Abstract
Mutations in human leucine-rich repeat kinase 2 (Lrrk2), a protein of yet unknown function, are linked to Parkinson's disease caused by degeneration of midbrain dopaminergic neurons. The protein comprises several domains including a GTPase and a kinase domain both affected by several pathogenic mutations. To elucidate the molecular interaction network of endogenous Lrrk2 under stoichiometric constraints, we applied QUICK (quantitative immunoprecipitation combined with knockdown) in NIH3T3 cells. The identified interactome reveals actin isoforms as well as actin-associated proteins involved in actin filament assembly, organization, rearrangement, and maintenance, suggesting that the biological function of Lrrk2 is linked to cytoskeletal dynamics. In fact, we demonstrate Lrrk2 de novo binding to F-actin and its ability to modulate its assembly in vitro. When tested in intact cells, knockdown of Lrrk2 causes morphological alterations in NIH3T3 cells. In developing dopaminergic midbrain primary neurons, Lrrk2 knockdown results in shortened neurite processes, indicating a physiological role of Lrrk2 in cytoskeletal organization and dynamics of dopaminergic neurons. Hence, our results demonstrate that molecular interactions as well as the physiological function of Lrrk2 are closely related to the organization of the actin-based cytoskeleton, a crucial feature of neuronal development and neuron function.
Parkinson's disease (PD)1 is the second most prevalent neurodegenerative disorder but the underlying pathogenesis still remains elusive. The human leucine-rich repeat kinase 2 (Lrrk2, OMIM 609007) gene has been identified as the most common causative gene of autosomal-dominant inherited and idiopathic PD (1). To date, more than 50 variants within the Lrrk2 locus have been described, seven of which are confirmed to be pathogenic (2). The clinical symptoms of Lrrk2 mutation carriers are indistinguishable from those of idiopathic PD patients, whereas the related neuropathology is pleomorphic, including α-synucleinopathy, tauopathy, and ubiquitin deposits or nigral neuronal loss solely (3, 4). Therefore, Lrrk2 might be central to neurodegenerative pathways associated with parkinsonism (5). The expression pattern of Lrrk2 comprises many regions of the central nervous system as well as some peripheral organs like heart, liver, lung, and kidney (4, 6). The protein localizes to various neuronal populations of the brain, primarily striatum, cortex, and hippocampus (7, 8) as well as to the dopaminergic (DA) neurons of the substantia nigra pars compacta (9, 10).
Lrrk2 encodes a large multidomain protein (∼286 kDa) belonging to the ROCO protein superfamily (4), comprising a characteristic combination of a ROC (Ras of complex protein) domain adjacent to a COR (C-terminal of ROC) domain (11), immediately followed by a kinase domain (mitogen-activated protein kinase kinase kinase). This catalytic region is flanked by other regulatory and protein-interaction domains (12). Although the domain structure of Lrrk2 is well known, the understanding of the physiological function and the impact of pathogenic mutations on the molecular mechanisms leading to PD still remain uncertain. In vitro studies demonstrated that Lrrk2 is both a functional kinase and GTPase, able to undergo autophosphorylation and perform phosphorylation of generic and putative physiological substrates (13–15). Some mutations have been shown to increase the protein's kinase activity (16–18) and to induce cellular toxicity in vitro, leading to the hypothesis that the kinase activity contributes to neurodegeneration (18, 19). However, divergent reports on the enzymatic activity of mutant Lrrk2 (20) and the distribution of point mutations throughout the sequence of Lrrk2 indicate that not just the kinase domain is crucial for the physiological function. It is rather likely that intra- and intermolecular protein-protein interactions (PPIs) inside as well as outside of the kinase domain are essential for the normal function. Hence, alterations in the PPIs may underlie the pathomechanisms by which mutant Lrrk2 causes parkinsonism (21). A key step in understanding the physiological function of Lrrk2 and associated PD pathology is placing Lrrk2 into biochemical pathways by the identification of its protein interaction partners.
Therefore, we systematically analyzed the Lrrk2 interactome using the QUICK (quantitative immunoprecipitation combined with knockdown) assay reported by Selbach and Mann (22). This approach assesses interactions between proteins at their endogenous levels and in their normal cellular environment by a combination of stable isotope labeling with amino acids in cell culture (SILAC) (23), RNA interference (RNAi), coimmunoprecipitation (co-IP), and quantitative mass-spectrometry (MS). By applying this screen in the murine NIH3T3 cell line we identified 36 proteins as robust Lrrk2 interactors whose majority are members of the actin family, actin-regulatory proteins, myosins, and proteins modulating their activity. To our knowledge, this is the first study reporting Lrrk2 protein interactions of the endogenous protein at its physiological expression level. We can show, that Lrrk2 cosediments with filamentous (F)-actin and affects the amount of F-actin in vitro, implying—in conjunction with the identified putative interactors—a function of Lrrk2 in the regulation of the actin cytoskeleton arrangement and/or dynamics. Significant morphological alterations of NIH3T3 cells upon lentiviral-mediated Lrrk2 knockdown raise further evidence for this hypothesis. Moreover, shRNA-based knockdown of Lrrk2 in primary ventral mesencephalic (VM) cells leads to impaired neurite outgrowth in developing DA neurons. These results indicate a functional association of Lrrk2 with the actin-based cytoskeleton, and suggest a role in regulating cellular morphology and structural plasticity.
EXPERIMENTAL PROCEDURES
Cloning of shRNA Expressing Lentiviral Vectors
For the lentiviral-mediated knockdown of Lrrk2, the previously reported second generation short hairpin RNAmir (shRNAmir) construct miB3 (24) was utilized. In addition, we designed a second, independent construct as described before (24), with the respective Lrrk2 target sequence sh-miB4 (AY792512 (2524–2544): 5′-AAAGTGCTCCGGTATCAGATG-3′) identified and selected using Ambion-web-based oligo-search software (siRNA Target Finder, http://www.ambion.com/techlib/misc/siRNA_finder.html). Both synthesized oligos (Metabion, Martinsried, Germany), consisting of 86 nucleotides coding for a 5′-pseudoBglII restriction site, a polymerase III-transcription start-site, a sense-oligo-loop-antisense-oligo, a transcription termination-site and a 3′-pseudoXbaI restriction site, were cloned into the pLVTH transfer vector as described elsewhere (25), resulting in pLVmiB3 (targeting within exon 1) (24) and pLVmiB4 (targeting within exon 20).
Viral Production
Recombinant lentiviruses were produced according to standard protocols as described before (25). In brief, subconfluent HEK293T cells were transiently cotransfected with 13 μg transfer vector, 13 μg pCMV-Gag/Pol dR8.92, 3 μg pRSV-Rev and 3.75 μg pMD2G by calcium phosphate precipitation and medium was changed after 16 h. Recombinant viruses were harvested 48 h post-transfection and concentrated 160-fold. Because all pLVTH vectors coexpress enhanced green fluorescent protein (GFP), titering of concentrated viral supernatants was performed by FACS analysis of transduced NIH3T3 cells. Therefore, cells were plated at a density of 1.5 × 105 cells per well on 6-well plates (Nunc, Rochester, NY) and serial dilutions of recombinant lentivirus were added 16 h later. The cells were cultured for 3 days, divided and 24 h later the percentage of GFP-positive cells was determined by FACS analysis. Wells containing less than 10% GFP-positive cells were used to calculate the amount of infectious units in NIH3T3 per microliter. The multiplicity of infection (MOI) represents the ratio of input infectious units calculated in NIH3T3 to the number of cells available for transduction.
NIH3T3 Cell Culture and Viral Transduction
NIH3T3 cells were maintained in DMEM (Invitrogen, Carlsbad, CA) containing 10% FBS (Invitrogen), 2 mm glutamine (Invitrogen), 50 U/ml penicillin, and 50 μg/ml streptomycin (Invitrogen) at 37 °C in a humidified atmosphere with 5% CO2 in air. For lentiviral transduction, cells were seeded at a density of 2.5 × 104 cells per well onto 6-well plates and transduced the next day with a MOI of five with LVTH and LVmiB3, respectively. Wildtype (wt) control cultures were prepared in parallel.
For the analysis of cell morphology, wt and transduced cells were divided and replated at 5%–10% density in DMEM/10% FBS on glass coverslips (Assistent, Sondheim, Germany) 5 days after transduction. At 4 h post-splitting, the cells were washed and serum starved with 0.5% FBS for 24 h.
QUICK Screen in NIH3T3 Cells
Endogenous Lrrk2 protein complex components were identified from NIH3T3 cells by using the QUICK approach. The assay and SILAC labeling were done essentially as described earlier (22, 23). Wt and LVmiB3-transduced NIH3T3 cells were grown for at least five passages in SILAC DMEM (PAA, Pasching, Austria) containing 10% dialyzed FBS (PAA) and either normal lysine and arginine or the heavy isotope labeled counterparts (Arg: 13C615N4; Lys: 13C6) (Silantes, Munich, Germany).
For immunoprecipitation, a rat-monoclonal Lrrk2—specific antibody (clone 1E11, produced in house by E. Kremmer (14, 24)) was covalently cross-linked to Protein G Sepharose 4 Fast Flow beads (GE Healthcare, Waukesha, WI) as outlined by Thermo Fisher Scientific (Waltham, MA) and described elsewhere (26). To this end, the matrix was equilibrated with 50 mm Na-borate buffer (pH 8.2) and the antibody was cross-linked at a density of 1 mg/ml resin using 25 mm dimethyl pimelimidate (Thermo Fisher Scientific) in cross-linking buffer (0.2 m triethanolamine, pH 8.2). After blocking the remaining active sites with 0.1 m ethanolamine (pH 8.2), the beads were washed twice with elution buffer (200 mm glycine, pH 2.5, 500 mm NaCl, 0.1% Nonidet P-40 (Roche, Penzberg, Germany)) to remove unbound antibody and then washed extensively with Na-borate buffer for use.
NIH3T3 cells were lyzed in ice-cold lysis buffer (30 mm Tris, pH 7.4, 150 mm NaCl, 10% glycerol, 1% Nonidet P-40, 1× Roche complete protease inhibitor mixture (Roche), 1× phosphatase inhibitor mixture 1 and 2 (Sigma, St. Louis, MO)). Extracts were kept on ice for 30 min with intermittent vortexing before they were centrifuged for 10 min at 16000 × g and 4 °C. The protein concentration of the lysates was determined by using the Bradford method (BioRad, Hercules, CA). Possible global changes in protein abundance induced by lentiviral-mediated RNAi were investigated by quantitative analysis of whole cell lysates.
Equal amounts of identically concentrated lysates derived from “heavy” and “light” SILAC labeled cells were precleared by incubation with Protein G Sepharose for 30 min at 4 °C under gentle agitation separately. For immunoprecipitation, precleared lysates were incubated with anti-Lrrk2-Protein G Sepharose matrix overnight at 4 °C with rotation. The matrices of the two separate immunoprecipitations were combined and washed three times with ice-cold HEPES buffer (20 mm HEPES, pH 7.5, 50 mm NaCl, 0.1% Nonidet P-40, 1× Roche complete protease inhibitor mixture, 1× phosphatase inhibitor mixture 1 and 2). Subsequently, protein complexes were eluted in Laemmli buffer (50 mm Tris, pH 6.8, 1% SDS, 10% glycerol, 100 mm β-mercaptoethanol, and bromphenol blue) for 15 min at 37 °C and gentle agitation. Both precipitated proteins and whole cell lysates were prefractionated by standard SDS-PAGE followed by tryptic in-gel cleavage as described earlier (27) before they were subjected to LC-MS/MS.
