline testing, which included fasting blood determinations, urine analysis, and flexible sigmoidoscopy with multiple mucosal biopsies. Subjects started a VLCD weight-loss program and were monitored weekly by the study physicians. The VLCD was continued until subjects had lost ≥8% of baseline weight, at which time the initial measurements were repeated. This degree of weight loss was chosen because a previous study showed that this would lower rectosigmoid proliferation in obese subjects by ≈40% (21). Subjects were counseled that the study required them to maintain their usual physical activity, and this was emphasized throughout the study.
Subjects consumed a reduced-calorie, Western-style diet that contained 50% of their calculated baseline caloric intake for 3 d before initiation of the VLCD. Subjects were fed a commercially available VLCD product (New Direction Program; Robard Corporation, Mount Laurel, NJ) that provided ≈782 kcal/d with an energy distribution of carbohydrate, fat, and protein of 25%, 25%, and 50%, respectively, and daily requirements of vitamins and minerals (Table 2). This VLCD provided a choice of shakes, soups, bars, and pudding in different flavors; subjects had 4 choices/d, consumed one item every 4 h, and drank ≥2 L water daily. Inpatients were monitored daily by the hospital staff, and outpatients were monitored weekly during clinic visits. Weekly blood tests for electrolytes and renal function tests were performed to monitor for complications that can occur with VLCD consumption.
TABLE 2.
Composition of the very-low-calorie diet1
| Nutrient | Daily intake | Percentage of energy |
| Energy (kcal) | 782 | 100 |
| Carbohydrates (g) | 48 | 25 |
| Fat (g) | 23 | 26 |
| Protein (g) | 96 | 49 |
The diet was fortified with vitamins and minerals and provided >100% of the Recommended Dietary Allowance on the basis of a 2000-kcal/d diet.
Procedures and sample collection
Fasting electrolytes, liver function tests, renal function tests, lipid profiles, and high-sensitivity C-reactive protein measurements were performed by the clinical chemistry laboratory at Memorial Sloan-Kettering Cancer Center. Aliquots of serum samples for cytokine measurements were stored at −80°C. Flexible proctosigmoidoscopy was performed between 0800 and 0900 after a 60-mL tap-water enema, and 12–14 rectosigmoid biopsies were obtained from 4 quadrants between 10 and 20 cm from the anal verge. Four biopsies for mucosal cytokine measurements were immediately frozen in liquid nitrogen; 6 biopsies for gene expression analysis were immersed in RNAlater (catalog no. 7024; Ambion) at 4°C for 24 h and frozen in liquid nitrogen. The remaining biopsies were frozen in optimum cutting temperature compound (OTC; International Medical Equipment, Chico, CA) and stored at −80°C for immunohistochemical analysis.
Sample analyses
Serum cytokines
Serum cytokines were measured in duplicate with the Human Proinflammatory 9-Plex MULTI-SPOT 96-well plate kit (catalog no. N05007A-1; Mesoscale Diagnostics, Gaithersburg, MD) (22) for IL-2, IL-8, IL-12p70, IL-1β, IL-6, and IL-10, TNF-α, and interferon-γ with a intraassay correlation ≤10% and interassay variance ≤10%.With the use of this multiplex assay, only IL-6 and -8 and TNF-α were detectable with a CV ≤10% and recovery rate ≥80%. Monocyte chemotactic protein (MCP)-1 was measured with a human Elisa kit (catalog no. DCP00; R&D Systems, Minneapolis, MN) with an intraassay correlation ≤8% and interassay variance ≤7%.
Mucosal cytokines
Two frozen mucosal biopsies were homogenized in RIPA buffer [25 mmol tris HCl/L (pH 7.6), 150 mmol NaCl/L, 1% NP-40, 1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate; catalog no. 89900; Pierce, Rockport, IL] with protease and phosphatase inhibitors at 4°C (catalog nos. P8340 and P5726; Sigma-Aldrich, Saint Louis, MO). The homogenate was centrifuged at 15,000 rpm for 20 min at 4°C, and the supernatant fluid was collected and stored at −80°C until further analysis. Mucosal cytokines were measured in duplicate with the Proinflammatory 9-Plex MULTI-SPOT 96-well plate kit (catalog no. N05007A-1; Mesoscale Diagnostics) as described for serum cytokines. Only mucosal IL-6, IL-8, IL-1β, and TNF-α were detectable with a CV ≤10% and recovery rate ≥80%. MCP-1 concentrations were measured as previously described. Mucosal protein concentrations were measured with the Pierce BCA assay kit (catalog no. 23225; Thermo Fischer, Waltham, MA) with Nanodrop technology (NanoDrop Technologies, Wilmington, DE). Mucosal cytokine concentrations are presented as picograms of each cytokine per milligram of mucosal protein. Samples with cytokine concentrations ≥2.5 SD from the mean were considered outliers and excluded from the final analysis.
