Abstract
The Karyopherin-β family of nuclear transport factors mediates the majority of nucleocytoplasmic transports. Although each of the 19 Karyopherin-βs transport unique sets of cargos, only three classes of nuclear localization and export signals, or NLSs and NESs, have been characterized. The short basic classical-NLS was first discovered in the 1980s and their karyopherin-bound structures first reported more than 10 years ago. More recently, structural and biophysical studies of Karyopherin-β2-cargo complexes led to definition of the complex and diverse PY-NLS. Structural knowledge of the leucine-rich NES is finally available more than ten years after the discovery of its recognition by the exportin CRM1. We review recent findings relating to how these three classes of nuclear targeting signals are recognized by their Karyopherin-β nuclear transport factors.
Introduction
The transport of proteins between the nucleus and the cytoplasm is signal-mediated: nuclear localization and export signals (NLSs and NESs) direct proteins in and out of the nucleus, respectively. The NPC allows ions, small molecules and many small proteins (<40 kDa) to diffuse between the nucleus and cytoplasm, but larger proteins require the assistance of nuclear transport factors to traverse the NPC. 19 Karyopherin-β (Kapβ; also known as Importin and Exportin) proteins in humans recognize NLS/NES, each functioning as distinct nuclear import, export or bidirectional transport factors (Table 1 [1]) [2,3]. Kapβs share similar molecular weights (90–150 kDa) and isoelectric points (pI = 4.0–5.0), low sequence identity (10–20%) and all contain 19–20 helical HEAT repeats arranged into superhelical or ring-like structures.
Table 1.
The Kapβ family of nuclear transport factors.
| Karyopherin-β subfamily [1] |
Human Karyopherin-βs |
S. Cerevisiae Karyopherin-βs |
Known Cargos | NLS/NES |
|---|---|---|---|---|
| IMB1 | Impβ (Kapβ1) | Kap95 | Proteins | Classical-NLS via Impα |
| IMB2 | Kapβ2 (Transportin) | Kap104 | Proteins | PY-NLS |
| IMB3 | Importin-5 (Kapβ3 or RanBP5) |
Kap121 (Pse1) | Proteins | ND |
| IMB4 | Importin-4 (RanBP4) | Kap123 | Proteins | ND |
| IMB5 | Importin-9 | Kap114 | Proteins | ND |
| IPO8 | Importin-7 (RanBP7), Importin- 8 (RanBP8) |
Kap119 (Nmd5), Kap108 (Sxm1) |
Proteins | ND |
| KA120 | Importin-11 | Kap120 | Proteins | ND |
| TNPO3 | Transportin-SR, Transportin-SR2 (Transportin-3 or TNPO3) |
Kap111 (Mtr10), Kap122 (Pdr6) |
Proteins | RS domains |
| XPO1 | CRM1 (Exportin-1) | CRM1 (Xpo1) | Proteins and RNAs |
Leucine-rich NES or classical-NES |
| XPO2 | CAS | Cse1 | Impα | Conformational; entire Impα |
| XPO4 | Exportin-4 | Proteins | ND | |
| XPO5 | Exportin-5 | Kap142 (Msn5) | RNA and proteins |
Conformational; entire pre- miRNA |
| XPO6 | Exportin-6 | Proteins | ND | |
| XPO7 | Exportin-7 (RanBP16) |
Proteins | ND | |
| XPOT | Exportin-t (Xpo-t) | Los1 | t-RNAs | Conformational; entire tRNA |
Each Kapβ recognizes a unique set of proteins or RNA, thus creating multiple transport pathways across the nuclear pore complex (NPC). Kapβs also bind nucleoporins (proteins in the NPC), targeting Kapβ-cargo complexes to the NPC for translocation. Kapβ-cargo interactions and transport directionality are regulated by the Ran GTPase nucleotide cycle [3]. RanGTP is concentrated in the nucleus, while RanGDP is concentrated in the cytoplasm. In import pathways, RanGTP and cargos bind Kapβs competitively, allowing cargo binding in the cytoplasm and RanGTP-mediated release in the nucleus. In contrast, in export pathways, RanGTP, cargos and Kapβs bind cooperatively for cargo binding in the nucleus and release in the cytoplasm as Ran-bound GTP is hydrolyzed.
The NLS (by extension, also the NES) was defined as a portion of nuclear transport cargo that is 1) necessary for import/export, 2) sufficient to import/export an unrelated protein, 3) able to bind transport factor directly and that 4) transport is abolished when the transport factor is disabled [4]. These criteria are generally useful until complications arise due to complexities in the cell, such as when transport is mediated by multiple signals and/or transport factors, signals are functional only when appropriately modified or if protein localization is also dictated by nuclear/cytoplasmic retention [5]. Many transport cargos are known for Importin-β (Impβ; also known as Karyopherin-β1), CRM1 (also known as Exportin1 or Xpo1) and Karyopherin-β2 (Kapβ2; also known as Transportin), but many fewer are known for the other 16 human Kapβs [6–10]. Correspondingly, classes of NLS recognized by Impβ, Kapβ2 and an NES for CRM1 have been identified but common properties of signals for the other Kapβs remain unknown. The first characterized nuclear transport signal is the short lysine-rich classical-NLS, which was discovered in the early 1980s and is recognized by Importin-α (Impα; also known as Karyopherin-α), which is an adaptor for Impβ [6,7]. More recently, structural analysis of Kapβ2-cargo complexes led to the discovery of a new class of NLS termed PY-NLS [8]. Finally, many export cargos contain leucine-rich NESs, which are recognized by export-Kapβ CRM1. Structures of the first CRM1-NES complex were reported in 2009 [11••,12••].
