Abstract
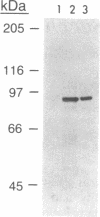
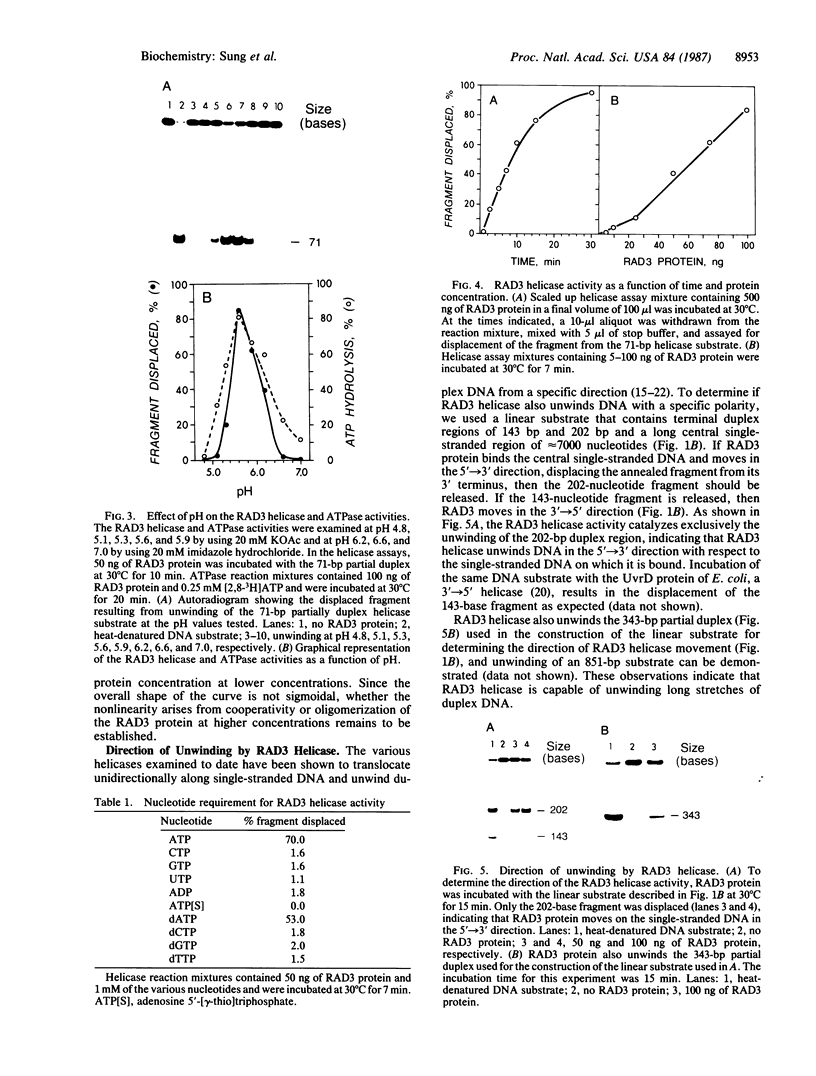
The Saccharomyces cerevisiae RAD3 gene, which is required for cell viability and excision repair of damaged DNA, encodes an 89-kDa protein that has a single-stranded DNA-dependent ATPase activity. We now show that the RAD3 protein also possesses a helicase activity that unwinds duplex regions in DNA substrates constructed by annealing DNA fragments of 71-851 nucleotides to circular, single-stranded M13 DNA. The DNA helicase activity is dependent on the hydrolysis of ATP, has a pH optimum of approximately 5.6, and is inhibited by antibodies raised against a truncated RAD3 protein produced in Escherichia coli. The RAD3 helicase translocates along single-stranded DNA in the 5'----3' direction. The direction of RAD3 helicase movement is consistent with the possibility that it unwinds DNA duplexes in advance of the replication fork during DNA replication.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Caron P. R., Kushner S. R., Grossman L. Involvement of helicase II (uvrD gene product) and DNA polymerase I in excision mediated by the uvrABC protein complex. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4925–4929. doi: 10.1073/pnas.82.15.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean F. B., Bullock P., Murakami Y., Wobbe C. R., Weissbach L., Hurwitz J. Simian virus 40 (SV40) DNA replication: SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc Natl Acad Sci U S A. 1987 Jan;84(1):16–20. doi: 10.1073/pnas.84.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geider K., Hoffmann-Berling H. Proteins controlling the helical structure of DNA. Annu Rev Biochem. 1981;50:233–260. doi: 10.1146/annurev.bi.50.070181.001313. [DOI] [PubMed] [Google Scholar]
- Golin J. E., Esposito M. S. Evidence for joint genic control of spontaneous mutation and genetic recombination during mitosis in Saccharomyces. Mol Gen Genet. 1977 Jan 18;150(2):127–135. doi: 10.1007/BF00695392. [DOI] [PubMed] [Google Scholar]
- Grippo P., Richardson C. C. Deoxyribonucleic acid polymerase of bacteriophage T7. J Biol Chem. 1971 Nov 25;246(22):6867–6873. [PubMed] [Google Scholar]
- Hartwell L. H., Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985 Jul;110(3):381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. R., Prakash S., Reynolds P., Polakowska R., Weber S., Prakash L. Isolation and characterization of the RAD3 gene of Saccharomyces cerevisiae and inviability of rad3 deletion mutants. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5680–5684. doi: 10.1073/pnas.80.18.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain I., Van Houten B., Thomas D. C., Abdel-Monem M., Sancar A. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6774–6778. doi: 10.1073/pnas.82.20.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R., Richardson C. C. Replication of duplex DNA by bacteriophage T7 DNA polymerase and gene 4 protein is accompanied by hydrolysis of nucleoside 5'-triphosphates. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1525–1529. doi: 10.1073/pnas.74.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad E. B. Method for the isolation of Escherichia coli mutants with enhanced recombination between chromosomal duplications. J Bacteriol. 1977 Apr;130(1):167–172. doi: 10.1128/jb.130.1.167-172.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- LeBowitz J. H., McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986 Apr 5;261(10):4738–4748. [PubMed] [Google Scholar]
- Masamune Y., Richardson C. C. Strand displacement during deoxyribonucleic acid synthesis at single strand breaks. J Biol Chem. 1971 Apr 25;246(8):2692–2701. [PubMed] [Google Scholar]
- Matson S. W. Escherichia coli helicase II (urvD gene product) translocates unidirectionally in a 3' to 5' direction. J Biol Chem. 1986 Aug 5;261(22):10169–10175. [PubMed] [Google Scholar]
- Matson S. W., George J. W. DNA helicase II of Escherichia coli. Characterization of the single-stranded DNA-dependent NTPase and helicase activities. J Biol Chem. 1987 Feb 15;262(5):2066–2076. [PubMed] [Google Scholar]