Immunohistochemistry
Six-micron sections were cut at −20°C onto plus slides (Fisher-Scientific, Waltham, MA) and stored at −80°C until assay. For immunohistochemical analyses, sections were quenched in ice-cold acetone and rehydrated in phosphate buffered saline (PBS). Sections were treated with purified mouse anti-human antibodies to cluster of differentiation (CD) 3 (T cell marker) at a 1:200 dilution (catalog no. 34740; BD Pharminogen, Franklin Lakes, NJ) and to CD163 (macrophage marker) at a 1:500 dilution (catalog no. BM4041; Novus Biologics, Littleton, CO). The secondary antibody, a biotin-labeled horse anti-mouse antibody at a 1:200 dilution was applied and followed by amplification with an avidin-biotin complex (1:100 of avidin and 1:100 of biotin; catalog no. 6102; Vector Laboratories, Burlingame, CA), and the signal was developed with chromogen-3-amino-9-ethylcarbazole (Sigma-Aldrich). The area of the sections and number of positive cells contained were counted manually with the National Institutes of Health Image J software (NIH IMAGE J 1.42; National Institutes of Health, Bethesda, MD) and reported as the number of positive cells per square millimeter. For each subject before and after the weight-loss regimen, all cells were counted in 3 separate sections >100 μm apart at a 10× magnification. Each section was analyzed in a random fashion by a single person blinded to their identity. A subset of sections were analyzed ≥3 times in a similar fashion, and the CV was <10%. Mucosal morphology was assessed by hematoxylin (Fisher Scientific, Waltham, MA) and eosin (Shandon, Pittsburg, PA) staining of sections by using a standard protocol. Data from only 8 subjects are presented for CD163 staining because the staining quality was inadequate for analysis in 2 subjects.
Gene expression
Total RNA was extracted from 2 rectosigmoid biopsy specimens by using the Trizol method (Invitrogen, Carlsbad, CA). RNA purification, quality assessment, and hybridization were performed as previously described (23). Biotin-labeled complementary RNA was hybridized onto Human HT-12v3 Ilumina expression chips (Illumina Inc, San Diego, CA). Samples from baseline and post–weight loss for each subject were analyzed on the same chip. The arrays were scanned by the Illumina BeadStation laser scanning imaging system (Illumina).
For determinations of real-time polymerase chain reactions (PCRs), 2 μg total RNA was used as a template for complementary DNA synthesis with a SuperScript-III First-Strand kit (Invitrogen). Complementary DNA was diluted in water, and amounts corresponding to 50 ng original RNA were used for quantitative gene expression by real-time PCR. SYBR Green PCR master mix (Applied Biosystems, Foster City, CA), primers from Sigma-Aldrich, and the ABI Prism7900 real-time PCR system (Applied Biosystems, Carlsbad, CA) were used for the real-time PCR. Six down-regulated and 4 up-regulated genes were assayed, which were normalized to the housekeeping gene GAPDH to adjust for the sample loading and efficiency of the reaction. Real-time PCR samples were assessed in duplicate, and messenger RNA concentrations were expressed as the log ratio of relative expression concentrations before and after treatment (see supplemental Table 1 under “Supplemental data” in the online issue for a list of real-time PCRs).
Analysis of the gene-expression microarray data
Gene-expression data from the microarrays were analyzed with the R software (version 2.1) and available packages from Bioconductor (http://www.bioconductor.org). Data were normalized by using robust spline normalization functions from the lumi package (Bioconductor). A classical quality control was performed. Expression concentrations with detection P values ≤ 0.05 were considered detectable. Probes with detectable expression in at least one sample and with SD >0.1 were used in further analysis. Expression values (log2 transformed) were modeled with the package limma (Bioconductor). Moderated paired t tests were used to test the significance of the effect of weight loss. P values were adjusted for multiple comparisons by using the Benjamini-Hochberg procedure. Genes with fold-changes ≥1.2 and P ≤ 0.05 were considered differentially expressed. Heat maps were generated by hierarchical clustering by using one Pearson r as the distance measure and complete linkage as the agglomeration procedure. The gene-expression data were deposited in the public domain (transcription profiling accession no. GSE20931).
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was conducted with the GSEA desktop application (http://www.broadinstitute.org/gsea) (24). Our analysis of gene expression before and after weight loss in each subject was performed in a paired fashion, and genes were ranked by their effect size (ie, the mean fold change for each subject). This rank-order list was used as the input to the GSEA. For each gene set, the enrichment score (ES) was calculated by using weighted Kolmogorov-Smirnov statistics to measure the proximity of the gene set to the top of the weight-loss effect ranked list. A high and positive ES indicated that the gene set or pathway was collectively up-regulated by the weight-loss intervention, whereas a negative ES indicated down-regulation. The significance (P value) of the observed ES was calculated by using simulations. Normalized ESs were used to compare analysis results across gene sets. The molecular signature database available at the Broad Institute, MIT (http://www.broadinstitute.org/gsea/msigdb), and gene sets created at the Rockefeller University from the data generated by IL-17 stimulation of keratinocytes (25, 26) were used to query the data. We used sets from 3 different collections from the GSEA website [ie, the curated canonical pathway database (C2), the Gene Ontology database (C5), and the transcription factor database (C3)]. Gene sets that were positively or negatively enriched with P < 0.05 and a false discovery rate (FDR) <5% were considered significant, except for the transcription factor gene sets for which an FDR <10% was considered significant.
Statistical analyses
Two-tailed paired t tests were used to compare anthropometric measurements, biochemical variables, serum and mucosal cytokine concentrations, and CD3 and CD163 cell counts at baseline and after weight loss. P ≤ 0.05 was considered significant. S-Plus (S+) version 8.1 for Windows software package (TIBCO Software Inc, Somerville, MA) was used for statistical analyses.
