Abstract
For mRNA to be transported from the nucleus to the cytoplasm, it must travel from the site of transcription through the nuclear interior to the nuclear pore. Studies in Saccharomyces cerevisiae have suggested a relationship between poly(A) RNA trafficking and myosin-like protein 1 (Mlp1p), a nuclear-pore associated protein that is homologous to the mammalian Tpr (translocated promoter region) protein [Kosova, B., Panté, N., Rollenhagen, C., Podtelejnikov, A., Mann, M., Aebi, U., and Hurt, E. (2000) J. Biol. Chem. 275, 343–350]. We identified a yeast two-hybrid interaction between the C-terminal globular domain of Mlp1p and Nab2p, a shuttling heterogeneous nuclear ribonucleoprotein that is required for mRNA export. Coimmunoprecipitation confirms that Nab2p also interacts with full-length Mlp1p and in vitro binding experiments show that Nab2p binds directly to the C-terminal domain of Mlp1p. In addition, our experiments reveal that the C-terminal domain of Mlp1p is both necessary and sufficient to cause accumulation of poly(A) RNA and Nab2p in the nucleus. We propose a model where Mlp1p acts as a checkpoint at the nuclear pore by interacting with export-competent ribonucleoprotein complexes through its C-terminal globular domain. This study identifies Nab2p as a heterogeneous nuclear ribonucleoprotein found in complex with Mlp1p and begins to delineate the path that mRNA travels from the chromatin to the nuclear pore.
Active genes are transcribed to pre-mRNA via RNA polymerase II and then pre-mRNA is processed within the nucleus to form mature mRNA transcripts (1). These processing events occur cotranscriptionally and include the addition of a 5′ 7-methylguanosine cap (2), the splicing of introns (3), and cleavage of the 3′ end followed by polyadenylation (2, 4). Fully processed transcripts are then exported from the nucleus to the cytoplasm where they can be translated into functional proteins at ribosomes (5, 6). Export of mature mRNA from the nucleus, which is coupled to mRNA processing, is an essential checkpoint in the regulation of gene expression (7). The detailed mechanism of export of mature mRNA transcripts from the nucleus is poorly understood; however, the complexes involved are beginning to be delineated (7).
Processing and export of mRNA from the nucleus are mediated by various RNA binding proteins, including heterogeneous nuclear ribonucleoproteins (hnRNPs), serine-arginine-rich proteins, and mRNA export factors (8, 9). The complement of RNPs on an mRNA transcript is dynamic throughout the life cycle of the transcript and it is likely that these RNPs serve as signals for proper processing and export. The RNPs present on a transcript are therefore candidates for markers identifying the maturation state of the mRNA transcript. Some of the hnRNPs shuttle between the nucleus and the cytoplasm, suggesting that they accompany the mRNA cargo from the nucleus to the cytoplasm (10–12). These shuttling hnRNPs interact with the cellular transport machinery to mediate the export of mRNP complexes from the nucleus through the nuclear pore complex (NPC). Ultimately, the NPC serves as the final nuclear checkpoint to ensure that only mature RNP complexes are exported from the nucleus.
The NPC is a large, proteinaceous channel embedded within the nuclear envelope that regulates all active transport between the nucleus and the cytoplasm (13, 14). This complex also extends into the cytoplasm and nucleoplasm through filament proteins that are thought to constitute the initial docking sites for import and export complexes (15). In mammalian cells, the translocated promoter region (Tpr) protein was originally identified as a component of the filaments that extend from the NPC into the nuclear interior (16, 17). Tpr is a 270-kDa protein consisting mostly of an α-helical coiled-coil structure with a carboxyl-terminal globular domain (18, 19). Recently, Tpr has been localized to both the nuclear basket of the NPC and discrete foci within the nucleus (20). It remains unclear whether Tpr is solely a structural component of the NPC or an NPC-associated protein that has functions both within the nucleus and at the NPC.
Possible functions for Tpr have been revealed through analysis of homologous proteins in various model systems. Tpr homologs have been found in Drosophila melanogaster (17), Xenopus laevis (16), and Saccharomyces cerevisiae (21). In S. cerevisiae, two homologs, Mlp1p (myosin-like protein 1) and Mlp2p (myosin-like protein 2), have been identified (21, 22). Although neither is essential for normal cell growth, studies have suggested that the proteins have a redundant function (21, 22). Both proteins are associated with the nuclear pore protein, Nic96p (23), and immuno-electron microscopy indicates that the proteins are localized to the nucleoplasmic side of the NPC with a distinct nuclear pool (21, 23). Insight into the function of Mlp1p came from the observation that overexpression of Mlp1p causes poly(A) RNA accumulation within the nucleus, as well as aggregation of Mlp1p within a chromatin-free, non-nucleolar region of the nucleus (21, 23). This phenotype has also been observed in mammalian cells on overexpression of Tpr (24). These observations suggest that Tpr/Mlp1p could play a role in mRNA metabolism.
