Abstract
The western clawed frog Xenopus tropicalis is an important model for vertebrate development that combines experimental advantages of the African clawed frog Xenopus laevis with more tractable genetics. Here we present a draft genome sequence assembly of X. tropicalis. This genome encodes over 20,000 protein-coding genes, including orthologs of at least 1,700 human disease genes. Over a million expressed sequence tags validated the annotation. More than one-third of the genome consists of transposable elements, with unusually prevalent DNA transposons. Like other tetrapods, the genome contains gene deserts enriched for conserved non-coding elements. The genome exhibits remarkable shared synteny with human and chicken over major parts of large chromosomes, broken by lineage-specific chromosome fusions and fissions, mainly in the mammalian lineage.
African clawed frogs (the genus Xenopus, meaning "strange foot") comprise more than twenty species of frogs native to Sub-Saharan Africa. The species Xenopus laevis was first introduced to the U.S. in the nineteen forties where a low-cost pregnancy test took advantage of the responsiveness of frogs to human chorionic gonadotropin(1). Since the frogs were easy to raise and had other desirable properties such as large eggs, external development, easily manipulated embryos and transparent tadpoles, X. laevis gradually developed into one of the most productive model systems for vertebrate experimental embryology(2).
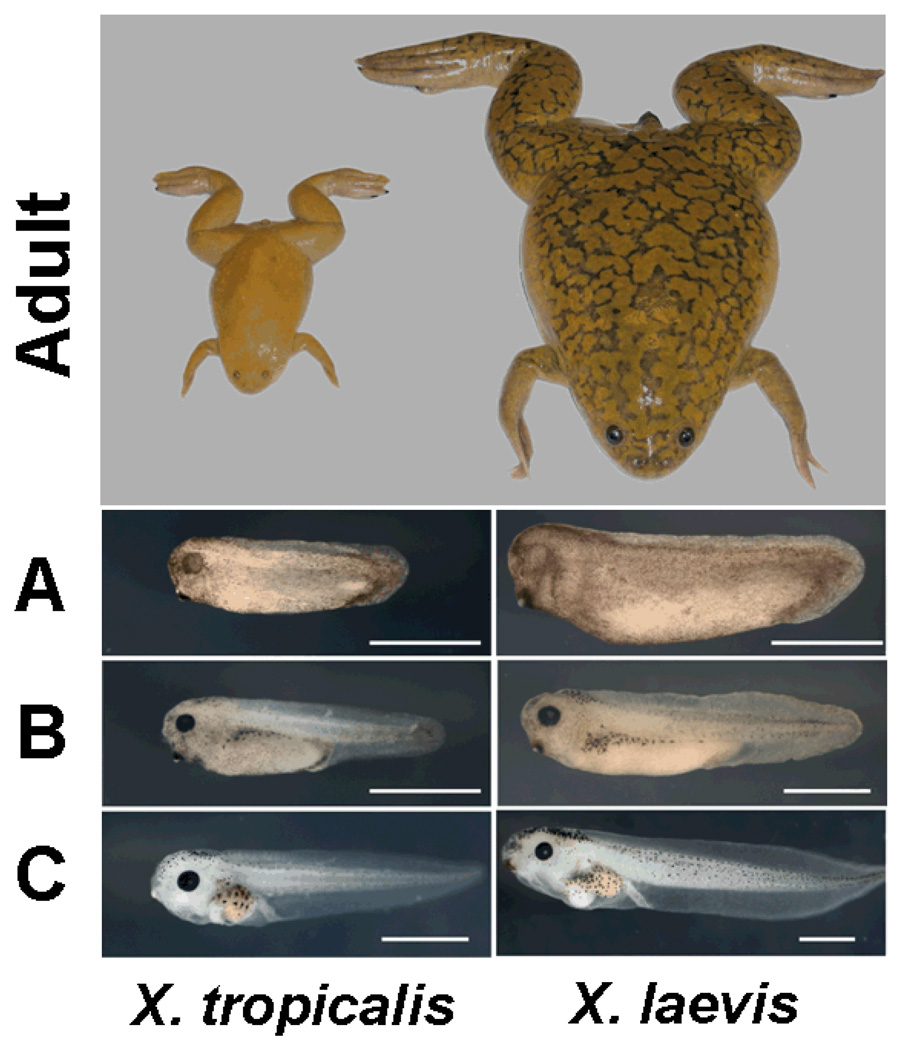
However, X. laevis has a large paleotetraploid genome with an estimated size of 3.1 billion bases (Gbp) on 18 chromosomes and a generation time of 1–2 years. In contrast, the much smaller diploid western clawed frog, X. tropicalis, has a small genome, about 1.7 Gbp on 10 chromosomes (3), matures in only 4 months and requires less space than its larger cousin. It is thus readily adopted as an alternative experimental subject for developmental and cell biology (Fig. 1).