RESULTS
Anthropometric and biochemical measurements
All 10 subjects successfully completed the study without significant adverse events in a mean of 46.5 ± 9.3 d. The VLCD resulted in a mean (±SD) decrease in body weight of 10.1 ± 1% (P < 0.01), decreased BMI of 10.2 ± 1.4% (P < 0.01), and reduction in waist circumference of 8.3 ± 4%, (P < 0.01). The weight-loss regimen also induced improvements in systolic and diastolic blood pressures, fasting blood sugar concentrations, total and LDL serum cholesterol and triglyceride concentrations, and total white blood cell counts (Table 3). Serum electrolyte concentrations, liver function tests, and renal function tests were not significantly altered.
TABLE 3.
Effect of diet-induced weight loss on anthropometric and biochemical measurements1
| Measurement | Baseline | After weight loss | Percentage change | P |
| Weight (kg) | 96 ± 12 | 86 ± 11 | 10 ± 1.4 | <0.01 |
| BMI (kg/m2) | 35 ± 3.5 | 31 ± 3 | 10 ± 1.4 | <0.01 |
| Waist circumference (cm) | 107 ± 11 | 97 ± 12 | 8.3 ± 4.4 | <0.01 |
| Systolic blood pressure (mm Hg) | 121 ± 8 | 107 ± 12 | 11.6 ± 7.7 | <0.01 |
| Diastolic blood pressure (mm Hg) | 79 ± 4 | 70 ± 6 | 11 ± 9.3 | <0.01 |
| Fasting blood glucose (mg/dL) | 95 ± 10 | 85 ± 6 | 9.7 ± 13.2 | 0.03 |
| Total cholesterol (mg/dL) | 187 ± 36 | 157 ± 26 | 15 ± 8.8 | <0.01 |
| LDL (mg/dL) | 110 ± 28 | 94 ± 19 | 13 ± 11 | 0.01 |
| Triglycerides (mg/dL) | 122 ± 45 | 93 ± 36 | 22 ± 18 | <0.01 |
| HDL (mg/dL) | 52 ± 9 | 45 ± 6 | 12 ± 14 | 0.02 |
| hs-CRP (mg/dL) | 0.8 ± 0.5 | 0.9 ± 0.7 | 1.4 ± 0.5 | 0.85 |
| WBC (1000/mm3) | 6.7 ± 1.3 | 5.6 ± 1.4 | 15.2 ± 18.1 | 0.02 |
All values are mean ± SDs; n = 10. hs-CRP, high-sensitivity C-reactive protein; WBC, peripheral white blood cell count.
Serum and mucosal cytokines
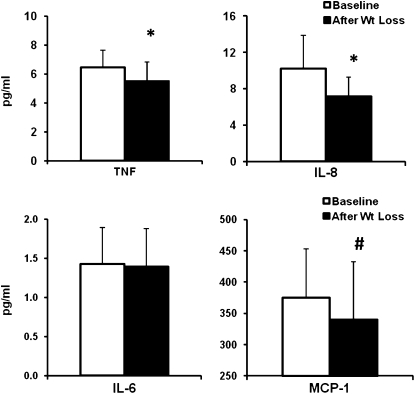
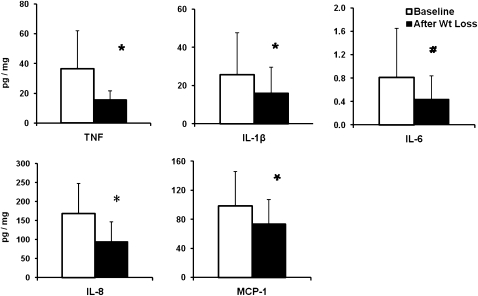
Diet-induced weight loss resulted in mean reductions in serum TNF-α concentrations of 15% and in IL-8 concentrations of 30% (both P < 0.05). There was also a trend toward decreased serum contents by 9% (P = 0.097) (Figure 1) but no change in concentrations of IL-6 and high-sensitivity C-reactive protein. Baseline serum MCP-1 concentrations were similar to those previously reported in obese individuals (27). In the rectosigmoid mucosa, concentrations of mucosal TNF-α decreased by 57% (P < 0.05), concentrations of IL-8 decreased by 44% (P < 0.05), concentrations of IL-1β decreased by 38% (P < 0.05), and concentrations of MCP-1 decreased by 25% (P < 0.05), and there was a trend toward decreased IL-6 concentrations by 45% (P = 0.06) (Figure 2). Data from one subject were excluded from the analysis because concentrations of all cytokines from her biopsies were >3 SDs from the mean of all subjects.
FIGURE 1.
Mean (±SD) changes (pg/mL) in serum cytokine and chemokine concentrations in 10 subjects before and after diet-induced weight loss. *P < 0.05, #P < 0.1. TNF, tumor necrosis factor; IL, interleukin; MCP, monocyte chemotactic protein.
FIGURE 2.
Mean (±SD) changes (pg/mg protein) in mucosal cytokines and chemokines in 9 subjects before and after diet-induced weight loss. *P < 0.05, #P < 0.1. TNF, tumor necrosis factor; IL, interleukin; MCP, monocyte chemotactic protein.
Immunohistochemistry
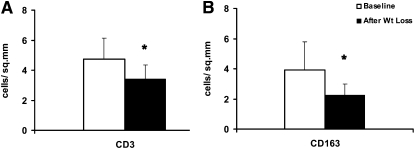
Standard hematoxylin and eosin staining of mucosal biopsies at the onset and end of the study showed no identifiable changes in total cellular content or mucosal morphology (data not shown). However, immunohistochemical staining showed a 28% decrease in CD3-positive T lymphocytes (P < 0.01) (n = 10) and a 42% decrease in CD163-positive macrophages (P < 0.05) (n = 8) (Figure 3) at the end of the study.
