Abstract
The structure and function of transfer RNA (tRNA) genes have been extensively studied for several decades, yet the general mechanisms controlling tRNA gene family evolution remain unclear, primarily because previous phylogenetics-based methods fail to distinguish between paralogs and orthologs that are highly similar in sequence. We have developed a system for identifying orthologs of tRNAs using flanking sequences to identify regions of conserved synteny and used it to annotate sets of orthologous tRNA genes across the 12 sequenced species of Drosophila. These data have allowed us to place the gains and losses of individual tRNA genes on each branch of the Drosophila tree and estimate rates of tRNA gene turnover. Our results show extensive rearrangement of the Drosophila tRNA gene complement over the last 60 My. We estimate a combined average rate of 2.18 ± 0.10 tRNA gene gains and losses per million years across the Drosophila lineage. We have identified 192 tRNAs that are ancestral to the genus, of which 157 are “core” tRNAs conserved in at least 11 of 12 extant species. We provide evidence that the core set of tRNA genes encode a nearly complete set of anticodons and have different properties from other “peripheral” tRNA genes, such as preferential location outside large tRNA clusters and higher sequence conservation. We also demonstrate that tRNA isoacceptor and alloacceptor changes by anticodon shifts have occurred several times in Drosophila, annotating 16 such events in functional tRNAs during the evolution of the genus.
Keywords: transfer RNA, genome evolution, noncoding RNA, tRNA identity, synteny map, gene duplication
Introduction
Transfer RNA (tRNA) genes are an important class of RNA genes that function to decode messenger RNA into protein sequences. tRNAs comprise some of the largest gene families in any organism, with several hundred functional tRNA genes predicted in most eukaryotes (reviewed in Griffiths-Jones [2007]). As with other noncoding RNA gene families, an accurate estimation of the functional tRNA gene content can be complicated by the widely varying numbers of pseudogenes found in some species, such as the rat (Gibbs et al. 2004). However, recent analysis of tRNA content in divergent species such as Drosophila and chicken suggest that the minimum functional tRNA gene set in metazoans is approximately 300 (Clark et al. 2007).
Because there are only 64 possible codons, the large number of tRNA genes present in eukaryotic genomes generates functional redundancy. Functionally equivalent tRNA genes that encode the same anticodon or isoacceptor group are typically transcribed from multiple loci in eukaryotic genomes (Long and Dawid 1980). Eukaryotic tRNA genes can be arranged in clusters that are often (Hayashi et al. 1980) but not always (Yen and Davidson 1980) homogeneous for a particular isoacceptor type. Previous studies in vertebrates have shown that tRNA gene clusters often contain several functionally equivalent loci with the same anticodon (Tang et al. 2009), suggesting that they have arisen by tandem gene duplication. However, clusters of tRNA genes with different anticodons suggest that members of some tRNA clusters may have more complex evolutionary histories (Tang et al. 2009). Despite these general observations about tRNA gene organization, little is known about the evolutionary mechanisms that determine the distribution and number of tRNA genes within eukaryotic genomes.
The fact that some clusters contain tRNA genes with different anticodon types raises the possibility that anticodon sequences may evolve after tandem gene duplication causing divergence in tRNA function. In vitro experiments have demonstrated that a single point mutation in an anticodon is sufficient to concurrently change tRNA amino acid identity and mRNA coupling capacity (Schulman and Pelka 1989; Saks et al. 1998). Moreover, tRNA functional shifts on an evolutionary timescale have been detected in metazoan mitochondrial genomes (Rawlings et al. 2003). In analysis of the 22 tRNA mitochondrial genes, Rawlings et al. (2003) found that leucine tRNA duplication and remolding events have occurred independently at least seven times within three major animal lineages. Anticodon switching in the nuclear genome of eukaryotes has not yet been investigated thoroughly, and the role that anticodon switching plays in determining tRNA gene family organization remains largely unexplored.
One of the key difficulties in studying tRNA gene family evolution is ambiguity in the homology relationships between loci across species. Sequence similarity between subsets of functionally equivalent tRNAs can be very high, and therefore, methods to infer homology based on the sequences of tRNAs themselves often cannot resolve orthologs from paralogs (Withers et al. 2006). Additionally, the absence of an ortholog from the data set because of gaps in the genome assembly or incomplete annotation may cause a paralogue to be falsely identified as the ortholog. Inclusion of additional information such as conservation of gene order and orientation in syntenic genomic regions can overcome this problem because orthologous genes will often be conserved in genomic location whether or not their sequences diverge significantly. Thus, use of local (or micro-) synteny maps, built from sequences flanking tRNA genes themselves, may hold the key to generating high-confidence sets of tRNA orthologs for evolutionary analysis.
Recent multispecies genome projects provide excellent material to study the dynamics of tRNA gene family evolution in eukaryotes. The Drosophila 12 genomes project has made available the genomes of a dozen species in the Drosophila genus together with their tRNA annotations (Clark et al. 2007). Spanning diverse habitats and ∼40 My of divergence, these 12 species vary considerably in their genome organization, morphology, ecology, and behavior. However, the most important aspects of the cellular, molecular, and developmental biology of these species are well conserved, including patterns of codon usage that are thought to correlate with tRNA abundance (Moriyama and Powell 1997; Vicario et al. 2007).
Here, we develop a synteny-based approach to study tRNA gene family evolution in 12 Drosophila genomes by mapping tRNA flanking regions to the D. melanogaster reference sequence. tRNA genes that map to the same region in D. melanogaster are assembled into orthologous sets of tRNA genes, from which we infer gains and losses of tRNA genes on each branch of the species tree. We quantify the level of turnover of tRNA genes across the genus Drosophila and propose the existence of core and peripheral sets of tRNA genes. Finally, we identify several cases of tRNA functional shifts by anticodon point mutations, demonstrating a greater than anticipated role for functional shifts in the evolution of eukaryotic tRNA gene organization.
