Abstract
One of the important questions in the field of virus research is about the balance between latent and lytic cycles of replication. Kaposi's sarcoma-associated herpesvirus (KSHV) remains predominantly in a latent state, with only 1–3% of cells supporting a lytic replication at any time. KSHV glycoprotein B (gB) is expressed not only on the virus envelope but also on the surfaces of the few cells supporting lytic replication. Using co-culture experiments, we determined that expression of KSHV gB on as few as 1–2% of human dermal microvascular endothelial cells resulted in a 10-fold inhibition of expression of ORF50, a viral gene critical for the onset of lytic replication. Also, we demonstrate that such a profound inhibitory effect of gB on the lytic cycle of virus replication is by repressing the ability of Egr-1 (early growth response-1) to bind and activate the ORF50 promoter. In general, virus-encoded late stage structural proteins, such as gB, are said to play major roles in virus entry and egress. The present report provides initial evidence supporting a role for membrane-associated gB expressed in a minimal number of cells to promote virus latency. These findings may have ramifications leading to a better understanding of the role of virus-encoded structural proteins not only in KSHV-related diseases but also in other viruses causing latent infections.
Keywords: DNA Viruses, Herpesvirus, Membrane, Signal Transduction, Viral Replication
Introduction
Kaposi's sarcoma-associated herpesvirus (KSHV)2 is a γ-herpesvirus that is etiologically associated with at least three types of cancers: Kaposi's sarcoma (KS), primary effusion lymphoma, and multicentric Castleman's disease (1). Similar to other herpesviruses, KSHV has two phases of replication: lytic and latent. In vivo, KSHV is known to remain predominantly in a latent state, with only 1–3% of cells supporting a lytic infection (2). Earlier studies have reported the ability of inflammatory cytokines and virus-encoded K1 and virus-encoded G protein-coupled receptor to inhibit lytic replication of KSHV (3–6). We now demonstrate that KSHV glycoprotein B (gB), one of the essential glycoproteins (7), is also involved.
KSHV gB is a lytic structural protein that is normally expressed on the envelope of mature virions but can be presented on the surface of cells supporting a lytic infection. A previous study conducted by our laboratory demonstrated that KSHV-infected cells supporting a lytic infection directed adhesion of cells. The mechanism of cell-cell adhesion, specifically in cells supporting a late stage of lytic replication, was mediated by gB instead of cellular integrins (8). This led us to suggest that gB expressed on the cell membranes may have a significant role in KS pathogenesis, particularly inducing cell survival signaling in cells apart from its role in virus binding, entry, and egress (7, 9, 10).
Cell survival is a crucial factor supporting virus infection and reactivation from latency (11). Activation of genes involved in cell survival is often regulated by NFκB signaling. KSHV infection activates NFκB signaling (12). Moreover, enhanced NFκB signaling is said to inhibit KSHV reactivation (4). Therefore, we determined the effects of gB on NFκB signaling using PCR arrays. Our results from the analysis of arrays demonstrated gB to significantly lower expression of egr-1 (early growth response-1) in target cells. Incidentally, gB expressed on 1–2% of cells could significantly inhibit Egr-1 expression in KSHV-infected target cells via autocrine and paracrine effects. Egr-1 is a nuclear protein with three consensus zinc finger sequences repeated in tandem that function as a transcription factor (13), regulating a variety of cellular functions (14). Earlier studies have demonstrated roles for proteins, such as the virus-encoded G protein-coupled receptor, expressed during the lytic cycle of replication (like gB), in mediating both autocrine and paracrine effects (15, 16) but primarily using transfection-based approaches. In the present report, for the first time, we used co-culture experiments to delineate the role of gB expressed on as few as 1–2% of cells to promote virus latency via altering Egr-1 expression. In the process, we describe an interesting pathophysiology initiated by gB expressed on a minimal number of cells.
EXPERIMENTAL PROCEDURES
Cells
HMVEC-d cells were propagated in EGMTM MV-microvascular endothelial cell medium (Clonetics) as per standard protocols (9). The passage numbers for HMVEC-d cells used in this study ranged between 5 and 9. 293 and BCBL-1 cells were cultured in DMEM and RPMI (Invitrogen) as per earlier studies (17, 18).
Plasmids
pCDNA3.1.CT-GFP-TOPO (pCDNA), gB/pCDNA3.1.CT-GFP-TOPO (gB/pCDNA), and gL/pCDNA are plasmids that encode full-length gB and gL, respectively (7, 19). Like gB, gL is also expressed independently on the cell membrane (19). Another plasmid, gB-RGA/pCDNA, encodes for full-length gB with a mutation to the RGD domain (RGD to RGA). All of these plasmids encode green fluorescent protein (GFP), which was used as a marker for sorting cells. We also used egr-1/pCDNA3.1(+) and gL/pCDNA3.1(+) vectors without the GFP tag for the short interfering RNA (siRNA) experiments (Fig. 2) and luciferase assay. Vectors pCMV and pCMV-MEK1 (20) were also used in this study.
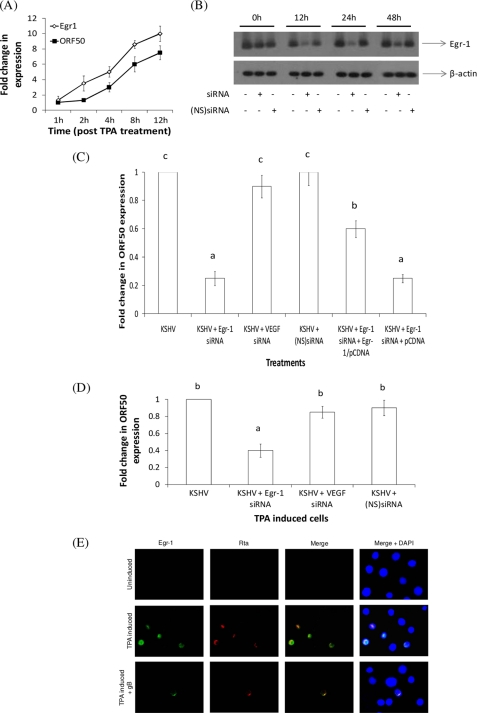
FIGURE 2.
Egr-1 plays a critical role in KSHV reactivation. A, effect of TPA-induced KSHV reactivation on egr-1 expression over a 12-h period. HMVEC-d cells were infected with KSHV for 48 h. These cells were then treated with TPA for different periods. At the end of incubation, the cells were lysed, and the expression of egr-1 and ORF50 was analyzed by qRT-PCR. Expression of genes in KSHV-infected cells was considered as 1-fold for comparisons. B, effect of transfecting HMVEC-d cells with siRNA specific to Egr-1. HMVEC-d cells were untransfected or were transfected either with specific siRNA or NS siRNA controls. After 0, 12, 24, and 48 h post-transfection, total RNA was isolated from cells and subjected to Northern blotting as per standard protocols (23). C, inhibition of Egr-1 by siRNA lowers ORF50 expression. siRNA-transfected HMVEC-d cells at the end of a 24-h incubation were infected with KSHV for 6 h and monitored for the expression of ORF50 by qRT-PCR. Expression of genes in untransfected cells was considered as 1-fold. D, inhibition of Egr-1 by siRNA lowers KSHV reactivation. HMVEC-d cells were infected with KSHV to establish a latent infection. These cells were untransfected or were transfected either with specific siRNA or NS siRNA controls. After 24 h post-transfection, cells were treated with TPA for 12 h prior to lysing the cells and monitoring the expression of ORF50 by qRT-PCR. Expression of genes in TPA-induced KSHV-infected cells was considered as 1-fold. In all of the above experiments, each bar denotes the average ± S.D. (error bars) of three experiments. The columns with different letters are statistically significant (p < 0.05) by least significant difference. E, Egr-1 is expressed in cells undergoing virus reactivation. Uninduced and TPA-induced BCBL-1 cells were sequentially stained with mouse anti-Egr-1 IgG and rabbit peptide IgGs to Rta, followed by incubating with anti-mouse-FITC and anti-rabbit TRITC, respectively, before examining under a fluorescent Nikon microscope (magnification ×100).
Antibodies
Antibodies to gB, anti-RGDgB-N1 (developed against RGD-containing peptide sequences in gB; amino acids 27–44), anti-gB-C (developed against non-RGD-containing peptide sequences in gB; amino acids 828–845) (9); anti-phospho-ERK1/2, anti-total ERK1/2, anti-actin, and monoclonal antibodies (15F7) to Egr-1 (Cell Signaling Technology, Beverly, MA) were used in this study. All of the above antibodies were made in rabbits. A mouse anti-Egr-1 monoclonal antibody purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) was used in the immunofluorescence assay.
