CONSPECTUS
FHA domains are protein modules that switch signals in diverse biological pathways by monitoring the phosphorylation of threonine residues of target proteins. As part of the effort to gain insight into cellular avoidance of cancer, FHA domains involved in the cellular response to DNA damage have been especially well characterized. The complete protein where the FHA domain resides and the interaction partners determine the nature of the signaling. Thus, a key biochemical question is: how do FHA domains pick out their partners from among thousands of alternatives in the cell? This Account discusses the structure, affinity, and specificity of FHA domains and the formation of their functional structure.
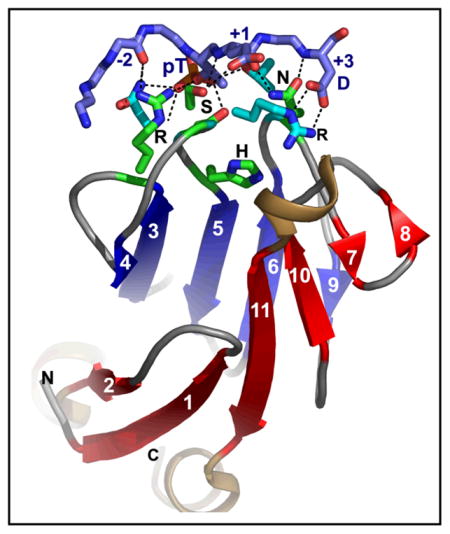
Although FHA domains share sequence identity at only five loop residues, they all fold into a β-sandwich of two β-sheets. The conserved Arg and Ser of the recognition loops recognize the phosphorylation of the Thr targeted. Side chains emanating from loops that join β-strand 4 with 5, 6 with 7, or 10 with 11 make specific contacts with amino acids of the ligand that tailor sequence preferences. Many FHA domains choose a partner in extended conformation, somewhat according to the residue three after the phosphoThr in sequence (pT+3 position). One group of FHA domains chooses a short carboxylate-containing side chain at pT+3. Another group chooses a long, branched aliphatic side chain. A third group prefers other hydrophobic or uncharged, polar side chains at pT+3. However, another FHA domain instead chooses on the basis of pT−2, pT−3, and pT+1 positions. An FHA domain from a marker of human cancer instead chooses a much longer protein fragment that adds a β-strand to its β-sheet and that presents hydrophobic residues from a novel helix to the usual recognition surface. This novel recognition site and more remote sites for the binding of other types of protein partners were predicted for the entire family of FHA domains by a bioinformatics approach.
The phosphopeptide-dependent dynamics of an FHA domain, SH2 domain, and PTB domain suggest a common theme: rigid, preformed binding surfaces support van der Waals contacts that provide favorable binding enthalpy. Despite the lack of pronounced conformational changes in FHA domains linked to binding events, more subtle adjustments may be possible. In the one FHA domain tested, phosphoThr peptide binding is accompanied by increased flexibility just outside the binding site and increased rigidity across the β-sandwich. The folding of the same FHA domain progresses through near-native intermediates that stabilize the recognition loops in the center of the phosphoprotein-binding surface; this may promote rigidity in the interface and affinity for targets phosphorylated on threonine.
Keywords: FHA domain, phosphorylation-dependent signaling, protein structure, molecular recognition, protein dynamics, protein folding
Introduction
Since phosphate is susceptible to nucleophilic attack, a good leaving group, and retained by cells due to its charge, it is widely exploited as a biological signal 1, 2. Phosphoesters are stable until meeting enzymes such as phosphatases that rapidly hydrolyze them 1. Phosphorylation and other post-translational modifications are monitored by non-covalent associations with a diverse array of protein modules that reversibly juxtapose protein partners in intracellular signaling cascades 3. Since threonine/serine phosphorylations are the most common protein kinase activities, several classes of protein adapters associate with proteins phosphorylated on hydroxyl-containing threonine residues 4. These adapters include FHA domains which assist in regulation of cell growth, division, differentiation, programmed cell death 4, DNA damage response, transcription of DNA to RNA, transport of cargo within cells, and protein degradation 5. FHA domains are found in more than 700 eukaryotic proteins such as kinases, phosphatases, motor proteins called kinesins, DNA-binding transcription factors, RNA-binding proteins, and metabolic enzymes. Identification of the domains in Forkhead transcription factors led to their naming as Forkhead-associated (FHA) domains 6. FHA domains are found in more than 500 bacterial proteins; some participate in injection of viral proteins into host cells, transmembrane transporters, and cell division 7. A unifying theme among these diverse roles is that FHA domains recognize threonine phosphorylation accompanying activation of protein Ser/Thr kinases.