It has been suggested that the effect of Mlp1p on poly(A) RNA localization is mediated by hnRNPs (23). Here we present the identification of the hnRNPs, Nab2p and Npl3p, found in complex with Mlp1p. Both Nab2p and Npl3p are shuttling hnRNPs in S. cerevisiae that are required for efficient poly(A) RNA export from the nucleus (11, 25, 26). Through a combination of in vivo and in vitro experiments, we demonstrate a direct interaction between the C-terminal globular domain of Mlp1p and Nab2p. Our analysis also reveals that expression of the C-terminal domain of Mlp1p (CT-Mlp1p) is both necessary and sufficient to cause poly(A) RNA and Nab2p to accumulate within the nucleus. Our results support the hypothesis that the effect of overexpression of Mlp1p on poly(A) RNA export from the nucleus is caused by retention of Nab2p and other hnRNPs in the nucleus. We propose a model where Mlp1p acts as a docking site at the NPC for export-competent RNP complexes. We further suggest that Mlp1p could act as a checkpoint at the NPC to ensure that only mature transcripts are exported from the nucleus.
Materials and Methods
Materials.
DNA manipulations were performed according to standard methods (27), and media were prepared by standard protocols (28). Yeast strains and plasmids used are described in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. Chemicals were obtained from Sigma, U.S. Biological (Swampscott, MA), or Fisher Scientific unless otherwise noted.
Yeast Two-Hybrid Screen.
The yeast two-hybrid reporter strain (EGY48) containing DNA binding domain (DBD)-NAB2 (pAC1101) was transformed with an S. cerevisiae genomic library cloned into the activation domain (AD) vector (pJG4-5) (Origene Technologies, Rockville, MD). The screen was conducted as described (29). Briefly, transformants were tested for a positive yeast two-hybrid interaction by galactose-dependent growth on media lacking leucine and expression of lacZ. The Nab2p and Mlp1p yeast two-hybrid interaction was confirmed by coexpressing either DBD-NAB2 and pJG4-5 vector, AD-MLP1 (pAC1195) and pEG202 vector, or DBD-NAB2 and AD-MLP1 in EGY48 cells. Cells were grown at 30°C on glucose or galactose minimal media, minimal media lacking leucine, or minimal media containing 200 μM 5-bromo-4-chloro-3-indolyl β-d-galactoside.
Immunoprecipitation.
For immunoprecipitation of myc-tagged proteins, NAB2-myc (pAC1126), NPL3-myc (pAC943), SRP1-myc (pAC963), or control vector (pAC2) was cotransformed with either CT-MLP1-GFP (pAC1197) or GFP alone (pAC1024) into WT (ACY192) yeast cells. Cultures were grown to saturation in 2% galactose minimal media and then harvested by centrifugation at 3,000 rpm. Cells were lysed and myc-tagged proteins were immunoprecipitated from protein extracts (8 mg total protein) with agarose-conjugated myc Ab (9E-10, Santa Cruz Biotechnology) (26). For RNase-treated samples, protein lysates were treated with 15 units (200 μg) RNase A for 30 min at 4°C (30). For immunoprecipitation of protein A-tagged Mlp1p, Mlp1p-protein A (ACY683) and WT (ACY192) cells were grown to log phase and collected by centrifugation. Cells were lysed in PBSMT (PBS/5 mM MgCl2/0.5% Triton X-100, pH 7.4) and protein A-tagged proteins were immunoprecipitated from protein extracts (30 mg total protein) by incubation overnight at 4°C with IgG Sepharose (Amersham Pharmacia). For all immunoprecipitation experiments, the bound fraction was washed three times with PBSMT and eluted from beads with loading buffer (125 mM Tris⋅HCl, pH 6.8/250 mM DTT/5% SDS/0.25% Bromophenol blue/25% glycerol). Lysates (30 μg total protein) and bound fractions were resolved by SDS/PAGE and analyzed by immunoblotting as described (31). Myc-tagged proteins were detected by using a 1:200 dilution of the 9E-10 mouse c-myc mAb (Santa Cruz Biotechnology). GFP-tagged proteins were detected by using a 1:5,000 dilution of anti-GFP rabbit polyclonal Ab (32). Protein A-tagged Mlp1p was detected by a 1:5,000 dilution of anti-IgG rabbit antibody (Jackson ImmunoResearch). Nab2p was detected by using a 1:50,000 dilution of a polyclonal Nab2p antibody (26).
In Vitro Binding Assay.
GST (pGEX4T-3) or GST-CT-MLP1 (pAC1340) was expressed in Escherichia coli DE3 cells. Cells were collected and lysed in 20 mM Tris, pH 8.0/0.5% Triton-X (buffer A) by sonication. Lysates were clarified by centrifugation and incubated with glutathione-Sepharose (Amersham Pharmacia) in buffer A for 2 h at 4°C. The beads were then washed with buffer A and increasing concentrations of NaCl (250–500 mM). Sepharose-bound GST or GST-CT-Mlp1p (6 μg) was incubated with 2 μg of purified His-Nab2p (26) or His-Srp1p control (33) at 4°C for 90 min. Unbound fractions were collected and the beads were washed four times with buffer A. Bound fractions were eluted with loading buffer and samples were analyzed by SDS/PAGE followed by Coomassie staining and immunoblotting with anti-GST and anti-His antibodies (Santa Cruz Biotechnology).
Microscopy.
For shuttling experiments, WT (ACY192) and rat7-1 (ACY194) cells expressing Nab2p-GFP (pAC719), ΔRGG-Nab2p-GFP (pAC980), or nuclear localization sequence (NLS)-nuclear export sequence (NES)-GFP (pAC213) were grown to log phase at 25°C and then shifted to 37°C or maintained at 25°C. For analysis of the effect of overexpression of Mlp1p on the localization of Nab2p, cells expressing Nab2p-GFP, ΔRGG-Nab2p-GFP, or NLS-NES-GFP and containing galactose-inducible FL-MLP1 (pAC1313), CT-MLP1 (pAC1196), ΔCT-MLP1 (pAC1315), or vector control (pAC19) were grown to log phase in minimal media with either 2% glucose or 2% galactose at 30°C. For all samples, cells were examined by using filters from Chroma Technology (Brattleboro, VT) and an Olympus BX60 epifluorescence microscope equipped with a Roper Scientific (Tucson, AZ) Quantix digital camera. All images were captured by using IP Lab spectrum software.
