SUMMARY
DNA ligase IV (LigIV) is critical for non-homologous end-joining (NHEJ), the major DNA double-strand break (DSB) repair pathway in human cells, and LigIV activity is regulated by XRCC4 and XLF (XRCC4-like factor) interactions. Here, we employ X-ray scattering (SAXS) data to characterize three-dimensional arrangements in solution for full-length XRCC4, XRCC4 in complex with LigIV tandem BRCT domains and XLF, plus the XRCC4·XLF·BRCT2 complex. XRCC4 forms tetramers mediated through a head-to-head interface and the XRCC4 C-terminal coiled-coil region folds back on itself to support this interaction. The interaction between XLF and XRCC4 is also mediated via head-to-head interactions. In the XLF·XRCC4·BRCT complex, alternating repeating units of XLF and XRCC4·BRCT place the BRCT domain on one side of the filament. Collective results identify XRCC4 and XLF filaments suitable to align DNA molecules and function to facilitate LigIV end joining required for DSB repair in vivo.
INTRODUCTION
DNA double strand breaks (DSBs) are a highly cytotoxic form of DNA damage, which if not repaired efficiently and/or accurately can lead to genome instability or cell death (Jackson and Bartek, 2009). DSBs are formed in response to naturally occurring cellular processes such as collapsed replication forks, as well as V(D)J and class switch recombination and are also induced by exposure to exogenous agents such as ionizing radiation (IR) and topoisomerase poisons (Lieber, 2008). In human cells, the major pathway for the repair of IR-induced DSBs is Non-Homologous End-Joining (NHEJ) (Lieber, 2010; Mahaney et al., 2009). The initial steps in NHEJ are detection of the DSB and protection of the DNA ends by the Ku70/80 heterodimer. This is followed by recruitment of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), which undergoes autophosphorylation that regulates pathway progression (Hammel et al., 2010). In subsequent steps, non-ligatable end groups and damaged termini are removed or repaired by processing enzymes that may include Artemis, polynucleotide kinase/phosphatase (PNKP), and/or DNA polymerases. The final step in NHEJ is DNA ligation, which is carried out by DNA ligase IV (LigIV) that exists in complex with the scaffolding protein XRCC4 (reviewed in (Lieber, 2010; Mahaney et al., 2009)). Ligation is regulated by XRCC4-like factor (XLF, also called Cernunnos), which interacts with XRCC4 (Ahnesorg et al., 2006; Buck et al., 2006).
LigIV is a member of the ATP-dependent DNA ligase family and functions exclusively in NHEJ (Ellenberger and Tomkinson, 2008). The C-terminal region of LigIV contains two breast and ovarian cancer susceptibility protein C-terminal (BRCT) domains separated by a short spacer (a tandem BRCT domain) that is required for interaction with XRCC4 (Critchlow et al., 1997). XRCC4 and LigIV form a stable complex and XRCC4 both stabilizes and stimulates the activity of LigIV (Grawunder et al., 1997; Modesti et al., 1999). XRCC4 also interacts with XLF (Ahnesorg et al., 2006; Buck et al., 2006), which stimulates the activity of LigIV towards non-compatible DNA ends in vitro (Gu et al., 2007; Tsai et al., 2007) by promoting re-adenylation of LigIV (Riballo et al., 2009). In addition, XRCC4 interacts with DNA (Modesti et al., 1999) and with the end-processing enzyme, PNKP (Koch et al., 2004). Thus, XRCC4 is a multifaceted protein that plays a critical and essential role in NHEJ. Defining the architectural and dynamic nature of XRCC4 interactions with its partner molecules is critical to understanding the regulation of the NHEJ complex and mechanisms of DSB repair.
XRCC4 is composed of a head domain (~ residues 1-119), an elongated alpha helical stalk (~residues 120-180) and a C-terminal region (~residues 180-334) of unknown function. It exists predominantly as a dimeric form that is mediated primarily by interaction of the two head domains (Junop et al., 2000). In addition, XRCC4 forms homo-multimers (dimers of dimers or tetramers) as well as higher order structures or filaments; however, the orientation of individual dimers within higher order multimers is a matter of debate (Callebaut et al., 2006; Dahm, 2008; Junop et al., 2000; Leber et al., 1998; Modesti et al., 2003; Recuero-Checa et al., 2009). The stalk domain of XRCC4 mediates both tetramerization and interaction with the BRCT domain of LigIV, and these interactions are mutually exclusive (Modesti et al., 2003; Sibanda et al., 2001; Wu et al., 2009). XRCC4 also interacts with DNA in a protein-concentration and DNA length-dependent manner (Modesti et al., 1999), and it has been suggested that XRCC4 filaments align DNA molecules to facilitate end-joining (Andres et al., 2007; Lu et al., 2007; Modesti et al., 1999).
XLF also consists of a globular head domain, an elongated coil-coil stalk and a disordered C terminal region; however, unlike XRCC4, the coiled-coil region doubles back on itself, positioning the C terminal region towards the head domains (Andres et al., 2007; Li et al., 2008). Also like XRCC4, XLF forms homodimers and tetramers (Andres et al., 2007; Callebaut et al., 2006; Hentges et al., 2006) through head domain/head domain interactions and binds DNA in a length and concentration dependent manner (Hentges et al., 2006; Lu et al., 2007). Mutational analysis suggests that the head domains of XRCC4 and XLF are required for the interaction between XLF and XRCC4 and with the XRCC4-LigIV complex (Andres et al., 2007; Deshpande and Wilson, 2007); yet, the structural arrangements within this complex are poorly understood.
The recent crystal structure of a truncated form of XRCC4 (residues ~1-203) in complex with the tandem BRCT domain of LigIV (Wu et al., 2009) provides important information on the interaction interface between both molecules. However, so far, only truncated forms of XRCC4 and XLF lacking their C-terminal domains have been successfully crystallized (Andres et al., 2007; Junop et al., 2000; Li et al., 2008), and only low-resolution (~30Å) EM negative stain reconstructions of full-length XRCC4 in complex with the tandem BRCT domain of LigIV have been obtained (Recuero-Checa et al., 2009). This likely reflects internal flexibility and/or conformational heterogeneity in the free or complexed state of full length XRCC4 and XLF, especially within the C terminal regions of both proteins, which have been predicted to be disordered (Junop et al., 2000; Li et al., 2008).
Such protein flexibility presents a major obstacle for classical structural methods such as X-ray crystallography, and alternative approaches are required to provide structural information on flexible molecules and dynamic complexes in solution. In recent years, small angle X-ray scattering (SAXS) has emerged as a fundamental tool for the study of biological molecules in solution (Putnam et al., 2007, Hura et al., 2009, Rambo and Tainer, 2010a). Here, we used SAXS to analyze the structure of full length XRCC4 alone and in complex with the LigIV tandem BRCT domains and with XLF. We generated truncations of XRCC4 to elucidate the structural properties of XRCC4 dimers and multimers as well as the functionality of its C-terminal region. We show that the XRCC4 C-terminus undergoes conformational changes upon BRCT binding and have characterized three-dimensional rearrangements of the XLF·XRCC4·BRCT complex. Combining information from known crystal structures with SAXS allowed us to construct a model for the three-dimensional atomic structures of the XLF·XRCC4·BRCT assembly. The resulting integrated information on assembly and conformational changes has specific physiological implications for the mechanism of XLF·XRCC4·Ligase IV in the ligation of DSBs.