Recognition of the classical-NLS by the Impα/β system
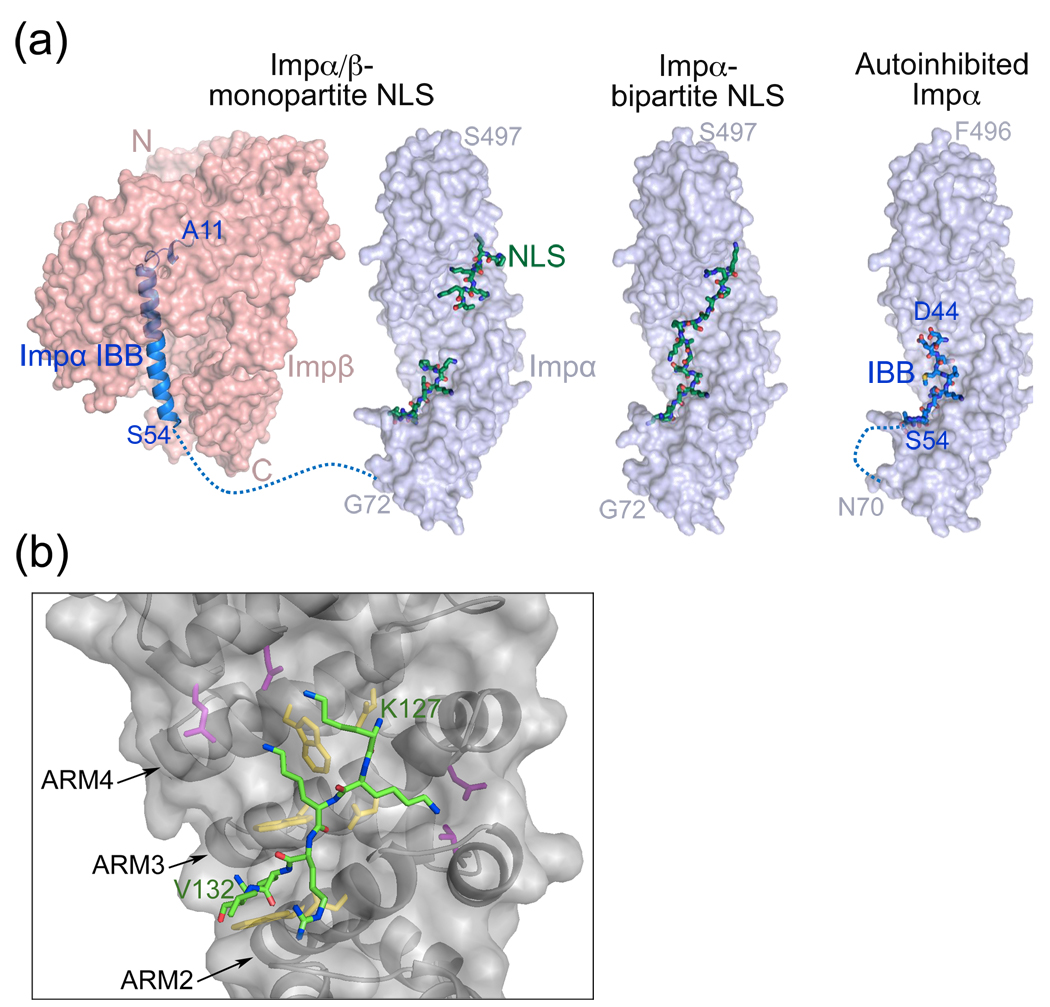
The first protein segments found to direct nuclear localization were identified in SV40 T antigen (PKKKRKV) and nucleoplasmin (AVKRPAATKKAGQAKKKKLD) [6,7]. These sequences were named nuclear localization signals or NLSs and they bind the armadillo (ARM) domain of Impα (Figures 1a and 1b) [2,3,13–15]. The N-terminal Impβ-binding or IBB domain of Impα in turn binds Impβ forming a ternary NLS-Impα/β complex (Figure 1a). The small sizes and relatively simple sequence patterns of these basic NLSs have facilitated identification of similar signals in many proteins. Since the discovery of other Kapβ pathways, the Impα/β system is now known as the classical pathway and its short basic signal the classical-NLS.
Figure 1.
Classical-NLS recognition by Impα/β. (a) Impβ (pink) binds to the IBB domain of Impα (blue ribbon); PDB ID 1QGK. Monopartite (1EJL) or bipartite (1EJY) classical-NLSs (green) bind to the Impα ARM domain (surface representation in blue). Full length Impα is autoinhibited. An NLS-like N-terminal segment (blue sticks) occupies the major NLS binding site on the ARM domain (blue surface) and prohibits binding of exogeneous NLS; PDB ID 1IAL. A dashed blue line represents the unstructured polypeptide chain that connects the Impα ARM domain to its IBB domain. (b) Interactions between the SV40 T antigen NLS (green) and the major binding site of Impα (grey; PDB ID 1BK6). Conserved tryptophan-asparagine pairs and acidic residues in Impα are colored yellow and magenta, respectively.
Structures of unliganded Impα, its NLS complexes and the Impβ-IBB structures as well as previous biochemical studies explained interactions of the three components [16–19]. A portion of the Impα IBB resembles a classical-NLS and binds in an intramolecular fashion to the Impα ARM domain, thus preventing binding with exogeneous NLS [17]. When Impβ binds the IBB, autoinhibition is relieved and the classical-NLS binding site exposed [18] (Figure 1).