Inhibitors
U0126 was purchased from Promega (Madison, WI) and used in this study.
Recombinant KSHV gB
Expression and purification of the recombinant KSHV gBΔTM (2106 bp; encoding amino acids 1–702 lacking the transmembrane and cytoplasmic domains) and gBΔTM-RGA proteins from the infected High-5 cells were done using nickel columns (PharMingen, Pasadena, CA) as per procedures described before (21). All of the reagents (including the above recombinant proteins) used in this study were prepared using endotoxin-free water. Additionally, the sample preparation was tested and determined free of contamination using an end point chromogenic Limulus amebocyte lysate assay (Charles River Laboratories, Charleston, SC).
Dose Optimization Assay for KSHV gBΔTM
Target cells were treated with different concentrations of gBΔTM at 37 °C in a V-bottom 96-well plate. After a 24-h incubation, the cells were analyzed for the expression of LDH, as an indicator of cell death. The LDH assay was performed using the CytoTox 96 non-radioactive kit (Promega) as per earlier studies (22).
PCR Arrays
We analyzed the manner by which gB could possibly alter signaling in endothelial cells using the NFκB signaling pathway PCR arrays. Briefly, cells cultured in growth medium were either left untreated or treated with 100 ng/ml gBΔTM at 37 °C. At the end of 2 and 4 h post-treatment, the cells were lysed, RNA was extracted, and cDNA was prepared (23). The cDNA was later used to analyze the profile of 84 genes regulated by NFκB signaling using the PCR array (catalog no. PAHS-025) as per the manufacturer's recommendations (SABiosciences, Frederick, MD).
Quantitative RT-PCR (qRT-PCR)
The qRT-PCR was performed using the synthesized cDNA in a 25-μl reaction volume to analyze the expression of ORF50, egr-1, and β-actin as per earlier protocols (24).
KSHV Infection of HMVEC-d Cells
HMVEC-d cells were infected as per earlier procedures (25). Briefly, monolayers of HMVEC-d cells were infected with a multiplicity of infection of 10 ORF73 units for 2 h (26). After incubation, the cells were washed three times in DMEM before incubating in growth medium for 48 h. This allowed virus to establish a latent infection of cells.
Co-culture Experiments
This assay was developed to analyze the effect of membrane-bound gB on the expression of egr-1 and ORF50 (virus reactivation). Two cell populations were used in this assay: the target and the effector cells. Target cells were HMVEC-d cells that were cultured as monolayers in 12-well plates. The effector cell population included 293 cells that were untransfected or were transfected with pCDNA, gB/pCDNA, or gL/pCDNA using the Nucleofector II device (27). We tested 293, HFF, HMVEC-d, and COS-1 cells as effector cells. We determined that 293 cells expressed gB on their cell surface at levels comparable with BCBL-1 cells that were induced with TPA for 48 h as measured by the mean fluorescent intensity (data not shown). Target cells were infected with KSHV as described above (26). These cells were washed twice in DMEM before trypsinizing them to obtain single cell suspension. The above single cell suspension of KSHV-infected target cells was mixed with 2, 5, 10, and 30% of effector cells that specifically expressed gB on their cell surface; seeded onto a 12-well plate; and incubated in growth medium at 37 °C. The gB- and gL-positive cells were sorted using a flow cytometer based on the expression of GFP. The time at which the mixture of cells was seeded onto 12-well plates was considered as 0 h. At different time points (1, 4, 8, 12, 24, 48, 72, 96, and 120 h) post-seeding, the cells were lysed, RNA was extracted, cDNA was prepared, and the expression of ORF50 was monitored by qRT-PCR using specific primers. The relative copy numbers of the ORF50 transcripts were calculated from the standard graph plotted using the Ct values for different dilutions of in vitro transcribed ORF50 (using ORF50/pGEM-T plasmid). These values were normalized to each other using the values of the β-actin control reactions. As reported earlier (28), the lowest limit of detection in the standard samples was 6–60 copies for the ORF50 gene. Expression of egr-1 in the above cells was monitored in the same lines.
Western Blotting
Equal amounts of protein were used in Western blotting experiments as per earlier studies (24). We used 60 μg (3 times more) of protein in this experiment for clarity of the bands. The bands were scanned, and the band intensities were assessed using the ImageQuaNT software program (Amersham Biosciences).
Silencing Egr-1 RNA Expression
Expression of Egr-1 was inhibited by the transfection of double-stranded RNA oligonucleotides as described previously (23, 29). The siRNAs used in this experiment were obtained from Dharmacon (Lafayette, CO) as the ON-TARGET plus Smart pool. The nonspecific (NS) siRNAs used were those described previously (29).
Immunofluorescence Assay for Examination of Egr-1 and Rta
Target cells were fixed for 10 min in ice-cold acetone and washed three times in phosphate-buffered saline (PBS). These cells were stained with mouse anti-Egr-1 monoclonal antibodies and rabbit peptide IgGs to Rta synthesized as per earlier studies (30) for 45 min at room temperature and incubated with goat anti-mouse FITC and goat anti-rabbit TRITC (Sigma) for 30 min at room temperature. Stained cells were washed in PBS and examined under a Nikon fluorescent microscope with appropriate filters.
In Vitro Transcription and Translation (IVT)
IVT of egr-1/pCDNA3.1(+) and gL/pCDNA3.1(+) was conducted as per earlier studies using the TNT-coupled rabbit reticulocyte lysate system (Promega).
Electromobility Shift Assay (EMSA) and Immunoprecipitated (IP) EMSA
IVT products of egr-1 or KSHV gL were evaluated by EMSA for DNA binding using a 25-bp DIG-labeled probe containing the ORF50 promoter (ORF50P) sequences (ORF50P probe). The probe encompasses the tttgacctgcgtgcgctctccggct sequence. 2.5 μl of IVT product and 50 ng/μl DIG-labeled probe were mixed in a binding buffer and allowed to incubate at room temperature for 30 min. Appropriate nonspecific probe at 1-μg amounts was used in the reaction. For supershift, IVT products were incubated with rabbit monoclonal antibodies to Egr-1 or nonspecific IgG at 37 °C for 30 min prior to the addition of the DIG-labeled probe. All samples were run on a 4% non- denaturing gel for ∼1.5 h and transferred to a PVDF membrane. The protein-DNA interaction was detected using the CSPD detection system (Roche Applied Science). The IP EMSA was conducted as per the procedures outlined in earlier studies (31) using 0.5 mg of protein from target cells, precleared with normal rabbit serum and protein A-Sepharose. In another set of experiments, we analyzed the ability of Egr-1 in the nuclear extracts to bind ORF50P by performing EMSA. Briefly, we used the annealed double-stranded DIG-labeled oligonucleotides containing the consensus binding site for Egr-1 on ORF50P as described above. EMSA was performed as per procedures outlined for testing IVT-synthesized Egr-1 using 4 μg of nuclear extract. For supershift assays, we incubated nuclear extracts with 1:50, 1:500, and 1:5000 dilutions of rabbit monoclonal antibodies to Egr-1 or nonspecific IgG at 37 °C for 30 min prior to the addition of the DIG-labeled probe and analyzing the protein-DNA interactions as per standard procedures.
Luciferase Assay
Target cells grown to 65% confluence were transiently co-transfected with 500 ng of pCDNA3.1(+), egr-1/pCDNA3.1(+), or gL/pCDNA3.1(+); 500 ng/well pGL3 or ORF50P (nucleotides 68,509–71,593; KSHV genome sequence NC_003409)/pGL3; and an internal control pRL-TK (Promega). Transfections were performed using the Nucleofector II device as per the recommendations of the manufacturer (Amaxa Inc., Gaithersburg, MD). At 48 h post-transfection, cells were harvested, and firefly and Renilla luciferase activities in cell extracts were recorded using the dual luciferase system. Luciferase activity was monitored using a Turner Systems Luminometer (Sunnyvale, CA) as in earlier studies (32). The relative luciferase activity was then calculated by normalizing ORF50P luciferase activity to control Renilla luciferase activity. The results were plotted as a percentage of the activity of the full-length promoter. In another set of experiments, 4 h prior to harvesting cells, the monolayers were incubated with 30% effector cells that were positive for gB expression or were transfected with empty vector. This was conducted to demonstrate the role of membrane-bound gB in regulating ORF50 activity.