Materials and Methods
tRNA Gene Sets
tRNA annotations and genome sequences for all 12 Drosophila species were obtained from the FlyBase 2008-07 release. FlyBase is presently missing some annotation of tRNA anticodons; 51 tRNAs were thus reclassified using tRNAscan-SE 1.23 (Lowe and Eddy 1997) with covariance analysis only mode (-C) for maximum sensitivity. Sequences annotated as pseudogenes by tRNAscan-SE were retained through all steps of the analysis.
Prior to mapping tRNAs from other genomes, the D. melanogaster genome was masked for repeats using RepeatMasker 3.2.7 (Smit et al. 1996–2004) and repeat libraries RM-20090120 (Jurka et al. 2005) configured to use WU-BlastN 2.0MP (04 May 2006) (Gish 1996–2004) with default parameters. Additionally, tRNA sequences in all Drosophila genomes were also masked to prevent mapping to paralogous D. melanogaster loci and to prevent inclusion of neighboring tRNA loci from query sequences in tRNA clusters. To assess the proportion of tRNA loci that are in repetitive DNA sequences, repeats in each Drosophila genome were annotated with RepeatMasker (Smit et al. 1996–2004) using species-specific repeat libraries generated by ReAS (Li et al. 2005) on the CAF1 assemblies (ftp://ftp.genomics.org.cn/pub/ReAS/drosophila/v2/consensus_fasta/).
Mapping tRNA Flanking Sequences
For each tRNA in all Drosophila genomes, a 10-kb region encompassing 5 kb from each flank (with the tRNA sequence masked) was searched against the D. melanogaster genome using WU-BlastN 2.0MP (4 May 2006) (Gish 1996–2004), with the hspsepSmax parameter (defining the maximum separation on the subject sequence of high-scoring pairs (HSPs) that are combined) set to the region length (10 kb) and an E value threshold of 10−10. As a balance between maximizing mapping success and minimizing multiple spurious mappings, 10 kb was chosen as the flanking region size (see supplementary materials, Supplementary Material online). A Drosophila tRNA locus was mapped to the D. melanogaster genome if two criteria are fulfilled: Blast HSPs are present from both sides of the query tRNA, and Blast HSPs are separated in the D. melanogaster genome by less than twice the length of the query sequence (i.e., 20 kb). Mappings to chromosome 4 in D. melanogaster were discarded because this chromosome arm has no tRNAs annotated, is composed mainly of heterochromatin and has a high frequency of repetitive sequences (Miklos et al. 1988; Bergman et al. 2006).
Each tRNA may map zero, one, or multiple times to D. melanogaster. The absence of a mapping indicates the flanking sequence of a tRNA has no orthologous location in D. melanogaster, and a single mapping implies one unique orthologous location. Multiple mappings show that flanking sequences map to regions of the D. melanogaster that are duplicated with respect to the query species. Most commonly, multiple mappings to the D. melanogaster genome occur for tRNAs located in clusters in D. melanogaster. In a small number of cases, multiple mappings to a single location in D. melanogaster are observed; these signify duplications in the query species with respect to D. melanogaster and again often occur in clusters. The size of mappings, defined as the distance between the HSPs in the D. melanogaster genome, is highly variable, ranging from high-resolution mappings of ∼70 bp to low-resolution mappings up to 20,000 bp. Low-resolution mappings are more common in species more distantly related to D. melanogaster.
Assembly of Ortholog Sets
Putatively orthologous tRNAs from each species were assembled into sets on the basis of overlapping coordinates in the D. melanogaster genome. If a tRNA gene is annotated in the D. melanogaster genome between the two mapped flanks, orthology with the D. melanogaster tRNA is inferred. Likewise, where tRNAs from multiple query species mapped to the same location in D. melanogaster but no tRNA is annotated in D. melanogaster itself, we assigned them as an orthologous set. Each orthologous set therefore consists of a group of tRNAs in Drosophila genomes that map to the same location in the D. melanogaster genome (fig. 1). Defining ortholog sets in this manner also allows us to propagate information (including the gene name) from the D. melanogaster genome to the entire set of orthologs.
FIG. 1.—
Mappings of Drosophila tRNA flanking regions to the D. melanogaster genome, visualized using the UCSC Genome Browser (Kent et al. 2002). Two sets of Thr:TGT orthologs are shown: the set on the right illustrates an ancestral tRNA with a loss in D. grimshawi and the set on the left illustrates a derived tRNA not present in the D. melanogaster genome that has been gained on the obscura group branch. The width of the mapped region shows the distance between HSPs of the two flanking regions of each query sequence; smaller intervals represent higher resolution mappings.
Preliminary lists of orthologs defined in this manner were then filtered by searching each tRNA against those it overlaps using BlastN to eliminate spurious ortholog matches. tRNAs that do not match another ortholog member with an e value <10−3 were removed from the list of overlaps.
Filtered overlap lists were then resolved into a single table of 1:1 orthologies for all 12 species. When a tRNA overlapped more than one mapping in another species, due to either low-resolution mappings or multiple mappings, orthology was preferentially assigned to a tRNA with the same identity and anticodon. This procedure is conservative with respect to our analysis of tRNA anticodon switches.
Some large tRNA clusters that vary in gene number across species produced complex sets of many:many mappings, which are particularly hard to resolve into 1:1 ortholog annotations. Accordingly, all ortholog sets were manually inspected, and 27 ambiguous cases involving multiple- or low-resolution mappings had members reassigned to alternative ortholog groups. The final ortholog table contained 753 rows, each representing a distinct ortholog set (see supplementary materials, Supplementary Material online).