RESULTS
The XRCC4 C-terminal region supports N-terminal domain interactions forming tetramers and filament-like structures
The XRCC4 high-resolution structure, spanning residues 1-203, displays a dimeric state with a tight interface between the N-terminal globular structure and an exposed coil-coil motif (Junop et al., 2000; Sibanda et al., 2001). However, the structure and function of the XRCC4 C-terminal region is unknown. To define the dynamic structure of full-length human XRCC4 and the role of the C-terminal domain, we employed SAXS (Putnam et al., 2007) combined with sample re-purification using size exclusion column chromatography (SEC) (Rambo and Tainer, 2010b). Pair distribution functions (P(r)) calculated from SAXS profiles of XRCC4 over 1-3 mg/ml indicate that the protein exists in a protein concentration dependent equilibrium of different oligomerization states in solution (Figure S1A). To characterize different oligomerization states, samples were re-purified prior to SAXS collection by a combination of SEC and multiangle light scattering (MALS), which allowed us to determine polydispersity and Mw for each fraction (Figure 1A-C). The maximal dimension (Dmax) of XRCC4 in the small molecular weight fraction (Mw MALS~90kDa, Mw SAXS~75kDa) was ~170Å (Figure 1C). Calculated envelopes show the position of the bulky head domains, while the wide protrusion most probably belongs to the XRCC4 C-terminal region (Figure 1D).
Figure 1. The XRCC4 C-terminal region supports the XRCC4-head to head interface.
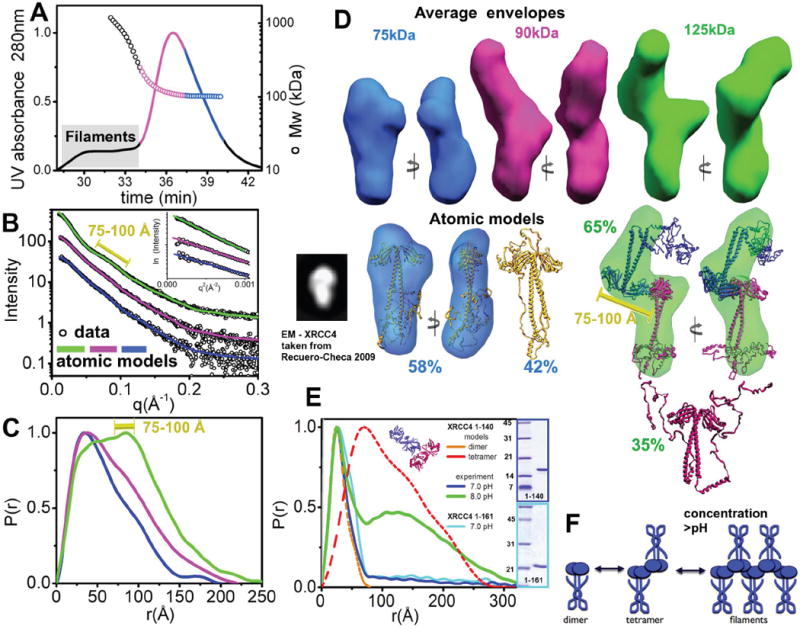
(A) SEC-MALS chromatograph of XRCC4 at 5.0 mg/ml. Solid lines represent the UV absorbance at 280nm and symbols represent molar mass vs. elution time. Magenta and blue regions represent the fractions collected for further SAXS measurement. Blue, SEC fraction of the XRCC4-dimer; magenta, the XRCC4 dimer-tetramer mixture at <1 mg/ml. Elution of filaments is indicated by the gray box.
(B) Experimental scattering profiles of the SEC fractions. Green, XRCC4 at 3.0 mg/ml without prior SEC preparation. This sample was filtrated with 1 MDa cut-off filter to eliminate filaments. Magenta and blue are as panel A. The theoretical scattering of the final MES models for XRCC4 (blue line, χ2=1.4), and a multi-component model of XRCC4 dimer-tetramer mixtures are shown (magenta line, χ2=1.4, 11% tetramer 89%.dimer; green line, χ2=1.8, 65% tetramer, 35% dimer). Inset - The Guinier plot with linear fit (violet line) in the limit qRG>1.3. The peak-distance at the 75-100Å interval is indicated (yellow bar).
(C) P(r) of XRCC4 assemblies computed from the experimental SAXS data shown in the same colors as panel B. The P(r) functions are normalized to unity at their maxima. The peak-distance at the 75-100Å interval is indicated (yellow bar).
(D) Top- Two views of the average SAXS envelopes of the XRCC4 assemblies colored as in panel B.
Middle - MES - Atomic models for XRCC4 dimers (gold) and tetramers (blue-red) along with their respective percentages. The major conformer is superimposed with the average envelope and rotated by 90°. The black inset shows the XRCC4 dimer described in a recent EM study (Recuero-Checa et al., 2009). The 75-100Å distance observed in the SAXS data is indicated (yellow bar).
(E) P(r) functions calculated for the SAXS curve of XRCC4 1-140 at 5.4 mg/ml and pH 7.0 (blue) or 8.0 (green) and XRCC4 1-161 at 5.4 mg/ml and pH 7.0 (light blue). Note the area at r>80Å corresponds to the residual amount of XRCC4 filaments formed through the head-to-head interaction. The theoretical P(r) for XRCC4 1-140 dimers and tetramers (cartoon model) are shown in orange and red, respectively. Inset - SDS-PAGE analysis of the XRCC4 1-140 and XRCC4 1-161 samples.
(F) Cartoon describing formation of XRCC4 at high protein concentration.
See also Figure S1.
To further examine the arrangement of the XRCC4 dimer and possible conformations of the C-terminal region, we employed rigid-body modeling (Pelikan et al., 2009) using rigid helices identified by secondary structure prediction (Figure S1B). Comparison of models with experimental data shows that the C-terminal region is in close contact with the coil-coil motif, with possible interactions with the N-terminal head domain (Figure 1D). Although the secondary structure prediction suggests that the C-terminal region of XRCC4 is disordered, a recent study using a de novo algorithm predicted that this region of mouse-XRCC4 exhibited globular characteristics (Recuero-Checa et al., 2009). However, an XRCC4 model with a globular C-terminus does not match the SAXS data (χ2~8), indicating that the globular conformation of the XRCC4 C-terminus likely does not exist in solution (Figure S1C). Although our SAXS data leads to a different model from that previously proposed (Recuero-Checa et al., 2009), the EM 2D projection of the mouse XRCC4 showed similar features to our SAXS reconstructions (Figure 1D). Notably, the folded back helical stalk indicated from our SAXS data is reminiscent of the structure of XLF (Andres et al., 2007; Li et al., 2008).
Given the predicted conformational disorder of the C-terminal region (Figure S1B), we reasoned that the coexistence of different conformations could contribute to the experimental SAXS profile. We therefore applied a Minimal Ensemble Search (MES) in which a genetic algorithm is used to identify the minimal ensemble of conformers required to best fit the experimental data (Bernado et al., 2007; Pelikan et al., 2009). Assembly of two MES-selected conformers slightly improved the fit to the experimental data (χ2=1.4 vs. χ2=1.8). Consistent with the earlier analysis, both conformers showed the C-terminal region folded back onto and twisted around the coil-coil motif (Figure S1CD).
We next examined the structural arrangements of XRCC4 tetramers in the collected higher molecular weight fraction (Figure 1A - magenta) and a filtrated sample, which had been passed through a filter without SEC (termed simple filtration, Figure 1BCD - green). Interestingly, SEC-MALS analysis of the higher molecular weight fraction showed a broad signal between 100 and >200 kDa suggesting polydispersity and indicating that the XRCC4 tetramer is in equilibrium with dimers and higher order oligomers. This is consistent with the previously determined high dissociation constant for XRCC4 tetramers (KD=22 μM) (Junop et al., 2000). The P(r) for the higher molecular weight fraction shows broadening with a distinct shoulder, indicating the presence of tetramers. The P(r) shoulder transforms into a maxima at r=75-100Å for the simply filtrated sample and indicates a larger fraction of tetramers and possible larger oligomers (Figure 1C). Calculated envelopes show the location of the XRCC4-dimer with the head domains in the central part of the envelope and the coil-coil motifs pointing outwards at the opposite extremity (Figure 1D). The regions corresponding to the coil-coil motif are more distinct for the simply filtrated samples (Figure 1BD), consistent with these samples containing a larger fraction of tetramers. These results demonstrate that the interaction of XRCC4-dimers within the tetramer is mediated through the head domains. The formation of larger oligomers observed at higher protein concentrations (>3.0mg/ml, Figure 1A and S1A) is consistent with repeated head-head complexation in the XRCC4 filaments (Figure 1E). Moreover, manual superposition of the XRCC4 tetrameric arrangement found in the crystal lattice (Junop et al., 2000) closely matches the SAXS envelopes (Figure 1D).