RESULTS
KSHV-soluble gB Lowers Egr-1 Expression
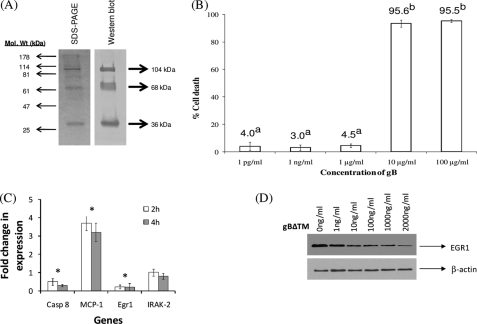
To determine the effect of KSHV gB on target cells, soluble gB protein lacking the transmembrane domain (gBΔTM) was first used. Coomassie staining of reduced gBΔTM protein yielded the expected three distinct bands at 36, 68, and 104 kDa, respectively (Fig. 1A). As suggested in the earlier studies (21), the 104-kDa protein probably represents the uncleaved gB precursor, and the 68-and 36-kDa proteins represent the cleaved and processed amino and carboxyl domains of the gBΔTM protein, respectively. Additional bands were not detected, indicating purity of the soluble protein. Treatment using rabbit anti-gB antibodies also led to the detection of three bands of the same molecular weight (Fig. 1A). The specificity of our antibodies was demonstrated by the lack of reactivity of normal rabbit sera and anti-gL antibodies (data not shown).
FIGURE 1.
A, expression and purification of KSHV gBΔTM in High-5 cells. The His-tagged gBΔTM from the cell pellets was purified using a nickel column. The purified protein samples under reduced conditions were resolved by 10% SDS-PAGE and stained by Coomassie Blue stain or analyzed by Western blotting using rabbit anti-gB antibodies. The three major bands of 36, 68, and 104 kDa are indicated by arrowheads. B, determining the cytotoxic effects of KSHV gBΔTM on HMVEC-d cells. Lactate dehydrogenase release as an indicator of percentage of cell death after treatment of HMVEC-d cells with different concentrations of gBΔTM is depicted. Data presented represent the average ± S.D. (error bars) of three experiments. The columns with different superscripts indicate statistical significance (p < 0.05) by least significant difference. C, KSHV gB suppresses egr-1. KSHV-infected target cells were either left untreated or treated with gBΔTM for 2 and 4 h, RNA was extracted, cDNA was synthesized, and analyzed by PCR arrays. Each bar denotes the average ± S.D. of three experiments. Asterisks above the columns denote the values to be statistically significant (p < 0.05) by least significant difference. D, KSHV gBΔTM induces a dose-dependent reduction in Egr-1 expression. KSHV-infected HMVEC-d cells were untreated or treated with different concentrations of gBΔTM for 2 h at 37 °C. At the end of incubation, the cells were lysed using gold lysis buffer, and lysate was resolved in a 10% SDS-PAGE, transferred to a PVDF membrane, and Western blotted using specific antibodies.
HMVEC-d cells that were treated with gBΔTM at concentrations of 1 μg/ml and below did not significantly induce cell death as monitored by the lactate dehydrogenase assay (Fig. 1B). These results were confirmed by the conventional trypan blue test. More than 95% of the target cells were found to be viable when the target cells were treated with gBΔTM at concentrations of 1 μg/ml or below (data not shown). Such non-toxic doses were used in this study. Comparable results were observed in 293, HFF, and primary effusion lymphoma cells (data not shown). Doses of about 1–4 μg/ml gBΔTM have been successfully used in functional assays (21, 33).
The manner by which gB could possibly alter NFκB signaling in KSHV-infected endothelial cells was analyzed using NFκB signaling pathway PCR arrays. When comparing untreated cells to cells that were treated with gBΔTM, the most significant changes observed were in the genes egr-1, MCP-1 (monocyte chemotactic protein-1), and caspase 8 (Fig. 1C). KSHV gBΔTM lowered expression of Egr-1 and caspase 8 mRNA in target cells. Incidentally, expression of MCP-1 was elevated in cells treated specifically with gBΔTM (Fig. 1C). Expression of IRAK-2 (Fig. 1C), β2-microglobulin, glyceraldehyde-3-phosphate dehydrogenase, and β-actin (data not shown) were not altered in response to treatment of cells with gBΔTM or in cells expressing gB on their cell surface, demonstrating the specificity of the data from the use of these arrays. These results were further confirmed by performing qRT-PCR using specific primers (data not shown).
Cell cycle plays a major role in allowing KSHV reactivation to occur (11, 34). Therefore, we performed the above experiment in KSHV-infected cells derived from G0/1 and S phases of cell cycle by the serum starvation method (18). Interestingly, egr-1 was significantly lowered in cells derived from both G0/1 and S phases (supplemental Figs. 1 and 2). The cells cultured in growth medium (Fig. 1C) responded in a manner similar to those in G0/1 phase compared with those in S phase. This could partly be because the majority of the HMVEC-d cells cultured in growth medium are in G0/1 phase (70–75%) followed by G2/M (13–16%) and S (5–8%) phase, respectively (supplemental Fig. 3).
The array data were confirmed by performing conventional Western blotting experiments. A dose-dependent inhibition of Egr-1 by gBΔTM in KSHV-infected target cells was observed (Fig. 1D). gBΔTM inhibited expression of Egr-1 when used at a concentration of ≥10 ng/ml. It did not alter the expression of β-actin at the concentrations tested (Fig. 1D). Nonspecific proteins GST and BSA (data not shown) did not have such an effect on Egr-1, confirming the specificity of the effect of gB. A similar effect on Egr-1 expression was noticed in BCBL-1 cells treated with increasing concentrations of soluble gBΔTM (data not shown). These results provide support for the ability of KSHV gB to inhibit the expression of Egr-1 at the transcriptional and translational levels in target cells.
Egr-1 Is Required for ORF50 Expression
KSHV Rta (replication and transcription factor) is encoded by ORF50, an immediate early gene that is responsible for the activation of several lytic genes (35). To date, a direct relationship between KSHV ORF50 and egr-1 has not been established. HMVEC-d cells supporting a latent KSHV infection were used to determine a possible association between the two. A significant increase in egr-1 expression was observed in KSHV-infected cells up to 12 h post-TPA treatment (Fig. 2A). Additionally, activation of ORF50 followed a similar increasing trend, indicating that TPA induced KSHV reactivation in these cells (Fig. 2A). In order to determine the necessity for Egr-1 in ORF50 expression, we monitored ORF50 expression in cells that were silenced for Egr-1 expression using specific double-stranded siRNA (36). The level of Egr-1 mRNA was significantly suppressed in HMVEC-d cells by siRNA specific to Egr-1 when compared with NS siRNA (Fig. 2B). An inhibition of 65 ± 8, 80 ± 4, and 55 ± 8% of Egr-1 mRNA was observed at 12, 24, and 48 h post-transfection.
KSHV replication follows a unique pattern in HMVEC-d cells (25). The lytic gene ORF50 is expressed as early as 2 h post-infection (PI), declining sharply by 24 h PI. This early expression of Rta is not sufficient to activate a lytic infection. KSHV is said to establish a latent infection 48 h PI. TPA treatment of KSHV-infected HMVEC-d cells 48 h PI led to expression of the full range of lytic genes (25). Thus, there are actually two distinct phases when ORF50 may be expressed in HMVEC-d cells: (i) during early stages of primary infection (TPA-independent) and (ii) when TPA is added to cells that are harboring virus DNA in a predominantly latent phase (TPA-dependent). Hence, we infected the above siRNA-transfected cells with KSHV (multiplicity of infection of 10 ORF73 units). After 6 h PI, ORF50 expression in the cells was monitored by qRT-PCR. This study monitored the expression of ORF50 during the early phase of primary infection. Cells transfected with siRNA specific to Egr-1 expressed reduced levels of ORF50 when compared with cells transfected with siRNA specific to VEGF or with NS siRNA or those left untransfected (Fig. 2C). This siRNA-specific decrease in the expression of ORF50 was significantly reversed when the cells were co-transfected with siRNA specific to egr-1 and 500 ng of egr-1/pCDNA3.1(+) plasmid compared with transfecting cells with siRNA specific to Egr-1 and 500 ng of pCDNA3.1(+) (Fig. 2C). Co-transfecting cells with gL/pCDNA3.1(+) did not reverse the inhibitory effect of siRNA specific to Egr-1 on ORF50 (data not shown). These results demonstrate the specific and crucial role for Egr-1 in the expression of Rta, the initiator of KSHV reactivation (37).
TPA-induced KSHV-infected cells that were transfected with siRNA specific to Egr-1 expressed reduced levels of ORF50 when compared with cells transfected with siRNA specific to VEGF or nonspecific NS siRNA or those left untransfected (Fig. 2D). These results suggest a key role for Egr-1 in KSHV reactivation.