Fig. 1.
Comparison of adults and tadpoles of X.tropicalis and X. laevis. Adult body length is 5 and 10 cm respectively. (A) tailbud (B) swimming tadpole (C) feeding tadpole. White bar indicates 1 mm.
As a group, amphibians are phylogenetically well-positioned for comparisons to other vertebrates, having diverged from the amniote lineage (mammals, birds, reptiles) some 360 million years ago. The comparison with mammalian and bird genomes also provides opportunity to examine the dynamics of tetrapod chromosomal evolution.
The X. tropicalis draft genome sequence described here was produced from ~7.6-fold redundant random shotgun sampling of genomic DNA from a seventh generation inbred Nigerian female. The assembly ((4), Tables S1–S3 and accession AAMC00000000) spans about 1.51 Gbp of scaffolds, with half of the assembled sequence contained in 272 scaffolds ranging in size from 1.56 to 7.82 Mb. Of known genes, 97.6% are present in the assembly, attesting to its near completeness in genic regions (4). Nearly two million Xenopus ESTs from diverse developmental stages and adult tissues complement the genome and enable studies of alternative splicing and identification of developmental stage- and tissue-specific genes (4).
Over a third of the frog genome consists of transposable elements (TEs), (Table S7), higher than the 9% TE density in the chicken genome (5) but comparable to the 40–50% density in mammalian genomes(6–7). Many families of frog TEs are more than 25% divergent from their consensus sequence, so like mammalian and bird TEs they have persisted for as long as 20–200 million years (5–6). This contrasts with the faster turnover observed in insects, nematodes, fungi, and plants (6, 8–9). Recently active TEs (1–5 Mya) are more common in frogs than in mammals or birds and are comparable with prevalence in fish, insects, nematodes, and plants. Among these is an unusually high diversity of very young families of L1 non-LTR retrotransposons, Penelope, and DIRS retrotransposons. In contrast to other vertebrates, most recognizable transposable elements (72%) are DNA transposons, rather than the retrotransposons that dominate other genomes (5–8, 10). Among these families(11–12), we identify Kolobok is a novel superfamily of DNA transposons. The genome also contains LTR retrotransposons of all major superfamilies, with higher diversity than in all other studied eukaryotes (Table S8). While most are ubiquitous, Copia, BEL, and Gypsy elements are not found in birds and mammals, suggesting that this subset became immobile after divergence from the amphibian lineage.
We estimate that the X. tropicalis genome contains 20,000 to 21,000 protein-coding genes using homology-based gene prediction methods and deep Xenopus EST and cDNA resources. These include orthologs of 79% of identified human disease genes (4). The genome contains 1,850 tandem expanded gene families with between 2 and 160 copies, accounting for nearly 24% of protein-coding loci. The largest expansion comprises tetrapod specific olfactory receptors (class II) occupying the first 1.7 Mb on scaffold_24. Other large expansions include protocadherins, bitter-taste receptors, and vomeronasal (pheromone) receptors (Table S9).