The monopartite classical-NLS
Classical-NLSs contain either one (monopartite classical-NLS) or two (bipartite classical-NLS) clusters of positively charged amino acids. Both NLS variants bind to the Impα ARM domain, which contains ten ARM repeats arranged as a cylindrical superhelix [16,20]. Highly conserved tryptophan-asparagine pairs in ARM repeats 2–4 define the major NLS binding site while a similar arrangement in ARM 7 and 8 constitutes the minor binding site. Monopartite-NLS conforms to the consensus sequence K-K/R-X-K/R (X is any amino acid) [4,21–23]. The signal binds Impα in extended conformation and interaction patterns are similar at both sites (Figure 1b) [16,20]. Asparagines of the Impα tryptophan-asparagine pairs hydrogen bond with the NLS backbone, tryptophans make hydrophobic interactions with the aliphatic side chains of lysines and arginines and peripheral polar and acidic residues make electrostatic interactions with charged NLS sidechains (Figure 1b) [16,20].
Besides the conserved basic residues, sequences flanking the basic clusters of classical-NLSs were also shown to make direct contact with the receptor and modulate Impα binding affinities [24]. In addition, post-translational modifications such as phosphorylation also modulate nuclear import rates. For example, phosphorylation of adjacent Ser111/112 N-terminal to the NLS of SV40 T antigen greatly enhances its nuclear import [25], while phosphorylation of the nearby Thr124 produces the opposite effect [26]. Fontes et al have further characterized the molecular and structural basis of phosphorylation in this NLS [24].
The bipartite classical-NLS
The bipartite classical-NLS contains a small N-terminal cluster of basic residues connected to the larger C-terminal cluster by a linker (e.g. AVKRPAATKKAGQAKKKKLD in nucleoplasmin), following a loose consensus sequence of (K/R)(K/R) X10–12 (K/R)3/5, where (K/R)3/5 represents three lysine or arginine residues out of five consecutive amino acids [27]. The C-terminal cluster binds the major NLS binding site of Impα while the N-terminal cluster binds the minor site [16,20]. Individual basic clusters bind Impα independently and combine to create a functional signal. For example, adding an N-terminal basic cluster to a non-functional monopartite classical-NLS increased binding affinity to result in a new functional bipartite NLS [22]. Interactions at both sites are similar to those for monopartite classical-NLSs and the basic clusters are structurally conserved [20,28]. In contrast, connecting linkers are structurally variable.
The bipartite classical-NLS linker is traditionally defined as 9–12 residues but recent studies show that up to 29 amino acids can be present in the linker of a functional bipartite NLS [29•]. These longer linkers are more dependent on specific amino acid sequences and strengths of the basic clusters [29•,30]. Although the linkers are tolerant to mutations, they also contribute to Impα binding affinity [30,31•]. In stronger NLSs, acidic residues and prolines are preferred in the linkers whereas hydrophobic and basic residues are more prevalent in linkers of weaker NLSs [30]. Optimization of Impα binding and import characteristics led to the design of two bipartite ‘super-NLSs’ that bound Impα with such high affinity as to inhibit its import activity [31•]. Linker residues do confer position-specific effects on import activity and thus the bipartite classical-NLS is a multipartite signal with energetic contributions from both basic clusters and linker.
Direct recognition of signals by Kapβs
Many proteins imported into the nucleus bind directly to a Kapβ. Sequences that bind Kapβs are diverse, encompassing both structural domains and linear epitopes. For example, Impβ binds directly and imports numerous proteins without involving Impα. Other than the helical Impβ-binding (IBB) domains of Impα and Snurportin 1 [18,32–34], most Impβ cargos show little sequence or structural similarity. Other than the structurally similar IBBs, cargos SREPB2 and the PTHRP segment are structurally diverse and bind at different sites on the Impβ [18,32–34]. Thus, general features among cargos that bind Impβ directly cannot be inferred at this time. The other 16 Kapβs are currently known to recognize small collections of diverse cargos, making it extremely difficult to identify or describe their NLSs or NESs. Nevertheless, the large sizes and low sequence identities of Kapβs coupled with identification of multiple cargo binding sites for several members [16], suggest that dozens of classes of nuclear targeting signals have yet to be discovered.
Nuclear import by Kapβ2
Kapβ2 is a prototypical Kapβ, which binds import cargos directly [35,36]. ~20 RNA binding proteins had been biochemically identified as Kapβ2 import cargos [35–40]. NLSs in hnRNP A1 (the 38-residue M9 sequence), HuR, TAP, hnRNP D and JKTPB showed little sequence similarity [35–39,41]. Such sequence diversity had prevented prediction of NLSs in other Kapβ2 cargos.