Comparison of Computational and Experimental tRNA Mappings
We compared the tRNA site orthologies inferred from our computational mapping for three species, D. melanogaster, D. pseudoobscura, and D. virilis, with those inferred from the polytene in situ hybridization data of Tonzetich et al. (1990). Tonzetich and colleagues reported the hybridization sites of seven tRNA genes [Arg-2(ACG), Lys-2(CTT), Ser-2b(GCT), Ser-7(AGA), Thr-3(TGT), Thr-4(CGT), Val-3b(CAC)] in four species: D. melanogaster, D. pseudoobscura, D. virilis, and D. hydei. By comparing these hybridization sites in terms of their linkage groups and label intensity, the authors inferred the likely orthologies of tRNA loci. To estimate how many of our computational mappings were supported by these experimental data, we reconciled the two data sets of inferred site orthologies. For a computational mapping to be supported, the experimental data must show an in situ hybridization signal on the expected chromosome arm, and the labeling intensities relative to number of tRNAs mapped to that site must be consistent with other labeling intensities for that gene and species.
tRNA Gains and Losses
Gains and losses of tRNA genes were placed on branches of the Drosophila tree automatically using an implementation of the Dollo parsimony method (Farris 1977). Each orthologous set represents a single gain on the tree, and therefore, a gain representing each tRNA set was placed on the most recent branch that would lead to all species represented in its set. Losses were then placed on subsequent branches leading to any species not represented in the set. Anticodon switches and pseudogenization events were also placed on branches by maximum parsimony.
The number of tRNAs expected to be conserved in at least 11 or 12 species given branch-specific turnover rates was determined by simulation. Fixing the observed numbers of gains and losses on each branch of the Drosophila tree, we simulated loss and gain of tRNAs on each successive branch by randomly subtracting tRNAs as losses before adding new tRNAs as gains. The simulation was repeated 1,000 times.
To find the 95% confidence intervals (CIs) for the average rate of loss and rate of gain plus loss across the Drosophila genus and separately on each of the 22 branches of the tree, we conducted a bootstrap analysis by sampling with replacement from the complete list of ortholog sets with their attached gain and loss events. CIs cannot be calculated for the average rate of gains using this method because gain events occur exactly one time on the tree and there is no variance in gains among ortholog sets.
Properties of tRNA Ortholog Sets
tRNAs located wholly within D. melanogaster intron boundaries of FlyBase 2008-07 gene annotations were classified as intronic. Clustering patterns were assessed by finding the number of D. melanogaster tRNAs with at least 1, 2, and 3 neighbors within 1,000 and 100,000 bases, respectively. For each definition, we assessed whether the proportion of clustered and not clustered tRNAs differed significantly between core and peripheral sets using a χ2 test. The complement of codons which the tRNAs of each group of ortholog sets could recognize and decode were assessed using the revised wobble base pairing rules (Guthrie and Abelson 1982) and the “superwobble” rule where necessary (Rogalski et al. 2008).
“Ancestral” tRNAs were defined as those present in at least one Sophophora and one Drosophila species. “Core” tRNAs were defined as those present in at least 11 of the 12 Drosophila species. Ortholog sets present in fewer than 11 species were considered “peripheral” tRNAs.
For each species, we measured the number of substitutions per site (uncorrected p-distance) for each gene with respect to the D. melanogaster ortholog, excluding tRNA introns. We tested core and peripheral genes separately, aligning the orthologous sequences using ClustalW 2.0.12 (Thompson et al. 1994) and counting nonidentical sites, excluding gaps. For each species, we tested the statistical significance of the difference in substitutions per site between core and peripheral sets using Mann–Whitney U tests.
Results
Mapping and Ortholog Assignment of Drosophila tRNA Genes
The number and properties of tRNAs mapped from each species to D. melanogaster are shown in table 1, and examples of ortholog sets with and without corresponding tRNA genes in D. melanogaster are shown in figure 1. Full mapping data in the form of GFF files are supplied in supplementary materials (Supplementary Material online). We observe that the proportion of tRNAs mapped to D. melanogaster and therefore located in regions of conserved synteny is generally high (>85%), even in the most divergent species of the Drosophila subgenus (D. mojavensis, D. virilis, and D. grimshawi). The only exceptions are the two species in the Sophophora subgenus with a significantly higher number of annotated tRNA genes, D. ananassae and D. willistoni, suggesting that the extra tRNAs present in these genomes are in regions without orthology to D. melanogaster. Indeed, both of these species have a high number of tRNA genes that are predicted to be pseudogenes or overlap species-specific repeats, and mapping rates for both pseudogenes and tRNAs that overlap repeats are in general low across species (table 1). This suggests that the elevated numbers of tRNAs annotated in D. ananassae and D. willistoni that do not map to the D. melanogaster genome may largely be due to pseudogenes created by recent proliferation of repeat sequences in these species. In total, 90% of all nonpseudogene tRNAs in 11 species map to the D. melanogaster genome. This indicates that our melanogaster-centric mapping approach at most misses only a small minority of orthologous clusters where the extended region is not present in D. melanogaster.
Table 1.