Since FHA domains are widely important in and representative of biological regulation, this Account focuses on chemical insights into how FHA domains work, starting with their structure, proceeding to specificity, and ending with how the functional structure may form. An FHA domain is organized into a β-sandwich of six-stranded and five-stranded β-sheets (Figure 1). Of the 120 to 140 amino acids therein, only five are identical in most FHA domains 6 (bold in Figure 2). Four of these are required for phosphoprotein interaction in one example 8, and lie along an edge of the five-stranded β-sheet in three contiguous loops (Figure 1) that contact phosphorylated partners. The central 4-5 recognition loop can be interrupted by a helix 9 (cyan in Figure 1). (The loops are numbered by the β-strands they connect). The adjacent 10-11 loop varies in length (top, middle of Figure 1). The FHA domains of Rad53 from baker’s yeast have helical insertions in loops distant from the phosphorecognition surface (Figure 1). Structures of FHA domains bound to pThr peptides 9–17 and affinities of pThr peptides for several, varied FHA domains 9, 10, 12, 13, 16–18 offer breadth of perspective on the structure and function of diverse FHA domains. General mechanisms shared by FHA domains have emerged: The typically low micromolar dissociation constants imply fast off-rates and depend heavily upon phosphorylation of the target threonine, which enables rapid and reversible switching and dissociation. The five conserved loop residues that support affinity for the phosphorylated threonine (pThr or pT) are surrounded by residues of the loops that differ among FHA domains in order to recognize the preferred sequences. These amino acid positions after or before the pThr are specified by the pT+X or pT−X nomenclature, respectively.
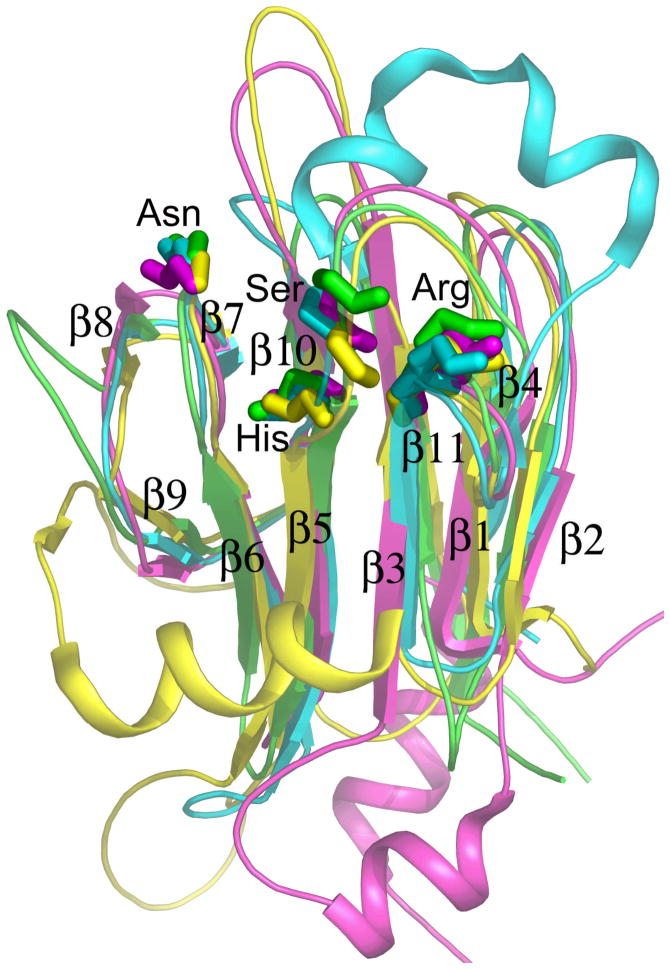
Figure 1.