To further examine this XRCC4-tetramer arrangement and the conformation of the C-terminal regions, we used rigid body modeling, as described above. The reconstructed atomic model gave an excellent match to the experimental data (χ2 =1.8), indicating a back-folded C-terminus similar to that observed in the dimeric state. A recently proposed model in which tetramers were formed via interactions between the coil-coil regions (Dahm, 2008; Junop et al., 2000; Modesti et al., 2003) does not agree with our experimental data, suggesting that the “coil-coil” tetramer does not exist in solution (Figure S1D).
To investigate the role of the C-terminal region in the formation of tetramers and filaments, we examined scattering results of XRCC4 truncations: XRCC4 1-140 and XRCC4 1-161. These constructs predominantly formed dimers in solution at protein concentrations approximately 4 times higher than the concentration at which the full length XRCC4 formed tetramers. Only a small proportion of the protein existed in the filament form at this concentration under standard buffer conditions (pH 7.0) (Figure 1E), and a higher proportion of filaments were observed at pH 8.0, as indicate by larger distances in the P(r) function (Figure 1E). A similar effect of pH was observed for full length XRCC4 (Figure S1E). These results indicate weak head-head interactions for the truncated XRCC4 and stabilization of this interaction by the C-terminal region in the full-length protein.
Together, our results are consistent with the C-terminal region of XRCC4 (residues 141-334) being important for XRCC4 tetramer formation. These analyses provide structural insights into full-length XRCC4, showing folding back of the disordered C-terminal region, similar to that observed in XLF, and a head-to-head interaction between XRCC4 dimers in tetramers and filaments that forms at high protein concentrations (Figure 1E and S1A).
LigIV tandem BRCT domains destabilize XRCC4 filaments
LigIV has the unique ability to catalyze ligation in NHEJ, and its function requires physical interaction with XRCC4 (Critchlow et al., 1997; Grawunder et al., 1997). Within the XRCC4·LigIV complex, interactions map to the central coil-coil region of XRCC4 and to the linker region between the two BRCT domains (Grawunder et al., 1998; Sibanda et al., 2001). The recent high-resolution structure of XRCC4 in complex with the LigIV BRCT domains reveals an extensive binding interface formed by a clamp-shaped helix-loop-helix motif spanning both BRCT1 and BRCT2 domains (Wu et al., 2009). To test the effects of the LigIV-BRCT domains on XRCC4 structural arrangements in solution, we combined XRCC4 with an excess of the tandem BRCT domain and examined scattering results of the re-purified XRCC4·BRCT assembly (Figure 2A).
Figure 2. The LigIV tandem BRCT domain destabilizes XRCC4 filaments.
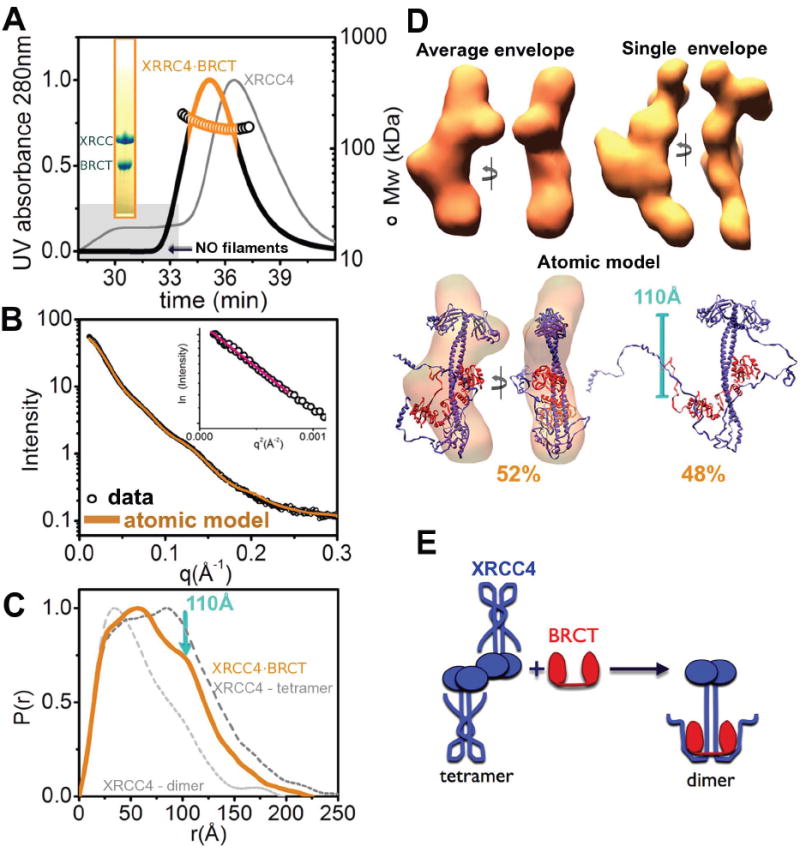
(A) SEC-MALS chromatograph of the XRCC4·BRCT complex (black) compared to XRCC4 alone (gray). Solid lines represent the UV at 280nm and symbols represent molar mass vs. elution time. The orange region represents the fraction collected for SAXS measurements. Note the absence of filaments for the XRCC4·BRCT complex (gray box). Inset - SDS-PAGE analysis of the peak fraction.
(B) Experimental scattering profiles for the collected XRCC4·BRCT fraction at ~1.0 mg/ml. The theoretical scattering (orange line, χ2=2.4) from the MES-model as shown in panel D. Inset - The Guinier plot with linear fit (red line).
(C) P(r) of the XRCC4·BRCT complex (orange) computed from the experimental SAXS data compared to the P(r) obtained for XRRC4-dimers and tetramers (light and dark gray, respectively). The P(r) functions are normalized to unity at their maxima. The average distance of the XRCC4 head domain to the BRCT domain (~110Å) is indicated.
(D) Top- Two views of the average and representative single SAXS envelopes of the XRCC4·BRCT complex.
Bottom - MES atomic models for XRCC 4·BRCT along with their respective percentages. The main conformer is superimposed on the average envelope and rotated by 90°. The distance observed in the SAXS data is indicated.
(E) Cartoon illustrating disruption of the XRCC4-tetramer upon BRCT complexation.
See also Figure S2.
MALS of the SEC peak fraction, in which both proteins co-eluted (Figure 2A), shows a low polydispersity peak with Mw~120 kDa, revealing the formation of an XRCC4·BRCT complex with a molar ratio of 2:1 (XRCC4:BRCT). The calculated P(r) function had a Dmax~220Å and differed from XRCC4 alone in the presence of a new maxima at r~110Å, which corresponds to the average distance between the head domains of XRCC4 and the BRCT domains (Figure 2CD). SAXS reconstructions reveal a protrusion in the central coil-coil region of XRCC4 and show BRCT complex formation. The broad-tail in the P(r) function (r=125-220Å) generates a slight bend in the extension relative to the central coil-coil region and is consistent with straightening and kinking of the XRCC4 coil-coil region observed upon BRCT complex formation (Wu et al., 2009). These SAXS results suggest further extension and stabilization of the C-terminal helices upon BRCT complex formation. The reconstructed atomic model closely matches the experimental data (Figure 2B) and shows displacement of the C-terminal helical stalks from the coil-coil region (Figure 2DE).