For verification of interaction partners, immunoprecipitation was conducted in separated settings as described above using cell lysates from non-SILAC labeled wt and LVmiB3-transduced NIH3T3 cells that were also cultured for at least five passages. Eluted proteins and cell lysates were subjected to SDS-PAGE and selected proteins were detected by Western blot analysis.
Liquid Chromatography (LC)-MS/MS and Data Analysis
LC-MS/MS analysis was performed on an Ultimate3000 nano high-pressure liquid chromatography (HPLC) system (Dionex, Sunnyvale, CA) coupled to a LTQ OrbitrapXL mass spectrometer (Thermo Fisher Scientific) by a nano spray ion source. Tryptic peptide mixtures were automatically injected and loaded at a flow rate of 30 μl/min in 95% buffer C (2% acetonitrile, 0.1% trifluoroacetic acid in HPLC grade water) and 5% buffer B (98% acetonitrile, 0.1% formic acid in HPLC grade water) onto a nano trap column (100 μm i.d. × 2 cm, packed with Acclaim PepMap100 C18, 5 μm, 100 Å, LC Packings). After 5 min, peptides were eluted and separated on the analytical column (75 μm i.d. × 15 cm, Acclaim PepMap100 C18, 3 μm, 100 Å, LC Packings) by a linear gradient from 5% to 40% of buffer B in buffer A (2% acetonitrile and 0.1% formic acid) at a flow rate of 300 nl/min over 140 min. Remaining peptides were eluted by a short gradient from 40% to 100% buffer B in 5 min. The eluting peptides were analyzed in the LTQ OrbitrapXL using XCalibur software (version 2.07). From the high resolution MS prescan with a mass range of 300 to 1500, the ten most intense peptide ions were selected for fragment analysis in the linear ion trap if they exceeded an intensity of at least 200 counts and if they were at least doubly charged. The normalized collision energy for CID was set to a value of 35 and the resulting fragments were detected with normal resolution in the linear ion trap. The lock mass option was activated and a background signal with a mass of 445.120020 was used as lock mass (28). Every ion selected for fragmentation, was excluded for 30 s by dynamic exclusion. All acquired spectra were processed and analyzed using the MaxQuant software (version 1.0.13.8) (29) and the mouse specific IPI database version 3.52 (number of sequences: 83947; http://www.maxquant.org/) in combination with Mascot (Matrix Science, Boston, MA, version 2.2). Trypsin/P was selected as enzyme for cleavage with two missed cleavages allowed. Fragment ion tolerance was set to 0.5 Da and the top six MS/MS peaks per 100 Da were used. A maximum of three labeled amino acids per peptide were allowed. The precursor ion tolerance was automatically set by the MaxQuant software. Cysteine carbamidomethylation was selected as fixed modification and methionine oxidation, phosphorylation on serine, threonine or tyrosine as well as protein acetylation were allowed as variable modifications. The peptide false discovery rate was set to 5%, the protein false discovery rate to 1%. All contaminants included in the database, with the exception of β-actin, as well as proteins that showed high variability (> 100%) were removed and only proteins, identified and quantified in at least two of five experiments, were considered for further analysis. If the identified peptide sequence set of one protein was equal to or contained the peptide set of another protein, these two proteins were grouped together by MaxQuant and not counted as independent protein hits. All details for the peptide and protein identifications and quantifications, including all parameters used for MaxQuant analysis are given in Supplemental File 1 for the QUICK assay and in Supplemental File 2 for the proteome comparison. MaxQuant normalized protein ratios were utilized for the downstream analyses and a p value of 0.05 was selected as threshold for significant enrichment of Lrrk2 complex components.
Protein Network Analysis
Protein-protein interactions were analyzed and visualized using Pathway Palette (30). Interaction networks were abstracted as graphs, where proteins are represented as nodes and their interactions as edges. Thereto, the genes corresponding to the significant enriched proteins within the QUICK assay (QUICK set) were uploaded to the Pathway Palette website (http://blaispathways.dfci.harvard.edu/Palette.html). In the case of protein hits with ambiguous assignment by the MaxQuant software, one gene was selected at random under the assumption that similar proteins share interaction partners. Known interactions between the proteins were retrieved from the Human Protein Reference Database (HPRD) (31) and the Biological General Repository for Interaction Datasets (BioGRID) (32). Interactions were evaluated as well as prioritized according to their “evidence,” referring to the experimental method used for the detection, provided by the database and using the following categorization: i) “BioGRID: Low-Throughput” or “HPRD: in vivo”, ii) “BioGRID: HTP/complex” or “HPRD: in vitro” and iii) “BioGRID: HTP/Pairwise” or “HPRD: yeast 2-hybrid.” To assess the physical cohesiveness of the resulting network, we implemented Physical Interaction Enrichment (PIE) analysis (33) using a custom Python script and a reference network consisting of the 29,377 mouse Entrez Gene IDs, with 51,460 known interactions between them, as represented in Pathway Palette. In total, a million randomly selected protein sets, having same size and node degree distribution as the QUICK set, were analyzed for the number of interactions within the sets to obtain a frequency distribution used for the calculation of physical cohesiveness (PIE score) and its significance. For a functional annotation of the Lrrk2-related network components, a pathway enrichment analysis was performed based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) (34) database using Pathway Palette. To further investigate the connectivity between the proteins of the QUICK set, interaction data from the HPRD and BioGRID database were used to construct an extended network by adding only first-order shared neighbors of the proteins, where two proteins from the QUICK set are linked by one interconnecting (intermediate) protein. Network complexity was reduced by removing edges between the first-order neighbors.
Affinity Purification of Lrrk2 and F-actin Cosedimentation Assay
Strep/Flag(SF)-TAP tagged wt Lrrk2 was transiently expressed in HEK293 cells and purified via the STREP-tag II as described earlier (35, 36). The Bradford assay was used to determine the protein concentration of the eluates, which were supplemented with 20% glycerol and stored at −80 °C prior to use.
Actin (Ca-ATP-G(globular)-actin) was purified from rabbit skeletal muscle according to Spudich and Watt (37), further purified over a Sephadex G-200 gel filtration column in G-buffer (5 mm Tris-HCl, pH 7.7, 0.1 mm CaCl2, 0.2 mm ATP, 0.2 mm dithiotreitol) and stored on ice. For F-actin cosedimentation assays, affinity purified SF-TAP tagged Lrrk2 in storage buffer (SB; desthiobiotin elution buffer, 20% glycerol) was added to a final concentration of 150 nm or 300 nm to 3 μm actin in G-buffer, supplemented with Roche complete protease inhibitor mixture. In all samples the combined volume of SB and protein in SB was always constant, i.e. 15 μl per 25 μl sample volume, thus ensuring constant buffer conditions. Polymerization was induced by the addition of KCl and MgCl2 to 100 mm and 2 mm, respectively. After incubation for 45 min at room temperature and 2 h at 4 °C, samples were subjected to centrifugation at 100,000 × g for 10 min at room temperature. The supernatants (S) were removed and pellets (P) were washed once with F-buffer (G-buffer containing 100 mm KCl and 2 mm MgCl2) before they were resuspended in Laemmli buffer. Equal amounts of both the pellets and supernatants from each reaction were analyzed by SDS-PAGE followed by Coomassie brilliant blue staining to mainly visualize actin or Western blotting using an anti-Flag antibody to show the presence of Lrrk2. To take into account any effects of the storage buffer on actin polymerization, samples containing only 3 μm actin and SB were included. A potential sedimentation of Lrrk2 irrespective of F-actin was monitored by incubating the protein at a concentration of 300 nm solely under actin polymerization buffer conditions. Protein intensities (I) on Coomassie gels and Western blots were quantified by densitometric analysis using ImageJ (National Institute of Mental Health (NIH), Bethesda, MD, USA; http://rsb.info.nih.gov/ij/) after background subtraction. The distribution of G- versus F-actin was determined from the intensity ratios I(S)/I(S)+I(P) and I(P)/I(S)+I(P), respectively, and expressed in percentages. To allow comparison over different Western blots, the signals for Lrrk2 intensities in the pellet fractions at different concentrations were normalized for each blot to the protein's intensity in the supernatant at 150 nm Lrrk2.
Primary VM Cultures and Viral Transduction
For the preparation of primary progenitor cultures of DA neurons all experiments were carried out in accordance with the European Community Council Directive (86/609/EEC) for care and use of laboratory animals. Primary embryonic mouse VM cells were isolated from embryos at gestation day 12.5 (E12.5) (animal breeding facility, Helmholtz Zentrum München, Munich, Germany), expanded and cultured as previously described (25). From day 7 in vitro (DIV) on, 10% FBS was added to the standard medium.
For the lentiviral transduction of VM cultures with LVTH, LVmiB3, and LVmiB4 at DIV1 a MOI of 0.5 was used. For a single experiment, each condition (wt, LVTH control, LVmiB3, and LVmiB4) was run in triplicate or quadruple wells.
Immunofluorescence
VM cultures and NIH3T3 cells were fixed with 4% paraformaldehyde (Sigma) in PBS for 15 min at 37 °C and subsequently washed with PBS for 10 min. After blocking in PBS containing 10% horse serum (Invitrogen) and 0.4% Triton X-100 (Sigma) for 1 h at room temperature, DA neurons were stained with mouse-monoclonal anti-tyrosine hydroxylase (TH) (clone LNC1, Millipore) diluted in PBS containing 2.5% horse serum and 0.1% Triton X-100 overnight at 4 °C. Fluorescent labeling was done by incubation with Alexa Fluor 568-conjugated donkey anti-mouse IgGs (Invitrogen) for 3 h at room temperature. For labeling of the actin cytoskeleton in NIH3T3 cells, coverslips were incubated with Alexa Fluor 568 phalloidin (Invitrogen) diluted in PBS containing 2.5% horse serum and 0.1% Triton X-100 for 90 min at room temperature. Nuclei were visualized by adding 4′,6-diamidino-2-phenylindol (Sigma).
Morphological Analysis of Cell Cultures
For the analysis of NIH3T3 cell morphology, fixed cells were immunostained for F-actin to visualize cell shape. Digital images from stained cultures were acquired using an Axioscope2 fluorescence microscope (Zeiss, Göttingen, Germany) equipped with an AxioCam HRc digital camera (Zeiss) and the AxioVision4 software package (Zeiss). Ten randomly selected fields per coverslip were imaged using a 10× objective and analyzed using ImageJ software. A binary mask was created by setting a threshold brightness that distinguished the fluorescent cells from the black background. Cell outlines were traced automatically and a single cell's area and perimeter of at least 500 cells per coverslip were measured. As a quantitative measure of cell shape, the ratio of perimeter to area (P:A-ratio) (38) of each cell was determined and the average P:A-ratio was calculated for each coverslip. Data obtained from all coverslips per experimental condition, with three coverslips per treatment group within one experiment, from four independent biological replications were pooled.
To quantify the neurite length of DA neurons in VM cultures at DIV9 and DIV14, digital images of 12 randomly selected fields per well from TH-immunostained cultures were captured using a 20× objective. Cultures were analyzed by manually tracing each individual process of a neuron from the cell body to the neurite tip using the NeuronJ plug-in of the ImageJ software. The length of neurites was calculated according to a calibrated scale using Excel software (Microsoft, Redmond, WA). Only individual processes without contacts to other neurites or cells were chosen for quantification. If a neurite branched, the longer of the branches was traced. Because it was not possible to distinguish between axons and dendrites in the cultures, all processes were considered equivalent and were referred to as neurites. To measure neurite length, a mean of 55 to 73 and 38 to 46 neurons per well was used for the analysis at DIV9 and DIV14, respectively. The obtained data, representing the total neurite length, were pooled from all wells per condition, three to four per experiment, over at least three independent experiments.