The fold of the FHA domain is illustrated for yeast RAD53 FHA1 11 (magenta) and FHA2 12 (yellow), human Chk2 9 (cyan), and plant kinase-associated protein phosphatase 25 (green). Side chains of five mostly conserved residues near the phospho-recognition surface are drawn.
Figure 2.
Structure-based alignment of sequences of FHA domains. Residues in the 3-4, 4-5, 6-7 and 10-11 recognition loops that contact phosphopeptide ligands are underlined. Mostly conserved residues are bold. Numbers inserted in the sequences count residues omitted from this display emphasizing recognition loops.
Determinants of PhosphoThr Peptide Affinity and Specificity
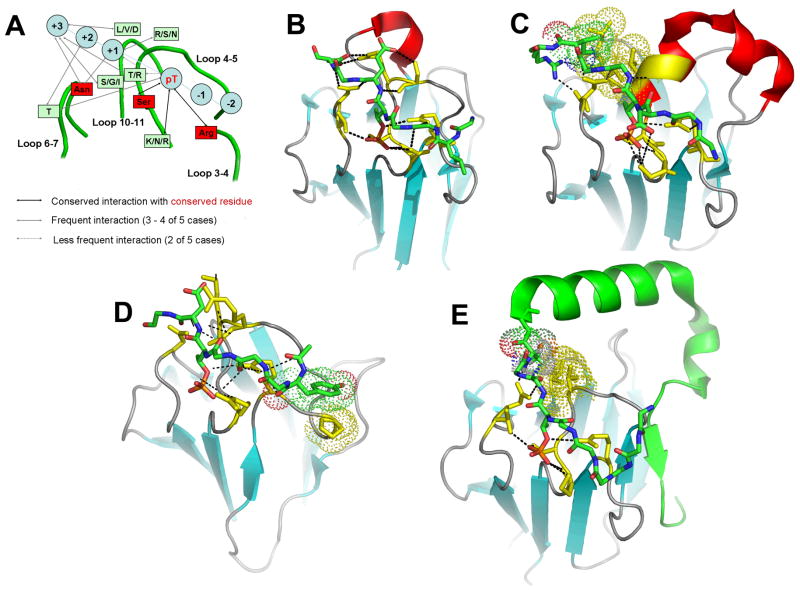
Residues and interactions shared by FHA domains for recognizing the phosphorylation of the threonine in a target peptide or protein ligand have been identified. Conserved in complexes of phosphopeptides with FHA domains are hydrogen bonds or salt bridges between the phosphate and the conserved Arg of the 3-4 loop, the conserved Ser of the 4-5 loop, and a Lys/Asn/Arg of the 4-5 loop (Figure 3A). The conserved Asn of the 6-7 loop often hydrogen bonds the pT+3 and pT+1 positions of peptide ligands (Figure 3A–C, E). The conserved Gly and His interact with each other and probably stabilize the architecture of the binding site.
Figure 3.
Comparison of structural details of phosphoThr peptide interactions with FHA domains. A) Summary of FHA domain interactions with peptides for Rad53 11-14 and Chk2 9. The structures of complexes in each panel represent: B) FHA1 of RAD53 (PDB code 1G6G) 13, C) Chk2 (1GXC) 9, D) PNK (1YJM) 17, and E) KI-67 (2AFF) 16. Each peptide is colored by atom: green for carbon, blue for nitrogen, and red for oxygen. FHA domain residues in contact with ligands are yellow. Electrostatic interactions and hydrogen bonds are shown with dotted lines. Hydrophobic interactions are represented with dots.
Given that phosphorylation of threonine is commonplace in cells and that five-residue protein sequences are enough to encode greater than 100,000 possible phosphoThr-containing sequences, what sequences does an FHA domain prefer to bind? Distinctive preferences in sequences of phosphoThr peptides from combinatorial libraries have been identified for several FHA domains 13. Both the FHA1 domain of Rad53p from baker’s yeast and the FHA domain of Cds1 from fission yeast selectively recognize a pT-V-x-D motif, particularly the negative charge of the Asp at the pT+3 position 13. Three FHA domains tested prefer a hydrophobic residue at pT+3: Ile by FHA2 of yeast Rad53p, Leu or Ile by human Mdc1, and Met or Tyr by Y127 from Mycobacterium tuberculosis 13. The FHA domain of KAPP from plants instead prefers Ser or Ala at pT+3 13.