To determine how the BRCT domain affects XRCC4 filament conformation, we incubated XRCC4 with different molar ratios of the BRCT domain, and SAXS data was collected without further purification. The narrowing of the P(r) function indicates accumulation of the XRCC4·BRCT complex and disruption of XRCC4 filaments (Figure S2). This straightening of the XRCC4 C-terminal region by BRCT interaction leads to displacement of C-terminal helical stalks from the coil-coil region and blocks stabilization of the head-head interface between two XRCC4-dimers (Figure 2E).
Dynamic nature of XLF·XRCC4 interactions
XLF is the most recently identified member of the NHEJ family of proteins. XLF and XRCC4 interact in solution with an affinity of 7.8μM and mutational analysis suggests that K63, K65, K99, of XRCC4 and L115 of XLF are involved in mediating a head-head interface (Andres et al., 2007); however, structural arrangements within the complex are unknown.
To examine the structure of the XRCC4·XLF complex, we first purified XLF 1-248 (referred to hereafter as XLF) in complex with the XRCC4 head domain (XRCC4 1-140). The SEC-MALS peak, where both proteins co-elute (Figure 3A), shows a broad signal between 100 and >200 kDa suggesting polydispersity and indicating that the XRCC4 1-140·XLF complex is dynamic in nature. We next examined scattering results from the fraction collected at the maxima of the SEC-MALS peak; however, repeated SEC of the collected peak fraction indicated that the complex underwent re-equilibration (Figure S3A). The SEC-MALS showed sample heterogeneity; however, distinct peaks at 80Å and 160Å in the SAXS profile, the P(r) function and the rod-like P(r) function with Dmax~200Å (Figure 3BC) allowed us to generate low-resolution models. However, SAXS envelopes calculated for two different concentrations of XLF·XRCC4 were slightly different (Figure S3D), likely reflecting heterogeneity of the sample. To further define the XLF·XRCC4 arrangement we carried out rigid-body modeling in combination with MES (Pelikan et al., 2009). Based on previous mutational analysis (Andres et al., 2007), we positioned XLF L115 into the XRCC4-head groove formed by helices α2, α3 and beta-sheets β6, β7, thus localizing all interaction-required residues (K63, K65, K99) (Figure 3D). We performed an exhaustive search for the best-fit conformation by rotating and translating the XLF molecule in close proximity to the XRCC4 groove. The best-fit model contained two XLF homodimers interacting with the XRCC4 grooves (χ2=4.1). However, a mixture of tetramer, hexamer, as well as octamer assemblies improved the match further, giving an excellent fit (χ2=1.9), consistent with the dynamic nature of the XLF·XRCC4 interaction (Figure 3DE, model 1). The best-fit model showed a tilt angle of ~45° between XRCC4 and XLF coil-coil regions (Figure 3DE). We also generated an alternative model 2 with a similar XRRC4·XLF interface but with 0° tilt angle between the coil-coil regions. The selected ensemble of oligomers model showed significantly worse fit (model 2, χ2= 2.7), suggesting that this arrangement is not accurate (Figure 3DE). We also tested the previously proposed model (Andres et al., 2007) in which the XLF head interacted with the lower region of the XRCC4 head domain. The calculated ensemble of this oligomeric state also gave a very poor fit (model 3, χ2=7.3) (Figure 3E).
Figure 3. Formation of XLF ·XRCC4 filaments.
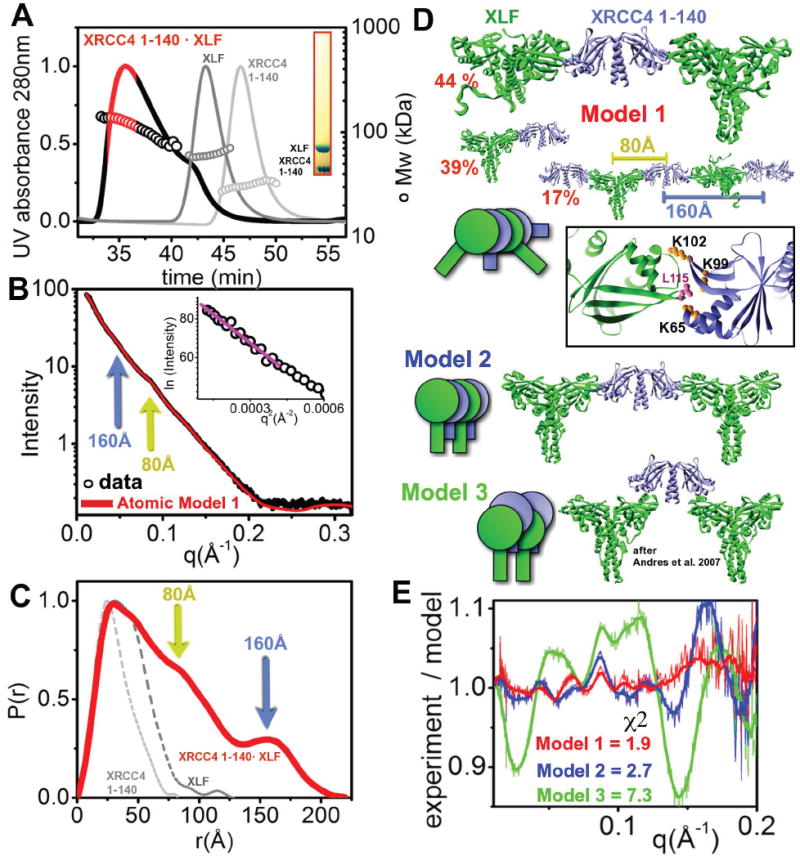
(A) SEC-MALS chromatograph of XLF·XRCC4 1-140 (black) compared to XLF and XRCC4 (1-140) (gray, light gray). Solid lines represent the UV at 280nm, and the symbols represent molar mass vs. elution time. The red region represents the fraction collected for SAXS measurement. Inset - SDS-PAGE analysis of the fraction.
(B) Experimental scattering profiles of the collected XLF·XRCC4 (1-140) fraction at 1.0 mg/ml. The theoretical scattering (red line) from the final MES-model shown in panel D (χ2=2.4). Two distinct spacings at 80Å and 160Å (yellow and blue arrow) are identical for experimental and theoretical profiles. Inset - The Guinier plot with linear fit (violet line).
(C) P(r) of XLF·XRCC4 1-140 (red) in comparison to XLF and XRCC4 1-140 (gray, and light gray respectively). The P(r) functions are normalized to unity at their maxima and distances at 80Å and 160Å are indicated.
(D) (top) Ensemble for atomic model 1 of XLF·XRCC4 1-140 along with respective percentages. The distances (80 and 160Å) observed in the SAXS data are indicated. Zoom-in of the reconstructed XLF·XRCC4 1-140 complex showing the interaction-interface and residues K65, K99 and K102 of XRRC4 and L115 of XLF. Model 2 and model 3 are shown as cartoons describing overall arrangements.
(E) Fit for the MES model 1 (shown in panel D), model 2 (tetramer 45%, hexamer 25 %, octamer 30%) and model 3 (tetramer 30%, hexamer 0 %, octamer 70%). The fits are shown as residuals.
See also Figure S3.
Consistent with previous results of mutational analysis (Wu et al., 2009), our model for the XRCC4·XLF head-head interface places XRCC4 residues K65 and K99 in direct interaction with XLF residue L115 (Figure 3D). Furthermore, no binding of the BRCT domain with XRCC4 1-140·XLF was observed (Figure S3), consistent with the BRCT domain interacting exclusively with the XRCC4 coil-coil motif which is not present in the XRCC4 1-140 construct.