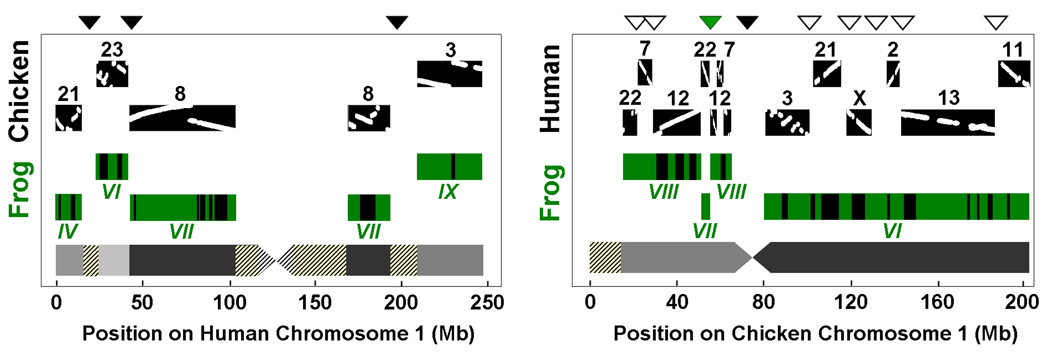
The X. tropicalis genome displays long stretches of gene colinearity with human and chicken (Fig. 2). Of the 272 largest scaffolds (totaling half the assembly) 267 show such colinearity (4). 60% of all gene models on these scaffolds can be directly associated with a human and/or chicken ortholog by conserved synteny. Patches of strict conserved colinearity are interrupted by large-scale inversions within the same linkage groups, and more rarely by chromosome breakage and fusion events, similar to the findings reported for human and chicken (Fig. 2, (5)) and in agreement with persistent conservation of linkage groups across chordates (13).
Figure 2.
Blocks of conserved tetrapod linkage for human (panel A) and chicken (panel B) chromosome 1 reveal fusions (solid black triangles) and break points (unfilled triangles) in amniotes. A total of three human fusions (panel A), seven human breaks (panel B), and one chicken fusion (panel B) is observed. The green triangle in panel B indicates the position of an apparent frog-specific break or ancestral amniote fusion. Grey areas indicate origin in different ancestral chromosomes. Shaded areas show larger regions with insufficient three-way synteny information. Detailed comparison of gene order in human and chicken reveals multiple large-scale inversions (dot plots on the black blocks). The green frog blocks consist of multiple scaffolds, 55 in panel A and 97 in panel B. Bars on the frog blocks show the location of scaffolds which do not contain markers from the linkage map, but have been predicted to associate with the linkage group by conserved synteny.
We uniquely placed 1,696 markers from the existing genetic map of X. tropicalis (http://tropmap.biology.uh.edu/map.html) onto a total of 691 scaffolds constituting more than 764 Mb of genomic sequence (4, 14). To identify lineage-specific fusion- and breakage-events within the mammals and sauropsids we identified blocks of conserved synteny between frog, human, and chicken. These blocks were detected using genomic probes comprising three-way orthologs between these tetrapods. 5,642 of these probes define conserved linkage blocks containing at least 15 genes and at least 2Mb of sequence (4, 14). The tetrapod ancestry of human and chicken chromosome 1 is outlined in Fig. 2. Remarkably, a core of more than 150 Mb of sequence spanning the centromere of human chr 1 (chicken chr 8, frog LG VII) has remained largely intact during ~360 million years of evolution since the tetrapod ancestor (Fig. 2A). Detailed shared synteny is interrupted by large-scale inversions, but gene order is frequently conserved over stretches of tens of Mb. Human chromosome 1 is seen to have grown by three lineage-specific mammalian fusions. In contrast, there are several mammalian-specific breakpoints (Fig. 2B). The genomic material on the entire q arm of chicken shows linkage conservation to frog LG VI while the human counterparts are scattered over regions of chromosomes 2, 3, 11, 13, 21, and X. The p arm indicates two mammalian breaks, suggesting that regions of chromosomes 7, 12, and 22 were once part of the same chromosome.
By extending this analysis to all human and chicken chromosomes we identified 22 human fusion and 21 fission events, versus only four fusions and one break in chicken. Clearly, the mammalian lineage has undergone considerably more rearrangement than the sauropsids, although the total chromosome count appears to have remained fairly constant. The segments analyzed here are distributed on 23 human and 22 chicken chromosomes, consistent with a derivation from 24 or 25 ancestral amniote chromosomes. Note that the chicken microchromosomes are unresolved by this analysis, preventing determination of the exact ancestral chromosome number. Both the vertebrate and eumetazoan ancestors have been suggested to have had about a dozen large chromosomes (13, 15). The current analysis indicates that the amniote ancestor had twice as many, suggesting substantial chromosome breakage on the amniotic stem.