Numbers of tRNAs, Pseudogenes, and tRNAs Annotated in Genome-Specific Repeats Are Compared across Drosophila Species
| Total tRNAs Annotated | tRNAs Annotated as Pseudogenes (% of total) | tRNAs Overlapping Repeats (% of total) | tRNAs Mapped (% of total) | Mapped Pseudogenes (% of mapped) | Mapped Repeat tRNAs (% of mapped) | |
| D. melanogaster | 297 | 4 (1) | 44 (15) | — | — | — |
| D. simulans | 268 | 2 (0.7) | 11 (4) | 255 (95) | 1 (0.4) | 4 (2) |
| D. sechellia | 312 | 13 (4) | 44 (14) | 295 (95) | 3 (1) | 11 (4) |
| D. yakuba | 380 | 52 (14) | 52 (14) | 340 (89) | 32 (9) | 10 (3) |
| D. erecta | 286 | 2 (0.7) | 6 (3) | 283 (99) | 2 (0.7) | 2 (0.7) |
| D. ananassae | 472 | 165 (35) | 181 (38) | 300 (64) | 37 (12) | 18 (6) |
| D. pseudoobscura | 295 | 1 (0.3) | 8 (3) | 262 (89) | 1 (0.4) | 2 (0.8) |
| D. persimilis | 306 | 1 (0.3) | 83 (27) | 259 (85) | 1 (0.4) | 24 (9) |
| D. willistoni | 460 | 164 (36) | 183 (40) | 241 (52) | 35 (15) | 22 (9) |
| D. mojavensis | 267 | 3 (1) | 7 (3) | 245 (92) | 2 (0.8) | 2 (0.8) |
| D. virilis | 279 | 1 (0.3) | 6 (2) | 246 (88) | 1 (0.4) | 2 (0.8) |
| D. grimshawi | 261 | 1 (0.3) | 23 (9) | 225 (86) | 1 (0.4) | 10 (4) |
Computational tRNA Orthology Mappings Are Consistent with Experimental Data
In order to assess the accuracy of our synteny-based ortholog detection method, we compared our computational mappings with previous experimental results from polytene in situ hybridization studies on seven tRNA genes [Arg-2(ACG), Lys-2(CTT), Ser-2b(GCT), Ser-7(AGA), Thr-3(TGT), Thr-4(CGT), Val-3b(CAC)] (Tonzetich et al. 1990) for the three species present in both data sets (D. melanogaster, D. pseudoobscura, and D. virilis). Out of 67 computational mappings for which hybridization data was available, 61 were supported (91%), with a further two explained by predicted losses on branches leading to D. melanogaster. Conversely, out of the 33 hybridization sites reported, computational mapping data provide support for 26 (79%). Thus, we conclude that our computational mappings are largely consistent with previous experimental data of Tonzetich et al. (1990), providing evidence that our computational method is accurately detecting orthologous tRNA genes.
tRNA Genes Have Undergone Substantial Flux in Drosophila
From the presence and absence of orthologous tRNAs in each species, we can infer the evolutionary history of that gene in the Drosophila genus, placing the gain and loss events on branches in the tree by parsimony (see Materials and Methods). Ortholog sets with more than two implied losses on the tree were manually inspected and a total of 27 ortholog groups were modified to achieve greater parsimony across all ortholog sets. We have also visualized mappings using custom tracks on the UCSC genome browser to refine our analysis (fig. 1).
Out of a total of 753 orthologous tRNA groups identified, only 192 (25%) were found to be ancestral to the Drosophila genus. These 192 genes have existed in locations of conserved synteny throughout the evolution of the Drosophila genus. Forty-seven (24%) of the ancestral tRNAs are present in all 12 extant species, and 110 tRNAs (57%) are core tRNA genes conserved in at least 11 of the 12 species. The remaining 82 ancestral and all nonancestral loci are peripheral tRNAs that are present in 10 or fewer species.
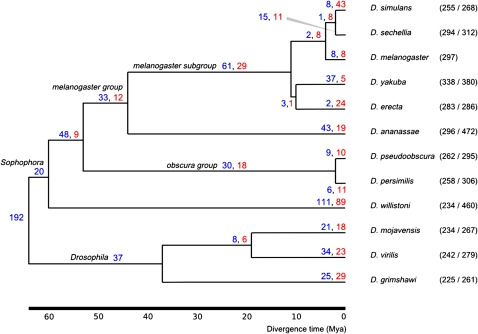
The numbers of tRNA gains and losses on each branch of the Drosophila phylogeny are shown in figure 2 (numbers of gains and losses) and figure 3 (rates of gains and losses). Considering only tRNAs from orthologous sets that are annotated in D. melanogaster, the combined rate of tRNA gene turnover (gains plus losses) within the Drosophila genus is 2.18 per million years (95% bootstrap CI: 2.08–2.28). The average rate of gains is 1.30 per million years, with 0.88 losses per million years (95% CI: 0.76–1.00). The higher rate of gain relative to loss could be due in part to the recent expansion of tRNA gene numbers by proliferation of tRNA-containing repeat sequences in D. willistoni and D. ananassae, some of which map to the D. melanogaster genome and are included in our rate estimates.
FIG. 2.—
Inferred tRNA gains (blue) and losses (red). The fraction of total annotated tRNAs with at least one mapping from each species to Drosophila melanogaster is shown in parentheses. Tree topology taken from Clark et al. (2007), with divergence times from Tamura et al. (2004).
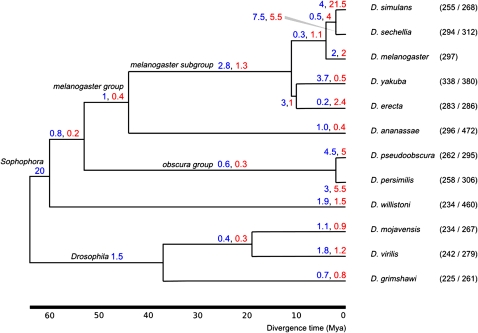
FIG. 3.—
Rates of tRNA gene gains (blue) and losses (red) per million years. Divergence times were derived from Tamura et al. (2004).
Some variation in the rate of tRNA gene gain and loss is observed on some branches. For example, a net gain and net loss of tRNAs are inferred on the lineages leading to D. yakuba and D. erecta, respectively. The high number of gains in the yakuba branch is in accordance with the elevated number of tRNAs annotated and the presence of unmapped tRNAs in known repeats in this genome (table 1). We also note that the branches leading to each member of the two pairs of species that have diverged most recently (simulans/sechellia and pseudoobscura/persimilis) show extraordinarily high rates of gain/loss, out of proportion with the most recent common ancestral branches leading to these lineages. We interpret these deviations from the long-term turnover rates as likely results of inaccuracies in the estimate of the divergence time of these branches or missing data from incomplete genome assemblies (see Discussion).