Next we examined full-length XRCC4 in complex with XLF. Similar to the complex between truncated XRCC4 (1-140) and XLF, SEC showed a broad signal, indicating that the XRCC4·XLF complex is also dynamic in nature (Figure 4A). Distinct peaks at 80Å and 160Å in the SAXS profile and P(r) function also indicate that complexes formed with full-length and truncated XRCC4 adopt a similar arrangement. Higher peak intensities in the higher protein concentration samples indicates the formation of longer oligomers that contain increasingly frequent repeat units at 80Å and 160Å (Figure 4C). Calculated SAXS envelopes reveal that the protrusion from the rod-like shape is consistent with the presence of the XRCC4 coil-coil region in the full length construct, however heterogeneity of the samples did not allow us to discern more detailed features (Figure S4A). Similar to the truncated complex, we performed MES analysis of the atomic models. Selected ensembles using a model in which the XRCC4·XLF-arrangement was identical to its truncated complex (Figure 4D) provided an excellent match (model 1, χ2=1.8) to the SAXS profile (Figure 4DE). The alternative model with parallel coil-coil regions gave a similar fit (model 2, χ2=1.7) with different ratios of oligomeric states in the selected ensemble (Figure S4B). The similarity between the fit of the two different models can be explained by the fact that the flexible XRCC4 C-terminus and the larger sample heterogeneity delivered SAXS profiles with fewer features. These profiles could be fitted with two different ensembles equally well (Putnam et al., 2007). However, the model ensemble in which the XLF head group interacted with the lower region of the XRCC4 head domain gave a poor fit (model 3, χ2=4.7) (Figure 4E).
Figure 4. Overall arrangement of the full length XRCC4·XLF filament.
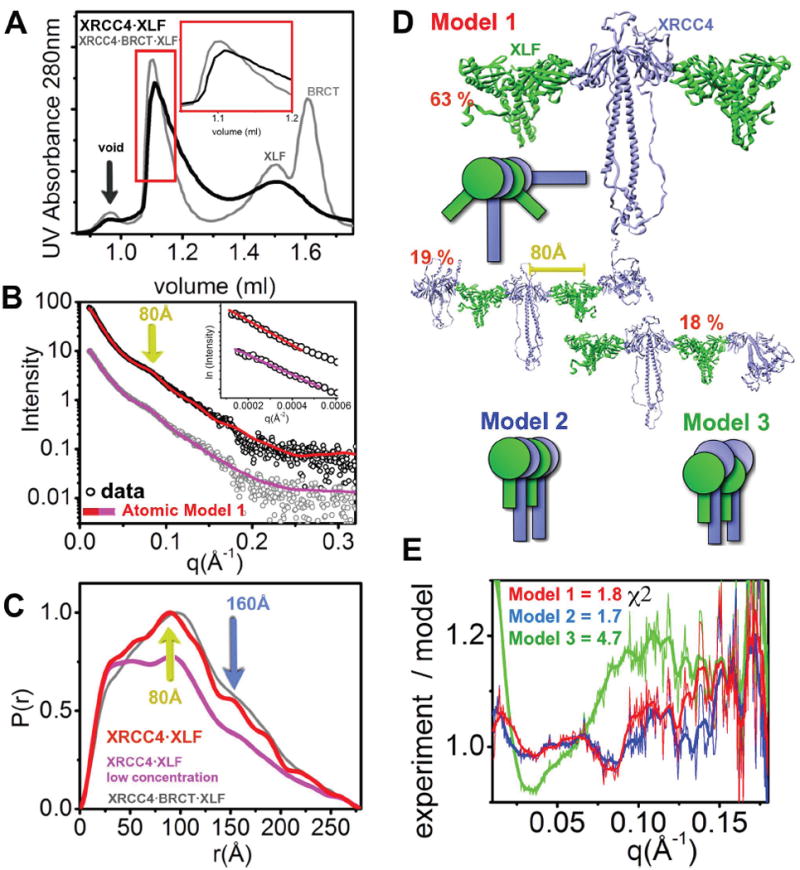
(A) SEC chromatograph of XLF·XRCC4 (black) compared to XLF·XRCC4·BRCT (gray). The red region represents the fraction collected for SAXS measurement.
(B) Experimental scattering profiles of the collected XLF·XRCC4 peak fraction at ~1.0 mg/ml (red) and after dilution to 0.5 mg/ml (violet). The theoretical scattering from the final MES-model shown in panel D is shown as a red (undiluted) or violet (diluted) line matching the experimental data (χ2=1.8, undiluted; χ2=1.3, diluted sample). Two distinct spacing at 80Å (yellow arrow) is identical for experimental and theoretical profiles. Inset - The Guinier plot with linear fit (violet line).
(C) P(r) of XLF·XRCC4 (red for undiluted and violet for dilution sample) in comparison to XRCC4·BRCT·XLF (gray). The P(r) functions are normalized to unity at their maxima. Two distinct distances at 80Å (yellow arrow) and 160Å (blue arrow) are indicated.
(D) Ensemble for atomic model 1 along with respective percentages. Model 2 and model 3 are shown as cartoons describing overall arrangements.
(E) Fits for the MES models shown in panel D and Figure S4B. The fits are shown as residuals.
See also Figure S4.
XLF tethers XRCC4·BRCT and forms an extended filament
To determine the structural arrangements of the XRCC4·XLF·BRCT assembly, we examined the complex of full-length XRCC4, XLF and BRCT after re-purification on SEC-MALS. The SEC-MALS peak, where all three proteins co-elute (Figure 5A), shows a broad signal between 150 and 200kDa similar to that observed for XRCC4·XLF and indicates the dynamic nature of the complex. Analysis of the SAXS profile obtained for the SEC peak fraction showed concentration dependence. The diluted fraction (1mg/ml) (Figure 5 BC - magenta) shows an anisotropic particle with a Dmax~280Å (Figure 5C).
Figure 5. Dynamic nature of the XLF·XRCC4·BRCT filament.
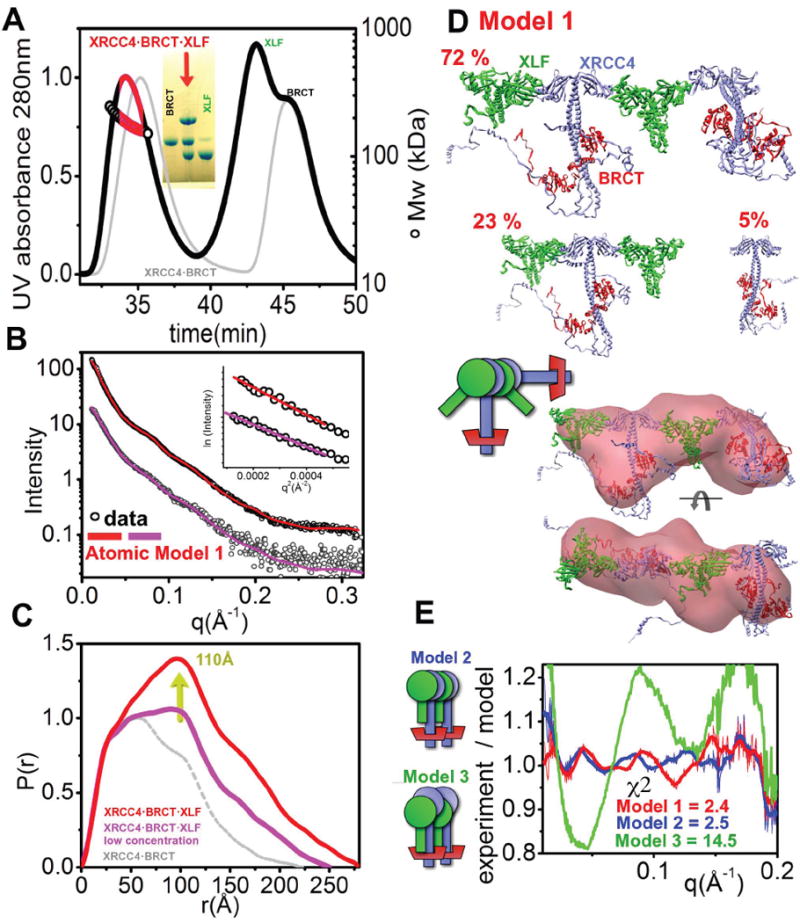
(A) SEC-MALS chromatograph of XLF·XRCC4·BRCT (black) in comparison to XRCC4·BRCT (gray). Solid lines represent the UV at 280nm, and the symbols represent molar mass. The red region represents the fraction collected for SAXS measurements. Inset - SDS-PAGE analysis of the peak-fractions.