FIGURE 3.
Mean (±SD) changes (number of cells/mm2) in mucosal T cells and macrophages before and after diet-induced weight loss. A: CD3-positive T cells (n = 10). B: CD163-positive macrophages (n = 8). *P < 0.05.
Microarray analyses
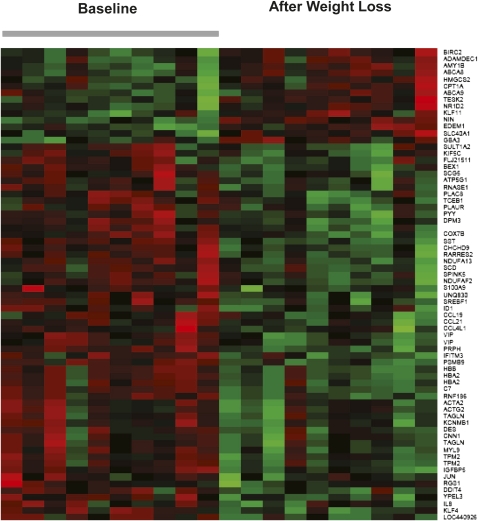
Microarray analyses revealed that diet-induced weight loss was accompanied by 1211 differentially expressed genes (with P ≤ 0.05) of which 70 genes (56 down-regulated and 14 up-regulated genes) had a fold-change ≥1.2. This differential pattern was strikingly shown in the heat map in Figure 4 (see supplemental Table 2 under “Supplemental data” in the online issue for the differential pattern shown in each gene). Genes involved in inflammation and cancer were down-regulated, including many genes associated with cytokines and chemokine signaling such as IL-8, CCL-21, CCL-19, and CCL4L1, as well as the proto-oncogenes JUN and FOS. Genes coding for molecules for which the circulating concentrations decreased with weight loss also were down-regulated, including peptide YY (PYY), vasoactive intestinal peptide (VIP), and somatostatin (SST).Genes that coded for gastrointestinal substrate transport, including ATP-binding cassette, subfamily A members 8 and 9 (ABCA8 and ABCA9), and carnitine palmitoyltransferase-I (CPT-1A) were up-regulated. Six down-regulated and 4 up-regulated genes were identified from the differentially expressed gene list for validation by real-time PCR, and gene-fold changes confirmed the microarray data findings (see supplemental Table 3 under “Supplemental data” in the online issue).
FIGURE 4.
Heat map of differentially expressed genes in rectosigmoid biopsies taken at baseline and after diet-induced weight loss in 10 individual subjects. The red color indicates a higher expression, and the green color indicates a lower expression.
Detecting changes in gene pathways by altered expression of single genes relies on relatively large changes and may be insensitive to many physiologically significant changes of lesser magnitude. A more sensitive approach has been GSEA, which examines many genes in a pathway for changes that may not, by themselves, be significant but are significant in the aggregate (28). In our analyses, we used gene sets curated in the canonical pathway database, in the Gene Ontology database, IL-17 pathway gene sets, and the transcription factor database from the GSEA website. In the canonical pathway database, 74 gene sets were down-regulated, and 24 gene sets were up-regulated (see supplemental Tables 4 and 5 under “Supplemental data” in the online issue); in the Gene Ontology database, 55 gene sets were down-regulated, and none were up-regulated (see supplemental Table 6 under “Supplemental data” in the online issue); and in the IL-17 gene sets, 6 gene sets were down-regulated and 2 gene sets were up-regulated (see supplemental Table 7 under “Supplemental data” in the online issue). Gene sets related to inflammation and cancer that were down-regulated are highlighted in Table 4 and include pathways activated by TNF-α, IL-6, IL-1, IL-17, chemokine and cytokine signaling pathways, prostaglandin synthesis pathways, and pathways involved in oxidative stress, glutathione metabolism, and apoptosis. Up-regulated mucosal gene pathways were involved in carbohydrate metabolism (see supplemental Table 5 under “Supplemental data” in the online issue). The reduction in oxidative stress pathways was supported by down-regulation of downstream gene targets FOS and JUN.
TABLE 4.