Using the observed frequencies for each branch, we simulated gain and loss events on the Drosophila phylogeny. Using the average number of genes present in at least 11 of 12 species after 1,000 simulations as a null distribution, we found that the number of core Drosophila tRNAs is larger than expected by chance (P < 0.001). This finding lends tentative support to the existence of a core tRNA set with a higher than usual level of syntenic conservation, as has been proposed in bacteria (Withers et al. 2006).
Core tRNAs Encode a Nearly Complete Set of Codons
The D. melanogaster tRNA complement comprises 44 different anticodons capable of decoding all 62 codons encoding amino acids. Drosophila melanogaster does not contain a CCC or CCG anticodon tRNA, which are expected to decode Gly(GGG) and Arg(CGG), respectively. Instead, tRNAGly(UCC) is expected to decode GGG and tRNAArg(UCG) to decode CGG, in accordance with the superwobble hypothesis (Rogalski et al. 2008). Intriguingly, the set of tRNA genes defined as core comprises 40 anticodons capable of decoding all codons except Asp(GAC)/(GAU), both decoded in melanogaster by tRNAAsp(GUC), and Trp(UGG), decoded by tRNATrp(CCA). Cognate tRNAs for these remaining codons have been mapped to D. melanogaster from other species. Both tRNAAsp(GUC) and tRNATrp(CCA) have breaks in syntenic conservation between Sophophora and Drosophila and between the melanogaster subgroup species and the rest of the genus. Within these divisions, little further syntenic variation is observed, indicating that even though these tRNAs fail to meet our requirements for core status they do show reasonable syntenic conservation in large sections of the genus.
Core tRNAs Are Underrepresented in Large Clusters
To address if core tRNAs are more or less likely to be found in clusters, we measured the mean number of core and peripheral tRNAs with a minimum of 1, 2, and 3 neighboring tRNAs within 1 kb and 100 kb. The results show that there is no significant difference between core and peripheral genes at both 1 kb and 100 kb distances if having just one neighboring tRNA gene defines a cluster (see table 2). However, increasing the number of genes required to define a cluster reveals that core and peripheral tRNA sets exhibit different clustering tendencies. Significant differences at P ≤ 0.05 (χ2) are observed for 1 kb with a minimum of two neighbors and for both distance parameters when requiring clusters to contain at least three neighbors. These observations suggest that core tRNAs are less likely than peripheral tRNAs to be located in large clusters.
Table 2.
The Proportions of Drosophila melanogaster Core and Peripheral tRNAs That Are Clustered
| Cluster Definition | Clusters | Clusters with Homogeneous Identity | Core tRNAs Clustered (%) | Peripheral tRNAs Clustered (%) | χ2, P Value |
| 1 kb, 1 neighbor | 71 | 49 | 65 (60) | 125 (66) | 0.15 |
| 1 kb, 2 neighbors | 26 | 14 | 23 (21) | 64 (33) | 0.01 |
| 1 kb, 3 neighbors | 10 | 3 | 5 (5) | 29 (15) | 0.003 |
| 100 kb, 1 neighbor | 58 | 34 | 92 (85) | 162 (86) | 0.45 |
| 100 kb, 2 neighbors | 30 | 11 | 67 (62) | 129 (68) | 0.14 |
| 100 kb, 3 neighbors | 23 | 7 | 51 (47) | 124 (66) | 0.001 |
Core tRNA Genes Are More Constrained in DNA Sequence
In order to test whether core tRNAs are more conserved at the sequence level than peripheral tRNAs, we compared substitutions in core and peripheral genes from each species against their D. melanogaster orthologs (see table 3). With the exception of D. willistoni, core tRNAs have fewer substitutions than peripheral tRNAs in each species tested. These differences are statistically significant or marginally significant (P < 0.1) for all species except D. ananassae, D. mojavensis, and D. virilis. Each of these three species diverged from D. melanogaster over 40 Ma, whereas D. mojavensis and D. virilis have low numbers of peripheral tRNAs with D. melanogaster orthologs, reducing the statistical power to reject the null hypothesis.
Table 3.
Comparison of Substitution Rates of Core and Peripheral tRNAs Having Drosophila melanogaster Orthologs
| Species | Core tRNAs with D. melanogaster Orthologs | Substitutions Per Site (core) | Peripheral tRNAs with D. melanogaster Orthologs | Substitutions Per Site (peripheral) | P Value |
| D. simulans | 103 | 0.002 | 136 | 0.005 | 0.09 |
| D. sechellia | 108 | 0.001 | 164 | 0.008 | 0.01 |
| D. yakuba | 108 | 0.001 | 174 | 0.005 | 0.04 |
| D. erecta | 107 | 0.001 | 163 | 0.006 | 0.04 |
| D. ananassae | 104 | 0.006 | 111 | 0.011 | 0.16 |
| D. pseudoobscura | 108 | 0.007 | 74 | 0.008 | 0.08 |
| D. persimilis | 108 | 0.006 | 74 | 0.009 | 0.01 |
| D. willistoni | 71 | 0.015 | 30 | 0.011 | 0.28 |
| D. mojavensis | 102 | 0.007 | 33 | 0.009 | 0.86 |
| D. virilis | 105 | 0.006 | 29 | 0.020 | 0.18 |
| D. grimshawi | 103 | 0.007 | 24 | 0.013 | 0.07 |
NOTE.—Substitution rates are shown with the number of tRNAs tested in both categories and the resultant P value (Mann–Whitney U test).