(B) Experimental scattering profiles of the collected XLF·XRCC4·BRCT fraction diluted to 1.0 mg/ml (violet) and the peak-fraction at ~3.0 mg/ml (red). The theoretical scattering from the final MES-models (shown in panel D) is shown as a solid line matching the experimental data (χ2=2.4 for the undiluted peak fraction, red; and χ2=1.3 for the diluted peak fraction, violet). Inset - The Guinier plot with linear fit (violet line).
(C) P(r) of XLF·XRCC4·BRCT calculated for the diluted fraction (violet) and undiluted peak-fraction (red) in comparison to XRCC4·BRCT (gray). The P(r) functions are normalized to unity at their first maxima r= 30Å. The average distance of the XRCC4 head domain to the BRCT domain (~110Å) is indicated by the yellow arrow.
(D) Top- MES-atomic models of XLF·XRCC4·BRCT calculated for the diluted sample shown along with their respective percentages.
bottom- Two views of the average SAXS envelope of undiluted XLF·XRCC4·BRCT complex. The envelope is superimposed on the main component of the MES model.
(E) Fit for the MES model 1, model 2 (tetramer 48%, hexamer 20 %, octamer 32%) and model 3 (tetramer 68%, octamer 15%, decamer 17%). The fits are shown as residuals.
See also Figure S5.
Next, we performed a validation of the XRCC4·BRCT·XLF atomic models using a similar approach to that described above. First, we built a model based on the described XLF·XRCC4 interface (Figure 3D) and the XRCC4·BRCT model (Figure 2D). The selected ensemble fit the data very well, in particular, the distinct spacing peak at q~0.075Å-1 (χ2=2.4) (Figure 5BE). The alternative model 2 gave a slightly worse fit (χ2=2.5) and model 3 fit the data poorly (χ2=14.5). The selected ensemble for the best fit model shows relatively little heterogeneity, with one component accounting for 72% of the entire ensemble (Figure 5D, model 1).
This atomic model yields an XRCC4·BRCT·XLF arrangement that can be superimposed with the SAXS envelope. The reconstructed envelope shows an elongated region, consistent with the presence of two XRCC4·BRCT complexes separated by a protrusion attributable to XLF (Figure 5D and S5). Together with analysis of the XRCC4·XLF complexes (Figure 3), this XRCC4·BRCT·XLF arrangement indicates that one XLF dimer tethers two XRCC4·BRCT complexes. The model also shows that XLF and XRCC4 are tilted relative to one other. Furthermore, XLF interacts with XRCC4 in a head-to-head fashion without introducing steric clashes with the LigIV-BRCT domain (Figure 4DE). The tilt angle between XRCC4·XLF may lead to an even larger separation of the two LigIV-BRCT tethered by a central XLF molecule. In this experimental model, an XLF dimer is predicted to bind two XRCC4 dimers and each XRCC4 dimer potentially may bind two dimers of XLF, thus generating filament-like assemblies containing alternating XLF and XRCC4 dimers. Our data shows that these filament-like structures are present in solution at high protein concentrations, mimicking the effective concentrations expected for proteins tethered at a DNA double-strand break.
DISCUSSION
LigIV is critical for DNA ligation in NHEJ, and its activity is regulated by interaction with both XRCC4 and XLF. However, the architectural and dynamic nature of the interaction between XRCC4 with XLF as well as the structure of disordered regions within the C-terminal regions of both XRCC4 and XLF has eluded elucidation by classical structural techniques including NMR, crystallography and EM. Here, we characterized three-dimensional rearrangements of full-length XRCC4, XRCC4 in complex with the tandem BRCT domains of LigIV and with XLF, as well as the XRCC4·XLF·BRCT complex in low-resolution models obtained from SAXS data. These results reveal important new insights into the structure and function of these XRCC4 complexes. First, our results show that XRCC4 tetramers are mediated through a head-to-head interface and that the C-terminal region of XRCC4 is folded back on itself and supports this interaction (Figures 1 and S1). This head-to-head arrangement is counter to proposed models in which two homodimers were suggested to interact through the coil-coil regions (Dahm, 2008; Junop et al., 2000; Modesti et al., 2003). The relatively high dimer- tetramer equilibrium (KD=22 μM) (Junop et al., 2000) and the fact that XRCC4 forms oligomers in a head-to-head fashion (Figure 1) indicate that XRCC4 can form long filaments in solution. XRCC4 filaments and higher order structures were observed at relatively high protein concentrations (approximately 3mg/ml or higher) and it is possible that high protein concentrations could promote non-specific protein-protein interactions. However, XRCC4 is a relatively abundant nuclear protein, and we speculate that filaments could form in the nucleus under conditions where protein crowding may occur (Richter et al., 2008), specifically at DSBs. We estimate that the concentration of XRCC4 in human cells is approximately 6-fold higher than that of LigIV (Mani et al, submitted), therefore a population of XRCC4 probably exists in cells that is not in complex with LigIV. We further propose that XRCC4 filaments may represent a transient storage form that dissociates into dimers upon interaction with the LigIV-BRCT domain (Figure 2DE). Indeed, the binding constant for XRCC4 tetramerization is weaker (KD 22 μM) (Figure 1E) than that of XRCC4 binding to the LigIV·BRCT domain (estimated KD~1 nM) (Modesti et al., 2003) and is reversible. Thus, the interaction of XRCC4 with LigIV will be dominant over tetramerization and shift the formation of filaments towards the dimeric form (Modesti et al., 2003). From our structural data, the BRCT domain probably promotes dissociation of XRCC4 filaments by the displacement of the XRCC4 C-terminus from the head region, which destabilizes the head-to-head interface (Figures 2 and S2).
Our overall model suggests that the C-terminal regions of XRCC4 are in close proximity to the XRCC4 head and stabilize the head-to-head interface of XRCC4 filaments. We propose that this arrangement is suitable to facilitate interaction with XRCC4 with DNA. XRCC4 interacts with DNA in a protein concentration and DNA length dependent manner (Modesti et al., 1999), and XRCC4 filaments may align along linear DNA molecules to facilitate DNA binding (Andres et al., 2007; Lu et al., 2007). Interestingly, the C-terminal region of XRCC4 is phosphorylated by DNA-PK in vitro (Lee et al., 2004; Yu et al., 2003) and DNA-PK phosphorylation disrupts binding to DNA in vitro (Modesti et al., 1999). Collectively these data support a model in which DNA is localized near the XRCC4 head/C-terminal region.