The extensive conserved synteny among tetrapods allows us to provisionally place frog scaffolds without genetic markers onto the linkage map. These are shown in Fig 2 as black bars within the blocks of conserved linkage with frog. A total of 170 large scaffolds containing about 200 Mb of sequence were assigned a linkage group in this manner. Such in silico inferred linkages will ultimately need to be verified experimentally, but have already proven useful in the positional identification and cloning of the gene responsible for the muzak mutation, which affects heart function (16).
The X. tropicalis genome exhibits extensive sequence conservation with other vertebrates, with the amphibian sequence filling a phylogenetic gap. Recognizable noncoding sequence conservation diminishes steadily with increasing evolutionary distance (Fig. S6). Frog genes adjacent to conserved non-coding sequences (CNS) are enriched or depleted in several gene ontology categories, including sensory perception of smell, response to stimulus, and regulation of transcription, among others (Table S16).
Gene deserts (defined as the top 3 percent of the longest intergenic regions) cover 17% of the genome and vary between 201 kbp and 1.2 Mbp. The 683 gene deserts contain almost 25% of CNSs. In mammalian genomes, these gene deserts have been found to harbor cis-regulatory elements(17).
The power of genome comparison and high-throughput transgenesis in Xenopus is illustrated in Fig. S7, where several mammalian-Xenopus CNS at the Six3 locus were assayed for enhancers regulating its eye- and forebrain-specific expression. The analysis suggests that frog-mammal comparisons may be more suitable than fish-mammal comparisons for identifying conserved cis-regulatory elements (see, e.g., CNS5 in Fig. S7).
Developmental pathways controlling early vertebrate axis specification were first implicated by work in Xenopus (2) but some interesting amphibian modifications can be found. For example, a Wnt ligand required for dorsal development, named Wnt11b in X. tropicalis, has been lost from mammals, but is found in the chick and zebrafish (as silberblick) (18). Despite its retention in these vertebrates, there is no evidence to support a maternal role in axis formation similar to Xenopus. Similarly a tbx16 homolog, vegT, is retained in frog, fish and chick, but is uniquely used in Xenopus for the establishment of the endoderm and mesoderm (19).
X. tropicalis also shows multiplications of genes deployed at the blastula and gastrula stages. For example, mammals have a single nodal gene, while X. tropicalis has more than 6. Synteny relationships reveal that nodal4 on scaffold 204 is orthologous to the single human nodal, while a cluster of more than 6 nodals on scaffold 34 is orthologous to the chicken nodal. Further analysis suggests that these two nodal loci arose in one of the whole-genome duplications at the base of vertebrate evolution and that the birds and mammals subsequently lost different nodal genes, while the lizard Anolis carolinensis has retained both copies (4).
The theme of duplication is reiterated by several transcription factors that act during gastrulation (4). The transcriptional activator siamois, expressed in the organizer, is triplicated locally in the genome; so far this gene is unique to the frog. The ventx genes are expressed at the same time, but opposite the organizer, and are present in six linked copies.
Conservation of the vertebrate immune system is highlighted by mammalian and Xenopus genome comparisons (20–21). While orthology is usually obvious, synteny has been an important tool to identify diverged genes. For example, a diverged CD8 beta retains proximity to CD8 alpha, and CD4 neighbors Lag3 and B protein. Similarly, an Interleukin2/21-like sequence was identified in a syntenic region between the tenr and centrin4 genes. The immunoglobulin repertoire provides further links between vertebrate immune systems. The IgW immunoglobulin was thought to be unique to shark/lungfish, but an orthologous IgD isotype in frog provides a connection between the fish and amniote gene families (22–23).
Unique antimicrobial peptides play an important role in skin secretions that are absent in birds, reptiles and mammals. Antimicrobial peptides (caerulein, levitide, magainin, PGLa/PYLa, PGQ, xenopsin), neuromuscular toxins (e.g. xenoxins) and neuropeptides (e.g. thyrotropin releasing hormone, TRH) (24) are secreted by granular glands and the first group represents an important defense against pathogens (25). Antimicrobial peptides are clustered in at least seven transcription units over 350 kbp on scaffold 811, with no intervening genes.