Anticodon Shifts Occur in Orthologous Eukaryotic Nuclear tRNA Genes
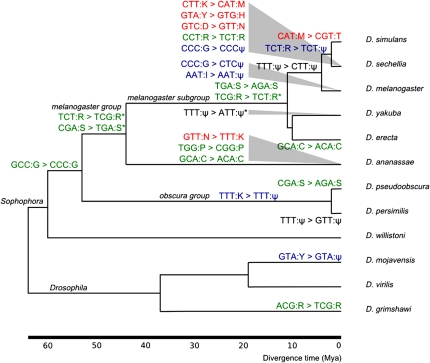
Having a complete set of orthologous tRNAs across multiple species has enabled the detection of several functional changes in nuclear tRNA genes. Sets of orthologs may contain the same functional anticodon sequences, different functional anticodon sequences (due to either isoaccepter or putative alloaccepter changes), or a mixture of functional and pseudogene predictions. Using a parsimony approach, we have detected a total of 22 changes in tRNA function (anticodon changes and pseudogenisations) involving 20 ortholog sets and placed these events on the Drosophila phylogeny (fig. 4). There are 11 cases of single base changes resulting in anticodon shifts in functional gene sequences that remain isoacceptors. Nine involve first anticodon bases, and the remaining two are third position changes (TCT:R → TCG:R and the reverse TCG:R → TCT:R). A further five mutations have resulted in putative alloacceptor changes. All involve single base mutations: 1 in the first base, 2 in the second, and 2 in the third anticodon position. There are six cases of functional genes becoming pseudogenes (pseudogenisations). Four anticodon shifts have equally parsimonious alternative branch placements (see supplementary materials, Supplementary Material online). Four anticodon shifts involve only nonfunctional pseudogenes. Thus, although tRNA identity is broadly conserved among ortholog sets, we do observe a low rate of anticodon shifting in orthologous tRNA genes across Drosophila species.
FIG. 4.—
Inferred tRNA identity changes placed on the Drosophila phylogeny by maximum parsimony. Isoacceptor anticodon changes are colored green, alloacceptor anticodon changes red, pseudogenisations blue, and anticodon changes in pseudogenes black. Pseudogenes are denoted by the symbol ψ. Note: GTA:Y → GTA:ψ on the D. mojavensis branch is both a pseudogenisation and anticodon flip. Asterisk indicates equally parsimonious alternative placements and/or changes (for details, see supplementary materials, Supplementary Material online).
Discussion
We have implemented a method for identifying orthologous sets of tRNA genes based on conserved microsynteny and analyzed the evolution of tRNA genes in 12 sequenced Drosophila genomes. We have used these ortholog sets to: 1) place inferred gain and loss events at their most parsimonious locations on the Drosophila phylogeny and estimate rates of tRNA gene turnover in Drosophila; 2) identify sets of ancestral, core, and peripheral sets of tRNA genes; 3) analyze the genomic features of a core and peripheral set of tRNA genes, and 4) detect functional changes among orthologs resulting from anticodon shifts and pseudogenisation.
Substantial tRNA Flux in the Drosophila Lineage
The results of the present study show in detail the extent to which tRNA gene families are in a state of flux in the Drosophila genus. Of the ∼300 tRNA genes present in each species, we identify 192 loci that were likely to be present in the ancestor of the genus Drosophila, yet only 47 of these ancestral loci are conserved in all 12 extant species. Over the long term, our results suggest that the rates of gain (1.30 gains per million years) and loss (0.88 losses per million years) are of the same order, and thus, the total number of tRNA genes across the Drosophila lineage over the last ∼40 My has been approximately constant despite ongoing gene gain and loss.
These rates of tRNA gene turnover in Drosophila are more than 2-fold higher than the rates calculated in a study of the tRNA evolution in bacteria (Escherichia coli, Shigella flexneri, and Salmonella typhimurium), which found average rates of 0.64 gains and 0.30 losses per million years (Withers et al. 2006). Taken at face value, the evidence suggests that tRNA gene turnover in prokaryotic genomes, with fewer tRNAs (∼100), is slower than in flies (∼300). Withers et al. (2006) derived rates from phylogenetic clustering analysis of tRNA gene sequences alone, which is likely to have a higher number of incorrect ortholog calls due to misclassification of paralogs with high sequence identity. The resulting overestimation of true orthologs would lead to an underestimate of the rate of tRNA gene gain and loss in bacteria. However, our results also underestimate the true rate of tRNA gene turnover in Drosophila because they do not consider tRNA genes that cannot be mapped by synteny to D. melanogaster. Assuming that all unmapped tRNAs represent gains at the tips of the tree gives an upper estimate of an additional 1.5 gains per million years. Based on these differences in turnover rate between bacteria and flies, we suggest that the size of a genomic tRNA gene complement, which impacts the redundancy in tRNA gene function, may influence the long-term evolutionary dynamics of tRNA gene turnover across species.
The combined rate of tRNA gene gain and loss is calculated as 2.18 ± 0.10 per million years. Over an average of 324 genes per species, we find 0.0067 gains and losses per gene per million years. The overall rate of protein-coding gene gain and loss in Drosophila has been estimated as 0.0012 gains and losses per gene per million years (∼14,000 genes per species) by probabilistic modeling of the variation in the size of gene families (Hahn et al. 2007). Notwithstanding the different methodologies, the available data suggest that tRNAs turnover ∼3–4 times faster than protein-coding genes in Drosophila. We interpret the relatively fast turnover rate for tRNAs to be facilitated by the high degree of functional redundancy in the tRNA gene complement in any single genome.