- Matson S. W., Richardson C. C. DNA-dependent nucleoside 5'-triphosphatase activity of the gene 4 protein of bacteriophage T7. J Biol Chem. 1983 Nov 25;258(22):14009–14016. [PubMed] [Google Scholar]
- Matson S. W., Tabor S., Richardson C. C. The gene 4 protein of bacteriophage T7. Characterization of helicase activity. J Biol Chem. 1983 Nov 25;258(22):14017–14024. [PubMed] [Google Scholar]
- Miller R. D., Prakash L., Prakash S. Genetic control of excision of Saccharomyces cerevisiae interstrand DNA cross-links induced by psoralen plus near-UV light. Mol Cell Biol. 1982 Aug;2(8):939–948. doi: 10.1128/mcb.2.8.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumovski L., Chu G., Berg P., Friedberg E. C. RAD3 gene of Saccharomyces cerevisiae: nucleotide sequence of wild-type and mutant alleles, transcript mapping, and aspects of gene regulation. Mol Cell Biol. 1985 Jan;5(1):17–26. doi: 10.1128/mcb.5.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumovski L., Friedberg E. C. A DNA repair gene required for the incision of damaged DNA is essential for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4818–4821. doi: 10.1073/pnas.80.15.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P., Higgins D. R., Prakash L., Prakash S. The nucleotide sequence of the RAD3 gene of Saccharomyces cerevisiae: a potential adenine nucleotide binding amino acid sequence and a nonessential acidic carboxyl terminal region. Nucleic Acids Res. 1985 Apr 11;13(7):2357–2372. doi: 10.1093/nar/13.7.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds R. J., Friedberg E. C. Molecular mechanisms of pyrimidine dimer excision in Saccharomyces cerevisiae: incision of ultraviolet-irradiated deoxyribonucleic acid in vivo. J Bacteriol. 1981 May;146(2):692–704. doi: 10.1128/jb.146.2.692-704.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Rupp W. D. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983 May;33(1):249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- Stahl H., Dröge P., Knippers R. DNA helicase activity of SV40 large tumor antigen. EMBO J. 1986 Aug;5(8):1939–1944. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Prakash L., Weber S., Prakash S. The RAD3 gene of Saccharomyces cerevisiae encodes a DNA-dependent ATPase. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6045–6049. doi: 10.1073/pnas.84.17.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. Template recognition sequence for RNA primer synthesis by gene 4 protein of bacteriophage T7. Proc Natl Acad Sci U S A. 1981 Jan;78(1):205–209. doi: 10.1073/pnas.78.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan M., Silver L. L., Nossal N. G. Bacteriophage T4 gene 41 protein, required for the synthesis of RNA primers, is also a DNA helicase. J Biol Chem. 1982 Oct 25;257(20):12426–12434. [PubMed] [Google Scholar]
- Wang T. S., Korn D. Reactivity of KB cell deoxyribonucleic acid polymerases alpha and beta with nicked and gapped deoxyribonucleic acid. Biochemistry. 1980 Apr 29;19(9):1782–1790. doi: 10.1021/bi00550a009. [DOI] [PubMed] [Google Scholar]
- Wilcox D. R., Prakash L. Incision and postincision steps of pyrimidine dimer removal in excision-defective mutants of Saccharomyces cerevisiae. J Bacteriol. 1981 Nov;148(2):618–623. doi: 10.1128/jb.148.2.618-623.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray W., Boulikas T., Wray V. P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981 Nov 15;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- Yarranton G. T., Gefter M. L. Enzyme-catalyzed DNA unwinding: studies on Escherichia coli rep protein. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1658–1662. doi: 10.1073/pnas.76.4.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung A. T., Mattes W. B., Oh E. Y., Grossman L. Enzymatic properties of purified Escherichia coli uvrABC proteins. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6157–6161. doi: 10.1073/pnas.80.20.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]