Fluorescence in Situ Hybridization (FISH).
The intracellular localization of poly(A) RNA was assayed by FISH (26). WT cells transformed with a galactose-inducible FL-MLP1 (pAC1313), CT-MLP1 (pAC1196), ΔCT-MLP1 (pAC1315), CT-MLP1-GFP (pAC1197), or vector control was grown in either 2% glucose or 2% galactose minimal media to log phase at 30°C. Cells were prepared as described (34). A digoxigenin-labeled oligo(dT) probe and either rhodamine- or FITC-conjugated antidigoxigenin Ab (1:200 dilution, Roche Molecular Biochemicals) were used to localize poly(A) RNA. Cells containing GFP-tagged proteins were simultaneously probed with a 1:500 dilution of anti-GFP Ab, which was detected with a FITC-conjugated anti-rabbit secondary Ab (1:1,000 dilution). Cells were also stained with 4′,6-diamidino-2-phenylindole-dihydrochloride (DAPI) to detect chromatin.
Results and Discussion
Identification of an Mlp1p–Nab2p Complex.
Nab2p is an essential shuttling hnRNP in S. cerevisiae that is required for efficient poly(A) RNA export (12, 26, 35). To gain insight into the function of Nab2p in poly(A) RNA metabolism, a yeast two-hybrid screen was conducted by using a LexA-DBD-Nab2 fusion protein (DBD-Nab2p). An S. cerevisiae genomic library fused to the VP16 AD was screened for Nab2p interacting proteins based on their ability to activate the LEU2 and lacZ reporter genes in the presence of DBD-Nab2p.
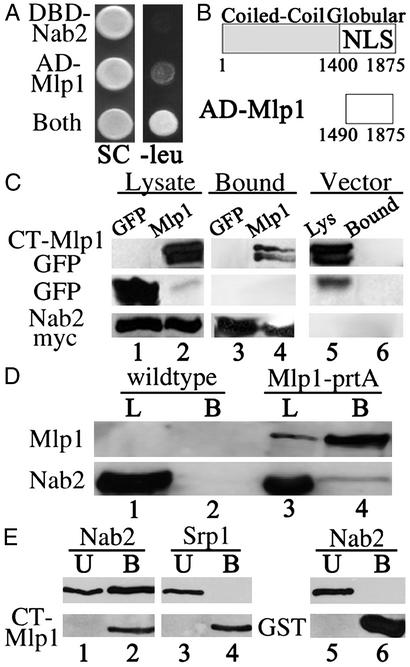
In this screen, we identified a yeast two-hybrid interaction between Nab2p and Mlp1p. Mlp1p is a 220-kDa protein consisting mostly of a coiled-coil α-helical structure and a globular C-terminal domain (22). Coexpression of AD-Mlp1p and DBD-Nab2p activates the LEU2 reporter, as indicated by growth on galactose minimal media lacking leucine (Fig. 1A). Neither AD-Mlp1p nor DBD-Nab2p alone could autoactivate the LEU2 reporter. Similar results were obtained for the lacZ reporter (data not shown). The domain of Mlp1p that interacts with Nab2p corresponds to the carboxyl-terminal globular domain of Mlp1p beginning at residue 1490 (Fig. 1B). Our results from the yeast two-hybrid analysis suggest that Nab2p and Mlp1p interact through the CT-Mlp1p.
Figure 1.
Nab2p directly interacts with CT-Mlp1p. (A) Cells expressing DBD-Nab2p and pJG4–5 vector, AD-Mlp1p and pEG202 vector, or DBD-Nab2p and AD-Mlp1p (Both) were grown on galactose synthetic complete (SC) or galactose plates lacking leucine (−leu). (B) Schematic of the domains and NLS (23) of Mlp1p and the fragment identified in the yeast two-hybrid screen (AD-Mlp1). Amino acid residues are designated below each schematic. (C) Immunoblot analysis of protein lysates containing either Nab2p-myc (lanes 1 and 2) or control vector (lane 5) and either GFP control (lane 1) or CT-Mlp1p-GFP (lanes 2 and 5). Lysates were immunoprecipitated with anti-myc Ab and bound fractions were probed with anti-GFP Ab (Top and Middle) or anti-myc Ab (Bottom) (lanes 3, 4, and 6). (D) Immunoblot analysis of protein lysates from WT (lane 1) or Mlp1p-protein A (lane 3) strains. Proteins were immunoprecipitated with IgG-Sepharose and bound fractions were probed simultaneously with anti-rabbit Ab and anti-Nab2p (lanes 2 and 4). (E) GST (lanes 5 and 6) and GST-CT-Mlp1p (lanes 1–4) were incubated with recombinant Srp1p (lanes 3 and 4) or Nab2p (lanes 1, 2, 5, and 6) and unbound (U) and bound (B) fractions were analyzed by immunoblotting.