In case of lentiviral transduced cultures, only GFP-positive cells and neurons were analyzed.
Camera lucida drawings for TH-immunoreactive (TH-ir) neurons were obtained from original images captured with a 20× objective. Representative, randomly selected neurons from each condition were manually traced using NeuronJ. Tracings were saved as a 16-bit TIFF file and converted utilizing ImageJ.
Dopaminergic Cell Counts
VM cultures were analyzed by fluorescence microscopy using an Axioscope2 fluorescence microscope. The number of DA neurons at DIV9 and DIV14 was determined by counting TH-ir cells under microscopical observation using a 20× objective within the culture wells. Cell counts are expressed as a percentage of the corresponding wt culture per 8-well chamber (equal to 81 mm2). Data obtained from all wells per treatment group of at least three independent biological replications were pooled.
mRNA Isolation from VM Cultures and Real-Time PCR (Semiquantitative)
Total RNA from DIV9 and DIV14 VM cultures was obtained using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to manufacturer's instructions. RNA integrity and quantity were determined with the RNA 6000 Nano Assay Kit on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Reverse transcription (RT) of RNA (1–2 μg per 20 μl reaction) was performed using the Omniscript RT Kit (Qiagen) and primed with an oligo-dT primer (Metabion). Semiquantitative real-time PCR was performed essentially as described (25), with previous published primer pairs for the detection of Lrrk2 and Oas1 (2′-5′-oligoadenylate synthetase) mRNA expression (24). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH)-forward (5′-GACAAAATGGTGAAGGTCGGTG-3′) and GAPDH-reverse (5′-AGGTCAATGAAGGGGTCGTTG-3′) primer were designed by GenBank mouse cDNA sequence analysis using VectorNTI software (Invitrogen). All primers were purchased from Metabion. Reactions were set up with 1 μl cDNA out of 20 μl RT-reaction, 1 μm of GAPDH and Oas1 primer or 10 μm of Lrrk2 primer and the QuantiTect SybrGreen PCR Kit (Qiagen). In addition, 1 mm MgCl2 was added to reactions with GAPDH primers. To rule out any DNA cross-contamination, blank samples were included. PCR cycling conditions were: start with 95 °C for 15 min, 40 cycles consisting of 94 °C for 15 s, 65 °C or 54 °C for 30 s and 72 °C for 30 s. Specificity of the amplification reaction was confirmed by melting curve analysis. Absolute expression levels were normalized to GAPDH expression within the same sample and the “mean normalized expression” was calculated (39). Data from at least three experiments were pooled and results are represented as expression changes relative to control.
Protein Preparation and Western Blot Analysis
For the preparation of protein extracts from VM cultures, cells were lysed in ice-cold lysis-buffer as described for NIH3T3 cells and equal amounts of total protein were subjected to Western blot analysis. To validate Lrrk2 interaction partners in NIH3T3 immunoprecipitates, 50% of the IP eluates and a lysate volume corresponding to 2% of the IP-input were used. Proteins were separated by standard SDS-PAGE and analyzed by Western blotting with chemiluminescent detection as reported (16) using the following primary antibodies: rat-monoclonal anti-Lrrk2 (clone 1E11), mouse-monoclonal anti-GAPDH (clone 6C5, Millipore), rabbit-polyclonal anti-GFP (Invitrogen), rabbit-polyclonal anti-myosin Id (Santa Cruz, Santa Cruz, CA), mouse-monoclonal anti-Arp3 (clone FMS338, Sigma) and mouse-monoclonal anti-tropomyosin (clone TM311, Sigma). In order to reprobe membranes, bound antibodies were removed by incubating the membranes in 62.5 mm Tris-HCl buffer, pH 6.8, containing 100 mm β-mercaptoethanol and 2% SDS for 20 min at 55 °C.
Western blot analysis of F-actin cosedimentation samples was based on IRDye infrared dye technology (Li-COR Biosciences, Lincoln, NE) on an Odyssey Infrared Imaging System (Li-COR Biosciences) following the manufacturer's guidelines. In brief, proteins separated by SDS-PAGE were electrophoretically transferred onto Hybond-C nitrocellulose membranes (GE Healthcare). The blots were blocked in 50% Odyssey buffer (Li-COR Biosciences) in PBS and subsequently incubated with rabbit-polyclonal anti-Flag antibody (Sigma) diluted in 50% Odyssey buffer in PBS containing 0.01% Tween20. For detection of the Flag-epitope, membranes were probed with an IRDye 800CW goat-polyclonal anti-rabbit secondary antibody (Li-COR Biosciences) and signals were detected using an Odyssey Infrared Imaging System.
Statistical Analysis
All data are expressed as means ± standard error of the mean (S.E.). The number of independent experiments is indicated by n. Multiple group analyses were made by one-way analysis of variance (ANOVA) followed by Tukey's or Holm-Sidak post-hoc test as well as by two-way ANOVA with Bonferroni's post-hoc analysis using GraphPad Prism 4 software (GraphPad Software Inc., San Diego, CA). Means were assumed to be significantly different when the p value for the null hypothesis was less than 0.05. The Chi-Square test (χ2) was applied to analyze the distributions of frequencies using Excel. Differences among the distributions were considered significant if p < 0.05. The p value of physical cohesiveness of the Lrrk2 interaction network was calculated as described (33).
RESULTS
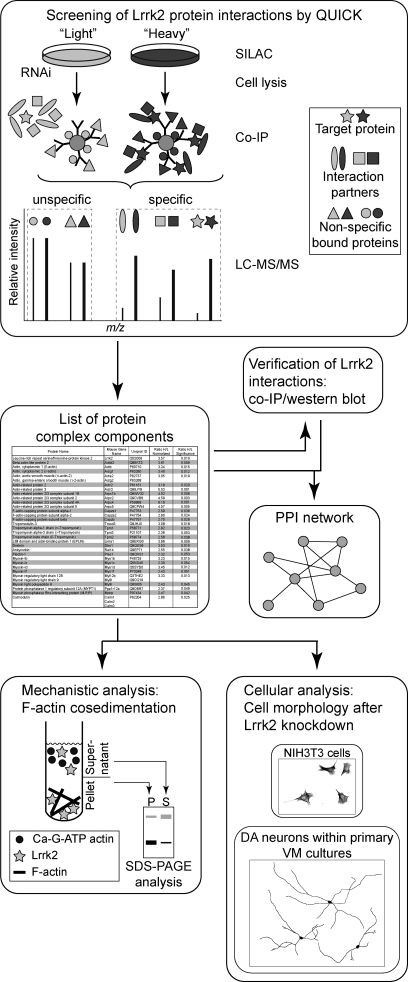
To screen for Lrrk2 PPIs we applied the QUICK approach (22) using NIH3T3 cells, resulting in a list of putative Lrrk2 complex components. A schematic overview of the experimental workflow followed in our study is presented in Fig. 1: a subset of the identified proteins was verified for its interaction with Lrrk2 by Western blot analysis following co-IP. Database-curated interaction data and interactions indicated by the experimental results were used to generate an Lrrk2 PPI network on the basis of the identified Lrrk2 complex partners. The result links Lrrk2 function to the actin-based cytoskeleton. Physiological relevance of the hypothesis was verified by both F-actin cosedimentation assays, determining the ability of Lrrk2 to bind F-actin and to modulate its assembly in vitro, and cellular assays, analyzing the morphology of NIH3T3 cells and DA neurons within primary VM cultures upon lentiviral-mediated knockdown of Lrrk2.
Fig. 1.
Schematic overview of the experimental procedure. The interactome of Lrrk2 was analyzed by QUICK using NIH3T3 cells. A list of putative Lrrk2 complex components was obtained and a selected subset was verified by Western blot analysis following co-IP. The identified Lrrk2 interacting proteins were mapped into a PPI network, integrating interaction data stored in the HPRD and BioGRID database as well as interactions identified through experimental results. The resulting protein network suggests a biological role of Lrrk2 within actin cytoskeletal dynamics. The physiological relevance of the hypothesis was analyzed through biochemical and cellular assays.
Lrrk2 Interacts with Components of the Actin Cytoskeleton
To identify Lrrk2 interacting proteins we conducted the QUICK approach (22), which combines SILAC, RNAi-induced knockdown, co-IP and quantitative MS to screen for endogenous PPIs.
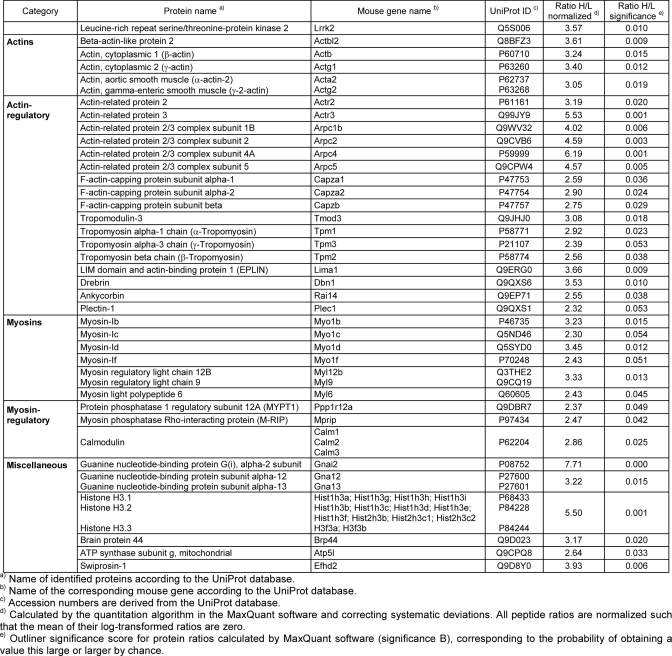
A prerequisite for the QUICK assay is the efficient depletion of the protein of interest within an appropriate cell type. We showed in a previous study that the expression of the Lrrk2-specific second generation shRNAmir construct miB3 in NIH3T3 cells results in an efficient Lrrk2 protein decline without induction of an immune response (24). Thus, corresponding viral supernatants were obtained and NIH3T3 cells were transduced with LVmiB3 1 day after plating. For the metabolic labeling with SILAC, wt and LVmiB3-transduced NIH3T3 cells were grown in “heavy” or “light medium” for at least five passages. Lrrk2 knockdown efficiency was monitored by Western blot analysis using the Lrrk2-specific antibody clone 1E11 (data not shown). To test whether the knockdown of Lrrk2 itself would lead to global changes in the proteome of NIH3T3 cells, whole cell lysates were prefractionated by SDS-PAGE, proteins were subjected to tryptic in-gel cleavage and the resulting peptides were analyzed by LC-MS/MS (Supplemental Table 1). SlLAC quantification did not reveal considerable alterations in the proteome after shRNA expression. Applying the QUICK approach, proteins were immunoprecipitated using a rat-monoclonal anti-Lrrk2 antibody and peptides from the pooled eluates were subjected to LC-MS/MS. Lrrk2 was significantly enriched 3.57-fold in the wt compared with the knockdown condition (p ≤ 0.05, Table I). In addition, we identified 36 proteins with significantly increased abundance ratios (p ≤ 0.05, Table I), which we considered as potential Lrrk2 interaction partners. Moreover, we found 355 proteins being equally abundant in both conditions and therefore considered to be nonspecific contaminants (Supplemental Table 2). By literature-based curation, the potential interactors were categorized according to their molecular function, with the majority of the identified proteins clustered into four groups and merely six of them possessing miscellaneous functions (group 5). Group 1 represents proteins belonging to the actin family, the second group includes actin-regulatory proteins that steer the formation of actin filaments and their organization in structural higher-order networks. Additionally, we identified proteins belonging to the myosin superfamily (group 3) and modulators of these actin-dependent motor proteins that are listed in group 4. Moreover, a KEGG pathway enrichment analysis was performed using Pathway Palette to obtain an unbiased indication of overrepresented cellular process categories in the set of Lrrk2 complex components. The analysis yielded the highest enrichment for proteins annotated to the “regulation of actin cytoskeleton” pathway (KEGG pathway entry #hsa04810), with the by far strongest significance of p < 10−10.35 (Supplemental Fig. 1). In addition, the proteins could be mapped to several other, not necessarily independent pathways whose significance did however not exceed 10−3.6 (“leukocyte transendothelial migration”).