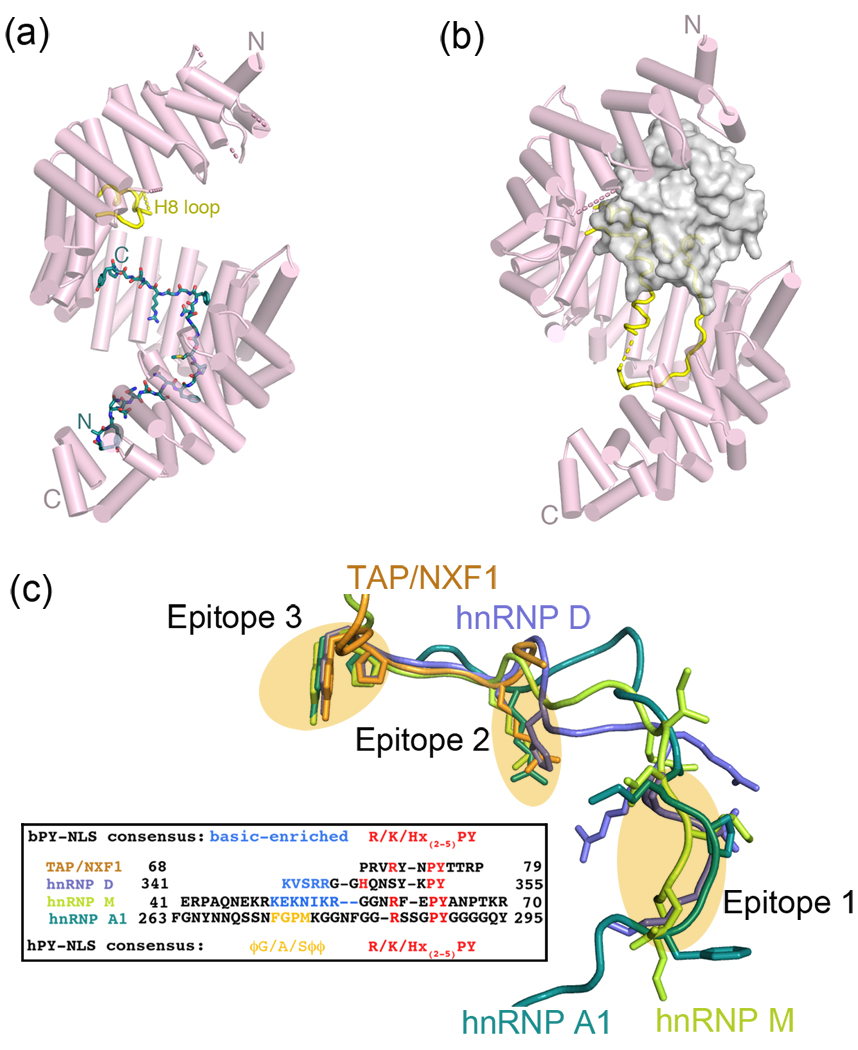
Numerous Kapβ2 structures now provide structural snapshots of this pathway (Figures 2a and 2b) [8,42–45]. Ran and cargo sites, in the N- and C-terminal arches of Kapβ2 respectively, are spatially distinct but are thermodynamically coupled by the 62-residue internal H8 loop, which occupies the cargo site and displaces cargo when Ran is bound [8,42,46] (Figure 2b).
Figure 2.
The Kapβ2 nuclear import pathway. (a) Crystal structure of Kapβ2 (pink) bound to the PY-NLS of hnRNP A1 (green; also known as the M9 sequence); PDB ID 2H4M. The internal loop in HEAT repeat 8 is mostly disordered and colored yellow. (b) Crystal structure of Kapβ2 (pink) bound to Ran•GppNHp (grey surface); PDB ID 1QBK. When Ran is bound, the H8 loop (yellow) is mostly ordered and binds to the PY-NLS binding site. (c) Structures of PY-NLSs from hnRNPs A1 (green; 2H4M), M (yellow-green; 2OT8), D (blue; 2Z5N) and mRNA export factor TAP/NXF1 (orange; 2Z5K) upon superposition of Kapβ2s. All four peptides spatially converge at epitopes 1, 2 and 3, which are connected by structurally variable linkers. PY-NLS sequences are shown in the box, with residues in epitope 1 shown in yellow (hydrophobic motif) and blue (basic motif) and conserved residues in epitopes 2 and 3 shown in red.
Diverse PY-NLS is defined by a collection of physical rules
Recent structural and biochemical studies revealed several common characteristics amongst the apparently disparate signals recognized by Kapβ2, unifying them into a new class of NLS termed PY-NLS [8]. Even though the 15–30 residue PY-NLSs are larger and more complex than the classical-NLSs, they bind Kapβ2 in similar extended conformation indicative of linear and rather long epitopes. However, unlike the classical-NLSs, the diverse PY-NLSs cannot be sufficiently described by a traditional consensus sequence. Instead, they are described by a collection of physical rules that include requirements for intrinsic structural disorder of a large peptide segment, overall basic character, and a set of sequence motifs [8].
PY-NLS consensus motifs
PY-NLS consensus motifs consist of a loose N-terminal hydrophobic or basic motif and a C-terminal RX2–5PY motif (Figure 2c). Composition of the N-terminal motifs divides the signal into hydrophobic (hPY-NLS) and basic subclasses (bPY-NLSs). The former contain consecutive predominantly hydrophobic residues that can be represented by the loose consensus of Φ1-G/A/S-Φ3-Φ4, where Φ is a hydrophobic residue (Figure 2c). The equivalent region in the bPY-NLS is a 4–20-residue segment that is enriched in basic residues. Kapβ2-bound PY-NLSs of hnRNP A1, hnRNP M, hnRNP D, JKTBP and TAP/NXF1 show structurally convergent consensus regions that are separated by linkers, highlighting the multipartite nature of the signal and confirming consensus designation of the motifs despite weak sequence similarity [43,45] (Figure 2c). The large, flat and mixed acidic/hydrophobic cargo-binding site of Kapβ2 allows recognition of the diverse PY-NLS sequences [43]. The hydrophobic motif of hPY-NLS and aliphatic portions of basic residues in bPY-NLS bind the hydrophobic patches while charged sidechains of the bPY-NLS bind the large acidic surface.