In our analysis of KSHV-infected BCBL-1 cells (another cell type that inherently carries KSHV DNA) under both uninduced and TPA-induced conditions, we determined that both Egr-1 and Rta are expressed in the nucleus of cells during virus reactivation (Fig. 2E). Egr-1 is a protein expressed only at a basal level under physiological conditions (38). The expression of both Egr-1 and Rta under natural (uninduced) or TPA-induced conditions could be reversed by treating with either soluble gB (Fig. 2E) or MAPK signaling inhibitor (U0126) but not by DMSO, the solvent for U0126 (data not shown). Similar results were observed in HMVEC-d and 293 cells (data not shown). In all (Figs. 1 and 2), these results demonstrate a direct link between the expression of egr-1 and ORF50 transcription and a possible role for KSHV gB to prolong the onset of a lytic infection by blocking egr-1 and ORF50 expression.
KSHV gB Expressed on a Minimal Number of Cells Is Sufficient to Regulate ORF50 and Egr-1 Expression
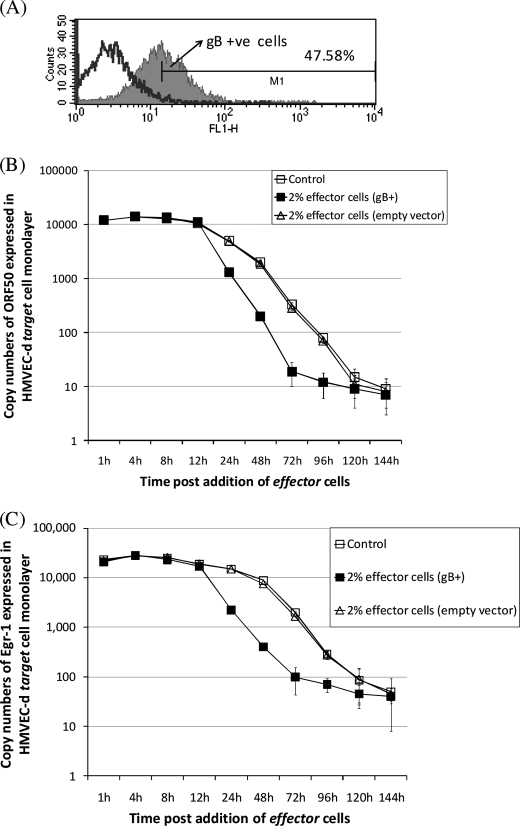
In order to test the ability of gB to promote virus latency under conditions similar to what may be observed in KS lesions, we developed an assay based on co-culturing cells. This involved the use of two different cell populations: target cells and effector cells. Effector cells expressing gB (gB+; Fig. 3A) on their surfaces significantly lowered ORF50 levels in KSHV-infected cells (Fig. 3B). ORF50 expression in KSHV-infected cells reached almost undetectable levels by 120 h. We found 2% effector cells expressing gB to reduce ORF50 to undetectable levels by 72 h (Fig. 3B). The results from using effector cells transfected with either empty vector (pCDNA) (Fig. 3B) or gL/pCDNA (data not shown) were comparable with the control (KSHV-infected target cells). ORF50 expression was inhibited to undetectable levels by 48 h itself when 30% gB+ effector cells were used compared with effector cells transfected with empty vector (data not shown). In this case, a sharp decline (up to 65%) in ORF50 expression was observed as early as 8–12 h postmixing. Our results also demonstrate that gB expressed on as few as 2% (Fig. 3C), 5%, 10%, or 30% (data not shown) of cells significantly lowered egr-1 expression compared with empty vector (Fig. 3C). There seems to be a direct correlation between the expression of egr-1 and ORF50 in response to signaling induced by membrane-bound gB in KSHV-infected cells. Silencing Egr-1 using specific siRNA lowers ORF50 expression (Fig. 2, C and D). We did not observe an additive effect by cell membrane-bound gB in lowering ORF50 expression in Egr-1-specific siRNA-transfected KSHV-infected target cells (data not shown). This further implicates gB in specifically lowering ORF50 expression via altering egr-1 expression. Our results also implicate a crucial role for the cell-cell contact to efficiently inhibit ORF50 and egr-1 expression. This is based on the fact that the supernatant obtained exclusively from the cultures of effector cells expressing gB on the cell surfaces failed to inhibit egr-1 or ORF50 expression levels in KSHV-infected target cells. Overall, we concluded that gB expressed on the cell surface of 1–2% of cells to significantly promote virus latency.
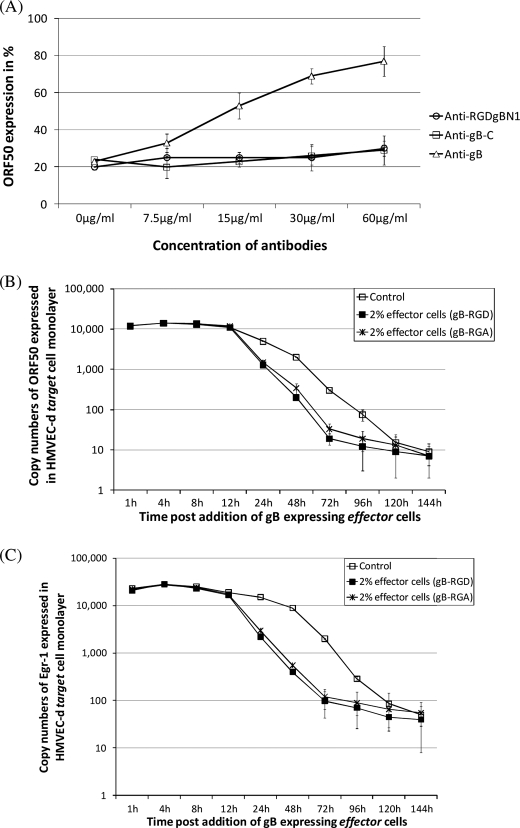
FIGURE 3.
KSHV gB expressed on a minimal number of cells lowers ORF50 and egr-1 expression. A, a FACScan flow cytometer was used to sort the gB- and GFP-positive effector cells (M1) from untransfected cells (unshaded area) as per laboratory procedures (8). B and C, KSHV-infected cells (control) were co-cultured with 2% effector cells expressing gB. At different time points, cells were lysed, and qRT-PCR was performed to monitor expression of ORF50 (B) and egr-1 (C) transcripts. Each point denotes the average ± S.D. (error bars) of three experiments.
Membrane-bound gB Blocks ORF50 Activity via the MAPK Signaling
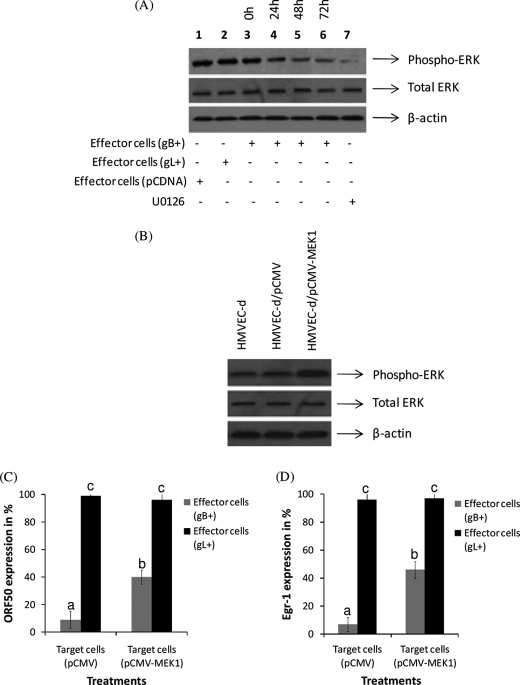
MAPK signaling is crucial for KSHV reactivation from latency (20, 39–41). Hence, we tested the effect of KSHV-encoded membrane-bound gB on ERK1/2 activity as measured by phosphorylation. We used the co-culture experiments as suggested in Fig. 3B to monitor the expression of phosphorylated ERK1/2. The level of ERK1/2 activity was significantly reduced in monolayer of HMVEC-d cells that were incubated with effector cells expressing gB on their cell surface (Fig. 4A, lanes 4–6) compared with effector cells expressing gL (Fig. 4, lane 2). Treating target cells with the MAPK inhibitor, U0126, for 24 h significantly lowered expression of the phosphorylated form of ERK1/2 (Fig. 4, lane 7). These results demonstrated the ability of KSHV membrane-bound gB expressed on a minimal number of cells to lower the MAPK signaling cascade that is crucial for the virus reactivation. Identical results were observed when KSHV-infected endothelial cells were treated with gBΔTM in the absence or in the presence of TPA (data not shown).
FIGURE 4.