To independently confirm the yeast two-hybrid interaction between the CT-Mlp1p and Nab2p, we performed coimmunoprecipitation by using a myc-tagged Nab2 protein (Fig. 1C). Protein extracts were generated from cells expressing either GFP or CT-Mlp1p-GFP and coexpressing Nab2p-myc or a vector control. Nab2p-myc was immunoprecipitated and bound fractions were analyzed with an anti-GFP Ab to determine whether Nab2p coimmunoprecipitates Mlp1p. Immunoblot analysis showed that CT-Mlp1p-GFP coimmunoprecipitates with Nab2p-myc, as indicated by the presence of CT-Mlp1p-GFP in the bound Nab2p-myc fraction (Fig. 1C, lane 4). This interaction does not depend on the presence of RNA as similar results were obtained when lysates were pretreated with RNase (data not shown). As specificity controls, the Nab2p-myc protein did not coimmunoprecipitate the control GFP protein (Fig. 1C, lane 3) nor did Mlp1p-GFP coimmunoprecipitate with a vector control (Fig. 1C, lane 6). This experiment confirms that Nab2p can interact with CT-Mlp1p.
To test whether Nab2p interacts with the full-length Mlp1 protein (FL-Mlp1p), a coimmunoprecipitation experiment was performed by using a yeast strain that expresses protein A-tagged Mlp1p. The expression and localization of the protein A-tagged Mlp1 protein is indistinguishable from endogenous Mlp1p (21). FL-Mlp1p was purified from yeast lysates by binding to IgG beads and the bound fraction was analyzed for copurification of Nab2p (Fig. 1D). Immunoblot analysis of the Mlp1p-bound fraction with a Nab2p antibody revealed an interaction between these two proteins (Fig. 1D, lane 4). Nab2p was not detected in an immunoprecipitation of lysates prepared from an untagged strain (Fig. 1D, lane 2). These experiments identify Nab2p as an hnRNP found in complex with Mlp1p.
The previous experiments establish that Nab2p associates with Mlp1p via the CT-Mlp1p, but do not address whether this is a direct interaction. To determine whether the interaction is direct, an in vitro binding assay was performed (Fig. 1E). In this experiment, GST control protein or GST-CT-Mlp1p expressed in E. coli was bound to glutathione-Sepharose and incubated with either purified recombinant Nab2p or a control protein, Srp1p, which does not interact with Mlp1p (23). Results indicate that Nab2p binds directly to CT-Mlp1p, as indicated by the presence of Nab2p in the bound CT-Mlp1p fraction (Fig. 1E, lane 2). Srp1p was detected only in the unbound CT-Mlp1p fraction, indicating that these two proteins do not interact directly, as reported (23) (Fig. 1E, lanes 3 and 4). Also, Nab2p does not nonspecifically interact with GST (Fig. 1E, lane 6). These results show that Nab2p binds directly to the CT-Mlp1p in vitro and suggest that Nab2p could directly interact with Mlp1p in vivo.
Characterization of the CT-Mlp1p.
Previous experiments have shown that overexpression of full-length Mlp1p causes accumulation of poly(A) RNA within the nucleus (23). Given the identification of a direct interaction between the CT-Mlp1p and Nab2p, an hnRNP, we hypothesized that overexpression of the CT-Mlp1p might block poly(A) RNA export by titrating and sequestering RNA-bound hnRNPs within the nucleus. We therefore tested whether overexpression of CT-Mlp1p is necessary and sufficient to inhibit poly(A) RNA export. For this experiment, we used constructs expressing FL-MLP1 (residues 1–1875) and CT-MLP1 (residues 1490–1875) under the control of the galactose inducible GAL1–10 promoter. As a control, we also created galactose-inducible ΔCT-MLP1 (residues 1–1489), which lacks the C-terminal globular domain. To assure that this control protein was targeted to the nucleus, we also fused a simian virus 40-NLS to ΔCT-MLP1 to create ΔCT-MLP1-NLS. Identical results were obtained for experiments conducted with either ΔCT-MLP1 or ΔCT-MLP1-NLS. Galactose-induced expression of all proteins was confirmed by immunoblotting (data not shown).
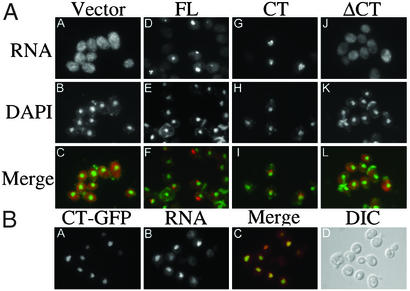
Expression of either FL-Mlp1p or CT-Mlp1p causes poly(A) RNA to accumulate within the nucleus (Fig. 2A D, E, G, and H). However, when ΔCT-Mlp1p is expressed, poly(A) RNA remains cytoplasmic, suggesting that the effect is mediated by CT-Mlp1p (Fig. 2AJ). Neither a vector control nor the Srp1p-negative control had any effect on poly(A) RNA localization (Fig. 2AA and data not shown). Closer observation revealed that on overexpression of CT-Mlp1p, poly(A) RNA accumulates outside of the DAPI-stained chromatin region (Fig. 2AI). This finding was also observed for overexpression of full-length Mlp1p, where poly(A) RNA accumulates in a chromatin-free region of the nucleus (Fig. 2AF) (23). We next tested whether poly(A)RNA colocalized with CT-Mlp1p. The CT-Mlp1p-GFP protein localized to the nucleus and formed distinct crescent shapes within the nucleus when overexpressed (Fig. 2BA). This localization is reminiscent of the localization of overexpressed full-length Mlp1p (21). Furthermore, the intranuclear poly(A) RNA accumulated in the same region as CT-Mlp1p-GFP (Fig. 2B A–C). Accumulation of poly(A) RNA within the nucleus correlated with expression of CT-Mlp1p-GFP, as determined by visualization of the GFP signal. These results indicate that expression of the CT-Mlp1p is both necessary and sufficient to alter the intracellular localization of poly(A) RNA. This finding supports our hypothesis that when Mlp1p is overexpressed sequestering of hnRNPs by CT-Mlp1p causes a poly(A) RNA export defect.