Negatively enriched gene sets related to inflammation and cancer in rectosigmoid biopsies before and after diet-induced weight loss in all 10 subjects1
| Name of gene set | Size | ES | NES | P | FDR |
| CYTOKINE_ACTIVITY | 72 | −0.51 | −2.17 | <0.001 | <0.001 |
| CHEMOKINE_RECEPTOR_BINDING | 34 | −0.65 | −2.30 | <0.001 | <0.001 |
| CHEMOKINE_ACTIVITY | 33 | −0.68 | −2.36 | 0.004 | <0.001 |
| INFLAMMATORY_RESPONSE | 108 | −0.46 | −2.02 | <0.001 | <0.001 |
| TNFALPHA_ALL_UP | 70 | −0.48 | −2.2 | <0.001 | <0.001 |
| TNFALPHA_30MIN_UP | 39 | −0.55 | −2.00 | <0.001 | 0.008 |
| CROONQUIST_IL6_STROMA_UP | 36 | −0.61 | −2.20 | 0.002 | 0.001 |
| IL6_FIBRO_UP | 42 | −0.50 | −1.82 | <0.001 | 0.009 |
| IL1 KC UP | 38 | −0.63 | −2.24 | <0.001 | <0.001 |
| IL17ANDTNF KC UP | 28 | −0.63 | −2.11 | <0.001 | <0.001 |
| IL17 GAFFEN | 30 | −0.56 | −1.95 | <0.001 | <0.001 |
| IL17 KC UP | 40 | −0.51 | −1.85 | <0.001 | 0.002 |
| ADDITIVE IL17 AND TNFA KC | 181 | −0.35 | −1.67 | <0.001 | 0.008 |
| EICOSANOID_SYNTHESIS | 15 | −0.67 | −1.88 | 0.004 | 0.027 |
| PROSTAGLANDIN_AND_LEUKOTRIENE_METABOLISM | 23 | −0.58 | −1.84 | 0.004 | 0.037 |
| ICOSANOID_METABOLIC_PROCESS | 15 | −0.7 | −1.99 | <0.001 | 0.042 |
| HSA00590_ARACHIDONIC_ACID_METABOLISM | 42 | −0.49 | −1.80 | <0.001 | 0.046 |
| GLUTATHIONE_METABOLISM | 30 | −0.65 | −2.24 | <0.001 | 0.001 |
| GLUTATHIONE_TRANSFERASE_ACTIVITY | 15 | −0.71 | −2.06 | <0.001 | 0.007 |
| ELECTRON_TRANSPORT_CHAIN | 87 | −0.66 | −2.86 | <0.001 | <0.001 |
| HSA00190_OXIDATIVE_PHOSPHORYLATION | 105 | −0.66 | −2.82 | <0.001 | <0.001 |
| PROTEASOME_DEGRADATION | 30 | −0.66 | −1.81 | 0.002 | 0.045 |
| UBIQUINONE_BIOSYNTHESIS | 14 | −0.66 | −2.02 | <0.001 | 0.001 |
| PROTEASOME | 17 | −0.66 | −2.04 | <0.001 | <0.001 |
Size, number of genes analyzed for each gene set; ES, enrichment score; NES, normalized ES; P, P value of ES; FDR, false discovery rate. Gene sets with P ≤ 0.005 and an FDR ≤5% are shown.
Importantly, the analysis of changes in transcription factor genes identified 38 down-regulated and 9 up-regulated transcription factor gene sets (see supplemental Tables 8 and 9 under “Supplemental data” in the online issue). These gene sets included down-regulated transcription factor gene sets for STAT3, STAT5A, STAT5B, NF-κB, ATF, CREB, and multiple serum response factors, which are recognized as very important in colorectal inflammation, proliferation, apoptosis, and oncogenesis, which are shown in Table 5.
TABLE 5.
Negatively enriched transcription factor gene sets related to inflammation and cancer in rectosigmoid biopsies before and after diet-induced weight loss in all 10 subjects1
| Name of gene set | Size | ES | NES | P | FDR |
| CCAWWNAAGG_V$SRF_Q4 | 63 | −0.53 | −2.13 | <0.001 | <0.01 |
| V$SRF_01 | 40 | −0.55 | −2.00 | <0.001 | <0.01 |
| V$SRF_Q5_01 | 165 | −0.41 | −1.94 | <0.001 | <0.01 |
| V$STAT3_01 | 16 | −0.62 | −1.82 | 0.005 | 0.01 |
| V$STAT5A_01 | 169 | −0.37 | −1.77 | <0.001 | 0.01 |
| V$STAT5B_01 | 171 | −0.37 | −1.74 | <0.001 | 0.02 |
| V$ATF_B | 133 | −0.35 | −1.63 | 0.001 | 0.06 |
| V$CREB_Q4_01 | 150 | −0.34 | −1.58 | 0.001 | 0.07 |
| V$NFKB_Q6_01 | 149 | −0.33 | −1.55 | 0.003 | 0.07 |
| GGGNNTTTCC_V$NFKB_Q6_01 | 91 | −0.35 | −1.52 | 0.005 | 0.08 |
Size, number of genes analyzed for each gene set; ES, enrichment score; NES, normalized ES; P, P value of ES; FDR, false discovery rate. Gene sets with P ≤ 0.005 and an FDR ≤10% are shown.
DISCUSSION
Epidemiologic studies indicated that obesity is accompanied by an increased risk of CRC, and intentional weight loss reduced this risk (2, 12). To explore potential mechanisms, we studied effects of diet-induced weight loss on the colorectal mucosa in obese women. A VLCD was used to induce a 10% weight loss, which resulted in reduced rectosigmoid mucosal cytokine and chemokine concentrations and fewer tissue macrophages and T cells. Gene-expression analysis showed down-regulation of multiple genes involved in inflammation and cancer, including genes involved in cytokine and chemokine signaling and the proto-oncogenes JUN and FOS. The gene-pathway analysis showed down-regulation of pathways activated by TNF-α, IL-6, IL-1, and IL-17 and pathways involved in chemokine and cytokine signaling, prostaglandin synthesis, oxidative stress, and glutathione metabolism. In addition, transcription factor pathways for STAT3, STAT5, NF-κB, ATF, CREB, and several serum response factors were down-regulated. These results strongly suggested that chronic inflammation was present in the colorectal mucosa of obese individuals and that diet-induced weight loss decreased inflammation and down-regulated inflammatory and cancer gene pathways.