Table I. Summary of Lrrk2 interacting proteins identified by quantitative immunoprecipitation combined with knockdown (QUICK). Proteins are categorized according to their molecular function. Those that could not be differentiated by the MaxQuant software are grouped within the same cell. Ratio H/L: ratio heavy to light labeled peptides.
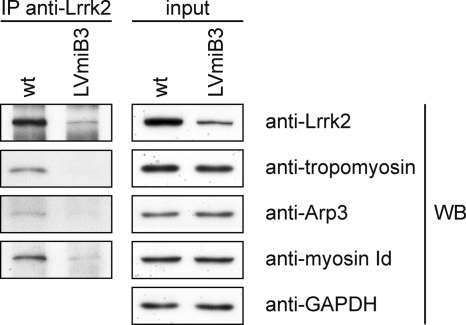
A selection of the identified Lrrk2 protein complex components was confirmed to interact with Lrrk2 by independent co-IP followed by Western blotting (Fig. 2), utilizing cell extracts from wt and LVmiB3-transduced NIH3T3 cells. The analysis with anti-myosin Id, anti-Arp3 and anti-tropomyosin revealed specific signals for wt cells compared with those of shRNA expressing NIH3T3, displaying no or weak bands, and thus confirming the results of the QUICK assay.
Fig. 2.
Verification of Lrrk2 interaction partners identified by QUICK using co-immunoprecipitation. Equal volumes of cell lysates with identical protein amounts from wt and LVmiB3-transduced NIH3T3 were immunoprecipitated using the anti-Lrrk2 antibody cross-linked to Protein G Sepharose. 50% of the immunoprecipitates and a lysate volume corresponding to 2% of the IP-input were separated by SDS-PAGE. Selected Lrrk2 protein complex components found through QUICK were verified by Western blotting using the indicated antibodies. Anti-GAPDH Western blotting assured equal protein loading of the input samples. IP, immunoprecipitation antibody; WB, Western blotting antibody.
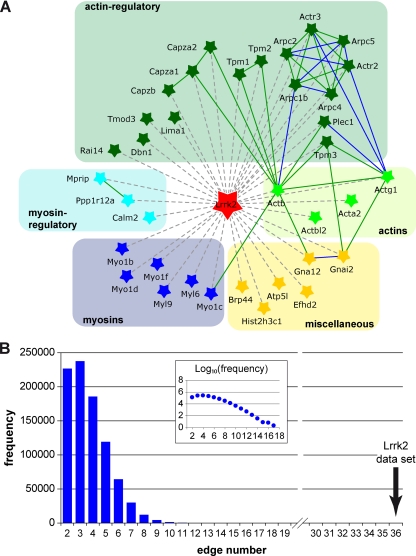
Interactions between the identified Lrrk2 complex partners were parsed and visualized using Pathway Palette in an evaluated and prioritized manner, applying interaction data stored in the HPRD and BioGRID database. Within the group of 37 specific interactors (nodes), the analysis identified 36 previously reported interactions (edges) between 20 individual nodes (Fig. 3A). To evaluate the physical cohesiveness of the inferred interaction network we applied PIE analysis. Fig. 3B shows the frequency distribution of edge numbers within the simulated protein sets (n = 1,000,000 simulated networks, set size 37). The analysis yielded a PIE score of 1.78 and a p value < 10−6 and thus reveals significant physical cohesiveness for the proteins identified within the QUICK assay, meaning that they interact with one another at a significantly higher frequency than a random set of proteins with the same node degree distribution. According to database searches for PPIs, interactions of proteins with Lrrk2 could not be automatically covered in the network. However, on the basis of the experimental results within both the QUICK assay and co-IP experiments, these associations were manually added, resulting in a Lrrk2 interaction network (Fig. 3A) that illustrates the close relationship between the identified proteins. To further analyze the connectivity between the Lrrk2 complex components, the network was expanded by linking pairs of proteins through interconnecting proteins using interaction data from the HPRD and BioGRID database, enabling the mapping of 10 additional proteins from the QUICK set into the network (Supplemental Fig. 2). Fidelity of the network was assessed through manual evaluation of all extensions in a literature-based fashion, with functional information extracted from peer-reviewed publications (Supplemental Table 3). The analysis revealed that the interconnecting neighbors predominantly comprise proteins associated with the actin cytoskeleton and/or affecting its modulation as well as myosins and their regulators. Thus, the extended network strengthens an inherent correlation among the identified Lrrk2 interacting proteins.
Fig. 3.
Network of Lrrk2 interacting proteins. Known interactions among the proteins identified by QUICK were analyzed and visualized by Pathway Palette. The obtained network displays significant physical cohesiveness. A, Out of the 36 proteins identified as specific Lrrk2 interactors, 20 had previously been described as connected through 36 interactions. Interactions of proteins with Lrrk2 were added manually, on the basis of their identification in the QUICK assay and, in part, verification by co-IP experiments. Proteins within the network are depicted by star-shaped nodes and colored according to their molecular function as classified in Table I. Solid lines denote known protein interactions from the HPRD and BioGRID database prioritized and qualified in an evidence-based fashion, referring to the PPI detection method, as reflected by a corresponding color-scheme of the edges: “BioGRID: Low-Throughput” or “HPRD: in vivo” type (green) and “BioGRID: HTP/Complex” or “HPRD: in vitro” type (blue). Manually added interactions are figured as dashed gray lines. Proteins are indicated by their gene names, with their full names shown in Table I. B, Distribution of interactions within a million randomly selected sets of proteins (set size 37) with the same node degree as those of the QUICK set. The graph insert represents the logarithmic depiction of interaction frequencies. The Lrrk2 data set gave significantly more interactions compared with the randomly selected sets.
Lrrk2 Binds F-actin and Affects its Polymerization In Vitro
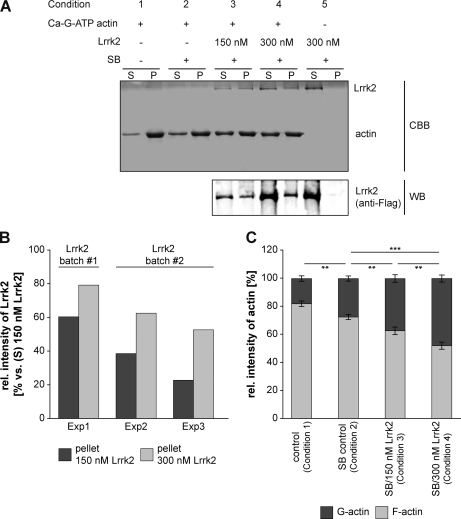
The finding that Lrrk2 interacted with actin isoforms and other cytoskeletal proteins suggested a possible association between Lrrk2 and F-actin. Therefore, we assessed F-actin binding of Lrrk2 in vitro in a cosedimentation assay using recombinant SF-TAP tagged Lrrk2 purified from HEK293 cells. After incubating a constant amount of actin (3 μm) with different concentrations of Lrrk2 (150 nm and 300 nm) under constant buffer conditions, actin was sedimented by centrifugation at steady-state level of polymerization. The pellets and supernatants, containing polymerized and unpolymerized actin, respectively, were analyzed by SDS-PAGE, followed by Coomassie brilliant blue staining (Fig. 4A, upper panel). (Co)sedimentation of Lrrk2 was visualized by Western blotting using an anti-Flag antibody (Fig. 4A, lower panel). Based hereupon, the amount of both Lrrk2 and actin present in the supernatants or pellets was quantified (Fig. 4B and C). In the absence of actin, Lrrk2 was only present in the supernatant, indicating that Lrrk2 did not self-aggregate forming pellets (Fig. 4A, condition 5). Coomassie brilliant blue staining following SDS-PAGE of the purified protein revealed the purity of the affinity preparation (Fig. 4A, upper panel, condition 5) as demonstrated in a previous study (36). In the presence of actin, Lrrk2 localized to the pellet fractions, visualized by Western blotting (Fig. 4A, lower panel, conditions 3 and 4; Fig. 4B). With increasing concentrations of Lrrk2 incubated with actin, we observed a concentration-dependent enhanced recovery of Lrrk2 with F-actin in the pellet fraction (Fig. 4B). Depending on the protein batch, the amount of Lrrk2 in the pellet increased between 1.3- and 2.3-fold using an added concentration of 300 nm versus 150 nm Lrrk2. Lrrk2 led furthermore to a significant increase in actin present in the supernatants, as shown by the quantification of Coomassie stained gels (Fig. 4A and C). Whereas in the control samples the majority of actin sedimented (Fig. 4A, upper panel, conditions 1 and 2; Fig. 4C), the addition of substoichiometric amounts of Lrrk2 decreased the amount of polymerized actin (Fig. 4A, upper panel, conditions 3 and 4; Fig. 4C). As demonstrated in Fig. 4C, the shift in the G-/F-actin ratio toward the monomeric form (in the supernatant) is statistically significant in the presence of increasing Lrrk2 (p < 0.01 and 0.001 versus SB buffer control, n = 4, one-way ANOVA, Holm-Sidak post-hoc test). At a concentration of 300 nm, Lrrk2 induced a ∼1:1 distribution of actin between supernatant (48.00 ± 2.54%) and pellet (52.00 ± 2.54%) (Fig. 4C).
Fig. 4.
Lrrk2 binds directly to F-actin and affects its polymerization/depolymerization in vitro. A, F-actin cosedimentation experiments were performed by incubating 3 μm Ca-G-ATP actin under polymerizing conditions with 150 nm (condition 3) or 300 nm (condition 4) SF-TAP tagged Lrrk2 purified from HEK293 cells in storage buffer (SB). Samples without Lrrk2 and SB (condition 1), with only SB (condition 2) or containing 300 nm Lrrk2 in the absence of actin (condition 5) served as controls. Samples with SB or Lrrk2/SB always contained a constant amount of the storage buffer components ensuring identical conditions. G-actin was allowed to polymerize until equilibrium was reached and sedimented by high speed centrifugation. The pellets (P) and supernatants (S) were analyzed by SDS-PAGE followed by Coomassie brilliant blue staining (upper panel) or Western blot using an anti-Flag antibody (lower panel). The results of a representative experiment are shown. Lrrk2 cosedimented with actin (condition 3 and 4), whereas in the absence of actin, Lrrk2 was only detected in the supernatant (condition 5). CBB: Coomassie brilliant blue; WB: Western blot. B, Relative amounts of Lrrk2 in the pellet upon incubation of 150 or 300 nm Lrrk2 with 3 μm actin for three experiments (Exp1–Exp3) using two different protein purifications (batch#1 and batch #2), based on quantification of Western blots as in A (lower panel). At the higher concentration, an increased amount of cosedimented Lrrk2 was consistently detected. C, Quantification of actin in the supernatant (G-actin) and pellet (F-actin) in the absence of SB (control, condition1), presence of SB (SB control, condition 2) or Lrrk2 (150 or 300 nm) in SB (condition 3 and 4). The addition of Lrrk2 led to a significant shift of actin from the pellet fraction to the supernatant (** p < 0.01, *** p < 0.001, n = 4, one-way ANOVA, Holm-Sidak post-hoc test). Whereas in Lrrk2 deficient samples the majority of actin was detected in the pellet fraction, a decrease in sedimented actin accompanied by a corresponding increase in unpolymerized actin in the supernatant was found in the presence of Lrrk2.