Physical properties that govern PY-NLS affinity
Mutagenesis and thermodynamic analysis revealed physical properties that govern PY-NLS binding affinity [47••]. The PY-NLS is composed of three linear epitopes that can each accommodate substantial sequence diversity (Figure 2c). For example, although tyrosine appears conserved in the C-terminal RX2–5PY motif, other amino acids especially hydrophobic ones are well tolerated in this position. Instead of the PY motif, cargos Nab2 and HuR have PL and PG dipeptides, respectively, [47••]. Interestingly, Kapβ2 binding partner ELYS which functions in NPC assembly likely has a PV dipeptide [48] and ciliary transport cargo KIF17 likely has a PL dipeptide in its ciliary localization signal (CLS) that also binds Kapβ2 [49]. The three PY-NLS epitopes are energetically modular and each epitope can contribute very differently to total binding energy in different PY-NLSs [47••]. Signal diversity can be achieved through combinatorial mixing of energetically weak and strong motifs while maintaining affinity appropriate for nuclear import function These concepts are illustrated by an extremely tight-binding peptide inhibitor that was designed by combining strong motifs from different PY-NLSs [43,47••]. Nuclear import function is lost as RanGTP can no longer dissociate the inhibitor.
PY-NLS prediction
The physical rules that describe the PY-NLS (structural disorder, positive charge and weak consensus motifs) were used together in a preliminary bioinformatics endeavor that predicted ~100 candidate Kapβ2 cargos [8]. 13 were previously known Kapβ2 cargos and many new PY-NLSs and several new cargos have been validated [8,47••]. However, narrow sequence patterns used in the initial search suggest that these predicted cargos are likely only a fraction of all PY-NLS-containing proteins [8]. Unfortunately, PY-NLS diversity will severely limit the usefulness of conventional sequence-based and comprehensive identification of the signal in genomes may be extremely challenging. The core problem is that binding energy is distributed across three epitopes in many different ways. Thus, simply relaxing sequence constraints will also increase “noise” and result in many wrong answers. However, the relatively small motif sizes and their energetic independence may allow the problem to be divided into manageable pieces. If binding energies for individual PY-NLS epitopes could be predicted accurately, one could empirically determine combinations that are functional. An approach that combines bioinformatics, structural modeling and prediction of binding energies may be a solution.
Nuclear export of the leucine-rich NES containing cargos by CRM1
The study of nuclear export signals or NESs has lagged behind that of nuclear import. There is only one known class of NES at this time. The first few identified NESs are short leucine-rich segments named leucine-rich NESs. Proteins harboring these NESs are recognized and exported by CRM1 [50–53].
CRM1-specific inhibitor Leptomycin B has facilitated identification and experimental validation of >200 broadly functioning cargos [54,55]. Leucine-rich NESs were usually identified by visual inspection of protein sequences and confirmed by in vivo nuclear export assay and/or in vitro CRM1 binding. Although early NESs in HIV Rev (LPPLERLTL) and protein kinase inhibitor A (LALKLAGLDI) are rich in leucines, many leucine-rich NESs contain other hydrophobic residues [9,10]. Therefore, the 10–15-residue signal is composed of three or four regularly spaced hydrophobic residues (consensus sequence of ϕ1-X2–3-ϕ2-X2–3-ϕ3-X-ϕ4, where ϕn represents L, V, I, F or M; and X can be any amino acid) [55–57]. The consensus describes a helical pattern and is quite ambiguous and will identify segments found in most helix-containing proteins. Like the PY-NLS, the leucine-rich NES is a complex and diverse signal that cannot be sufficiently described by a consensus sequence alone. Despite its non-specific pattern, the NES consensus describes only ~40% of known NESs [55,58•]. In order to more accurately describe NES sequences, Kosugi et al. generated a large number of NES peptides in a random peptide library screen and delineated multiple distinct consensus sequences, which now collectively describe more than 86% of known functional NESs [58•].
CRM1-NES structures