How does an FHA domain achieve specificity for its partners, from among thousands of alternative phosphoThr sequences it might encounter in the cell?
As we consider what might adjust the specificity of FHA domains, we next examine shared and varying structural features among complexes of FHA domains with phosphopeptides. The residues FHA domains use to recognize the pT+3 of ligands vary. Some yeast FHA domains use an Arg to recognize Asp at pT+3 in the peptide (see Conspectus showing Rad53 FHA1), while others select the Ile or Leu at pT+3 with a hydrophobic pocket formed by the 4-5, 6-7, and 10-11 loops. FHA domains usually show weaker selection at the −4 to +2 positions 13. Contacts between +1 through +3 positions of the phosphopeptide and the 6-7 loop or 10-11 loop of the FHA domain are frequent (Figure 3A). We now examine particular structures of FHA domains with pThr peptides bound, in order to point out interactions that may generalize to other FHA complexes and then proceed in each case to distinctive interactions that may tailor the specificity of that FHA domain.
Aspartate Preference at pT+3
Rad53p regulates the DNA damage response in baker’s yeast 19. In addition to a kinase domain, it comprises FHA1 and FHA2 domains required for DNA damage-dependent Rad53 activation 20. Its target protein of Rad9 is phosphorylated in response to DNA damage and interacts with the C-terminal FHA domain of Rad53 21. The interaction of FHA1 of Rad53 and a Rad9p-derived phosphoThr peptide features an extensive network of hydrogen bonds that align and order the peptide binding loops 13. A hydrogen bond between the phosphoThr and the Thr in the 6-7 loop of Rad53 FHA1 (Figure 3B) supplements the conserved contacts with the phosphate (Figure 3A). Side chains of the conserved Arg, conserved Asn, and Asn86 donate hydrogen bonds to main chain carbonyl oxygen atoms at pT+1, pT−2, and pT−4 positions of the peptide 13 (Conspectus and Figure 3B). The conserved Asn accepts an H-bond from the amide group of Asp at pT+3. These backbone interactions could be widely shared among FHA domain interactions with phosphorylated partners to lend affinity. FHA1 of Rad53 makes other contacts that tailor its distinctive specificity. The carboxylate group of Asp at pT+3 in the peptide forms a salt bridge to the guanidinium moiety of Arg83 in the 4-5 loop of FHA1, providing affinity 13 (Conspectus and Figure 3B). Rad53 FHA1 binds to Rad9-derived pThr peptides, containing pTxxD sequences, by contacting pT, pT+1 and pT+2 peptide positions using Ser and Arg from the 4-5 loop and Thr and Asn from the 6-7 loop 11.(Numbering of secondary structure in NMR structures of Rad53 domains is one less than here and other structural reports). Recognition of pThr and Asp at pT+3 are keys to the binding affinity and specificity, while pT+1 and pT+2 residues appear to fine tune recognition.
Preferences for Branched Aliphatics at pT+3
FHA2 of Rad53p forms an extensive hydrogen bonding network with a pThr peptide ligand using residues from the 3-4, 4-5, 6-7, and 10-11 loops 12. The backbone carbonyl oxygen of an Asn in the 4-5 loop forms an H-bond with the amide proton at pT+1. A Thr OγH from the 6-7 loop makes a hydrogen bond with the carboxylate group of Glu at pT+2 12. Extensive hydrophobic interactions occur between a Thr methyl group from the 6-7 loop and Ile of the 10-11 loop of FHA2 and the peptide’s pThr methyl group, Glu alkyl group at pT+2, and Leu at pT+3 12. The hydrophobic contacts account for FHA2’s preference for Ile or Leu at the pT+3 position 13.