Our model also reveals the importance of the XRCC4 head domain for tetramer/filament formation. Interestingly, XRCC4 lacking the N-terminal 60-100 amino acids, which encompasses the head domains, fails to complement the radiation sensitivity defect of XRCC4-deficient cells in vivo (Grawunder et al., 1998; Leber et al., 1998) even though this region is not required for interaction with LigIV (Grawunder et al., 1998). Together, these studies suggest that XRCC4 has LigIV independent functions in the cell that are mediated by the head domains of XRCC4. We propose that head domain-mediated tetramerization and filament formation aligns DNA filaments and that this function of XRCC4 is required for DSB repair in vivo.
XRCC4 also interacts with XLF. The overall structure of the human XLF homodimer resembles that of the XRCC4 homodimer except that it has a shorter coil-coil region followed by a turn that reverses the direction of the polypeptide, positioning it towards the head domain (Andres et al., 2007; Li et al., 2008). We show that the XRCC4 C-terminal region adopts a similar folded back structure that reaches towards the head domain. The high conformational disorder of this C-terminal region and its possible low affinity for the coil-coil region could explain displacement of the C-terminus from the coil-coil region upon BRCT binding. From our structural data, we propose a model for the XRCC4·XLF interface that is mediated via a head-to-head interaction with a relatively small, buried interface (Figure 3). The atomic model of the XRCC4·XLF assembly gives a excellent match to the SAXS data and indicates that the conserved α2, α3 and β6, β7 sheets of XRCC4 interact with the conserved region of XLF between the β6 and β7 sheets (Figure 3), as proposed from mutational analysis (Andres et al., 2007; Malivert et al., 2010). The major difference between their model and our SAXS based model is in the orientation of the coiled coil domains of each homodimer (Figure 3F). The XLF β6, β7 patch (containing L115) functions as a key in the XRCC4 cavity formed by the conserved helices α2, α3 and sheets β6, β7 (containing interaction-required residues K63, K65, K99). In our structure, the tilt angle of ~45° between the XRCC4 dimers bound to XLF molecule allows for filament formation without introducing steric clashes upon binding of LigIV through the BRCT domain (Figure 4DE). This model not only satisfies our SAXS data and mutational analyses (Andres et al., 2007; Malivert et al., 2010), but also allows the BRCT C-terminus to interact with XLF weakly as previously reported (Deshpande and Wilson, 2007). This arrangement of XLF and XRCC4·BRCT places the BRCT domain on one side of the filament by alternating repeated units of XLF and XRCC4·BRCT. The advantage of the centered XLF and the tilt angle in between two external XRCC4 dimers is that this arrangement positions the LigIV-BRCT domain a considerable distance from the head domains/filament (Figure 4ED). Since the LigIV catalytic domain is tethered to the BRCT domains by a long, flexible linker (Recuero-Checa et al., 2009; Watts and Brissett, 2010), our data support a model in which the XRCC4·XLF filament acts in a cooperative manner as a DNA “holder” (Lu et al., 2007) and that the catalytic domain of LigIV reaches over to the DSB through its flexible linker (Perry et al., 2010). Although the recent EM study shows minimal flexibility between XRCC4 and LigIV in the XRCC4·LigIV complex (Recuero-Checa et al., 2009), we propose that the intrinsic motion of LigIV is persistent in solution under physiological conditions. To test the dynamic character of catalytically active LigIV, the conformation of the LigIV catalytic domain should change during DNA ligation process such that the BRCT domain may provide a scaffold allowing the catalytic domain to interact with the XRCC4·XLF - DNA “holder” while allowing certain structural flexibility. Furthermore, in support of XRCC4·XLF - DNA “holder” model as discussed above, XRCC4·XLF can interact with unusually long DNA molecules (Lu et al., 2007) and formation of such a filaments would be predicted to stabilize binding of long DNA substrates.
EXPERIMENTAL PROCEDURES
Protein purification
Proteins were cloned, expressed in bacteria and purified as described in Supplementary Material. Purified proteins used in this study were full-length XRCC4 (aa 1-344), XRCC4 1-140, XRCC4 1-161, XLF 1-248 and DNA LigIV 618-911.
Size-Exclusion Chromatography (SEC) and Multi-Angle laser Light Scattering (MALS)
MALS experiments were performed in-line after SEC Chromatographic separations as described in Supplementary Material. The light scattering experiments were used to perform analytical scale chromatographic separations for mass determination of the principle peaks in the SEC analysis.
Data collection and evaluation
SAXS data were collected at the ALS beamline 12.3.1 (SIBYLS) LBNL Berkeley, California (Hura et al., 2009). Wavelength λ=1.0 Å and the sample-to-detector distances set to 1.5 m resulting in scattering vectors, q, ranging from 0.001 Å-1 to 0.32 Å-1. The scattering vector is defined as q = 4π sinθ/λ, where 2θ is the scattering angle. All experiments were performed at 20°C and data was processed as described (Hura et al., 2009). The experimental SAXS data for different protein concentrations were investigated for aggregation using Guinier plots (Guinier and Fournet, 1955). The radius of gyration RG was derived by the Guinier approximation I(q) = I(0) exp(-q2RG2/3) with the limits qRG<1.3. The program GNOM (Svergun, 1992) was used to compute the pair-distance distribution functions, P(r). This approach also provided the maximum dimension of the macromolecule, Dmax. The overall shapes were restored from the experimental data using the program DAMMIF with P1 symmetry operator. (Franke and Svergun, 2009). Ten bead models obtained for each SAXS profile were averaged by DAMAVER (Volkov and Svergun, 2003) to construct the averaged model representing general structural features. Bead models were converted to volumetric format.
Atomic model building
In our rigid body modeling strategy BILBOMD, molecular dynamics (MD) simulations were used to explore conformational space adopted by XRCC4 C-terminus. The crystal structures of XRCC4 (PDBid: 1fu1) and the XRCC4·BRCT complex (PDBid: 3ii6) were used to construct the initial atomic models. The initial models were constructed by connecting XRCC4 or XRCC4·BRCT with predicted helical stacks identified by secondary structure prediction (Figure S1B). These atomic models were used as templates for rigid body modeling. For each registered conformation, the theoretical SAXS profile and the corresponding fit to the experimental data were calculated using the program FoXS (Schneidman-Duhovny et al., 2010). The flexible C-terminus in the XRCC4 and XRCC4·BRCT structures was represented by an ensemble containing 2 different conformations. The scattering from such an ensemble is readily computed by averaging the individual scattering patterns from the conformers. To select an appropriate minimal ensemble from pool of ~10000 MD - generated conformations, Minimal Ensemble Search (MES) was employed (Pelikan et al., 2009). The scattering curves from all the structures in the MD – pool were first pre-computed and the subsequent genetic algorithm-selection operators were performed using these patterns and not the structures (Bernado et al., 2007). The final model was calculated to best fit the experimental curve I(q)experiment minimizing the discrepancy χ2 between the experimental and calculated multiconformational profile. Due to the dynamic character of XRCC4, XRCC4·XLF and XRCC4·BRCT·XLF the samples represented an equilibrium of the different oligomerization states and MES was used to identify the fraction ratios of the components required to best fit the experimental data (Pelikan et al., 2009). The multicomponent scattering I(q) from such a minimal ensemble was computed by averaging the individual scattering profiles from conformers:
I1,2,3…N (q) were the scattering profiles from the single oligomers and the momentum transfer. A MES based search was used to select an appropriate ensemble of oligomers from a pool of all generated oligomers (homodimer, tetramer, hexamer, octamer, decamer). The final model achieved the best fit to the experimental curve I(q)experiment by minimizing the discrepancy χ2 between the experimental and calculated multi-component curve. Comparison of the structural properties of the selected oligomers (two for XRCC4; three for XRCC4·XLF and XRCC4·XLF·BRCT) in the ensemble subset allowed us to distinguish the degree of heterogeneity of the experimental system.