Apparent Variation in Rates of tRNA Gain and Loss Across Lineages
Despite evidence for a long-term pattern of stability in the number of tRNA genes across the genus Drosophila, there is evidence that the tRNA gene complements of closely related species may be very different. Some lineages, such as D. ananassae, D. yakuba, and D. willistoni, have a much higher tRNA gene number than the average, indicating a higher rate of gain than loss on these lineages. However, the majority of extra tRNA genes above the average of the genus are predicted to be pseudogenes (Clark et al. 2007). Thus, short-term increases in gene number may not resolve into longer term growth of the tRNA complement in these lineages if pseudo-tRNAs are ultimately deleted, like most nonfunctional DNA in Drosophila (Petrov et al. 1996). In fact, these transient bursts of tRNA gene number may simply reflect other genomic processes that create new repetitive DNA, as D. ananassae, D. yakuba, and D. willistoni genomes also have some of the highest transposable element activity and repeat content of the sequenced species (Clark et al. 2007). Consistent with this, the majority of excess tRNA genes in D. ananassae and D. willistoni do not map to orthologous regions present in the D. melanogaster genome, and large fractions of them are located in annotated repeats.
Among loci that are mappable to orthologous regions, we find that the two most recently diverged species pairs (D. simulans/D. sechellia and D. pseudoobscura/D. persimilis) each show very high rates of tRNA gain and loss postspeciation. One reason for these elevated rates may be the shortness of the branches themselves, where erroneous underestimation of the divergence time may lead to overestimation of the rate of gene gain and loss. Another reason for higher estimated rates of gain or loss on the lineages is that these genome assemblies have low sequencing coverage (D. sechellia and D. persimilis) (Clark et al. 2007) or are a mosaic of several different low coverage assemblies (D. simulans) (Begun et al. 2007). Assembly gaps resulting from low coverage may lead to incorrect parsimony assignments and hence to artificially elevated numbers of losses and gains for the two species pairs involving D. sechellia and D. persimilis. Gaps in the genome assembly also provide a plausible explanation for the high rate of loss inferred on the D. simulans branch. The strongest evidence for difference in the short-term rate of tRNA gain appears to be for D. yakuba, whose genome is sequenced to deep coverage (Clark et al. 2007) and whose tRNA complement maps well to the D. melanogaster genome. Thus, apparent short-term variation in the rate of tRNA gene gain and loss may overall be better explained by repeat-driven expansion of pseudo-tRNAs and genome assembly artifacts rather than real deviation from the long-term process of steady-state tRNA gene number with ongoing turnover.
Evidence for a Core Set of tRNAs in Drosophila
Although it is clear that there is a high level of turnover among the Drosophila tRNAs, we detect a significantly larger number of core tRNA ortholog groups that are conserved in 11 or 12 species than would be expected by chance. We believe that this observation reflects unequal rates of tRNA turnover for a core set of highly conserved tRNAs in the Drosophila genomes, supporting previous evidence from studies in E. coli (Withers et al. 2006) and in vertebrates (Tang et al. 2009). In general, core tRNAs have fewer substitutions than peripheral genes from the same species, suggesting higher selective constraint on these loci. Moreover, the group of D. melanogaster tRNAs conserved in at least 11 species is able to decode all but two codons. This suggests that the core set may be able to function to a large extent independently of other tRNA loci and are supplemented by a set of more peripheral tRNA genes that turnover more rapidly. Finally, the mechanism for increased conservation of a subset of tRNAs may in part be related to their genomic environment because the most widely conserved tRNAs are less likely to be located in large clusters.
Impact of tRNA Anticodon Shifts
The present study is the first large-scale multispecies investigation of changes in tRNA function in nuclear genomes. For the most part, we observed broad conservation in tRNA identity among ortholog sets across Drosophila species. Nevertheless, we did observe 22 changes in tRNA function, 16 of which involve mutations in anticodons. The potential evolutionary importance of tRNA functional shifts is demonstrated by the reassignment 170 Ma of the CUG codon of fungal Candida and Debaryomyces species from tRNALeu(CAG) to tRNASer(CAG) (Ohama et al. 1993). Previous work in animal mitochondria (Rawlings et al. 2003) and the nuclear genomes of cow (Tang et al. 2009) and mouse (Coughlin et al. 2009) have provided evidence for switches in tRNA function. However, these studies are unable to show exactly which sets of orthologous genes have been involved in the switches using sequence similarity-based approaches.
Although tRNAs with the same anticodon tend to cluster together, nearly half of clusters in D. melanogaster contain two or more anticodons (table 2). We show that anticodon shifts have occurred several times in Drosophila, providing an explanation for the origin of tRNA clusters with different anticodon sequences and a potential mechanism for genomes to meet the changing demands of codon usage. Patterns of codon usage bias in the genus Drosophila appear to be conserved, with the exception of the D. willistoni lineage, which shows a dramatic reduction in codon bias (Bergman et al. 2002; Powell et al. 2003; Clark et al. 2007; Vicario et al. 2007). We do not find any direct evidence for tRNA functional shifts playing a role in the evolution of codon usage bias in D. willistoni, as we have not detected any anticodon shifts on this long branch. However, many D. willistoni tRNAs do not have assigned orthologs in the other 11 species, and these unmapped loci may be more prone to functional changes. It therefore remains possible that the very high number of tRNA loci on this lineage and the high level of gene turnover observed may have permitted a shift in codon usage. However, as other lineages such as D. ananassae also have elevated numbers of tRNA genes, shifts in codon usage do not appear to be directly coupled with high tRNA gene numbers in Drosophila.
Conclusions
We have compiled sets of orthologs of the tRNAs of 12 Drosophila genomes using an approach based on microsynteny marked by conserved flanking regions. Synteny approaches provide reliable orthology calls in multigene families with high sequence identity. We show that tRNAs of Drosophila have a high rate of turnover, but a subset are particularly conserved in both sequence and synteny, which we argue represent a core set of tRNAs. tRNAs in the core set are able to recognize nearly all codons and are preferentially located outside large clusters. We find evidence for tRNA functional changes by anticodon shifts, suggesting a mechanism to explain how clusters of tRNAs with different anticodons arise and how the tRNA gene complement may respond to the changing demands of codon usage during evolution.