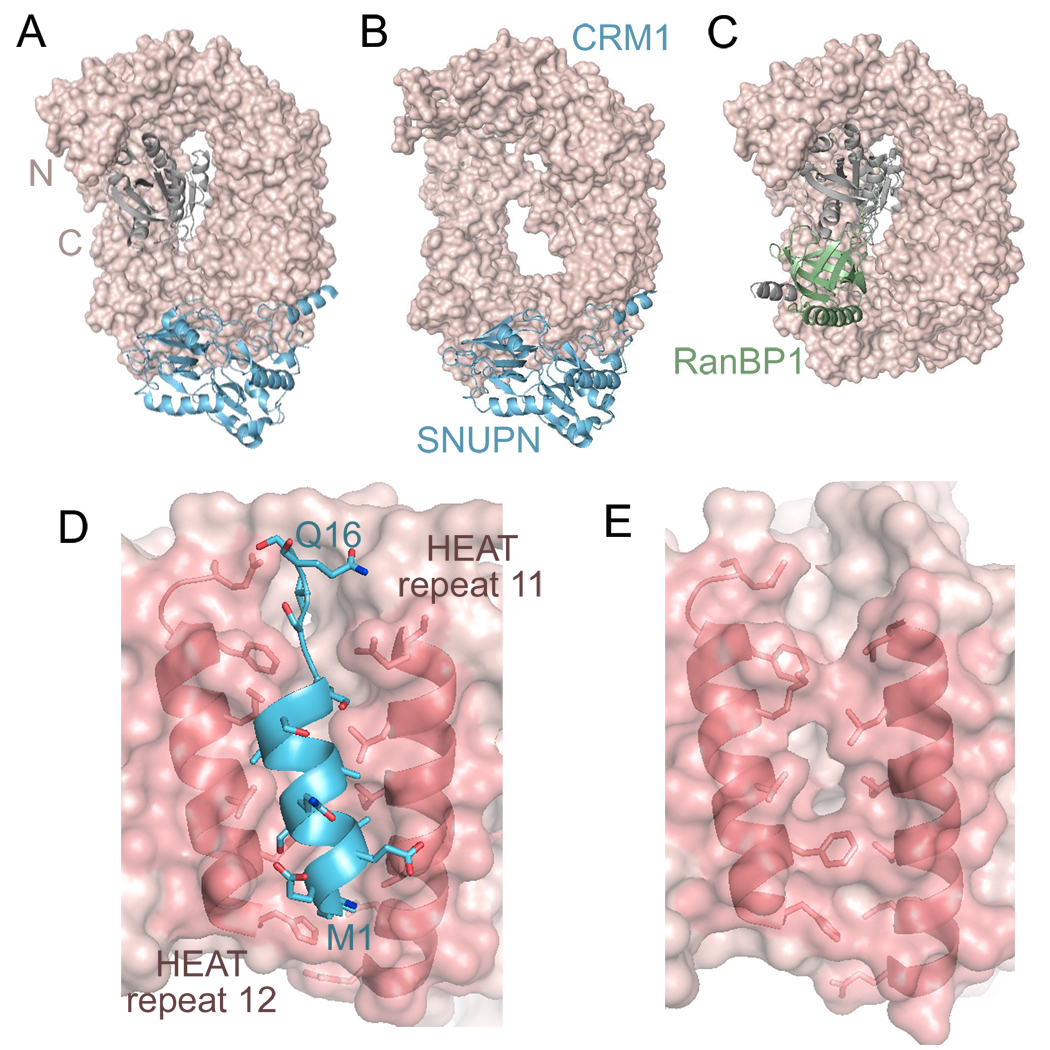
Structures of CRM1 bound to cargo Snurportin 1 (SNUPN), with and without RanGTP have recently been solved [11••,12••,59•] (Figures 3a and 3b). With the exception of the Ran binding regions, both CRM1 structures are virtually identical. CRM1 has 20 HEAT repeats that are arranged into a ring (Figures 3a–c). RanGTP binds inside the ring, contacting H1-H9, its C-terminus and the long β-hairpin loop of H9 [12••] (Figure 3a). Unlike other karyopherins, which all seem to bind cargos using their concave surfaces, SNUPN binds to the convex side of CRM1 [11••,12••].
Figure 3.
Structures of CRM1 complexes. The structure of CRM1 (pink, surface representation) bound with cargo SNUPN (light blue ribbon) with (a, PDB ID 3GJX) and without (b, PDB ID 3GB8) RanGTP (grey). (c) The structure of CRM1 (pink) bound with RanGTP (grey) and RanBP1 (green; PDB ID 3M1I). (d) The leucine-rich NES of SNUPN (light blue ribbon) bound to the hydrophobic groove of CRM1 (pink). The helices of HEAT repeats 11 and 12 are shown as ribbons and sidechains that line the hydrophobic groove are shown. (e) Rearrangement of the hydrophobic groove in the CRM1-RanGTP-RanBP1 complex. Helices of HEAT repeats 11 and 12 are shown as in (d). Helix movement resulted in narrower or ‘closed’ hydrophobic groove, which is incompatible with NES binding.
SNUPN binds CRM1 in a multipartite manner through its N-terminus, nucleotide binding domain (NBD) and C-terminal tail [11••,12••]. The first 16 residues of SNUPN contain a leucine-rich NES [11••], which binds to a hydrophobic groove at HEAT repeats H11 and H12 of CRM1 (Figure 3d). The N-terminal location and the 30-residue loop that follows allow the NES to protrude away from the rest of SNUPN and be accessible for CRM1-binding. The NES forms a short 3-turn amphiphatic helix, followed by three residues in extended conformation. Hydrophobic side-chains are aligned for placement into the hydrophobic groove, leaving polar residues exposed on the opposite side. Interaction between the NBD and CRM1 is mostly electrostatic, with the NBD basic surface binding to an acidic CRM1 surface that is adjacent to the NES-binding groove.
Leptomycin B alkylates Cys 528 of human CRM1, which is located in the NES-binding groove [11••,12••]. The inhibitor likely occupies the hydrophobic groove and prevents NES binding. Binding of Ran effector RanBP1 to CRM1 and RanGTP also displaces the NES. Koyama and Matsuura showed that RanBP1 binding shifts the internal H9 loop of CRM1 to the concave side of the NES-binding groove (H11 and H12), driving helices there together and closing the hydrophobic groove to dissociate the NES [60•] (Figures 3d and 3e). It is unclear if the NES groove is similarly closed in unliganded CRM1.
Physical characteristics of the leucine-rich NES
The helical structure of the leucine-rich NES reflects its consensus sequence and may be an important and prevalent physical characteristic of the signal. Interestingly, the most active NES sequences identified in an export activity assessment study bear the helical consensus of ϕ1-X3-ϕ2-X2-ϕ3-X-ϕ4 [57]. The same sequence pattern was found to be the most abundant natural NESs [58•]. However, the CRM1 groove likely also accommodates non-helical NESs, as a few of the new consensus sequences by Kosugi et al are not helical patterns [58•]. High proline content of the well-known NES from HIV Rev (LPPLERLTL) suggests that this particular signal is either a severely distorted helix or adopts an extended conformation. Additional structures of leucine-rich NESs bound to CRM1 will be required to map the repertoire of NES conformations that are recognized by CRM1.
The ambiguous NES consensus, prevalence of the sequence patterns in the genome and the high likelihood that hydrophobic sequences are found in the interior of proteins all suggest that many identified and predicted leucine-rich NESs may be false positives. It is critical that a nuclear targeting signal be accessible, within the full-length cargo protein, for interactions with its Kapβ. Regions surrounding NESs tend to be over-represented by polar and acidic residues, perhaps reflecting protein surface locations [55,58•]. Leucine-rich NESs located at protein termini, in structurally disordered regions or flanked by long loops, are more likely to be accessible to CRM1.