Protein kinase Chk2, mammalian homolog of Rad53p and Cds1 checkpoint kinases from yeasts, is phosphorylated and activated in response to DNA damage, resulting in checkpoint activation and cell cycle arrest 22. Like FHA2 of Rad53, Chk2 FHA selectively binds pT peptides containing Ile or Leu at pT+3. Hydrophobic residues are preferred at the pT−2, pT+1, and pT+2 positions 9. The peptide HFDpTYLI with this sequence pattern packs against the 3-4, 4-5, 6-7, and 10-11 loops of the Chk2 FHA domain 9. The pThr γ-methyl group makes van der Waals contacts with the conserved Asn of the 6-7 loop, as well as with Thr, Tyr, and conserved Ser of the 4-5 loop 9. A Lys in the 4-5 loop forms a salt bridge with the phosphate group and selects for Asp at pT−1.
Changing Preference at pT+3
Some FHA domains violate trends in recognition of the key pT+3 position of the peptide ligand 20. While FHA1 of Rad53p prefers Asp at pT+3 11, 13, it can switch specificity to bind a peptide with a physiological recognition site with Ile at pT+3 14,20. FHA1 interacts similarly with the phosphate of these differing peptides, forming a salt bridge with the conserved Arg of the 3-4 loop and hydrogen bond with the conserved Asn of the 4-5 loop 14. FHA1 uses Arg from the 4-5 loop to form a salt bridge with Asp at pT+3 of the first peptide. By contrast, FHA1 employs hydrophobic interactions with the pT+3 position of the physiological peptide sequence; the Ile at pT+3 makes hydrophobic contacts with a Gly and Val from the 10-11 loop of FHA1 14 (Figure 2, 3A). Moreover, FHA1 makes additional hydrophobic contacts with the physiological site at its pT+1, pT+2, and pT+4 positions 14. Thus, FHA1 can recognize pT+3 with electrostatics or alternatively can recognize pT+1 through pT+4 with hydrophobic contacts 14.
A Preference for Polar or Hydrophobic Group at pT+3
When the kinase-interacting FHA domain (KI-FHA) from plant kinase-associated protein phosphatase recognizes several trans-phosphorylated receptor-like kinase domains 18, 23, it attenuates signaling through receptor-like protein kinases at the cytoplasmic membrane 24. A principal, but not lone, site of KI-FHA binding in vitro was found in BRI1-Associated Kinase-1 (BAK1) at its Thr546 in region XI near the C-terminal end of the kinase domain 18. This site was initially suggested by pThr peptides from receptor-like kinase domains that bind KI-FHA. The favorable enthalpy and the hydrophobic effect from contacts with these pThr peptides account for their 8 to 30 μM KD values for KI-FHA 18. The sequence preferences of KI-FHA for pThr peptides 13 are shared with the Thr546 binding site in the BAK1 kinase 18, namely the preference for Ser at pT+3 and Gln at pT+1. KI-FHA from KAPP is unique in having a long and mobile 8-9 loop that joins the other four loops in recognition of phosphopartners 18, 25, 26.
Choosing Residues N-terminal to pThr
An FHA domain with distinctive determinants of specificity has been elucidated 17. Mammalian polynucleotide kinase (PNK) plays a key role in the repair of DNA with either single-strand breaks or double-strand DNA breaks 17. PNK acts as a 5′-kinase/3′-phosphatase to create 5′-phosphate/3′-hydroxyl termini required for ligation during repair. PNK is recruited to repair complexes through interactions between its N-terminal FHA domain and phosphorylated components. PNK is notable in not needing contact with the pT+3 position 17. Instead, its FHA domain depends more upon pT−2, pT−3, and pT+1 positions of peptide ligands 27, contrasting other well-studied FHA domains. The equivalent of β-strand 4 in other FHA domains is missing from PNK, but the flanking “3-4” and “4-5” recognition loops remain and form the interface with a peptide ligand 17. Several electrostatic interactions between a negatively charged pT peptide ligand and the very positively charged surface of the FHA domain of PNK are critical for the micromolar affinity 17. The second Arg in its “4-5” loop (Figure 2) interacts with the phosphate and Asp at pT−3 of the ligand (Figure 3D). The preceding Arg and Lys in the “4-5” loop (Figure 2) interact with the peptide’s Glu at pT−2 and Asp at pT+1. Hydrogen bonds are present between the conserved Asn and pT+1 backbone and between the conserved Arg and pT−2 backbone. Tyr at pT−4 stacks with a Pro from PNK (at right in Figure 3D) 17.