Supplementary Material
Supplementary materials are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
The authors thank Matthew Ronshaugen, Antonio Marco, and Aaron Webber for useful discussion and comments on the manuscript and Dave Ardell for motivating the investigation of tRNA functional switching. This work was funded by a Biotechnology and Biological Sciences Research Council studentship to H.H.R. and by a University of Manchester Fellowship to S.G.J.
References
- Begun DJ, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman CM, et al. Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol. 2002;3:RESEARCH0086. doi: 10.1186/gb-2002-3-12-research0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman CM, Quesneville H, Anxolabehere D, Ashburner M. Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biol. 2006;7:R112. doi: 10.1186/gb-2006-7-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Coughlin DJ, Babak T, Nihranz C, Hughes TR, Engelke DR. Prediction and verification of mouse tRNA gene families. RNA Biol. 2009;6:195–202. doi: 10.4161/rna.6.2.8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris JS. Phylogenetic analysis under Dollo's law. Syst Zool. 1977;26:77–88. [Google Scholar]
- Gibbs RA, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Gish W. 1996–2004. WU-BLAST [Internet]. St. Louis: Washington University [cited 2008 April 24]. Available from: http://blast.wustl.edu. [Google Scholar]
- Griffiths-Jones S. Annotating noncoding RNA genes. Annu Rev Genomics Hum Genet. 2007;8:279–298. doi: 10.1146/annurev.genom.8.080706.092419. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Abelson J. Organization and expression of tRNA genes in Saccharomyces cerevisiae. 1982 The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression, Cold Spring Harbor Monograph Archive. p. 487–528. [Google Scholar]
- Hahn MW, Han MV, Han SG. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 2007;3:e197. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, et al. Hybridization of tRNAs of Drosophila melanogaster to polytene chromosomes. Chromosoma. 1980;76:65–84. doi: 10.1007/BF00292227. [DOI] [PubMed] [Google Scholar]
- Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, et al. ReAS: recovery of ancestral sequences for transposable elements from the unassembled reads of a whole genome shotgun. PLoS Comput Biol. 2005;1:e43. doi: 10.1371/journal.pcbi.0010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long EO, Dawid IB. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos GL, Yamamoto MT, Davies J, Pirrotta V. Microcloning reveals a high frequency of repetitive sequences characteristic of chromosome 4 and the beta-heterochromatin of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1988;85:2051–2055. doi: 10.1073/pnas.85.7.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama EN, Powell JR. Codon usage bias and tRNA abundance in Drosophila. J Mol Evol. 1997;45:514–523. doi: 10.1007/pl00006256. [DOI] [PubMed] [Google Scholar]
- Ohama T, et al. Non-universal decoding of the leucine codon CUG in several Candida species. Nucleic Acids Res. 1993;21:4039–4045. doi: 10.1093/nar/21.17.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA, Lozovskaya ER, Hartl DL. High intrinsic rate of DNA loss in Drosophila. Nature. 1996;384:346–349. doi: 10.1038/384346a0. [DOI] [PubMed] [Google Scholar]
- Powell JR, Sezzi E, Moriyama EN, Gleason JM, Caccone A. Analysis of a shift in codon usage in Drosophila. J Mol Evol. 2003;57(Suppl 1):S214–S225. doi: 10.1007/s00239-003-0030-3. [DOI] [PubMed] [Google Scholar]
- Rawlings TA, Collins TM, Bieler R. Changing identities: tRNA duplication and remolding within animal mitochondrial genomes. Proc Natl Acad Sci U S A. 2003;100:15700–15705. doi: 10.1073/pnas.2535036100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski M, Karcher D, Bock R. Superwobbling facilitates translation with reduced tRNA sets. Nat Struct Mol Biol. 2008;15:192–198. doi: 10.1038/nsmb.1370. [DOI] [PubMed] [Google Scholar]
- Saks ME, Sampson JR, Abelson J. Evolution of a transfer RNA gene through a point mutation in the anticodon. Science. 1998;279:1665–1670. doi: 10.1126/science.279.5357.1665. [DOI] [PubMed] [Google Scholar]
- Schulman LH, Pelka H. The anticodon contains a major element of the identity of arginine transfer RNAs. Science. 1989;246:1595–1597. doi: 10.1126/science.2688091. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. RepeatMasker Open-3.0 [Internet] 1996–2004. Institute for Systems Biology [cited 2009 June 01]. Available from: http://www.repeatmasker.org. [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- Tang DT, Glazov EA, McWilliam SM, Barris WC, Dalrymple BP. Analysis of the complement and molecular evolution of tRNA genes in cow. BMC Genomics. 2009;10:188. doi: 10.1186/1471-2164-10-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonzetich J, Hayashi S, Grigliatti TA. Conservatism of sites of tRNA loci among the linkage groups of several Drosophila species. J Mol Evol. 1990;30:182–188. doi: 10.1007/BF02099944. [DOI] [PubMed] [Google Scholar]
- Vicario S, Moriyama EN, Powell JR. Codon usage in twelve species of Drosophila. BMC Evol Biol. 2007;7:226. doi: 10.1186/1471-2148-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers M, Wernisch L. dos Reis M. Archaeology and evolution of transfer RNA genes in the Escherichia coli genome. RNA. 2006;12:933–942. doi: 10.1261/rna.2272306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen PH, Davidson N. The gross anatomy of a tRNA gene cluster at region 42A of the D. melanogaster chromosome. Cell. 1980;22:137–148. doi: 10.1016/0092-8674(80)90162-2. [DOI] [PubMed] [Google Scholar]