Obesity is accompanied by elevated circulating concentrations of insulin and IGF-1, and changes in the insulin-IGF axis are believed to be important in CRC pathogenesis in obesity through the PIK3/Akt pathway (29) Insulin is mitogenic only at supraphysiologic concentrations through the activation of the IGF-1 pathway (30). Physiologic concentrations of IGF-1 are growth promoting and mitogenic, and increased concentrations of IGF-1, when adjusted for concentrations of its binding protein, are associated with a modest increase in CRC risk (31). Leptin concentrations also increase in obesity, and leptin has growth-promoting, mitogenic, and antiapoptotic properties in colon cancer cell lines but not in the normal epithelium (32) In the Adenomatous polyposis coli (APCmin) mouse, leptin did not promote CRC or xenograft growth in mice (33). However, obesity is recognized as a chronic inflammatory state with increased serum cytokine concentrations, including those of TNF-α, IL-6 and -8, and MCP-1 (27, 34), and evidence of inflammation in adipose tissue, liver, skeletal muscle, and coronary arteries (18). In our study, weight loss significantly reduced circulating concentrations of TNF-α, IL-8, and MCP-1. Increased concentrations of obesity are accompanied by progressively higher concentrations of serum cytokines and increased risk of colorectal neoplasia (27, 35). This relation is supported by evidence that cytokines have a procarcinogenic effect in colorectal tissues (20). Reducing systemic TNF-α activity (36) and knocking out NF-κB in myeloid cells (37) decreased colonic tumors in mouse colitis models. Thus, this diet regimen–induced reduction in systemic cytokines might contribute to lowering colon neoplasia in obese individuals.
The current study showed that the diet-induced weight loss reduced rectosigmoid mucosal concentrations of the inflammatory mediators TNF-α, IL-6, IL-8, IL-1β, and MCP-1. In confirmation, the expression of gene pathways regulated by TNF-α and IL-6 and -1β also decreased. In mouse models of colitis-induced CRC, the inhibition of activated gene pathways downstream of TNF-α and IL-6 decreased CRC. We also observed a decreased expression of transcription factor pathways implicated in colorectal inflammation, CRC, and other types of cancer, including STAT3, NF-κB, ATF, CREB, and STAT5 (38). STAT3 activation up-regulates inflammatory cytokines such as IL-6, IL-1β, IL-8, MCP-1, and COX-2 (39). NF-κB induces the expression of many inflammatory mediators and is a core transcription factor in the immune response. Both STAT3 and NF-κB signaling are recognized as pathways involved in inflammation-induced carcinogenesis (38). STAT3 and STAT5 can expand regulatory T cells, which inhibit anti-tumor immunity and promote tumor progression (40). An analogous pathway of inflammation that leads to increased risk of liver cancer in obesity was recently published (41). Thus, diet-induced weight reduction down-regulates core transcription factors that are crucial in inflammation and carcinogenesis and thereby may reduce risk of CRC. The expression of gene pathways involved in prostaglandin synthesis were also lower. In human obesity and mouse obesity models, the increased expression of the COX2 gene is a major regulator of colonic prostaglandin synthesis. Caloric restriction has been shown to reduce colonic COX2 expression in mouse obesity accompanied by decreased numbers of aberrant crypt foci (11).
The current study showed that our experimental regimen reduced rectosigmoid mucosal T cell and macrophage numbers accompanied by decreased expression of IL-8, CCL19, CCL20, CCL4L1, and gene pathways associated with chemokine activity. These gene pathways are involved in mucosal leukocyte accumulation, and their activity is increased in the colonic mucosa of patients with inflammatory bowel disease during acute exacerbations and down-regulated during remissions (42).
Oxygen free-radical formation and increased oxidative stress link colonic inflammation and cancer formation (20). Obesity enhances systemic oxidative stress, which, through intermediary pathways, enhances the expression of proinflammatory and cell cycle genes that can lead to tissue inflammation and DNA damage and result in loss of tumor suppressor functions that are characteristic of CRC (43). Western diet–consuming obese mice have shown enhanced oxidative stress as well as inflammation in the colorectal mucosa, (9) and calorie restriction has reduced oxygen free-radical formation and DNA damage in rodent liver and heart tissue (44, 45). Our study subjects showed down-regulation of pathways involved with oxidative phosphorylation and FOS and JUN genes that activate the transcription of proinflammatory and cell proliferative mediators and can promote several human cancers (19). Thus, obesity creates an environment in the colon that promotes the initiation and progression of CRC, and calorie restriction has an opposite effect by reducing oxygen free-radical formation and decreasing oxidative stress.
The current experimental model was chosen as a proof-of-principle study for several reasons. Our experience with similar translational studies has shown that endpoint data obtained in the same individual obtained before and after an intervention provided optimum and reproducible information. A weight loss of over 8% of initial weight was used because a previous study showed that this would greatly lower proliferation rates in humans (21). Previous experience permitted us to study only 10 subjects because the research staff of the Rockefeller University Hospital has provided remarkable nutritional and environmental control in intervention studies. One limitation is that the protocol did not include control subjects such as obese individuals fed an isoenergetic diet by using VLCD products. As a result, it is not possible to determine whether the observed improvements in the inflammatory and carcinogenesis pathways were changes in energy balance, adiposity, or diet composition. However, unpublished data (P Holt, 2010) using dramatically differing diets have not shown similar degrees of changes in mucosal cytokines or gene array pathway analysis. Irrespective of the precise factors responsible, our analysis showed a marked reduction in inflammatory and colon carcinogenesis endpoints with the intervention, which indicated that obesity must be accompanied by increases in these pathways. We recognize that the diet can alter luminal microbiota diversity, which might influence epithelial cell signaling (46) and, thus, contribute to some of the changes that we observed.