Binding Sites for Phosphoproteins
PhosphoThr peptides have been essential for studies of associations with FHA domains. Phosphoprotein fragments, however, offer more physiological relevance, but with challenges in obtaining suitable phosphorylation and solubility.
Similarity of Binding of Globular Phosphoprotein and PhosphoPeptide
BRI1 is a plant receptor Ser/Thr kinase that is critical for brassinosteroid hormonal signaling and development in plants 28. Once BRI1 is activated, it is attenuated by kinase-associated protein phosphatase (KAPP) that uses its KI-FHA domain to recognize the activated kinase domain 18. The surface of KI-FHA that binds the 43 kDa kinase domain of BRI1 was mapped by NMR. The BRI1 recognition surface of KI-FHA is essentially same as that for the pThr peptides, involving the 3-4, 4-5, 6-7, 8-9 and 10-11 loops of KI-FHA 18 (Figure 4). The affinity of GST-BRI1 for KI-FHA appears to be moderate, like it is for pThr peptides 18. Consequently, results on peptide interactions with KI-FHA may offer some value in representing interactions with receptor-like kinases 18, 25, 26.
Figure 4.
Receptor kinase binding site (panel B) on KI-FHA corresponds to pThr peptide binding site (panel A). Residues with NMR peaks most shifted by phosphorylated partner are colored red and those shifted less yellow. In A), the ligand is the pT546 peptide from plant kinase BAK1. In B), the partner is the BRI1 kinase domain from plants fused to GST 18.
Examples of Novel Motifs Added to Interface
Unlike FHA domains discussed above, Ki67FHA lacks affinity for short phosphopeptides. Ki67 is a cancer marker found in the nucleus. The FHA domain at its N-terminus binds human NIFK in a mitosis- and phosphorylation-dependent manner 29. The structure of the FHA domain of human Ki67 bound to a phosphorylated 44-residue fragment of NIFK is a unique physiological complex 16. The affinity of the 44-residue hNIFK phosphopeptide for human Ki67FHA is the highest affinity yet demonstrated between an FHA domain and a phosphorylated partner. To gain affinity for Ki67FHA, NIFK must undergo sequential phosphorylation of Thr238 by cyclin-dependent kinase 1 in order for glycogen synthase kinase 3 to phosphorylate the Thr234 that Ki67FHA recognizes 16. When this phosphorylated 44-residue NIFK fragment binds to Ki67FHA, it forms a helix and a β-strand that adds to the edge of the β-sheet of Ki67FHA to form an extended interface that buries 1,450 Å2 16. The structure of the recognition surfaces of Ki67FHA is mostly preformed, except that the 1-2, 5-6 and 6-7 loops adjust in conformation when the 44-mer phosphopeptide binds 16. Three regions of Ki67FHA (Figure 3E) interact with the 44-residue phosphopeptide to provide more contacts and a KD of 77 nM 16, which is about 100-fold greater affinity than typical of short pThr peptides. Three sites contribute to the high affinity between NIFK226–2693P and Ki67FHA (Figure 3E). Contributing most to affinity is the phosphorylated central threonine (T234) of the NIFK peptide that interacts with the well-characterized pThr binding surface. The phosphate of T234 accepts H-bonds donated by the conserved Arg from the 3-4 loop, Lys and conserved Ser from the 4-5 loop, and a partly conserved Thr from the 6-7 loop. The backbone of the peptide pT+1 and pT+3 positions form hydrogen bonds with the backbone and a side chain, respectively, of the conserved Asn from the 6-7 loop of Ki67FHA. Next in importance for affinity are NIFK residues 260–264 that form a novel β-strand (green arrow in Figure 3E) hydrogen bonded anti-parallel to β-strand 4 16. Finally, the 4-5 and 10-11 loops of Ki67FHA dock with the α-helix formed in the hNIFK peptide once bound. A Phe, Leu, and Arg (side chains not shown) on one side of this helix from NIFK fit into a hydrophobic patch formed by the 4-5 loop and 10-11 loop of KI67FHA (Figure 3E), likely both stabilizing the helix and enhancing affinity 16.