KSHV membrane-bound gB lowers ERK1/2 activity. A, KSHV-infected cells (control) were co-cultured with 2% effector cells expressing gB, gL, or empty vector. At different time points, cells were lysed with gold lysis buffer, proteins were resolved in a 10% SDS-PAGE, and the expression of the phosphorylated form of ERK1/2 was monitored by Western blotting. Representational data showing the effect of gL and pCDNA at the 72 h time point (lanes 1 and 2) are depicted. B, transfection of HMVEC-d cells with pCMV-MEK1 induced elevated ERK activity. HMVEC-d cells were untransfected (lane 1) or transfected with pCMV (lane 2) or pCMV-MEK1 (lane 3). After 48 h of transfection, the cells were lysed, and the proteins were resolved by SDS-PAGE. The blots were probed for phospho-ERK1/2, total ERK1/2, and β-actin by Western blotting. KSHV membrane-bound gB regulates ORF50 (C) and egr-1 (D) activity via modulating ERK1/2 activity. The above cells were infected with KSHV. KSHV-infected cells (control) were co-cultured with 2% effector cells expressing gB or gL. At the end of 48 h, the cells were lysed, and qRT-PCR was performed to monitor expression of ORF50 (C) and egr-1 (D). Expression of ORF50 and egr-1 is presented as a percentage when compared with the effector cells that were transfected with empty vector. Each point denotes the average ± S.D. (error bars) of three experiments. The columns with different letters are statistically significant (p < 0.05) by least significant difference.
Additionally, we tested the effect of transient transfection of HMVEC-d cells with a vector encoding MEK1 on the ability of membrane-bound gB to lower ORF50 activity by co-culture experiments. A 4-fold-enhanced ERK1/2 activity in cells transfected with pCMV-MEK1 was noted when compared with both untransfected cells and those that were transfected with empty vector (Fig. 4B). We observed a significant drop in the ability of membrane-bound gB expressed on the 2% effector cells to lower ORF50 (Fig. 4C) and egr-1 (Fig. 4D) activity in cells overexpressing ERK1/2 activity. As expected, comparable results were observed when the cells were treated with U0126 (data not shown). Based on these results, we concluded that membrane-bound gB mediates its ultimate effect on egr-1 > ORF50 activity via the MAPK signaling pathway.
Effect of gB on ORF50 and Egr-1 Expression is Arg-Gly-Asp (RGD)-independent
KSHV gB has been demonstrated to mediate virus attachment and entry of target cells in an RGD-dependent manner (9, 21, 42). Therefore, we performed studies to determine if the RGD domain in gB had a role in regulating egr-1 and ORF50 expression. Accordingly, we analyzed the ability of antibodies to the RGD domain (anti-RGDgB-N1) in gB to block gB-mediated ORF50 suppression in KSHV-infected target cells. Earlier studies have documented anti-RGDgB-N1 to block the function of gB that assists in the entry of KSHV into target cells (9). For this purpose, we used 30% gB+ effector cells because they lowered ORF50 expression in a short period (4–8 h postmixing) of time (data not shown). Treatment of the effector cells expressing gB on their surface with antibodies to gB blocked the ability of gB to inhibit ORF50 expression (Fig. 5A). However, anti-RGDgB-N1 and anti-gB-C (antibodies developed against a non-RGD-containing peptide sequence) failed to significantly lower the ability of membrane-bound gB to inhibit ORF50 expression in target cells (Fig. 5A). Also, treatment of KSHV-infected target cells with gBΔTM-RGA (soluble form of gB with an RGA domain instead of an RGD) lowered ORF50 expression in cells (data not shown).
FIGURE 5.
KSHV gB modulates expression of egr-1 in an RGD-independent manner. A, 30% effector cells expressing gB were incubated with different concentrations of different antibodies for 1 h at 37 °C prior to incubating them with the target cells. At the end of 4 h of incubation, the cells were lysed, and ORF50 was monitored by qRT-PCR. The effect of antibodies on gB-repressed ORF50 expression is presented as a percentage in comparison with target cells + effector cells transfected with empty vector (considered as 100%). Data represent the average ± S.D. (error bars) of three experiments. In another set of experiments, KSHV-infected cells (control) were co-cultured with 2% effector cells expressing gB or gB-RGA. At different time points, cells were lysed, and qRT-PCR was performed to monitor expression of ORF50 (B) and egr-1 (C). Each point denotes the average ± S.D. of three experiments.
To further confirm the above results, we performed the co-culture experiments using 2% gB+ effector cells. In this experiment, we tested the effect of expressing wild type gB (gB-RGD) over gB carrying a mutation to the RGD domain (gB-RGA) on ORF50 and egr-1 expression in KSHV-infected target cells. gB-RGA expressed on the cell surface of 2% of effector cells was able to lower expression of both ORF50 and egr-1 in KSHV-infected cells in a manner similar to gB-RGD (Fig. 5, B and C). These results suggest that membrane-bound gB alters the expression of ORF50 in an RGD-independent manner.
KSHV gB Inhibits Egr-1 Binding to the ORF50P
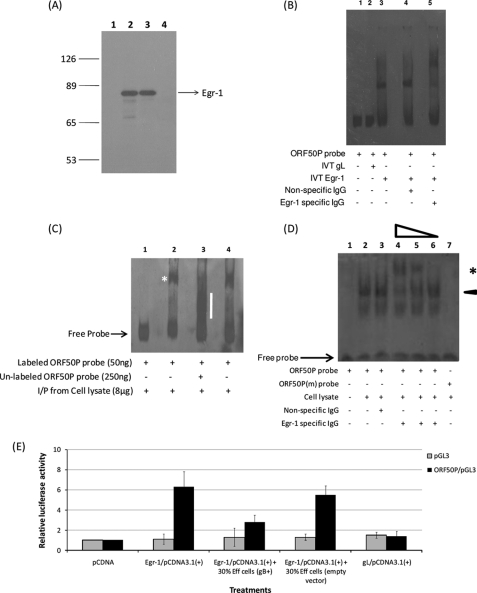
Our results implicate gB in suppressing ORF50 expression by modulating Egr-1 expression (Figs. 1–5). Egr-1 protein is a transcription factor capable of regulating the transcription of several genes (13, 43). Egr-1 is said to bind a GC-rich DNA template (such as GCGC(G/T)GGGCG, GCGGGGGCG, and CGCCCATGC) on the promoter and initiate gene transcription (43). Such GC-rich Egr-1 binding sequences have been identified by us in the promoter region of KSHV ORF50. Hence, we conducted experiments to determine if Egr-1 bound ORF50P sequences. EMSAs were performed using DIG-labeled specific probes and in vitro transcribed and translated Egr-1 proteins. The IVT of egr-1/pCDNA3.1 plasmid resulted in the generation of [35S]methionine-labeled specific protein of about ∼78 kDa (Fig. 6A, lane 2). IVT of empty vector did not produce any protein of the predicted molecular size (Fig. 6A, lane 1). These results were further confirmed by immunoprecipitation using specific rabbit monoclonal antibodies (15F7) to Egr-1 (Cell Signaling, Danvers, MA). Rabbit anti-Egr-1 antibodies specifically immunoprecipitated the IVT product of egr-1/pCDNA3.1(+) (Fig. 6A, lane 3). Rabbit anti-gB antibodies did not immunoprecipitate Egr-1 (Fig. 6A, lane 4). These results confirmed the authenticity of the clone. We tested if this IVT synthesized Egr-1-bound ORF50P. Our results demonstrated that the IVT-synthesized Egr-1 bound the ORF50P probes specifically and formed a discrete band (Fig. 6B, lane 3) compared with IVT gL (Fig. 6B, lane 2). This band (Egr-1/ORF50P probe) was supershifted to higher molecular weights by antibodies to Egr-1 (Fig. 6B, lane 5) but not by control IgGs (Fig. 6B, lane 4), indicating that this complex was indeed specifically formed by Egr-1 interacting with ORF50P probe. Similar results were obtained using IP Egr-1 proteins from nuclear lysates of KSHV-infected cells (Fig. 6C, lane 2). Furthermore, excess unlabeled ORF50P probes were able to restrict binding of IP Egr-1 proteins to labeled probes, demonstrating the specificity of this binding (Fig. 6C, lane 3). Finally, we performed gel shift assays to demonstrate the ability of Egr-1 obtained from within the nucleus (whole nucleus extract) of KSHV-infected cells to bind ORF50P (Fig. 6D). Nuclear extracts from KSHV-infected cells (obtained 6 h PI) had a strong ORF50P probe-protein complex band (Fig. 6D, lane 2). This ORF50P probe-protein complex band was specifically supershifted when incubated with antibodies to Egr-1 (Fig. 6D, lanes 4–6) compared with nonspecific IgGs (Fig. 6D, lane 3). There was a dose response in supershift with increasing concentrations of the antibodies to Egr-1 (Fig. 6D, lanes 4–6). Also, there was no binding of the Egr-1 protein to the ORF50P(m) probe, which has a mutation in the putative Egr-1 binding site (Fig. 6D, lane 7), further demonstrating the specificity of this interaction. The above results confirm the ability of Egr-1 to specifically bind ORF50P.