Figure 2.
Characterization of the CT-Mlp1p. (A) Cells expressing a galactose-inducible vector (A–C), FL-Mlp1p (D–F), CT-Mlp1p (G–I), or ΔCT-Mlp1p (J–L) were grown in galactose and poly(A) RNA was visualized by fluorescence in situ hybridization analysis. Corresponding DAPI staining and merged images are shown. DAPI, green; poly(A) RNA, red; merge, yellow. (B) Cells expressing a galactose-inducible CT-Mlp1p-GFP protein were grown in galactose and CT-Mlp1p-GFP and poly(A) RNA were visualized by indirect immunofluorescence (A) and fluorescence in situ hybridization analysis (B). Corresponding merged (C) and differential interference contrast (DIC) (D) images are shown. GFP, green; poly(A) RNA, red; merge, yellow. (Magnification: ×100.)
Overexpression of Mlp1p Inhibits Cell Growth.
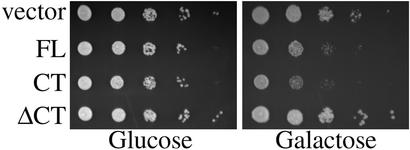
Because poly(A) RNA export from the nucleus is an essential cellular process, we hypothesized that blocking poly(A) RNA export would inhibit cell growth. To test this hypothesis, WT cells containing a vector control or plasmids expressing FL-MLP1, CT-MLP1, or ΔCT-MLP1 were grown to saturation in glucose media, where expression is not induced, and then serially spotted onto glucose or galactose plates (Fig. 3). On galactose plates where Mlp1p is overexpressed, cells expressing FL-Mlp1p or CT-Mlp1p grew more slowly than the vector control or cells expressing ΔCT-Mlp1p. Thus, overexpression of the CT-Mlp1p inhibits cell growth, which is consistent with a decrease in the efficiency of poly(A) RNA export from the nucleus.
Figure 3.
Overexpression of Mlp1p inhibits cell growth. WT cells containing a galactose-inducible vector control, FL-MLP1 (FL), CT-MLP1 (CT), or ΔCT-MLP1 (ΔCT) were grown to saturation in glucose, serially diluted, and spotted onto glucose (Left) or galactose (Right) minimal media plates.
Mlp1p Blocks Nab2p Export from the Nucleus.
Our results indicate that the CT-Mlp1p directly interacts with Nab2p and that overexpression of this domain causes accumulation of poly(A) RNA within the nucleus. We have proposed that the inhibition of poly(A) RNA export is caused by titration of RNA-bound Nab2p/hnRNPs by Mlp1p. If this model is correct, overexpression of the CT-Mlp1p should also block the export of Nab2p from the nucleus.
Nab2p is a shuttling protein (12, 26, 36) with a nuclear steady-state localization (35). We predict that Mlp1p overexpression should sequester Nab2p in the nucleus; however, this localization would be indistinguishable from the WT steady-state nuclear localization of Nab2p. Therefore to analyze Nab2p export, it was first necessary to generate a mutant of Nab2p that still shuttles, but has a more cytoplasmic steady-state localization than WT Nab2p due to a slowed rate of nuclear import. With such a mutant, nuclear accumulation caused by inhibition of Nab2p export could be readily observed as a shift from the cytoplasm to the nucleus.
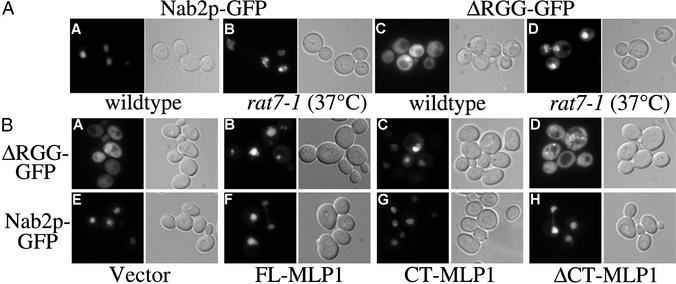
A slower importing mutant of Nab2p was created by deleting the RGG domain of Nab2p, the domain that binds the Nab2p import receptor, Kap104p (12, 37). In WT cells at steady state, full-length Nab2p-GFP is localized within the nucleus (Fig. 4AA) (35). In contrast, ΔRGG-Nab2p-GFP is localized throughout the cell (Fig. 4AC). The localization of ΔRGG-Nab2p-GFP throughout the cell could be caused by (i) lack of import into the nucleus or (ii) a decrease in the rate of import such that Nab2p import is now slower than export. To distinguish between these possibilities, we examined ΔRGG localization in cells where protein export is blocked. If the ΔRGG protein never enters the nucleus, a block in protein export should not affect its steady-state localization. In contrast, if the ΔRGG protein can enter the nucleus, then blocking export would result in nuclear accumulation of the ΔRGG protein.