Conclusions
Even though 19 different Kapβs mediate nuclear transport of proteins, only three classes of nuclear targeting signals have been characterized. The Impα/β heterodimer recognizes the monopartite and bipartite classical-NLSs, Kapβ2 recognizes the PY-NLS and CRM1 recognizes the leucine-rich NES. The monopartite classical-NLS is a compact signal with a well-defined sequence pattern but the bipartitie classical-NLS is larger and more complex. The PY-NLS and the leucine-rich NES are very diverse signals that cannot be sufficiently described by traditional sequence patterns but are instead described by collections of physical rules. One of the most important requirements for a nuclear targeting signal is its accessibility within the cargo protein, indicating that the signal either lacks secondary structure or it is located in a disordered or flexible region. Dozens of nuclear targeting signals have yet to be discovered and many are likely poorly defined in sequence. The concept of defining nuclear targeting signals with a series of weakly restrictive rules may be generally true.
Acknowledgements
We thank Bostjan Kobe, Anita Corbett, Dirk Görlich and Thomas Güttler for discussions. This work is funded by the National Institutes of Health (R01-GM069909 and GM007062), Welch Foundation (I-1532), Leukemia and Lymphoma Society Scholar award and UT Southwestern Endowed Scholars Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.O'Reilly AJ, Field MC. Evolution of the karyopherin-β family of nucleocytoplasmic transport factors. doi: 10.1371/journal.pone.0019308. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran EJ, Bolger TA, Wente SR. SnapShot: nuclear transport. Cell. 2007;131:420. doi: 10.1016/j.cell.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 3.Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 4.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suel KE, Chook YM. Kap104p imports the PY-NLS-containing transcription factor Tfg2p into the nucleus. J Biol Chem. 2009;284:15416–15424. doi: 10.1074/jbc.M809384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dingwall C, Sharnick SV, Laskey RA. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell. 1982;30:449–458. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- 7.Kalderon D, Richardson WD, Markham AF, Smith AE. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 8.Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 10.Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 11. Dong X, Biswas A, Süel K, Jackson L, Martinez R, Gu H, Chook YM. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature. 2009;458:1136–1141. doi: 10.1038/nature07975.•• Structure of the CRM1-SNUPN complex and discovery of the leucine-rich NES in SNUPN.
- 12. Monecke T, Guttler T, Neumann P, Dickmanns A, Gorlich D, Ficner R. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science. 2009;324:1087–1091. doi: 10.1126/science.1173388.•• Structure of the CRM1-SNUPN-RanGTP complex.
- 13.Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 14.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 15.Conti E, Izaurralde E. Nucleocytoplasmic transport enters the atomic age. Curr Opin Cell Biol. 2001;13:310–319. doi: 10.1016/s0955-0674(00)00213-1. [DOI] [PubMed] [Google Scholar]
- 16.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 17.Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat Struct Biol. 1999;6:388–397. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- 18.Cingolani G, Petosa C, Weis K, Muller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 19.Moroianu J, Blobel G, Radu A. The binding site of karyopherin alpha for karyopherin beta overlaps with a nuclear localization sequence. Proc Natl Acad Sci U S A. 1996;93:6572–6576. doi: 10.1073/pnas.93.13.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fontes MR, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J Mol Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- 21.Chelsky D, Ralph R, Jonak G. Sequence requirements for synthetic peptide-mediated translocation to the nucleus. Mol Cell Biol. 1989;9:2487–2492. doi: 10.1128/mcb.9.6.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodel MR, Corbett AH, Hodel AE. Dissection of a nuclear localization signal. J Biol Chem. 2001;276:1317–1325. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- 23.Yang SN, Takeda AA, Fontes MR, Harris JM, Jans DA, Kobe B. Probing the specificity of binding to the major nuclear localization sequence-binding site of importin-alpha using oriented peptide library screening. J Biol Chem. 2010;285:19935–19946. doi: 10.1074/jbc.M109.079574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fontes MR, Teh T, Toth G, John A, Pavo I, Jans DA, Kobe B. Role of flanking sequences and phosphorylation in the recognition of the simian-virus-40 large T-antigen nuclear localization sequences by importin-alpha. Biochem J. 2003;375:339–349. doi: 10.1042/BJ20030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubner S, Xiao CY, Jans DA. The protein kinase CK2 site (Ser111/112) enhances recognition of the simian virus 40 large T-antigen nuclear localization sequence by importin. J Biol Chem. 1997;272:17191–17195. doi: 10.1074/jbc.272.27.17191. [DOI] [PubMed] [Google Scholar]
- 26.Jans DA, Ackermann MJ, Bischoff JR, Beach DH, Peters R. p34cdc2-mediated phosphorylation at T124 inhibits nuclear import of SV-40 T antigen proteins. J Cell Biol. 1991;115:1203–1212. doi: 10.1083/jcb.115.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dingwall C, Laskey RA. Nuclear targeting sequences: a consensus? Trends Biol. Sci. 1991;16:178–181. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 28.Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure Fold Des. 2000;8:329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 29. Lange A, McLane LM, Mills RE, Devine SE, Corbett AH. Expanding the definition of the classical bipartite nuclear localization signal. Traffic. 2010;11:311–323. doi: 10.1111/j.1600-0854.2009.01028.x.• Examination of the linker lengths in bipartite classical-NLS.