FIGURE 6.
KSHV gB inhibits Egr-1 binding to ORF50P. A, IVT of the egr-1 gene and specificity of the rabbit anti-Egr-1 monoclonal antibodies. The pCDNA3.1 (lane 1) and egr-1/pCDNA3.1 (lane 2) were transcribed and translated in vitro using rabbit reticulocyte lysates and [35S]methionine. The IVT products were resolved in a 10% SDS-polyacrylamide gel and autoradiographed. Lanes 3 and 4, immunoprecipitation of IVT products from the egr-1/pCDNA3.1 plasmid by anti-Egr-1 (lane 3) and anti-gB antibodies (lane 4). B, IVT Egr-1 binds ORF50P. EMSA was performed with a DIG-labeled ORF50P probe and IVT-synthesized Egr-1. Shown is IVT-synthesized Egr-1 binding to the ORF50P probe (lane 3) compared with that of IVT gL (lane 2). The specificity of the Egr-1-ORF50P was confirmed by incubating the reaction mixture with either nonspecific IgG (lane 4) or monoclonal antibodies to Egr-1 (lane 5). Lane 1 contains the DNA probe only. C, IP Egr-1 from the nuclear extracts of KSHV-infected HMVEC-d cells bound ORF50P. IP EMSA was done with nuclear extracts from KSHV-infected HMVEC-d cells 12 h post-TPA treatment. Nuclear protein (0.5 mg) from KSHV-infected cells was precleared with normal rabbit serum and protein A-Sepharose at 4 °C for 1 h. The supernatant was incubated sequentially with anti-Egr-1 or rabbit preimmune IgG and protein A-Sepharose beads at 4 °C overnight. The complexes were dissociated from beads and used in an EMSA performed with DIG-labeled probes specific to ORF50P (see “Experimental Procedures”). The specificity of the binding was confirmed by incubating the DNA-protein complex with 250 ng of unlabeled ORF50P probe. D, nuclear extracts from KSHV-infected cells had a strong and specific ORF50P-protein complex band. EMSA was performed using 4 μg of nuclear extract obtained from the above KSHV-infected cells (6 h PI) using DIG-labeled ORF50P probes (lanes 1–6) or ORF50P(m) probes (lane 7). An arrowhead denotes the ORF50P probe-protein complex (lane 2). Incubating nuclear extracts with 1:50 (lane 4) and 1:500 (lane 5) dilutions of antibodies to Egr-1 supershifted the ORF50P probe-protein complex compared with incubating the extracts with either a 1:5000 dilution of antibodies to Egr-1 (lane 6) or a 1:50 dilution of nonspecific IgG (lane 3). An asterisk marks the supershifted band. Lane 1, DNA probe only. E, overexpression of Egr-1 activates full-length ORF50P. HMVEC-d cells were cultured to 65% confluence in 6-well plates. The cells were co-transfected with a combination of at least one construct taken from the following groups: (i) 500 ng of pGL3 (empty vector) or ORF50P/pGL3; (ii) 500 ng of pcDNA3.1(+), egr-1/pCDNA3.1(+), or gL/pCDNA3.1(+); and (iii) 500 ng of control vector pRL-TK (27). Forty-four hours post-transfection, the cells were incubated with 30% effector (Eff) cells expressing gB or empty vector for another period of 4 h at 37 °C. At the end of the incubation, cells were lysed, and luciferase activity was measured. Firefly luciferase activity from ORF50P/pGL3 was normalized to the corresponding Renilla luciferase activity. The -fold activation of ORF50P/pGL3 by pCDNA was considered as 1-fold (mean ± S.E. (error bars); n = 3).
To further authenticate the role of gB in inhibiting Egr-1-mediated transcription of ORF50, transient transfection assays were performed. We observed an enhanced ORF50P (nucleotides 68,509–71,593) activity (as measured by the luciferase activity) when the cells were transfected with plasmid encoding Egr-1 (egr-1/pCDNA3.1) compared with an empty vector (pCDNA3.1) or gL/pCDNA3.1 (Fig. 6E). However, incubating the above cells with 30% gB+ effector cells (Fig. 6E) or 30% gB-RGA effector cells (data not shown) led to a dramatic reduction in Egr-1-mediated ORF50P activity. We also observed a similar response in ORF50P (nucleotides 68,674–71,596) (44) activity due to endogenously overexpressing Egr-1 (data not shown). Taken together, these results further implicate a role for Egr-1 in gB-mediated repression of ORF50 expression (promoting virus latency).
DISCUSSION
This study was conducted in order to better understand the role of cell membrane-bound (and not envelope-associated) KSHV-encoded gB in viral pathogenesis, specifically on its ability to promote latency. Our results identified soluble gB as significantly inhibiting Egr-1 expression in cells (Fig. 1, C and D, and supplemental Figs. 1 and 2). Interestingly, gBΔTM did not alter the expression of interferons (IFN-α, β, and γ) as monitored by the PCR arrays and qRT-PCR (data not shown). This is a critical piece of information because earlier studies have demonstrated human cytomegalovirus gB to activate IFN-responsive pathways (45) and that IFNs are known to inhibit replication of several viruses, including HIV and influenza virus (46, 47). Egr-1 (also known as Zif268, NGFI-A, and Krox24) is a transcription factor that is involved in diverse biological functions, including growth, proliferation, and differentiation (14). The role of Egr-1 in KSHV pathogenesis has not been previously established. Therefore, we chose to elaborate on its function in the present study.
Egr-1 regulates expression of several viral genes and plays a crucial role in the replication of different viruses (48–52). Egr-1 was elevated in KSHV-infected cells that were treated with TPA, an agent used to induce KSHV reactivation (Fig. 2, A and E). Inhibiting expression of cellular Egr-1 significantly lowered KSHV reactivation as measured by the expression of virally encoded ORF50 (Fig. 2, C and D). KSHV Rta (encoded by ORF50) is the switch carried by the virus to initiate virus reactivation (37). Multiple pathways involving a variety of transcription factors, including autoactivation by Rta, have been shown either to enhance or inhibit ORF50 gene transcription (20, 44, 53–60). Such a repertoire of different and, to some extent, redundant activating factors or pathways makes for an efficient mechanism to control one of the key features of virus replication, specifically reactivation during different stages of pathogenesis as well as under different cellular environments. All of the factors that have been described to date and are known to regulate the expression of ORF50/Rta can be classified as cellular proteins, nuclear-associated virus-encoded proteins, or chemicals. For the first time, we describe a role for the physiologically relevant number of infected cells expressing a virus-encoded late structural protein, gB, on their cell membranes to mediate ORF50/Rta expression.
The physiological relevance of these findings was confirmed by analyzing the effect of gB expressed by 1–2% of cells on KSHV-infected cells during the course of a primary infection of endothelial cells. Our results confirmed the ability of gB expressed on a minimal number of cells to significantly lower both ORF50 (Fig. 3B) and egr-1 (Fig. 3C) expression and thereby hasten the process of virus latency via both autocrine and paracrine effects. This is an interesting observation because earlier studies have defined a crucial role for MAPK signaling (Ras > Raf > MEK > Erk) in Ets-1-induced virus reactivation (20, 39–41). Treating cells with TPA or any form of stress leads to an increase in the expression of Ras > Raf > MEK > ERK > Ets-1 signaling. Ets-1 is a transcription factor that plays an important role in cell proliferation, angiogenesis, and immune response (61, 62). It also binds the egr-1 promoter sequence and up-regulates the expression of Egr-1 (48, 63). Moreover, MAPK signaling has been shown to regulate Egr-1 expression in virus-infected cells (64). Because gB was able to inhibit Egr-1 expression, we predicted that it would also lower MAPK signaling. We observed membrane-bound gB expressed by 1–2% of cells to inhibit phospho-ERK1/2 expression in KSHV-infected cells (Fig. 4A). A similar effect of soluble gB on MAPK signaling was observed (data not shown). Moreover, the ability of gB to lower ORF50 (Fig. 4C) and egr-1 (Fig. 4D) expression in KSHV-infected cells overexpressing MEK1/2 (Fig. 4B) was significantly quenched. Taken together, we concluded that KSHV gB lowers Egr-1 expression by inhibiting the upstream MAPK signaling activity.