Figure 4.
Overexpression of CT-Mlp1p causes Nab2p accumulation within the nucleus. (A) GFP signal was visualized in WT cells (A and C) and rat7-1 cells shifted to 37°C (B and D) expressing Nab2p-GFP (A and B) or ΔRGG-Nab2p-GFP (C and D). Corresponding differential interference contrast images are shown. (B) GFP signal was visualized in WT cells that express a vector control (A and E), FL-Mlp1p (B and F), CT-Mlp1p (C and G), or ΔCT-Mlp1p (D and H) and coexpress ΔRGG-Nab2p-GFP (A–D) or Nab2p-GFP (E–H). Corresponding differential interference contrast images are shown. (Magnification: ×100.)
A protein export assay was performed by using the rat7-1 temperature-sensitive mutation of the Nup159p/Rat7p nucleoporin, which inhibits protein export from the nucleus (38). After a shift to the nonpermissive temperature (37°C) where protein export is blocked, ΔRGG-Nab2p-GFP accumulates within the nucleus of rat7-1 cells (Fig. 4AD). This result indicates that ΔRGG-Nab2p-GFP is, in fact, able to shuttle into and out of the nucleus, but is imported more slowly than the full-length Nab2 protein. Further analysis of this mutant has revealed that the ΔRGG-Nab2p protein can functionally replace the endogenous full-length Nab2 protein in vivo (41), indicating that ΔRGG-Nab2p can still perform the essential function of Nab2p. These experiments establish that the ΔRGG-Nab2p mutant can be used as a tool to analyze cellular effects on Nab2p export.
To test whether overexpression of Mlp1p affects Nab2p export, we analyzed the localization of ΔRGG-Nab2p-GFP in cells overexpressing Mlp1p (Fig. 4B). Cells containing the galactose-inducible FL-MLP1, CT-MLP1, ΔCT-MLP1, or vector control and coexpressing either Nab2p-GFP, ΔRGG-Nab2p-GFP, or an NLS-NES-GFP control protein were grown in either glucose or galactose and the GFP signal was monitored in live cells. When either FL-Mlp1p or CT-Mlp1p is overexpressed, ΔRGG-Nab2p-GFP accumulates within the nucleus showing that Nab2p export is blocked (Fig. 4B B and C). However, when ΔCT-Mlp1p is expressed, ΔRGG-Nab2p-GFP remains cytoplasmic, suggesting that the effect is mediated by the CT-Mlp1p (Fig. 4BD). Neither a vector control nor the Srp1p negative control had any effect on ΔRGG localization (Fig. 4AA and data not shown). The cytoplasmic steady-state localization of an NLS-NES-GFP shuttling reporter protein is not affected by Mlp1p overexpression, indicating that overexpression of Mlp1p does not globally affect all protein export (Fig. 6, which is published as supporting information on the PNAS web site). Nab2p-GFP remains within the nucleus in all cells, indicating that there is no apparent effect on Nab2p import (Fig. 4B E–H). The localization of all GFP-tagged proteins in cells grown in glucose, where Mlp1p expression is repressed, was indistinguishable from the localization in WT cells (data not shown). These results indicate that expression of the CT-Mlp1p blocks Nab2p export from the nucleus.
Analysis of Mlp1p–hnRNP Complexes.
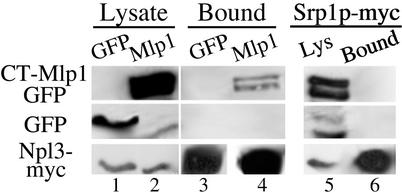
To better understand the role that Mlp1p plays in RNP export, we sought to identify other proteins in complex with Mlp1p. We therefore extended our immunoprecipitation analysis to another hnRNP, Npl3p, and a non-hnRNP shuttling nuclear control protein, Srp1p (39). Protein extracts from cells expressing either a GFP control protein or CT-Mlp1p-GFP and coexpressing Npl3p-myc or Srp1p-myc were immunoprecipitated with myc Ab. Immunoblot analysis of bound fractions indicated that Npl3p-myc coimmunoprecipitated CT-Mlp1p-GFP (Fig. 5, lane 4), whereas the control Srp1p-myc protein did not (Fig. 5, lane 6), indicating that the interactions observed are not caused by nonspecific interactions with all nuclear proteins or the myc tag. The Npl3p-myc protein also did not coimmunoprecipitate the control GFP (Fig. 5, lane 3). As with Nab2p, the interaction between Npl3p and Mlp1p is not sensitive to RNase treatment (data not shown). This result suggests that Mlp1p associates with multiple hnRNPs and possibly multiple RNP complexes.
Figure 5.
CT-Mlp1p coimmunoprecipitates Npl3p. Immunoblot analysis of protein lysates containing Npl3p-myc (lanes 1 and 2) or Srp1p-myc (lane 5) and GFP control (lane 1) or CT-Mlp1p-GFP (lanes 2 and 5). Proteins were immunoprecipitated with anti-myc Ab and bound fractions were probed with anti-GFP Ab (Top and Middle) or anti-myc Ab (Bottom) (lanes 3, 4, and 6).
Our work identifies hnRNPs physically associated with Mlp1p in vivo. The fact that Mlp1p is a nuclear pore-associated protein suggests that Mlp1p–RNP complexes identified represent RNP complexes at later stages of mRNA processing that are export competent. Recently, several RNA binding proteins in D. melanogaster have been shown to colocalize with Tpr during conditions of heat shock (40), suggesting that these interactions and functions may be conserved. Identification of other proteins associated with Mlp/Tpr may help to delineate the RNP signals present on mature mRNA transcripts ready for export from the nucleus.