Overall, our results demonstrate an association of Lrrk2 to F-actin. Moreover, under the in vitro conditions used, the G-actin/F-actin balance at equilibrium is altered in favor of G-actin indicating that Lrrk2 affects actin polymerization.
Effect of Lentiviral-Mediated Knockdown of Lrrk2 on NIH3T3 Morphology
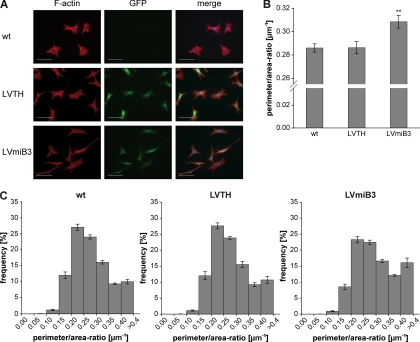
The actin cytoskeleton plays a fundamental role in eukaryotic cells and is a major determinant of cell morphology. The assembly and disassembly of filamentous actin structures provide a driving force for dynamic processes within the cell. Fibroblasts are commonly used to study the mechanisms of actin cytoskeleton reorganization (40, 41). Therefore, we used NIH3T3 cells to investigate whether Lrrk2 depletion affects the cellular morphology. Wt, LVTH- and LVmiB3-transduced NIH3T3 cells were plated on glass coverslips and serum-starved for 24 h. Upon silencing of Lrrk2 the cells displayed a change in morphology in terms of elongation and narrowing of the cell body, whereas the wt and LVTH-transduced controls had a typical fibroblast-like morphology (Fig. 5A). As a quantitative measure of cell shape, we determined the ratio of perimeter to area (P:A-ratio) (38) and calculated both the average P:A-ratio (Fig. 5B) and its frequency distribution (Fig. 5C) for the different conditions. Frequency distribution analysis confirmed the immunocytochemical observations. The P:A-ratio in all three conditions ranged from 0.05 μm−1 to greater than 0.40 μm−1 with the majority of cells falling in the range between 0.20 μm−1 and 0.35 μm−1 (Fig. 5C). However, the distribution of the P:A-ratio revealed a shift toward higher values in the Lrrk2 knockdown cultures (Fig. 5C), indicating that a significant proportion of the cells showed cell body elongation and narrowing. The proportions of cells in the ranges of 0.15–0.2 μm−1 and 0.2–0.25 μm−1 were significantly reduced in the shRNA-treated cultures (p < 0.001 versus wt, n = 4, two-way-ANOVA, Bonferroni post-hoc test; wt: 12.06 ± 1.0% and 27.07 ± 1.0%; LVTH: 12.00 ± 1.3% and 27.59 ± 0.9%; LVmiB3: 8.53 ± 0.8% and 23.28 ± 1.0%), whereas the percentage of cells with a ratio greater than 0.4 μm−1 was significantly increased (p < 0.001 versus wt, n = 4, two-way ANOVA; wt: 10.07 ± 0.7%, LVTH: 10.68 ± 1.1% and LVmiB3: 16.07 ± 1.5%). This resulted in an increase in the average P:A-ratio of NIH3T3 cells expressing the miB3 silencing construct from 0.286 ± 0.004 μm−1 and 0.286 ± 0.005 μm−1 for wt and LVTH-control cells, respectively, to 0.309 ± 0.005 μm−1 for LVmiB3 cells (p < 0.01 versus wt, n = 4, one-way ANOVA, Tukey's post-hoc test) (Fig. 5B). These results denote that NIH3T3 cells undergo marked alterations in cell morphology upon Lrrk2 knockdown.
Fig. 5.
Knockdown of Lrrk2 alters the morphology of NIH3T3 cells. NIH3T3 cells were plated onto 6-well plates and transduced the following day with lentiviral vectors encoding Lrrk2 targeting shRNA (miB3) or the transfer vector LVTH alone as indicated. Five days after transduction, cells were replated at a density of 5%–10% on glass coverslips, serum-starved for 24 h and analyzed. A, To visualize cell shape, cells were immunostained for F-actin. LVmiB3-transduced NIH3T3 cells displayed cell body elongation and narrowing. Scale bar: 50 μm. B, To quantify the observed alterations in cell shape the perimeter to area ratio (P:A-ratio) was determined. Silencing of Lrrk2 resulted in a significant increase of this ratio (** p < 0.01 versus wt, n = 4, one-way ANOVA, Tukey‘s post-hoc test). C, Relative frequency distributions of P:A-ratios for wt, LVTH- or LVmiB3-transduced NIH3T3 cells. Ratios were grouped into consecutive bins increasing by 0.05 μm−1, ranging from 0.05 to > 0.4 μm−1, and were calculated as percentage of cells with a given ratio. The knockdown of Lrrk2 led to a shift of the P:A-ratio toward higher values, denotative for an increase in the amount of cells exhibiting an enhanced P:A-ratio.
Lentiviral-Mediated Knockdown of Lrrk2 in Primary VM Cultures Results in a Significant Decrease in Dopaminergic Neurite Length
On the basis of the pathological association between Lrrk2 and PD and the importance of the actin cytoskeleton in neuronal morphogenesis (42), we investigated the role of endogenous Lrrk2 in neuronal cells. Therefore, we applied the lentiviral-mediated knockdown of Lrrk2 by shRNA expression to primary VM cultures. In addition to miB3 we designed a second, independent shRNAmir construct on the basis of the naturally expressed microRNA-30 precursor, miB4.
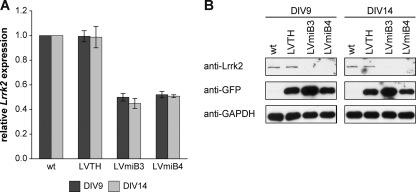
Primary VM cultures derived from E12.5 mouse embryos were transduced with lentiviral vectors encoding the Lrrk2 targeting shRNAs (LVmiB3 and LVmiB4) or the empty vector (LVTH) 1 day after plating and were analyzed at DIV9 or DIV14. Transduction of the VM cultures with LVmiB3 and LVmiB4 resulted in a significant decrease of Lrrk2 mRNA levels relative to wt cultures (p < 0.001, one-way ANOVA) (Fig. 6A) and led to a relative Lrrk2-expression ratio of 0.50 ± 0.03 (LVmiB3, n = 5) and 0.52 ± 0.03 (LVmiB4, n = 3) at DIV9 and 0.45 ± 0.04 (LVmiB3, n = 5) and 0.51 ± 0.01 (LVmiB4, n = 4) at DIV14 (Fig. 6A). The decline in Lrrk2 mRNA levels after expression of the two silencing constructs was accompanied by a corresponding reduction in Lrrk2 protein content (Fig. 6B). We showed in a previous study that the expression of miB3 in cortical cultures does not activate a shRNA-mediated interferon response (24). Likewise, we observed no meaningful induction of Oas1 expression in LVmiB3- and LVmiB4-transduced primary VM cultures (data not shown) hence excluding an immune response, providing accordance with published RNAi guidelines (43, 44).
Fig. 6.
Knockdown of Lrrk2 in primary VM cultures. VM tissue was isolated from E12.5 mouse embryos and cells were plated onto 8-well chamber slides. Cultures were transduced at DIV1 with LVTH, LVmiB3 and LVmiB4 and analyzed at DIV9 or DIV14. A, Semiquantitative real-time PCR displayed reduced Lrrk2 mRNA levels in primary VM cultures transduced with the two independent shRNA-constructs LVmiB3 and LVmiB4. B, Western blot analysis revealed that the transduction with LVmiB3 and LVmiB4 resulted in decreased Lrrk2 protein levels at both time points. GFP and GAPDH Western blots served as controls for transduction efficiency and protein load, respectively.
For the phenotypic analysis of shRNA expressing primary VM cultures, cells were fixed, dopaminergic neurons were visualized by TH-immunohistochemistry and examined by fluorescence microscopy. The transduction with lentiviral vectors encoding Lrrk2 targeting shRNAs or the empty vector alone resulted in a transduction efficiency of TH-ir neurons of about 90%, determined at both DIV9 and DIV14 (data not shown). First, we quantified the amount of TH-ir cells relative to nontransduced wt cultures. At DIV9 no significant alterations in the relative number of TH-ir neurons was observed (p > 0.05 versus wt, n = 4–5, one-way ANOVA; wt: 99.28 ± 1.20%, LVTH: 102.12 ± 3.64%; LVmiB3: 102.16 ± 3.22% and LVmiB4: 97.34 ± 8.37%) (Fig. 7A). As recently as at DIV14, transduction of the cultures with either LVmiB3 or LVmiB4 caused a significant decrease in relative cell counts to 78.38 ± 4.18% and 74.92 ± 5.40%, respectively (p < 0.01 versus wt, n = 3–5, one-way ANOVA) (Fig. 7A). The relative amount of TH-ir neurons in the LVTH control expressing cultures revealed a reduction to 89.46 ± 5.79%, which was not statistically significant (p > 0.05 versus wt, n = 4, one-way ANOVA).
Fig. 7.
Lentiviral-mediated knockdown of Lrrk2 in primary VM cultures leads to an impaired neurite outgrowth of developing DA neurons. Lrrk2 expression in primary VM cultures was depleted by lentiviral delivery of two independent silencing constructs (LVmiB3 and LVmiB4). Non-transduced (wt) or LVTH-transduced cells served as negative controls and cultures were analyzed at DIV9 or DIV14. A, Cell counts of TH-ir cells per well. No differences in the amount of DA neurons was evident at DIV9, whereas the knockdown of Lrrk2 resulted in a decrease in TH-ir cell counts at DIV14 (** p < 0.01 versus wt DIV14, n = 3–5, one-way ANOVA, Tukey‘s post-hoc test). B, Quantification of neurite length of TH-ir neurons. Knockdown of Lrrk2 led to a significant decrease in the average neurite length at DIV9 and DIV14 (*** p < 0.001 versus wt DIV9, n = 4–5, +++ p < 0.001 versus wt DIV14, n = 3–5, one-way ANOVA, Tukey‘s post-hoc test). C, Corresponding frequency distributions of neurite length for the different treatment groups (wt, LVTH, LVmiB3, and LVmiB4) at DIV9 and DIV14. Absolute neurite lengths were grouped into 40 μm bins of increasing size and the percentage of DA neurites of given length was calculated. The expression of miB3 and miB4 resulted at both time points in a shifted distribution toward shorter processes compared with wt and LVTH-control cultures, reflecting an increase in the number of shorter neurites and a decrease of longer neurites.