KSHV interacts with integrins on the cell membrane via the RGD motif present in gB, which facilitates virus entry into cells (9). Therefore, we analyzed if the RGD motif in gB had a role in transducing signals crucial in regulating Egr-1 expression. Our results implicate gB expressed on the cell membranes as repressing ORF50 and egr-1 expression via an RGD-independent method of signaling (Fig. 5). Taken together, our data implicate a yet to be defined motif in gB crucial for lowering the expression of Egr-1. Such an RGD-independent gB signaling was reported in recent studies that analyzed the role of gB in virus egress and KS pathogenesis (65).
Recent studies have demonstrated the ability of Egr-1 to bind the EBV-encoded ZTA promoter and initiate virus reactivation (48, 52). Interestingly, KSHV Rta is the functional equivalent of EBV ZTA (66). KSHV Rta activates KSHV early lytic genes, including virus-encoded interleukin 6 (IL-6) and polyadenylated nuclear RNA, and a late gene, small viral capsid antigen (67). It is considered as the switch that triggers virus reactivation. Here, we showed the ability of Egr-1 to bind and drive the ORF50P activity (Fig. 6, B–E). Incidentally, Egr-1-driven promoter activity in HMVEC-d cells was significantly lowered by the expression of the membrane-bound gB and not gL (Fig. 6E). Membrane-bound gB lacking the RGD motif was also able to significantly lower the Egr-1-enhanced ORF50P activity (data not shown). Taken together, we established that gB alters Egr-1 > KSHV ORF50 expression by inhibition of MAPK signaling pathways via an RGD-independent manner.
We maintain that gB expressed on the surface of a minimal number of cells plays a critical role in promoting virus latency or delaying the onset of KSHV reactivation. We are completely aware of the fact that the membrane-bound gB is only part of the complicated and well orchestrated machinery that promotes latency. Furthermore, we suspect that Egr-1 holds a pivotal position to mediate this effect. Proteins encoded by different viruses have also demonstrated the ability to alter KSHV reactivation from latency. Specifically, EBV has been shown to inhibit KSHV lytic replication through the activity of LMP-1 (latent membrane protein-1) (68). The mechanism for LMP-1 function is via inhibition of Egr-1 expression. The relationship between the onset of AIDS and KSHV-related diseases further supports a role for Egr-1 during virus reactivation. HIV-1 infection is a cofactor for AIDS-related KS development, and it has been demonstrated to induce KSHV reactivation (69). The effect of HIV infection may be directed by the activity of the Tat protein. However, the specific function for the HIV Tat protein to stimulate lytic KSHV infection is still being debated (70, 71). Moreover, recent evidence suggests that the Tat protein may inhibit Egr-1 expression in target cells (72). Thus, it is possible that the Tat protein may be able to inhibit lytic KSHV infection and promote tumor progression in a manner similar to KSHV gB. Apart from AIDS-KS, there are other forms of KS, like the classical KS, endemic KS, and transplantation-associated KS, where there is an absence of the HIV-1 Tat protein. Under such circumstances, we hypothesize a crucial role for KSHV-encoded gB expressed on the cell surface to regulate viral reactivation via modulating the MAPK > Egr-1 signaling pathway. In the case of AIDS-KS, we predict that the role of gB in KS pathogenesis would be more of an additive effect to that of HIV Tat.
The idea that the virus-encoded protein (gB) that is expressed during the lytic phase of infection actively suppresses reactivation in adjacent infected cell populations is a particularly appealing concept because such a mechanism would help to prevent a cascade of reactivation that would potentially destroy the latently infected cell population. A self-limiting reactivation pathway is conducive to sustaining a predominantly latent infection that is typical of KSHV, EBV, and other γ-herpesviruses. For the first time, using the co-culture experiments, we have provided evidence to demonstrate the ability of membrane-bound gB expressed in as few as 1–2% of cells to significantly promote latency through both autocrine and paracrine effects. We strongly believe that there is a role for soluble proteins in the form of inflammatory cytokines/growth factors in mediating the paracrine effects of gB. In general, virus-encoded late stage structural proteins, such as gB, are said to play major roles in virus entry and egress (9, 65). These findings will add a new perspective to understanding the role of such structural glycoproteins in modulating virus reactivation and pathogenesis. Future studies will be aimed at further delineating the role of Egr-1 in Rta expression and reactivation, the mechanism by which gB mediates its paracrine effect, and interactions between the specific domain on gB and the host cell receptor molecules critical to regulate Egr-1 expression. A special emphasis on the role of MCP-1 (Fig. 1C) in mediating the paracrine effects of gB is already under way. Further studies in this area may lead to a better understanding of KSHV biology with respect to glycoproteins and their role in regulating virus reactivation.
Acknowledgments
We thank Dr. Adrian Whitehouse (Institute of Molecular and Cellular Biology University of Leeds, UK) for kindly providing the plasmid encoding the full-length ORF50 promoter. We also thank Mr. A. M. Huxley and Dr. Adrian Reber (Centers for Disease Control, Atlanta, GA) for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health, NIBIB, Grants R21EB006483 (to S. M. A.) and 5F31CA132560-02 (to O. F. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- KSHV
- Kaposi's sarcoma-associated herpesvirus
- KS
- Kaposi's sarcoma
- gB
- glycoprotein B
- gL
- glycoprotein L
- HMVEC-d
- human dermal microvascular endothelial
- pCDNA
- pCDNA3.1.CT-GFP-TOPO
- qRT-PCR
- quantitative RT-PCR
- IVT
- in vitro transcription and translation
- TRITC
- tetramethylrhodamine isothiocyanate
- IP
- immunoprecipitated
- DIG
- digoxigenin
- PI
- post-infection
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- NS
- nonspecific
- ORF50P
- ORF50 promoter.
REFERENCES
- 1.Hamden K. E., Whitman A. G., Ford P. W., Shelton J. G., McCubrey J. A., Akula S. M. (2005) Leukemia 19, 18–26 [DOI] [PubMed] [Google Scholar]
- 2.Chen L., Lagunoff M. (2005) J. Virol. 79, 14383–14391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milligan S., Robinson M., O'Donnell E., Blackbourn D. J. (2004) J. Virol. 78, 2591–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye F. C., Zhou F. C., Xie J. P., Kang T., Greene W., Kuhne K., Lei X. F., Li Q. H., Gao S. J. (2008) J. Virol. 82, 4235–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon M., Cesarman E., Boshoff C. (2006) Blood 107, 277–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee B. S., Paulose-Murphy M., Chung Y. H., Connlole M., Zeichner S., Jung J. U. (2002) J. Virol. 76, 12185–12199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akula S. M., Pramod N. P., Wang F. Z., Chandran B. (2001) Virology 284, 235–249 [DOI] [PubMed] [Google Scholar]
- 8.Dyson O. F., Oxendine T. L., Hamden K. E., Ford P. W., Akula S. M. (2008) Cell Microbiol. 10, 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akula S. M., Pramod N. P., Wang F. Z., Chandran B. (2002) Cell 108, 407–419 [DOI] [PubMed] [Google Scholar]
- 10.Subramanian R., D'Auvergne O., Kong H., Kousoulas K. G. (2008) J. Virol. 82, 7144–7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryan B. A., Dyson O. F., Akula S. M. (2006) J. Gen. Virol. 87, 519–529 [DOI] [PubMed] [Google Scholar]
- 12.An J., Sun Y., Sun R., Rettig M. B. (2003) Oncogene 22, 3371–3385 [DOI] [PubMed] [Google Scholar]
- 13.Christy B., Nathans D. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 8737–8741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiel G., Cibelli G. (2002) J. Cell. Physiol. 193, 287–292 [DOI] [PubMed] [Google Scholar]
- 15.Cannon M., Philpott N. J., Cesarman E. (2003) J. Virol. 77, 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi Y. B., Nicholas J. (2010) Virology 397, 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ford P. W., Hamden K. E., Whitman A. G., McCubrey J. A., Akula S. M. (2004) Cancer Biol. Ther. 3, 876–881 [DOI] [PubMed] [Google Scholar]