Previous work has suggested a role for Nab2p in regulating poly(A) tail length and proper mRNA export (25, 26). Nab2p may bind pre-mRNA transcripts during polyadenylation and remain bound to the transcript to indicate that proper polyadenylation has occurred. It is possible that Nab2p serves as the primary contact between Mlp1p and the RNP complex. It is also possible that Mlp1p could make multiple contacts with various proteins in the RNP complex. The only common motif shared between Nab2p and Npl3p is the characteristic RGG domain; however, in the case of Nab2p this domain does not appear to be responsible for the interaction with Mlp1p (Fig. 4B and unpublished data). Further investigation will be necessary to test whether Mlp1p interacts directly with multiple hnRNPs or whether the interactions occur through Nab2p.
Our results also show that the effect of overexpression of Mlp1p on poly(A)RNA export is mediated by the C-terminal domain, presumably through direct interactions with RNP complexes. This conclusion is supported by our finding that expression of the C-terminal domain also blocks poly(A) RNA and Nab2p export in a Δmlp1Δmlp2 background (data not shown). Our hypothesis is that overexpression of Mlp1p titrates and sequesters hnRNPs, and consequently mRNA, within the nucleus through interactions with its C-terminal domain. This work provides support for the hypothesis that Nab2p, possibly in conjunction with other hnRNPs, is the functional link between Mlp1p and mRNA.
CT-Mlp1p shares 17% identity (39% similarity) with the C-terminal domain of the mammalian Tpr protein. This analysis identifies potential conserved residues that could mediate the interaction between Tpr/Mlp and hnRNPs. It should also be noted that Mlp1p and Mlp2p share 22% identity (32% similarity) within the C-terminal domain. Mlp1p and Mlp2p have similar domain structures and cellular localization (21–23), but there is no evidence for a relationship between Mlp2p and mRNA export (23). Whether these proteins are solely redundant homologs or have independent functions is still unknown.
This work has important implications for understanding mRNA processing and export and the function of Mlp/Tpr. The data presented in this article in conjunction with the fact that neither MLP1 nor MLP2 is essential lead us to propose that Mlp/Tpr could act as a checkpoint for quality control of mRNA processing. When present, Mlp/Tpr may interact with RNPs on export-competent transcripts and permit exit from the nucleus. However, if the proper complement of RNPs was not present, the complex would be retained within the nucleus for further processing. Consistent with this model, genetic and physical interactions between the Mlp proteins and RNA export factors have also been identified (F. Stutz, personal communication). In the absence of Mlp/Tpr, this checkpoint would be bypassed and RNP complexes would be nondiscriminately exported, regardless of whether the proper RNP complement was present. This model could account for the fact that neither MLP1 nor MLP2 is essential, as most checkpoint proteins are not essential under normal conditions. Further investigation of Δmlp1Δmlp2 mutant yeast will be necessary to test this model. In addition, with the identification of two hnRNPs in complex with Mlp1p, experiments can now be designed to better understand how the Mlp–RNP interaction contributes to mRNA export from the nucleus.
Supplementary Material
Acknowledgments
We thank P. Fanara, E. Griffis, M. Powers, S. T. Warren, M. T. Harreman, S. W. Leung, and K. A. Marfatia for their helpful experimental and manuscript suggestions. We also acknowledge M. P. Rout and P. A. Silver for reagents used in this study. This work was supported in part by a National Institutes of Health grant (to A.H.C.). D.M.G. is a member of the Graduate Program in Biochemistry, Cell and Developmental Biology and was funded by Emory University as an Emory University Minority Graduate Fellow. C.P.J. was sponsored by the Emory University Scientific Undergraduate Research Experience Program funded by the Howard Hughes Medical Institute.