- 30.Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J Biol Chem. 2009;284:478–485. doi: 10.1074/jbc.M807017200. [DOI] [PubMed] [Google Scholar]
- 31. Kosugi S, Hasebe M, Entani T, Takayama S, Tomita M, Yanagawa H. Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity-based profiling. Chem Biol. 2008;15:940–949. doi: 10.1016/j.chembiol.2008.07.019.• Design of peptide inhibitors that bind Impα with extremely high affinity.
- 32.Cingolani G, Bednenko J, Gillespie MT, Gerace L. Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta. Mol Cell. 2002;10:1345–1353. doi: 10.1016/s1097-2765(02)00727-x. [DOI] [PubMed] [Google Scholar]
- 33.Lee SJ, Sekimoto T, Yamashita E, Nagoshi E, Nakagawa A, Imamoto N, Yoshimura M, Sakai H, Chong KT, Tsukihara T, Yoneda Y. The structure of importin-beta bound to SREBP-2: nuclear import of a transcription factor. Science. 2003;302:1571–1575. doi: 10.1126/science.1088372. [DOI] [PubMed] [Google Scholar]
- 34.Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G. Molecular basis for the recognition of snurportin 1 by importin beta. J Biol Chem. 2008;283:7877–7884. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- 35.Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 36.Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin beta2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci U S A. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawamura H, Tomozoe Y, Akagi T, Kamei D, Ochiai M, Yamada M. Identification of the nucleocytoplasmic shuttling sequence of heterogeneous nuclear ribonucleoprotein D-like protein JKTBP and its interaction with mRNA. J Biol Chem. 2002;277:2732–2739. doi: 10.1074/jbc.M108477200. [DOI] [PubMed] [Google Scholar]
- 38.Truant R, Kang Y, Cullen BR. The human tap nuclear RNA export factor contains a novel transportin-dependent nuclear localization signal that lacks nuclear export signal function. J Biol Chem. 1999;274:32167–32171. doi: 10.1074/jbc.274.45.32167. [DOI] [PubMed] [Google Scholar]
- 39.Fan XC, Steitz JA. HNS a nuclear-cytoplasmic shuttling sequence in HuR. Proc Natl Acad Sci U S A. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siomi MC, Eder PS, Kataoka N, Wan L, Liu Q, Dreyfuss G. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki M, Iijima M, Nishimura A, Tomozoe Y, Kamei D, Yamada M. Two separate regions essential for nuclear import of the hnRNP D nucleocytoplasmic shuttling sequence. Febs J. 2005;272:3975–3987. doi: 10.1111/j.1742-4658.2005.04820.x. [DOI] [PubMed] [Google Scholar]
- 42.Chook YM, Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature. 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- 43.Cansizoglu AE, Lee BJ, Zhang ZC, Fontoura BM, Chook YM. Structure-based design of a pathway-specific nuclear import inhibitor. Nat Struct Mol Biol. 2007;14:452–454. doi: 10.1038/nsmb1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cansizoglu AE, Chook YM. Conformational heterogeneity of karyopherin beta2 is segmental. Structure. 2007;15:1431–1441. doi: 10.1016/j.str.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imasaki T, Shimizu T, Hashimoto H, Hidaka Y, Kose S, Imamoto N, Yamada M, Sato M. Structural Basis for Substrate Recognition and Dissociation by Human Transportin 1. Molecular Cell. 2007;28:57–67. doi: 10.1016/j.molcel.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 46.Chook YM, Jung A, Rosen MK, Blobel G. Uncoupling Kapbeta2 substrate dissociation and ran binding. Biochemistry. 2002;41:6955–6966. doi: 10.1021/bi012122p. [DOI] [PubMed] [Google Scholar]
- 47. Suel KE, Gu H, Chook YM. Modular organization and combinatorial energetics of proline-tyrosine nuclear localization signals. PLoS Biol. 2008;6:e137. doi: 10.1371/journal.pbio.0060137.•• Energetic organization of the PY-NLS.
- 48.Lau CK, Delmar VA, Chan RC, Phung Q, Bernis C, Fichtman B, Rasala BA, Forbes DJ. Transportin regulates major mitotic assembly events: from spindle to nuclear pore assembly. Mol Biol Cell. 2009;20:4043–4058. doi: 10.1091/mbc.E09-02-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, Hammond JW, Truong YN, Margolis B, Martens JR, Verhey KJ. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol. 2010 doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 51.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M. CRM1 is responsible for intracellular transport meditted by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 52.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 53.Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 54.Hutten S, Kehlenbach RH. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 2007;17:193–201. doi: 10.1016/j.tcb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 55.la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 56.Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henderson BR, Eleftheriou A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res. 2000;256:213–224. doi: 10.1006/excr.2000.4825. [DOI] [PubMed] [Google Scholar]
- 58. Kosugi S, Hasebe M, Tomita M, Yanagawa H. Nuclear export signal consensus sequences defined using a localization-based yeast selection system. Traffic. 2008;9:2053–2062. doi: 10.1111/j.1600-0854.2008.00825.x.• new consensus sequences for the leucine-rich NES from random peptide library screen and mutagenesis.
- 59. Dong X, Biswas A, Chook YM. Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nat Struct Mol Biol. 2009;16:558–560. doi: 10.1038/nsmb.1586.• Mechanism of SNUPN dissociation from CRM1 in the cytoplasm.
- 60. Koyama M, Matsuura Y. An allosteric mechanism to displace nuclear export cargo from CRM1 and RanGTP by RanBP1. EMBO J. 2010;29:2002–2013. doi: 10.1038/emboj.2010.89.• Structure of the CRM1-RanGTP-RanBP1 complex.