- 18.Dyson O. F., Ford P. W., Chen D., Li Y. Q., Akula S. M. (2009) J. Cell Mol. Med. 13, 1920–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naranatt P. P., Akula S. M., Chandran B. (2002) Arch. Virol. 147, 1349–1370 [DOI] [PubMed] [Google Scholar]
- 20.Ford P. W., Bryan B. A., Dyson O. F., Weidner D. A., Chintalgattu V., Akula S. M. (2006) J. Gen. Virol. 87, 1139–1144 [DOI] [PubMed] [Google Scholar]
- 21.Wang F. Z., Akula S. M., Sharma-Walia N., Zeng L., Chandran B. (2003) J. Virol. 77, 3131–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw A. M., Braun L., Frew T., Hurley D. J., Rowland R. R., Chase C. C. (2000) Virology 268, 159–166 [DOI] [PubMed] [Google Scholar]
- 23.Hamden K. E., Ford P. W., Whitman A. G., Dyson O. F., Cheng S. Y., McCubrey J. A., Akula S. M. (2004) J. Virol. 78, 13381–13390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dyson O. F., Bryan B. A., Lambert P. J., Ford P. W., Akula S. M. (2007) Intervirology 50, 245–253 [DOI] [PubMed] [Google Scholar]
- 25.Krishnan H. H., Naranatt P. P., Smith M. S., Zeng L., Bloomer C., Chandran B. (2004) J. Virol. 78, 3601–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naranatt P. P., Akula S. M., Zien C. A., Krishnan H. H., Chandran B. (2003) J. Virol. 77, 1524–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ralph J. A., Zocco D., Bresnihan B., Fitzgerald O., McEvoy A. N., Murphy E. P. (2007) Am. J. Pathol. 170, 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ford P. W., Hamden K. E., Whitman A. G., Bryan B. A., Chintalgattu V., McCubrey J. A., Dyson O. F., Akula S. M. (2005) Virus Res. 114, 172–176 [DOI] [PubMed] [Google Scholar]
- 29.Akula S. M., Ford P. W., Whitman A. G., Hamden K. E., Bryan B. A., Cook P. P., McCubrey J. A. (2005) Blood 105, 4516–4522 [DOI] [PubMed] [Google Scholar]
- 30.Wang S. E., Wu F. Y., Fujimuro M., Zong J., Hayward S. D., Hayward G. S. (2003) J. Virol. 77, 600–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamaluddin M., Choudhary S., Wang S., Casola A., Huda R., Garofalo R. P., Ray S., Brasier A. R. (2005) J. Virol. 79, 15302–15313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chappell W. H., Green T. D., Spengeman J. D., McCubrey J. A., Akula S. M., Bertrand F. E. (2005) Cell Cycle 4, 1389–1395 [DOI] [PubMed] [Google Scholar]
- 33.Sharma-Walia N., Naranatt P. P., Krishnan H. H., Zeng L., Chandran B. (2004) J. Virol. 78, 4207–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McAllister S. C., Hansen S. G., Messaoudi I., Nikolich-Zugich J., Moses A. V. (2005) J. Virol. 79, 2626–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukac D. M., Kirshner J. R., Ganem D. (1999) J. Virol. 73, 9348–9361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogishima T., Shiina H., Breault J. E., Terashima M., Honda S., Enokida H., Urakami S., Tokizane T., Kawakami T., Ribeiro-Filho L. A., Fujime M., Kane C. J., Carroll P. R., Igawa M., Dahiya R. (2005) Oncogene 24, 6765–6772 [DOI] [PubMed] [Google Scholar]
- 37.Sun R., Lin S. F., Staskus K., Gradoville L., Grogan E., Haase A., Miller G. (1999) J. Virol. 73, 2232–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gius D., Cao X. M., Rauscher F. J., 3rd, Cohen D. R., Curran T., Sukhatme V. P. (1990) Mol. Cell. Biol. 10, 4243–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen A., Brodie C., Sarid R. (2006) J. Gen. Virol. 87, 795–802 [DOI] [PubMed] [Google Scholar]
- 40.Xie J., Ajibade A. O., Ye F., Kuhne K., Gao S. J. (2008) Virology 371, 139–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu F., Harada J. N., Brown H. J., Deng H., Song M. J., Wu T. T., Kato-Stankiewicz J., Nelson C. G., Vieira J., Tamanoi F., Chanda S. K., Sun R. (2007) PLoS Pathog. 3, e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garrigues H. J., Rubinchikova Y. E., Dipersio C. M., Rose T. M. (2008) J. Virol. 82, 1570–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao X., Mahendran R., Guy G. R., Tan Y. H. (1993) J. Biol. Chem. 268, 16949–16957 [PubMed] [Google Scholar]
- 44.Wilson S. J., Tsao E. H., Webb B. L., Ye H., Dalton-Griffin L., Tsantoulas C., Gale C. V., Du M. Q., Whitehouse A., Kellam P. (2007) J. Virol. 81, 13578–13586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyle K. A., Pietropaolo R. L., Compton T. (1999) Mol. Cell. Biol. 19, 3607–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palm M., Garigliany M. M., Cornet F., Desmecht D. (2010) Vet. Res. 41, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hou W., Wang X., Ye L., Zhou L., Yang Z. Q., Riedel E., Ho W. Z. (2009) J. Virol. 83, 3834–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang Y., Lee H. H., Chen Y. T., Lu J., Wu S. Y., Chen C. W., Takada K., Tsai C. H. (2006) J. Virol. 80, 7748–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen S. H., Yao H. W., Chen I. T., Shieh B., Li C. (2008) J. Clin. Invest. 118, 3470–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romagnoli L., Sariyer I. K., Tung J., Feliciano M., Sawaya B. E., Del Valle L., Ferrante P., Khalili K., Safak M., White M. K. (2008) Virology 375, 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silva P. N., Soares J. A., Brasil B. S., Nogueira S. V., Andrade A. A., de Magalhães J. C., Bonjardim M. B., Ferreira P. C., Kroon E. G., Bruna-Romero O., Bonjardim C. A. (2006) Biochem. J. 398, 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zalani S., Holley-Guthrie E., Kenney S. (1995) J. Virol. 69, 3816–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan H., Xie J., Ye F., Gao S. J. (2006) J. Virol. 80, 5371–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deng H., Young A., Sun R. (2000) J. Gen. Virol. 81, 3043–3048 [DOI] [PubMed] [Google Scholar]
- 55.Gwack Y., Byun H., Hwang S., Lim C., Choe J. (2001) J. Virol. 75, 1909–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Curreli F., Cerimele F., Muralidhar S., Rosenthal L. J., Cesarman E., Friedman-Kien A. E., Flore O. (2002) J. Virol. 76, 5208–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang S. E., Wu F. Y., Chen H., Shamay M., Zheng Q., Hayward G. S. (2004) J. Virol. 78, 4248–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bottero V., Sharma-Walia N., Kerur N., Paul A. G., Sadagopan S., Cannon M., Chandran B. (2009) Virology 392, 34–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harrison S. M., Whitehouse A. (2008) FEBS Lett. 582, 3080–3084 [DOI] [PubMed] [Google Scholar]
- 60.Li Q., Zhou F., Ye F., Gao S. J. (2008) Virology 379, 234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhat N. K., Thompson C. B., Lindsten T., June C. H., Fujiwara S., Koizumi S., Fisher R. J., Papas T. S. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 3723–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hashiya N., Jo N., Aoki M., Matsumoto K., Nakamura T., Sato Y., Ogata N., Ogihara T., Kaneda Y., Morishita R. (2004) Circulation 109, 3035–3041 [DOI] [PubMed] [Google Scholar]
- 63.Robinson L., Panayiotakis A., Papas T. S., Kola I., Seth A. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7170–7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cai Y., Liu Y., Zhang X. (2006) Virology 355, 152–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Subramanian R., Sehgal I., D'Auvergne O., Kousoulas K. G. (2010) J. Virol. 84, 1704–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao W., Tang Y., Lin S. F., Kung H. J., Giam C. Z. (2003) J. Virol. 77, 3809–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun R., Lin S. F., Gradoville L., Yuan Y., Zhu F., Miller G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10866–10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu D., Coleman T., Zhang J., Fagot A., Kotalik C., Zhao L., Trivedi P., Jones C., Zhang L. (2007) J. Virol. 81, 6068–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ganem D. (1998) Curr. Clin. Top. Infect. Dis. 18, 237–251 [PubMed] [Google Scholar]
- 70.Harrington W., Jr., Sieczkowski L., Sosa C., Chan-a-Sue S., Cai J. P., Cabral L., Wood C. (1997) Lancet 349, 774–775 [DOI] [PubMed] [Google Scholar]
- 71.Varthakavi V., Smith R. M., Deng H., Sun R., Spearman P. (2002) Virology 297, 270–280 [DOI] [PubMed] [Google Scholar]
- 72.Darbinian N., Darbinyan A., Czernik M., Peruzzi F., Khalili K., Reiss K., Gordon J., Amini S. (2008) J. Cell. Physiol. 216, 128–134 [DOI] [PMC free article] [PubMed] [Google Scholar]