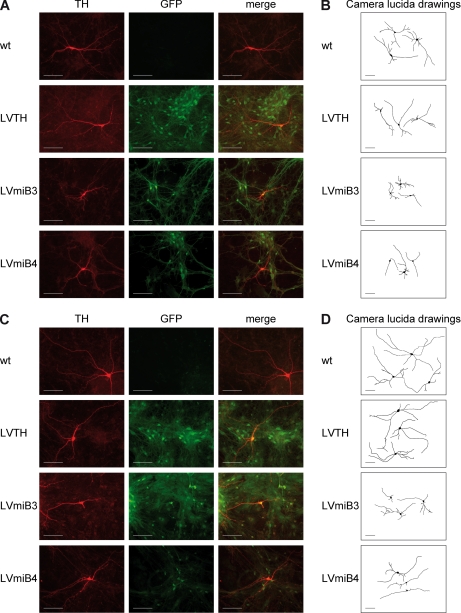
Similar to NIH3T3 cells, we analyzed whether the depletion of Lrrk2 had an effect on the morphology of DA neurons. Fig. 8 shows representative images and camera lucida drawings of untreated wt, LVTH control and Lrrk2 knockdown (LVmiB3 and LVmiB4) cultures at DIV9 and DIV14. Knockdown of Lrrk2 led to shortened neurites as early as at DIV9 (Fig. 8A and B), which was even more pronounced at DIV14 (Fig. 8C and D). Quantitative measurements of the neurite length at DIV9 confirmed the immunocytochemical observations and revealed that the transduction with LVmiB3 led to a significant shift in the neurite length distribution (p < 0.001 versus wt, χ2) to shorter neurites (Fig. 7C). Expression of the LVmiB4 construct led also to a shift toward lower value, although it was not significant (p > 0.05 versus wt, χ2). However, the population of neurites with a length of 40 μm or shorter increased significantly compared with wt cultures (p < 0.001, n = 4–5, two-way ANOVA; wt: 24.04 ± 0.71%, LVmiB4: 33.29 ± 1.01%). Furthermore, the relative amount of processes with a length between 40–80 μm and 80–120 μm decreased from 22.68 ± 0.91% and 22.39 ± 0.81% (wt) to 19.51 ± 1.24% and 19.55 ± 0.89% (LVmiB4) (p < 0.05 versus wt, n = 4–5, two-way ANOVA). The alterations in neurite length distributions were reflected in a significant decrease in the average length of neurite extensions of TH-ir neurons expressing shRNAs targeting Lrrk2 over wt controls (p < 0.001 versus wt, n = 4–5, one-way ANOVA) (Fig. 7B). The outgrowth declined from 90.60 ± 0.98 μm in wt cultures to 58.92 ± 0.58 μm in LVmiB3-transduced and 82.20 ± 1.89 μm in LVmiB4-transduced cultures. In the LVTH control no alterations in the frequency distribution and average neurite length (91.54 ± 1.34 μm) were observed. Neurons cultivated until DIV14 possessed on average significantly longer processes than at DIV9 (p < 0.001 DIV9 versus DIV14, n = 3–5, two-way ANOVA), regardless of whether they expressed shRNA constructs or not (Fig. 7B). However, the average neurite length of LVmiB3- or LVmiB4-transduced TH-ir neurons at DIV14 was still significantly decreased compared with the wt or LVTH condition (p < 0.001 versus wt, n = 3–5, one-way ANOVA; wt: 122.03 ± 1.71 μm, LVTH: 120.45 ± 2.81 μm, LVmiB3: 69.73 ± 1.03 μm, LVmiB4: 103.65 ± 2.35 μm) (Fig. 7B). The corresponding neurite length distributions displayed a shifting to lower values, which was significant for both Lrrk2 targeting shRNA constructs (p < 0.001 LVmiB3 versus wt, p < 0.01 LVmiB4 versus wt, χ2) (Fig. 7C). Thus, depletion of Lrrk2 in primary VM cultures resulted in an impaired outgrowth of developing DA neurites.
Fig. 8.
TH-ir neurons expressing Lrrk2 mRNA targeting shRNAs. Primary VM progenitor cultures were transduced with lentiviruses encoding Lrrk2 silencing shRNA-constructs (LVmiB3 and LVmiB4) or containing the transfer vector (LVTH) alone as indicated. (A and C) DA neurons were visualized using immunofluorescent labeling for tyrosine hydroxylase (TH) (left columns) at DIV9 (A) or DIV14 (C). GFP fluorescence (center columns) indicated successfully transduced neurons whereas the combination of TH-immunostaining and GFP fluorescence (right panel; merge) demonstrated the efficient transduction of DA neurons. (B and D) Camera lucida drawing of representative, randomly selected GFP-positive TH-ir cells at DIV9 (B) and DIV14 (D). Drawings were obtained by manually tracing of the neuron in NeuronJ. TH-ir neurons in miB3- and miB4-expressing cultures exhibited shorter neurites compared with non-transduced (wt) or LVTH-transduced neurons at DIV9 and DIV14. Scale bar: 50 μm.
DISCUSSION
Since the first linkage of Lrrk2 to PD in 2004 (4, 6) much attention has been drawn on the enzymatic activity of Lrrk2 rather than the physiological role of the protein within intact cells and tissues. Thus, knowledge about the endogenous function of Lrrk2 and Lrrk2-mediated signaling pathways is still limited. Studies on the analysis of Lrrk2 protein interactions published so far are based on either yeast 2-hybrid screens and/or the overexpression of the tagged protein (13, 21, 45–47). However, several drawbacks are linked to these approaches (22). To identify specific interactors of Lrrk2, we applied QUICK (22) and identified 36 potential Lrrk2 complex partners (Table I) whose vast majority is functionally linked to the actin cytoskeleton, its rearrangement, dynamics, and maintenance as indicated by both literature-based curation and KEGG pathway enrichment analysis. Filamentous actin superstructures are essential for a wide variety of cell properties and function in the generation and maintenance of cell morphology and polarity, in endocytosis and intracellular trafficking, in cell division, contractility and motility, including neuronal outgrowth (48). Spatial organization and dynamic assembly of different actin filamentous structures is controlled by a large number of actin-regulatory proteins performing specific recognition and/or catalytic yet coordinated functions (49–51). Beside some actin isoforms, we found a number of proteins that are known to modulate actin dynamics (49, 52–57) as Lrrk2 interactors. In addition, we also identified members of the myosin I and II family and modulators of these actin-dependent motor proteins. Myosins generally use actin filaments as tracks along which they move, thereby promoting contractility or cell motility and vesicular transport (58), but their involvement in the assembly/disassembly (59, 60) and remodeling of actin filaments (61) has also been shown.
The assembled PPI network based on the putative Lrrk2 complex partners identified in the study (Fig. 3A) proved to be significantly enriched in previously validated interactions, as curated by the HPRD and BioGRID database (Fig. 3B). The significant physical cohesiveness of the Lrrk2 complex partners reflects their biological coherence and functional interdependence thereby reinforcing the validity of the remaining, novel Lrrk2 interactions implied by the experimental results. Thus, the inferred Lrrk2 PPI network, which links Lrrk2 to proteins that are involved in the assembly of actin-based structures, indicates that the function of Lrrk2 is related to the actin cytoskeleton and its dynamic regulation. This is further supported by the extension of the network through the incorporation of first-order shared neighbors, revealing a high degree of connectivity between the Lrrk2 interacting proteins by intermediate proteins (Supplemental Fig. 2). A functional evaluation of the extensions in a literature-based fashion demonstrated that the interconnecting proteins are predominantly closely associated with the actin cytoskeleton, its modulation and/or the actin-dependent myosins (Supplemental Table 3), affirming the network's fidelity. Therefore, Lrrk2 protein complex partners and consequently Lrrk2 itself can be mapped into an evolutionary and functionally conserved PPI network regulating the actin-based cytoskeleton. This is indicative for a role of Lrrk2 in actin-based biological processes and in line with previous studies that suggest an association of Lrrk2's biological function with the actin cytoskeleton (62–64). Although the study by Habig and colleagues reports that the knockdown of Lrrk2 results in the differential expression of genes involved in actin cytoskeleton signaling in SH-SY5Y (63), the knockdown does not affect the proteome of NIH3T3 cells as shown here. However, these findings are not mutually exclusive as investigations on the transcriptome and proteome analyze two different levels of a biological system. Divergent mRNA and protein expression levels have often been described, being most likely the result of posttranscriptional regulatory mechanisms (65, 66). Thus, beside integrating genomic and proteomic data sets, their meaningful interpretation requires functional annotation and consideration in the context of biological pathways and functional processes (66). Accordingly, Habig et al. as well as our study ascribe the same cellular role to Lrrk2 based on their experimental findings, namely its association with actin cytoskeleton-based mechanisms.
The suggested functional connection of Lrrk2 with the actin cytoskeleton is further emphasized by the ability of affinity purified Lrrk2 to associate with F-actin as demonstrated in vitro (Fig. 4). Moreover, under the stoichiometric conditions tested in vitro, Lrrk2 decreased the amount of polymerized actin at steady state, thus affecting the G-actin/F-actin ratio in favor of the monomer. These data may indicate that Lrrk2 itself possesses at least a basal regulatory activity on actin polymerization as a direct modulator. In the in vitro context, it might have either monomer sequestration or, given its substoichiometric activity, capping activity. Further analysis is needed to clarify the mechanism underlying the observed effects of Lrrk2 on the G-/F-actin ratio. Given the cellular abundance of actin, it also needs consideration that the Lrrk2/actin stoichiometry used in vitro (1/10 to 1/20) may be higher than one can expect in vivo, although local Lrrk2 to actin ratios in cells may be sufficiently high at actin filaments or, considering the observed association of Lrrk2 to membranous structures (67, 68), close to membranes. It cannot also be excluded that a modified form of Lrrk2 (e.g. phosphorylated) is more active in exerting its modulating function within the cell. Alternatively, in conjunction with its identified interactors Lrrk2 may act as passive user of the microfilament system by scaffolding/cooperating with the actin-regulatory and motor proteins. Taken together, Lrrk2 can be placed in the protein network whose mutual interactions govern the spatiotemporal localization and activity of the actin cytoskeleton.
As the physiological function of the actin cytoskeleton comprises the generation and maintenance of cell morphology, alterations in cell shape are a consequence of modifications in the organization of cytoskeletal components. We demonstrated that the depletion of Lrrk2 in NIH3T3 cells by RNAi leads to significant morphological alterations (Fig. 5), reflecting a general perturbation of normal cytoskeletal organization. Given that Lrrk2-associated PD is characterized by the degeneration of DA neurons in the substantia nigra pars compacta (3) as in idiopathic PD, we assayed the physiological relevance of Lrrk2 in DA neurons. The depletion of Lrrk2 in primary VM cultures derived from embryonic mice resulted in a decrease of the average neurite length in developing DA neurons due to an impaired neurite outgrowth (Figs. 7 and 8). Although it was reported that off-target effects following the expression of shRNAs in rat hippocampal cultures can alter the neuronal morphology by innate antiviral response pathways (69) this option can be excluded as the delivery of shRNA expressing constructs in our approach lacked the induction of Oas1 expression. Moreover, Bauer et al. (24) demonstrated that the induction of an interferon response due to the delivery of shRNAs is associated with a significant decline of neurons, rarefaction of neurites as well as fragmented and condensed nuclei. In contrast, the neurites of DA neurons expressing shRNA targeting Lrrk2 mRNA displayed outgrowth that, though diminished compared with the wt, continued from DIV9 to DIV14 (Fig. 7B). This reveals further that the neurites were rather subjected to reduced outgrowth ability than to a retraction process. Additionally, the relative amount of DA neurons in our study remained unaltered upon Lrrk2 silencing at DIV9. A physiological relevance of Lrrk2 on neurite outgrowth was also demonstrated by others. Gillardon recently showed that neurons derived from Lrrk2-deficient mouse ES cells also exhibit significantly shorter neurites than wt neurons (70). However, a converse effect of Lrrk2 on neurite length was observed after transduction of rat cortical neurons with Lrrk2 shRNA vectors that led to an increase in neurite length (71). This discrepancy is difficult to reconcile. One point that might contribute to these conflicting results is the difference in cell culture namely primary progenitor VM cultures and ES cell derived neurons from mouse versus cortical cultures derived from P1 rat brains. Besides the two different animal models that were used, the developmental stage of the neurons represented by the cellular systems also differs. Both the progenitor VM cultures and the ES cell derived neurons mimic the prenatal period of neuronal development whereas the cortical cultures represent the postnatal phase.