In conclusion, our study shows that diet-induced weight loss in obese women reduced colorectal mucosal inflammation as shown by decreased inflammatory cytokines, inflammatory cells, down-regulated inflammatory and cancer gene pathways, and transcription factor gene sets that enhance inflammation and CRC risk. Thus, obesity appeared to be accompanied by a low-grade inflammatory state in the colorectal mucosa, and diet-induced weight loss decreased such inflammation. Because colorectal inflammation is an important cofactor for colorectal carcinogenesis, such an inflammatory state may contribute to enhanced CRC risk in obesity; weight loss counteracts this effect and may thereby lower CRC risk.
Supplementary Material
Acknowledgments
We thank Jan L Breslow for his advice and support, Jeanne Walker for her close supervision of study volunteers, James G Krueger and Artemis Khatcherian for help with immunohistochemistry, and Juana Gonzalez for technical help with the Mesoscale multiplex assay at the Clinical and Translational Science Center immunology core facility.
The authors’ responsibilities were as follows—SP: study concept and design, data acquisition, data analysis and interpretation, and drafting of the manuscript; LMN: study design and VLCD management; MS-F: statistical analyses; and PRH: study concept and design, data interpretation, and drafting of the manuscript. None of the authors had a conflict of interest.
REFERENCES
- 1.Renehan AG, Tyson M, Egger M, Heller RF, Zwahlen M. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet 2008;371:569–78 [DOI] [PubMed] [Google Scholar]
- 2.Larsson SC, Wolk A. Obesity and colon and rectal cancer risk: a meta-analysis of prospective studies. Am J Clin Nutr 2007;86:556–65 [DOI] [PubMed] [Google Scholar]
- 3.Flegal KM, Carroll MD, Ogden CL, Curtin LR. Prevalence and trends in obesity among US adults, 1999-2008. JAMA 2010;303:235–41 [DOI] [PubMed] [Google Scholar]
- 4.Mokdad AH, Serdula MK, Dietz WH, Bowman BA, Marks JS, Koplan JP. The spread of the obesity epidemic in the United States, 1991-1998. JAMA 1999;282:1519–22 [DOI] [PubMed] [Google Scholar]
- 5.Kuczmarski RJ, Flegal KM, Campbell SM, Johnson CL. Increasing prevalence of overweight among US adults. The National Health and Nutrition Examination Surveys, 1960 to 1991. JAMA 1994;272:205–11 [DOI] [PubMed] [Google Scholar]
- 6.Gravaghi C, Bo J, Laperle KM, et al. Obesity enhances gastrointestinal tumorigenesis in Apc-mutant mice. Int J Obes (Lond) 2008;32:1716–9 [DOI] [PubMed] [Google Scholar]
- 7.Weber RV, Stein DE, Scholes J, Kral JG. Obesity potentiates AOM-induced colon cancer. Dig Dis Sci 2000;45:890–5 [DOI] [PubMed] [Google Scholar]
- 8.Newmark HL, Yang K, Lipkin M, et al. A Western-style diet induces benign and malignant neoplasms in the colon of normal C57Bl/6 mice. Carcinogenesis 2001;22:1871–5 [DOI] [PubMed] [Google Scholar]
- 9.Erdelyi I, Levenkova N, Lin EY, et al. Western-style diets induce oxidative stress and dysregulate immune responses in the colon in a mouse model of sporadic colon cancer. J Nutr 2009;139:2072–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy BS, Wang CX, Maruyama H. Effect of restricted caloric intake on azoxymethane-induced colon tumor incidence in male F344 rats. Cancer Res 1987;47:1226–8 [PubMed] [Google Scholar]
- 11.Raju J, Bird RP. Energy restriction reduces the number of advanced aberrant crypt foci and attenuates the expression of colonic transforming growth factor beta and cyclooxygenase isoforms in Zucker obese (fa/fa) rats. Cancer Res 2003;63:6595–601 [PubMed] [Google Scholar]
- 12.Parker ED, Folsom AR. Intentional weight loss and incidence of obesity-related cancers: the Iowa Women's Health Study. Int J Obes Relat Metab Disord 2003;27:1447–52 [DOI] [PubMed] [Google Scholar]
- 13.Sandhu MS, Dunger DB, Giovannucci EL. Insulin, insulin-like growth factor-I (IGF-I), IGF binding proteins, their biologic interactions, and colorectal cancer. J Natl Cancer Inst 2002;94:972–80 [DOI] [PubMed] [Google Scholar]
- 14.Kim S, Keku TO, Martin C, et al. Circulating levels of inflammatory cytokines and risk of colorectal adenomas. Cancer Res 2008;68:323–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole BF, Logan RF, Halabi S, et al. Aspirin for the chemoprevention of colorectal adenomas: meta-analysis of the randomized trials. J Natl Cancer Inst 2009;101:256–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arber N, Eagle CJ, Spicak J, et al. Celecoxib for the prevention of colorectal adenomatous polyps. N Engl J Med 2006;355:885–95 [DOI] [PubMed] [Google Scholar]
- 17.Kim S, Baron JA, Mott LA, et al. Aspirin may be more effective in preventing colorectal adenomas in patients with higher BMI (United States). Cancer Causes Control 2006;17:1299–304 [DOI] [PubMed] [Google Scholar]
- 18.Zeyda M, Stulnig TM. Obesity, inflammation, and insulin resistance–a mini-review. Gerontology 2009;55:379–86 [DOI] [PubMed] [Google Scholar]