Supplementary Material
Acknowledgments
We thank the Berkeley Lab Advanced Light Source and SIBYLS beamline staff at 12.3.1 for aiding solution scattering data collection and members of the Lees-Miller laboratory for comments. This work was supported by the National Institutes of Health (NIH) Structural Cell Biology of DNA Repair Machines P01 grant CA92584 (JAT/SPLM) as well as operating grant 69139 from the Canadian Institutes of Health Research (SPLM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS. Crystal Structure of Human XLF: A Twist in Nonhomologous DNA End-Joining. Mol Cell. 2007;28:1093–1101. doi: 10.1016/j.molcel.2007.10.024. [DOI] [PubMed] [Google Scholar]
- Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Malivert L, Fischer A, Mornon JP, Revy P, de Villartay JP. Cernunnos interacts with the XRCC4 x DNA-ligase IV complex and is homologous to the yeast nonhomologous end-joining factor Nej1. J Biol Chem. 2006;281:13857–13860. doi: 10.1074/jbc.C500473200. [DOI] [PubMed] [Google Scholar]
- Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- Dahm K. Role and regulation of human XRCC4-like factor/cernunnos. J Cell Biochem. 2008;104:1534–1540. doi: 10.1002/jcb.21726. [DOI] [PubMed] [Google Scholar]
- Deshpande RA, Wilson TE. Modes of interaction among yeast Nej1, Lif1 and Dnl4 proteins and comparison to human XLF, XRCC4 and Lig4. DNA Repair (Amst) 2007;6:1507–1516. doi: 10.1016/j.dnarep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annu Rev Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shpae determination in small-angle scattering. J Appl Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- Grawunder U, Zimmer D, Kulesza P, Lieber MR. Requirement for an interaction of XRCC4 with DNA ligase IV for wild-type V(D)J recombination and DNA double-strand break repair in vivo. J Biol Chem. 1998;273:24708–24714. doi: 10.1074/jbc.273.38.24708. [DOI] [PubMed] [Google Scholar]
- Gu J, Lu H, Tsai AG, Schwarz K, Lieber MR. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence. Nucleic Acids Res. 2007;35:5755–5762. doi: 10.1093/nar/gkm579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinier A, Fournet F. Small angle scattering of X-rays. New York: Wiley Interscience; 1955. [Google Scholar]
- Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges P, Ahnesorg P, Pitcher RS, Bruce CK, Kysela B, Green AJ, Bianchi J, Wilson TE, Jackson SP, Doherty AJ. Evolutionary and functional conservation of the DNA non-homologous end-joining protein, XLF/Cernunnos. J Biol Chem. 2006;281:37517–37526. doi: 10.1074/jbc.M608727200. [DOI] [PubMed] [Google Scholar]
- Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, 2nd, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junop MS, Modesti M, Guarne A, Ghirlando R, Gellert M, Yang W. Crystal structure of the Xrcc4 DNA repair protein and implications for end joining. Embo J. 2000;19:5962–5970. doi: 10.1093/emboj/19.22.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CA, Agyei R, Galicia S, Metalnikov P, O’Donnell P, Starostine A, Weinfeld M, Durocher D. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. Embo J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber R, Wise TW, Mizuta R, Meek K. The XRCC4 gene product is a target for and interacts with the DNA-dependent protein kinase. J Biol Chem. 1998;273:1794–1801. doi: 10.1074/jbc.273.3.1794. [DOI] [PubMed] [Google Scholar]
- Lee KJ, Jovanovic M, Udayakumar D, Bladen CL, Dynan WS. Identification of DNA-PKcs phosphorylation sites in XRCC4 and effects of mutations at these sites on DNA end joining in a cell-free system. DNA Repair (Amst) 2004;3:267–276. doi: 10.1016/j.dnarep.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Li Y, Chirgadze DY, Bolanos-Garcia VM, Sibanda BL, Davies OR, Ahnesorg P, Jackson SP, Blundell TL. Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ. Embo J. 2008;27:290–300. doi: 10.1038/sj.emboj.7601942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Pannicke U, Schwarz K, Lieber MR. Length-dependent binding of human XLF to DNA and stimulation of XRCC4.DNA ligase IV activity. J Biol Chem. 2007;282:11155–11162. doi: 10.1074/jbc.M609904200. [DOI] [PubMed] [Google Scholar]
- Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiationinduced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malivert L, Ropars V, Nunez M, Devret P, Miron S, Faure G, Guerois R, Mornon JP, Revy P, Charbonnier JB, et al. Delineation of the XRCC4 interacting region in the globular head domain of cernunnos/XLF. J Biol Chem. 2010 doi: 10.1074/jbc.M110.138156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesti M, Hesse JE, Gellert M. DNA binding of Xrcc4 protein is associated with V(D)J recombination but not with stimulation of DNA ligase IV activity. Embo J. 1999;18:2008–2018. doi: 10.1093/emboj/18.7.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesti M, Junop MS, Ghirlando R, van de Rakt M, Gellert M, Yang W, Kanaar R. Tetramerization and DNA ligase IV interaction of the DNA double-strand break repair protein XRCC4 are mutually exclusive. J Mol Biol. 2003;334:215–228. doi: 10.1016/j.jmb.2003.09.031. [DOI] [PubMed] [Google Scholar]
- Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JJ, Cotner-Gohara E, Ellenberger T, Tainer JA. Structural dynamics in DNA damage signaling and repair. Current opinion in structural biology. 2010 doi: 10.1016/j.sbi.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr Opin Struct Biol. 2010a;20:128–37. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Improving small-angle X-ray scatteringdata for structural analyses of the RNA world RNA. 2010b;16(3):638–466. doi: 10.1261/rna.1946310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recuero-Checa MA, Dore AS, Arias-Palomo E, Rivera-Calzada A, Scheres SH, Maman JD, Pearl LH, Llorca O. Electron microscopy of Xrcc4 and the DNA ligase IV-Xrcc4 DNA repair complex. DNA Repair (Amst) 2009;8:1380–1389. doi: 10.1016/j.dnarep.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Riballo E, Woodbine L, Stiff T, Walker SA, Goodarzi AA, Jeggo PA. XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic acids research. 2009;37:482–492. doi: 10.1093/nar/gkn957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Nessling M, Lichter P. Macromolecular crowding and its potential impact on nuclear function. Biochim Biophys Acta. 2008;1783:2100–2107. doi: 10.1016/j.bbamcr.2008.07.017. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, Yu X, Chen J, Songyang Z. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. J Biol Chem. 2003;278 doi: 10.1074/jbc.C300407200. [DOI] [PubMed] [Google Scholar]
- Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, Blundell TL, Pellegrini L. Crystal structure of an Xrcc4-DNA ligase IV complex. Nat Struct Biol. 2001;8:1015–1019. doi: 10.1038/nsb725. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Sali A. FoXS: A Web server for Rapid Computation and Fitting of SAXS Profiles. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq461. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI. Determination of the regularization Parameter in Indirect- Transform Methods using perceptual criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci U S A. 2007;104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts FZ, Brissett NC. Linking up and interacting with BRCT domains. DNA Repair (Amst) 2010;9:103–108. doi: 10.1016/j.dnarep.2009.10.010. [DOI] [PubMed] [Google Scholar]
- Wu PY, Frit P, Meesala S, Dauvillier S, Modesti M, Andres SN, Huang Y, Sekiguchi J, Calsou P, Salles B, Junop MS. Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Mol Cell Biol. 2009;29:3163–3172. doi: 10.1128/MCB.01895-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Wang W, Ding Q, Ye R, Chen D, Merkle D, Schriemer D, Meek K, Lees-Miller SP. DNA-PK phosphorylation sites in XRCC4 are not required for survival after radiation or for V(D)J recombination. DNA Repair (Amst) 2003;2:1239–1252. doi: 10.1016/s1568-7864(03)00143-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