Besides its critical role for a wide range of fundamental cellular processes, actin-based cytoskeletal dynamics is crucial for the development of the nervous system. Migration of neurons as well as extension of neurites relies on an organized actin polymerization (72), actin-regulatory proteins and myosins (50). The effects of Lrrk2 knockdown observed in cellulo may be supported by the identified interactors as well as by the link between Lrrk2 and actin filaments found in vitro, collectively indicative for an impact of Lrrk2 on cytoskeleton integrity mediated by variations in actin-based dynamics. Lrrk2 being placed in a functional network of proteins required for a proper actin network formation during neurite outgrowth by the in vitro studies and the concomitant observed decrease in neurite length of developing DA neurons after Lrrk2 knockdown moreover suggest that Lrrk2 acts as a modulator of actin dynamics during neurite extension.
Neurite outgrowth is the result of numerous extracellular stimuli targeting the actin cytoskeleton of each individual neuron. Small Rho GTPases are essential components that link these stimuli to actin but their neuronal downstream effectors binding to actin or responsible for scaffolding are largely unknown (73). Lrrk2 being a GTPase dependent kinase, harboring both protein interaction domains as well as enzymatic domains, may be involved in such processes and serves as a scaffold protein within signaling pathways controlling the dynamic actin cytoskeleton organization. Several LRR-containing proteins and members of the ROCO protein superfamily participate in the functional assembly of the actin network (53, 74–77) making Lrrk2 not the first member of this family to be associated with components of the cytoskeleton.
Clearly, further research has to determine the physiological role of Lrrk2 in actin-based cytoskeleton processes. Because Lrrk2 is a multi-domain protein, comprising in addition to its GTPase and kinase activity several protein interaction domains that are likely to facilitate various PPIs, different biochemical functions can be accomplished by Lrrk2. This view is supported by several studies demonstrating that Lrrk2 is involved in various, not necessarily independent, physiological processes including kinase signaling, vesicular transport, regulation of microtubules, cytoskeleton assembly and the chaperone system (78). The proteomic approach taken here renders new insights on how Lrrk2 molecularly interacts with the cytoskeletal machinery controlling actin turnover. A direct influence on actin filament dynamics is observed in vitro although in cells Lrrk2 may also, and maybe mainly, act as connector or scaffold for associated actin-regulatory and motor proteins and thereby regulating both actin filament assembly and stability. Indeed, on the cellular level, loss of Lrrk2 influences the generation and/or maintenance of cell morphology in non-neuronal cells and interferes with neurite outgrowth as well as neurite maintenance of DA neurons. Consistent with previous studies (63, 64) our results suggest a biological function of Lrrk2 in actin-related processes, whose impairment may underlie PD-associated neurodegeneration.
Acknowledgments
We thank Stephanie Schöffmann, Felix von Zweydorf, and Davy Waterschoot for excellent technical assistance, Dr Hakan Sarioglu, Silke Becker, and Sandra Helm from the Core Facility Proteomics, Helmholtz Zentrum München, for MS analysis and Dr Andrea Huber-Brösamle for helpful discussions.
Footnotes
* This work was supported by the NGFN2 SMP Proteomics (FKZ: 01GR0449, subproject 9), NGFN-Plus (FKZ: 01GS08140, subproject 12), the German Federal Ministry of Education and Research (BMBF: QuantPro, FKZ: 0316865A and DYNAMO, FKZ: 0315513A), the Initiative and Networking Fund of the Helmholtz Association (Helmholtz Alliance for Mental Health in an Aging Society) (HelMA, FKZ: HA-215, topic 3 WP11), the EU grant SYSCILIA (HEALTH-F5–2010-241955), FWO grant G.0441.10 (to CA), BOF-grant 01J04806 (to MVT), the Dana-Faber Cancer Institute (to JAM and MA) and the National Human Genome Research Institute (P50HG004233; to JAM and MA).
 This article contains supplemental Files 1–2, Figs. 1–2 and Tables 1–3.
This article contains supplemental Files 1–2, Figs. 1–2 and Tables 1–3.
1 The abbreviations used are:
- PD
- Parkinson's disease
- ANOVA
- analysis of variance
- co-IP
- co-immunoprecipitation
- DA
- dopaminergic
- DIV
- days in vitro
- F-actin
- filamentous actin
- G-actin
- globular actin
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- GFP
- green fluorescent protein
- Lrrk2
- leucine-rich repeat kinase 2
- Oas1
- 2′-5′-oligoadenylate synthetase
- P:A ratio
- perimeter to area ratio
- PIE
- physical interaction enrichment
- PPI
- protein-protein interaction
- QUICK
- quantitative immunoprecipitation combined with knockdown
- RNAi
- RNA interference
- SB
- storage buffer
- shRNA
- short hairpin RNA
- SILAC
- stable isotope labeling by amino acids in cell culture
- TH
- tyrosine hydroxylase
- TH-ir
- TH-immunoreactive
- VM
- ventral mesencephalon
- wt
- wildtype.
REFERENCES
- 1. Elbaz A. (2008) LRRK2: bridging the gap between sporadic and hereditary Parkinson's disease. Lancet Neurol. 7, 562–564 [DOI] [PubMed] [Google Scholar]
- 2. Paisán-Ruiz C. (2009) LRRK2 gene variation and its contribution to Parkinson disease. Hum. Mutat. 30, 1153–1160 [DOI] [PubMed] [Google Scholar]
- 3. Wszolek Z. K., Pfeiffer R. F., Tsuboi Y., Uitti R. J., McComb R. D., Stoessl A. J., Strongosky A. J., Zimprich A., Müller-Myhsok B., Farrer M. J., Gasser T., Calne D. B., Dickson D. W. (2004) Autosomal dominant parkinsonism associated with variable synuclein and tau pathology. Neurology 62, 1619–1622 [DOI] [PubMed] [Google Scholar]
- 4. Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R. J., Calne D. B., Stoessl A. J., Pfeiffer R. F., Patenge N., Carbajal I. C., Vieregge P., Asmus F., Müller-Myhsok B., Dickson D. W., Meitinger T., Strom T. M., Wszolek Z. K., Gasser T. (2004) Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44, 601–607 [DOI] [PubMed] [Google Scholar]
- 5. Taylor J. P., Mata I. F., Farrer M. J. (2006) LRRK2: a common pathway for parkinsonism, pathogenesis and prevention? Trends Mol. Med. 12, 76–82 [DOI] [PubMed] [Google Scholar]
- 6. Paisán-Ruíz C., Jain S., Evans E. W., Gilks W. P., Simón J., van der Brug M., López de Munain A., Aparicio S., Gil A. M., Khan N., Johnson J., Martinez J. R., Nicholl D., Carrera I. M., Pena A. S., de Silva R., Lees A., Martí-Massó J. F., Pérez-Tur J., Wood N. W., Singleton A. B. (2004) Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron 44, 595–600 [DOI] [PubMed] [Google Scholar]
- 7. Higashi S., Moore D. J., Colebrooke R. E., Biskup S., Dawson V. L., Arai H., Dawson T. M., Emson P. C. (2007) Expression and localization of Parkinson's disease-associated leucine-rich repeat kinase 2 in the mouse brain. J. Neurochem. 100, 368–381 [DOI] [PubMed] [Google Scholar]
- 8. Taymans J. M., Van den Haute C., Baekelandt V. (2006) Distribution of PINK1 and LRRK2 in rat and mouse brain. J. Neurochem. 98, 951–961 [DOI] [PubMed] [Google Scholar]
- 9. Han B. S., Iacovitti L., Katano T., Hattori N., Seol W., Kim K. S. (2008) Expression of the LRRK2 gene in the midbrain dopaminergic neurons of the substantia nigra. Neurosci. Lett. 442, 190–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Higashi S., Biskup S., West A. B., Trinkaus D., Dawson V. L., Faull R. L., Waldvogel H. J., Arai H., Dawson T. M., Moore D. J., Emson P. C. (2007) Localization of Parkinson's disease-associated LRRK2 in normal and pathological human brain. Brain Res. 1155, 208–219 [DOI] [PubMed] [Google Scholar]
- 11. Bosgraaf L., Van Haastert P. J. (2003) Roc, a Ras/GTPase domain in complex proteins. Biochim. Biophys. Acta 1643, 5–10 [DOI] [PubMed] [Google Scholar]
- 12. Mata I. F., Wedemeyer W. J., Farrer M. J., Taylor J. P., Gallo K. A. (2006) LRRK2 in Parkinson's disease: protein domains and functional insights. Trends Neurosci. 29, 286–293 [DOI] [PubMed] [Google Scholar]
- 13. Gandhi P. N., Chen S. G., Wilson-Delfosse A. L. (2009) Leucine-rich repeat kinase 2 (LRRK2): a key player in the pathogenesis of Parkinson's disease. J. Neurosci. Res. 87, 1283–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gloeckner C. J., Schumacher A., Boldt K., Ueffing M. (2009) The Parkinson disease-associated protein kinase LRRK2 exhibits MAPKKK activity and phosphorylates MKK3/6 and MKK4/7, in vitro. J. Neurochem. 109, 959–968 [DOI] [PubMed] [Google Scholar]
- 15. Qing H., Wong W., McGeer E. G., McGeer P. L. (2009) Lrrk2 phosphorylates alpha synuclein at serine 129: Parkinson disease implications. Biochem. Biophys. Res. Commun. 387, 149–152 [DOI] [PubMed] [Google Scholar]
- 16. Gloeckner C. J., Kinkl N., Schumacher A., Braun R. J., O'Neill E., Meitinger T., Kolch W., Prokisch H., Ueffing M. (2006) The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum. Mol. Genet. 15, 223–232 [DOI] [PubMed] [Google Scholar]
- 17. West A. B., Moore D. J., Biskup S., Bugayenko A., Smith W. W., Ross C. A., Dawson V. L., Dawson T. M. (2005) Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. U.S.A. 102, 16842–16847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. West A. B., Moore D. J., Choi C., Andrabi S. A., Li X., Dikeman D., Biskup S., Zhang Z., Lim K. L., Dawson V. L., Dawson T. M. (2007) Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum. Mol. Genet. 16, 223–232 [DOI] [PubMed] [Google Scholar]
- 19. Smith W. W., Pei Z., Jiang H., Dawson V. L., Dawson T. M., Ross C. A. (2006) Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat. Neurosci. 9, 1231–1233 [DOI] [PubMed] [Google Scholar]
- 20. Greggio E., Cookson M. R. (2009) Leucine-rich repeat kinase 2 mutations and Parkinson's disease: three questions. ASN Neuro. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ding X., Goldberg M. S. (2009) Regulation of LRRK2 stability by the E3 ubiquitin ligase CHIP. PLoS One 4, e5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Selbach M., Mann M. (2006) Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK). Nat. Methods 3, 981–983 [DOI] [PubMed] [Google Scholar]
- 23. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 24. Bauer M., Kinkl N., Meixner A., Kremmer E., Riemenschneider M., Förstl H., Gasser T., Ueffing M. (2009) Prevention of interferon-stimulated gene expression using microRNA-designed hairpins. Gene Ther. 16, 142–147 [DOI] [PubMed] [Google Scholar]
- 25. Bauer M., Szulc J., Meyer M., Jensen C. H., Terki T. A., Meixner A., Kinkl N., Gasser T., Aebischer P., Ueffing M. (2008) Delta-like 1 participates in the specification of ventral midbrain progenitor derived dopaminergic neurons. J. Neurochem. 104, 1101–1115 [DOI] [PubMed] [Google Scholar]
- 26. Liu W. Z., Boucias D. G., McCoy C. W. (1995) Extraction and characterization of the insecticidal toxin hirsutellin A produced by Hirsutella thompsonii var. thompsonii. Exp. Mycol. 19, 254–262 [DOI] [PubMed] [Google Scholar]