- 19.Klaunig JE, Kamendulis LM, Hocevar BA. Oxidative stress and oxidative damage in carcinogenesis. Toxicol Pathol 2010;38:96–109 [DOI] [PubMed] [Google Scholar]
- 20.Coussens LM, Werb Z. Inflammation and cancer. Nature 2002;420:860–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinbach G, Heymsfield S, Olansen NE, Tighe A, Holt PR. Effect of caloric restriction on colonic proliferation in obese persons: implications for colon cancer prevention. Cancer Res 1994;54:1194–7 [PubMed] [Google Scholar]
- 22.Chowdhury F, Williams A, Johnson P. Validation and comparison of two multiplex technologies, Luminex and Mesoscale Discovery, for human cytokine profiling. J Immunol Methods 2009;340:55–64 [DOI] [PubMed] [Google Scholar]
- 23.Protiva P, Cross HS, Hopkins ME, et al. Chemoprevention of colorectal neoplasia by estrogen: potential role of vitamin D activity. Cancer Prev Res (Phila) 2009;2:43–51 [DOI] [PubMed] [Google Scholar]
- 24.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007;23:3251–3 [DOI] [PubMed] [Google Scholar]
- 25.Nograles KE, Zaba LC, Guttman-Yassky E, et al. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br J Dermatol 2008;159:1092–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaffen SL. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol 2009;9:556–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim CS, Park HS, Kawada T, et al. Circulating levels of MCP-1 and IL-8 are elevated in human obese subjects and associated with obesity-related parameters. Int J Obes (Lond) 2006;30:1347–55 [DOI] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer 2008;8:915–28 [DOI] [PubMed] [Google Scholar]
- 30.Giovannucci E, Michaud D. The role of obesity and related metabolic disturbances in cancers of the colon, prostate, and pancreas. Gastroenterology 2007;132:2208–25 [DOI] [PubMed] [Google Scholar]
- 31.Wei EK, Ma J, Pollak MN, et al. A prospective study of C-peptide, insulin-like growth factor-I, insulin-like growth factor binding protein-1, and the risk of colorectal cancer in women. Cancer Epidemiol Biomarkers Prev 2005;14:850–5 [DOI] [PubMed] [Google Scholar]
- 32.Fenton JI, Hord NG, Lavigne JA, Perkins SN, Hursting SD. Leptin, insulin-like growth factor-1, and insulin-like growth factor-2 are mitogens in ApcMin/+ but not Apc+/+ colonic epithelial cell lines. Cancer Epidemiol Biomarkers Prev 2005;14:1646–52 [DOI] [PubMed] [Google Scholar]
- 33.Aparicio T, Kotelevets L, Tsocas A, et al. Leptin stimulates the proliferation of human colon cancer cells in vitro but does not promote the growth of colon cancer xenografts in nude mice or intestinal tumorigenesis in Apc(Min/+) mice. Gut 2005;54:1136–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fain JN. Release of interleukins and other inflammatory cytokines by human adipose tissue is enhanced in obesity and primarily due to the nonfat cells. Vitam Horm 2006;74:443–77 [DOI] [PubMed] [Google Scholar]
- 35.Adams KF, Leitzmann MF, Albanes D, et al. Body mass and colorectal cancer risk in the NIH-AARP cohort. Am J Epidemiol 2007;166:36–45 [DOI] [PubMed] [Google Scholar]
- 36.Popivanova BK, Kitamura K, Wu Y, et al. Blocking TNF-alpha in mice reduces colorectal carcinogenesis associated with chronic colitis. J Clin Invest 2008;118:560–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greten FR, Eckmann L, Greten TF, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 2004;118:285–96 [DOI] [PubMed] [Google Scholar]
- 38.Grivennikov SI, Karin M. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev, 2010;21:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 2009;9:798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol 2007;7:41–51 [DOI] [PubMed] [Google Scholar]
- 41.Park EJ, Lee JH, Yu GY, et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 2010;140:197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banks C, Bateman A, Payne R, Johnson P, Sheron N. Chemokine expression in IBD. Mucosal chemokine expression is unselectively increased in both ulcerative colitis and Crohn's disease. J Pathol 2003;199:28–35 [DOI] [PubMed] [Google Scholar]
- 43.Klaunig JE, Kamendulis LM. The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol 2004;44:239–67 [DOI] [PubMed] [Google Scholar]
- 44.Gredilla R, Barja G, Lopez-Torres M. Effect of short-term caloric restriction on H2O2 production and oxidative DNA damage in rat liver mitochondria and location of the free radical source. J Bioenerg Biomembr 2001;33:279–87 [DOI] [PubMed] [Google Scholar]
- 45.Gredilla R, Sanz A, Lopez-Torres M, Barja G. Caloric restriction decreases mitochondrial free radical generation at complex I and lowers oxidative damage to mitochondrial DNA in the rat heart. FASEB J 2001;15:1589–91 [DOI] [PubMed] [Google Scholar]
- 46.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology 2009;136:65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.