Abbreviations
- NPC
nuclear pore complex
- RNP
ribonucleoprotein
- hnRNP
heterogeneous nuclear RNP
- DAPI
4′,6-diamidino-2-phenylindole-dihydrochloride
- NLS
nuclear localization sequence
- NES
nuclear export sequence
- DBD
DNA binding domain
- AD
activation domain
- Tpr
translocated promoter region
- Mlp1p
myosin-like protein 1
- CT-Mlp1p
C-terminal domain of Mlp1p
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Cramer P, Srebrow A, Kadener S, Werbajh S, de la Mata M, Melen G, Nogues G, Kornblihtt A R. FEBS Lett. 2001;498:179–182. doi: 10.1016/s0014-5793(01)02485-1. [DOI] [PubMed] [Google Scholar]
- 2.Shatkin A J, Manley J L. Nat Struct Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 3.Hastings M L, Krainer A R. Curr Opin Cell Biol. 2001;13:302–309. doi: 10.1016/s0955-0674(00)00212-x. [DOI] [PubMed] [Google Scholar]
- 4.Akker S A, Smith P J, Chew S L. J Mol Endocrinol. 2001;27:123–131. doi: 10.1677/jme.0.0270123. [DOI] [PubMed] [Google Scholar]
- 5.Maquat L E, Carmichael G G. Cell. 2001;104:173–176. doi: 10.1016/s0092-8674(01)00202-1. [DOI] [PubMed] [Google Scholar]
- 6.Zenklusen D, Stutz F. FEBS Lett. 2001;498:150–156. doi: 10.1016/s0014-5793(01)02482-6. [DOI] [PubMed] [Google Scholar]
- 7.Reed R, Hurt E. Cell. 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- 8.Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 9.Reed R, Magni K. Nat Cell Biol. 2001;3:E201–E204. doi: 10.1038/ncb0901-e201. [DOI] [PubMed] [Google Scholar]
- 10.Michael W M. Trends Cell Biol. 2000;10:46–50. doi: 10.1016/s0962-8924(99)01695-5. [DOI] [PubMed] [Google Scholar]
- 11.Flach J, Bossie M, Vogel J, Corbett A H, Jinks T, Willins D A, Silver P A. Mol Cell Biol. 1994;14:8399–8407. doi: 10.1128/mcb.14.12.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aitchison J D, Blobel G, Rout M P. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 13.Rout M P, Aitchison J D. J Biol Chem. 2001;276:16593–16596. doi: 10.1074/jbc.R100015200. [DOI] [PubMed] [Google Scholar]
- 14.Wente S R. Science. 2000;288:1374–1377. doi: 10.1126/science.288.5470.1374. [DOI] [PubMed] [Google Scholar]
- 15.Panté N, Aebi U. Science. 1996;273:1729–1732. doi: 10.1126/science.273.5282.1729. [DOI] [PubMed] [Google Scholar]
- 16.Cordes V C, Reidenbach S, Rackwitz H R, Franke W W. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimowska G, Aris J P, Paddy M R. J Cell Sci. 1997;110:927–944. doi: 10.1242/jcs.110.8.927. [DOI] [PubMed] [Google Scholar]
- 18.Byrd D A, Sweet D J, Panté N, Konstantinov K N, Guan T, Saphire A C, Mitchell P J, Cooper C S, Aebi U, Gerace L. J Cell Biol. 1994;127:1515–1526. doi: 10.1083/jcb.127.6.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bangs P L, Sparks C A, Odgren P R, Fey E G. J Cell Biochem. 1996;61:48–60. doi: 10.1002/(sici)1097-4644(19960401)61:1<48::aid-jcb7>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Frosst P, Guan T, Subauste C, Hahn K, Gerace L. J Cell Biol. 2002;156:617–630. doi: 10.1083/jcb.200106046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strambio-de-Castillia C, Blobel G, Rout M P. J Cell Biol. 1999;144:839–855. doi: 10.1083/jcb.144.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolling R, Nguyen T, Chen E Y, Botstein D. Mol Gen Genet. 1993;237:359–369. doi: 10.1007/BF00279439. [DOI] [PubMed] [Google Scholar]
- 23.Kosova B, Panté N, Rollenhagen C, Podtelejnikov A, Mann M, Aebi U, Hurt E. J Biol Chem. 2000;275:343–350. doi: 10.1074/jbc.275.1.343. [DOI] [PubMed] [Google Scholar]
- 24.Bangs P, Burke B, Powers C, Craig R, Purohit A, Doxsey S. J Cell Biol. 1998;143:1801–1812. doi: 10.1083/jcb.143.7.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hector R E, Nykamp K R, Dheur S, Anderson J T, Non P J, Urbinati C R, Wilson S M, Minvielle-Sebastia L, Swanson M S. EMBO J. 2002;21:1800–1810. doi: 10.1093/emboj/21.7.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green D M, Marfatia K A, Crafton E B, Zhang X, Cheng X, Corbett A H. J Biol Chem. 2002;277:7752–7760. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 28.Adams A, Gottschling D E, Kaiser C A, Stearns T. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 29.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 30.Shen E C, Stage-Zimmermann T, Chui P, Silver P A. J Biol Chem. 2000;275:23718–23724. doi: 10.1074/jbc.M002312200. [DOI] [PubMed] [Google Scholar]
- 31.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seedorf M, Damelin M, Kahana J, Taura T, Silver P A. Mol Cell Biol. 1999;19:1547–1557. doi: 10.1128/mcb.19.2.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fanara P, Hodel M R, Corbett A H, Hodel A E. J Biol Chem. 2000;275:21218–21223. doi: 10.1074/jbc.M002217200. [DOI] [PubMed] [Google Scholar]
- 34.Wong D H, Corbett A H, Kent H M, Stewart M, Silver P A. Mol Cell Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson J T, Wilson S M, Datar K V, Swanson M S. Mol Cell Biol. 1993;13:2730–2741. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duncan K, Umen J G, Guthrie C. Curr Biol. 2000;10:687–696. doi: 10.1016/s0960-9822(00)00527-3. [DOI] [PubMed] [Google Scholar]
- 37.Lee D C, Aitchison J D. J Biol Chem. 1999;274:29031–29037. doi: 10.1074/jbc.274.41.29031. [DOI] [PubMed] [Google Scholar]
- 38.Krebber H, Taura T, Lee M S, Silver P A. Genes Dev. 1999;13:1994–2004. doi: 10.1101/gad.13.15.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solsbacher J, Maurer P, Bischoff F R, Schlenstedt G. Mol Cell Biol. 1998;18:6805–6815. doi: 10.1128/mcb.18.11.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimowska G, Paddy M R. Exp Cell Res. 2002;276:223–232. doi: 10.1006/excr.2002.5525. [DOI] [PubMed] [Google Scholar]
- 41. Marfatia, K. A., Crafton, E. B., Green, D. M. & Corbett, A. H. (2003) J. Biol. Chem., in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.