- 27. Gloeckner C. J., Boldt K., Ueffing M. (2009) Strep/FLAG tandem affinity purification (SF-TAP) to study protein interactions. Curr. Protoc. Protein Sci. Chapter 19, Unit 19. 20 [DOI] [PubMed] [Google Scholar]
- 28. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 29. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 30. Askenazi M., Li S., Singh S., Marto J. A. (2010) Pathway Palette: a rich internet application for peptide-, protein- and network-oriented analysis of mass spectrometry data. Proteomics 10, 1880–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mishra G. R., Suresh M., Kumaran K., Kannabiran N., Suresh S., Bala P., Shivakumar K., Anuradha N., Reddy R., Raghavan T. M., Menon S., Hanumanthu G., Gupta M., Upendran S., Gupta S., Mahesh M., Jacob B., Mathew P., Chatterjee P., Arun K. S., Sharma S., Chandrika K. N., Deshpande N., Palvankar K., Raghavnath R., Krishnakanth R., Karathia H., Rekha B., Nayak R., Vishnupriya G., Kumar H. G., Nagini M., Kumar G. S., Jose R., Deepthi P., Mohan S. S., Gandhi T. K., Harsha H. C., Deshpande K. S., Sarker M., Prasad T. S., Pandey A. (2006) Human protein reference database–2006 update. Nucleic Acids Res. 34, D411–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stark C., Breitkreutz B. J., Reguly T., Boucher L., Breitkreutz A., Tyers M. (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34, D535–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sama I. E., Huynen M. A. (2010) Measuring the physical cohesiveness of proteins using Physical Interaction Enrichment (PIE). Bioinformatics [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kanehisa M., Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gloeckner C. J., Boldt K., Schumacher A., Roepman R., Ueffing M. (2007) A novel tandem affinity purification strategy for the efficient isolation and characterisation of native protein complexes. Proteomics 7, 4228–4234 [DOI] [PubMed] [Google Scholar]
- 36. Gloeckner C. J., Boldt K., von Zweydorf F., Helm S., Wiesent L., Sarioglu H., Ueffing M. (2010) Phospho-peptide analysis reveals two discrete clusters of phosphorylation in the N-terminus and the Roc domain of the Parkinson-disease associated protein kinase LRRK2. J Proteome Res. 9, 1738–1745 [DOI] [PubMed] [Google Scholar]
- 37. Spudich J. A., Watt S. (1971) The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 38. Chevallier J., Koop C., Srivastava A., Petrie R. J., Lamarche-Vane N., Presley J. F. (2009) Rab35 regulates neurite outgrowth and cell shape. FEBS Lett. 583, 1096–1101 [DOI] [PubMed] [Google Scholar]
- 39. Simon P. (2003) Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 19, 1439–1440 [DOI] [PubMed] [Google Scholar]
- 40. Castets M., Schaeffer C., Bechara E., Schenck A., Khandjian E. W., Luche S., Moine H., Rabilloud T., Mandel J. L., Bardoni B. (2005) FMRP interferes with the Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Hum. Mol. Genet. 14, 835–844 [DOI] [PubMed] [Google Scholar]
- 41. Vardouli L., Moustakas A., Stournaras C. (2005) LIM-kinase 2 and cofilin phosphorylation mediate actin cytoskeleton reorganization induced by transforming growth factor-beta. J. Biol. Chem. 280, 11,448–11,457 [DOI] [PubMed] [Google Scholar]
- 42. Luo L. (2002) Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu. Rev. Cell Dev. Biol. 18, 601–635 [DOI] [PubMed] [Google Scholar]
- 43. Editorial in Nature Cell Biology (2003) Whither RNAi? Nat. Cell Biol. 5, 489–490 [DOI] [PubMed] [Google Scholar]
- 44. Cullen B. R. (2006) Enhancing and confirming the specificity of RNAi experiments. Nat. Methods 3, 677–681 [DOI] [PubMed] [Google Scholar]
- 45. Gandhi P. N., Wang X., Zhu X., Chen S. G., Wilson-Delfosse A. L. (2008) The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. J. Neurosci. Res. 86, 1711–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ko H. S., Bailey R., Smith W. W., Liu Z., Shin J. H., Lee Y. I., Zhang Y. J., Jiang H., Ross C. A., Moore D. J., Patterson C., Petrucelli L., Dawson T. M., Dawson V. L. (2009) CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 2897–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sancho R. M., Law B. M., Harvey K. (2009) Mutations in the LRRK2 Roc-COR tandem domain link Parkinson's disease to Wnt signalling pathways. Hum. Mol. Genet. 18, 3955–3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pollard T. D., Cooper J. A. (2009) Actin, a central player in cell shape and movement. Science 326, 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. dos Remedios C. G., Chhabra D., Kekic M., Dedova I. V., Tsubakihara M., Berry D. A., Nosworthy N. J. (2003) Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 83, 433–473 [DOI] [PubMed] [Google Scholar]
- 50. Ishikawa R., Kohama K. (2007) Actin-binding proteins in nerve cell growth cones. J Pharmacol Sci 105, 6–11 [DOI] [PubMed] [Google Scholar]
- 51. Lambrechts A., Van Troys M., Ampe C. (2004) The actin cytoskeleton in normal and pathological cell motility. Int. J. Biochem.. Cell Biol. 36, 1890–1909 [DOI] [PubMed] [Google Scholar]
- 52. Cooper J. A. (2002) Actin dynamics: tropomyosin provides stability. Curr. Biol. 12, R523–525 [DOI] [PubMed] [Google Scholar]
- 53. Fischer R. S., Fowler V. M. (2003) Tropomodulins: life at the slow end. Trends Cell Biol. 13, 593–601 [DOI] [PubMed] [Google Scholar]
- 54. Fontao L., Geerts D., Kuikman I., Koster J., Kramer D., Sonnenberg A. (2001) The interaction of plectin with actin: evidence for cross-linking of actin filaments by dimerization of the actin-binding domain of plectin. J. Cell Sci. 114, 2065–2076 [DOI] [PubMed] [Google Scholar]
- 55. Ishikawa R., Hayashi K., Shirao T., Xue Y., Takagi T., Sasaki Y., Kohama K. (1994) Drebrin, a development-associated brain protein from rat embryo, causes the dissociation of tropomyosin from actin filaments. J. Biol. Chem. 269, 29,928–29,933 [PubMed] [Google Scholar]
- 56. Maul R. S., Song Y., Amann K. J., Gerbin S. C., Pollard T. D., Chang D. D. (2003) EPLIN regulates actin dynamics by cross-linking and stabilizing filaments. J. Cell Biol. 160, 399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mullins R. D., Heuser J. A., Pollard T. D. (1998) The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. U.S.A. 95, 6181–6186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Winder S. J., Ayscough K. R. (2005) Actin-binding proteins. J. Cell Sci. 118, 651–654 [DOI] [PubMed] [Google Scholar]
- 59. Haviv L., Gillo D., Backouche F., Bernheim-Groswasser A. (2008) A cytoskeletal demolition worker: myosin II acts as an actin depolymerization agent. J. Mol. Biol. 375, 325–330 [DOI] [PubMed] [Google Scholar]
- 60. Osherov N., May G. S. (2000) In vivo function of class I myosins. Cell Motil. Cytoskeleton 47, 163–173 [DOI] [PubMed] [Google Scholar]
- 61. Vicente-Manzanares M., Ma X., Adelstein R. S., Horwitz A. R. (2009) Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10, 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Alegre-Abarrategui J., Christian H., Lufino M. M., Mutihac R., Venda L. L., Ansorge O., Wade-Martins R. (2009) LRRK2 regulates autophagic activity and localises to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet 18, 4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Häbig K., Walter M., Poths S., Riess O., Bonin M. (2008) RNA interference of LRRK2-microarray expression analysis of a Parkinson's disease key player. Neurogenetics 9, 83–94 [DOI] [PubMed] [Google Scholar]
- 64. Jaleel M., Nichols R. J., Deak M., Campbell D. G., Gillardon F., Knebel A., Alessi D. R. (2007) LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem. J. 405, 307–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tian Q., Stepaniants S. B., Mao M., Weng L., Feetham M. C., Doyle M. J., Yi E. C., Dai H., Thorsson V., Eng J., Goodlett D., Berger J. P., Gunter B., Linseley P. S., Stoughton R. B., Aebersold R., Collins S. J., Hanlon W. A., Hood L. E. (2004) Integrated genomic and proteomic analyses of gene expression in Mammalian cells. Mol. Cell. Proteomics 3, 960–969 [DOI] [PubMed] [Google Scholar]
- 66. Waters K. M., Pounds J. G., Thrall B. D. (2006) Data merging for integrated microarray and proteomic analysis. Brief Funct. Genomic Proteomic 5, 261–272 [DOI] [PubMed] [Google Scholar]
- 67. Biskup S., Moore D. J., Celsi F., Higashi S., West A. B., Andrabi S. A., Kurkinen K., Yu S. W., Savitt J. M., Waldvogel H. J., Faull R. L., Emson P. C., Torp R., Ottersen O. P., Dawson T. M., Dawson V. L. (2006) Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann. Neurol. 60, 557–569 [DOI] [PubMed] [Google Scholar]
- 68. Hatano T., Kubo S., Imai S., Maeda M., Ishikawa K., Mizuno Y., Hattori N. (2007) Leucine-rich repeat kinase 2 associates with lipid rafts. Hum. Mol. Genet. 16, 678–690 [DOI] [PubMed] [Google Scholar]
- 69. Alvarez V. A., Ridenour D. A., Sabatini B. L. (2006) Retraction of synapses and dendritic spines induced by off-target effects of RNA interference. J. Neurosci. 26, 7820–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gillardon F. (2009) Leucine-rich repeat kinase 2 phosphorylates brain tubulin-beta isoforms and modulates microtubule stability–a point of convergence in Parkinsonian neurodegeneration? J. Neurochem. 110, 1514–1522 [DOI] [PubMed] [Google Scholar]
- 71. MacLeod D., Dowman J., Hammond R., Leete T., Inoue K., Abeliovich A. (2006) The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron 52, 587–593 [DOI] [PubMed] [Google Scholar]
- 72. Meyer G., Feldman E. L. (2002) Signaling mechanisms that regulate actin-based motility processes in the nervous system. J. Neurochem. 83, 490–503 [DOI] [PubMed] [Google Scholar]
- 73. Luo L. (2000) Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1, 173–180 [DOI] [PubMed] [Google Scholar]
- 74. Abysalh J. C., Kuchnicki L. L., Larochelle D. A. (2003) The identification of pats1, a novel gene locus required for cytokinesis in Dictyostelium discoideum. Mol. Biol. Cell 14, 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bosgraaf L., Russcher H., Smith J. L., Wessels D., Soll D. R., Van Haastert P. J. (2002) A novel cGMP signalling pathway mediating myosin phosphorylation and chemotaxis in Dictyostelium. EMBO J. 21, 4560–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Davy D. A., Ball E. E., Matthaei K. I., Campbell H. D., Crouch M. F. (2000) The flightless I protein localizes to actin-based structures during embryonic development. Immunol. Cell Biol. 78, 423–429 [DOI] [PubMed] [Google Scholar]
- 77. Kuo J. C., Lin J. R., Staddon J. M., Hosoya H., Chen R. H. (2003) Uncoordinated regulation of stress fibers and focal adhesions by DAP kinase. J. Cell Sci. 116, 4777–4790 [DOI] [PubMed] [Google Scholar]
- 78. Anand V. S., Braithwaite S. P. (2009) LRRK2 in Parkinson's disease: biochemical functions. FEBS J. 276, 6428–6435 [DOI] [PubMed] [Google Scholar]