X. tropicalis occupies a key phylogenetic position among previously sequenced vertebrate genomes, namely amniotes and teleost fish. Given the utility of the frog as a genetic and developmental biology system and the large and increasing amounts of cDNA sequence from the pseudo-tetraploid X. laevis, the X. tropicalis reference sequence is well poised to advance our understanding of genome and proteome evolution in general, and vertebrate evolution in particular.
Supplementary Material
Acknowledgments
This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396. This research was supported in part by the Intramural Research Program of the NIH, National Library of Medicine, and by a grant to R.K.W. from the National Human Genome Research Institute (NHGRI U01 HG02155) with supplemental funds provided by the National Institute of Child Health and Human Development. We thank Richard Gibbs and Steve Scherer of the Human Genome Sequencing Center, Baylor College of Medicine, for their contributions to SSLP identification and mapping.
References and Notes
- 1.Hogben L, Gordon C. J. Exp. Biol. 1930;7 [Google Scholar]
- 2.Brown DD. J Biol Chem. 2004 Oct 29;279:45291. doi: 10.1074/jbc.X400008200. [DOI] [PubMed] [Google Scholar]
- 3.Tymowska J. Cytogenet Cell Genet. 1973;12:297. doi: 10.1159/000130468. [DOI] [PubMed] [Google Scholar]
- 4.SOM. 2010 [Google Scholar]
- 5.I. C. G. S. Consortium. Nature. 2004 Dec 9;432:695. [Google Scholar]
- 6.Lander ES, et al. Nature. 2001 Feb 15;409:860. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Waterston RH, et al. Nature. 2002 Dec 5;420:520. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 8.Kapitonov VV, Jurka J. Genetica. 1999;107:27. [PubMed] [Google Scholar]
- 9.Kapitonov VV, Jurka J. Proc Natl Acad Sci U S A. 2003 May 27;100:6569. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.RiceConsortium. Nature. 2005 Aug 11;436:793. [Google Scholar]
- 11.Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. [Google Scholar]
- 12.Kapitonov VV, Jurka J. DNA Cell Biol. 2004 May;23:311. doi: 10.1089/104454904323090949. [DOI] [PubMed] [Google Scholar]
- 13.Putnam NH, et al. Science. 2007 Jul 6;317:86. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 14.SOD. 2010 [Google Scholar]
- 15.Woods IG, et al. Genome Res. 2005 Sep;15:1307. doi: 10.1101/gr.4134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abu-Daya A, Sater AK, Wells DE, Mohun TJ, Zimmerman LB. Dev Biol. 2009 Dec 1;336:20. doi: 10.1016/j.ydbio.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Science. 2003 Oct 17;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 18.Garriock RJ, Warkman AS, Meadows SM, D'Agostino S, Krieg PA. Dev Dyn. 2007 May;236:1249. doi: 10.1002/dvdy.21156. [DOI] [PubMed] [Google Scholar]
- 19.Kofron M, et al. Development. 1999 Dec;126:5759. doi: 10.1242/dev.126.24.5759. [DOI] [PubMed] [Google Scholar]
- 20.Du Pasquier L, Schwager J, Flajnik MF. Annu Rev Immunol. 1989;7:251. doi: 10.1146/annurev.iy.07.040189.001343. [DOI] [PubMed] [Google Scholar]
- 21.Robert J, Ohta Y. Dev Dyn. 2009 Jun;238:1249. doi: 10.1002/dvdy.21891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohta Y, Flajnik M. Proc Natl Acad Sci U S A. 2006 Jul 11;103:10723. doi: 10.1073/pnas.0601407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y, et al. Proc Natl Acad Sci U S A. 2006 Aug 8;103:12087. doi: 10.1073/pnas.0600291103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreil G. Skin Secretions of Xenopus Laevis. In: Tinsley HR, editor. The Biology of Xenopus. Oxford: The Zoological Society of London; 1996. [Google Scholar]
- 25.LAR Rollins-Smith LK, Houston CJLE, Woodhams DC. Antimicrobial peptide defenses in amphibian skin Integrative and Comparative Biology. vol. 45. 2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.