Another unique behavior is that the yeast Dun1 FHA domain preferentially binds a sequence having two phosphoThr residues from the Rad53 SQ/TQ cluster domain; this activates the DUN1-dependent transcriptional response to DNA damage 36. The conserved Arg of the 3-4 loop and conserved Ser of the 4-5 loop, which in other FHA domains recognize a single phosphoThr, instead reorient in Dun1 for recognizing the first and second phosphoThr residues, respectively, from the bound peptide 36.
Other Proposed Protein-Binding Sites
Functional surfaces of FHA domains were predicted 25 by a bioinformatics method 30. The approach predicts β-strand 4 to be a functional surface, consistent with the structure of KI67FHA where β-strand 4 is paired with the extended hNIFK peptide 16 (at right in Figure 3E). Since the bioinformatics prediction applies to the entire protein family of FHA domains 30, β-strand 4 may turn out to contribute to protein recognition events of other FHA domains as well. The predicted functional surfaces include the phoshoprotein binding surface, broad and variable in properties among FHA domains, and a recognition surface most distant from the phoshoprotein binding site 25. This patch is centered about Ile157 of Chk2 (at bottom of Figure 3C), includes residues from β-strand 9 and the 5-6 loop 25, and is the site of the I157T mutation in Li-Fraumeni cancer patients that disrupts binding of BRCA1, CDC25A, and p53 (proteins critically important in cancer) 9. Another functional surface around β-strands 7 and 10 was predicted by the bioinformatics. At this site the crystalline FHA domain of human CHFr dimerizes such that β7/8 (a hairpin in all other FHA domain structures) straightens out into single β-strand that H-bonds with the β7/8 strand from the other chain of the dimer 31. This swaps strands 9–11 into a β-sandwich with strands 1–7 from the other chain. In KI-FHA, β-strand 8 lacks measurable structural stability 32 and is mobile on the millisec scale 26. If low stability and mobility of β-strand 8 generalize to other FHA domains, it could account for β-strand 8 being malleable enough to straighten out in crystals of segment-swapped CHFr.
Dynamics and Energetics of pThr Peptide Binding
15N NMR relaxation of KI-FHA from KAPP indicates that its residues corresponding to those most important for the affinity and specificity of FHA domains are rigid on the fast timescale of picoseconds in KI-FHA 26. The conserved Gly, Arg, Ser, His, and Asn (Figure 1) are rigid in both free and bound states 26. Of five residues of KI-FHA likely to form the subsite for the pT+3 residue, a Leu and conserved Asn of the 6-7 loop and Gly and Thr of the 10-11 loop are rigid 26. The fifth, a Glu of the 4-5 loop, becomes rigid only once the pThr peptide binds. The rigidity of these recognition residues confirms that key recognition loops are pre-formed in KI-FHA. Why might this be? The intrinsically high backbone rigidity of these key residues should diminish free energy costs of loss of conformational entropy upon binding. The rigidity of these key recognition residues is likely to promote enthalpically favorable Van der Waals contacts between the pThr peptide and KI-FHA domain. Rigidity promoting such energetically favorable contacts between phosphopeptide and protein was proposed for the recognition of pTyr peptides by an SH2 domain 33 and a PTB domain 34. Consistent with that outlook, favorable enthalpy of association provides at least half of the free energy of binding of pThr peptides to KI-FHA 18. Since a rigid, preformed binding site conducive to enthalpically favorable contacts with phosphopeptide is shared among the SH2, PTB, and FHA domains tested, this behavior might generalize to other FHA domains as well.
Binding of a pThr peptide increases rigidity of KI-FHA globally. The average rigidity of the backbone on the fast timescale is increased with pThr peptide bound 26. In fact, sites of binding-enhanced rigidity are observed with the β-sandwich in all four of the β-hairpins (those with blue coloration in Figure 5) 26. The nanosecond fluctuations enriched in its loops are quenched once the phosphopeptide is bound. Perhaps the binding-increased rigidity of KI-FHA reflects the favorable enthalpy of association 18 that might result from improved internal packing and hydrogen bonding 26.
Figure 5.
Dynamics changes in KI-FHA upon phosphoThr peptide binding, derived from 15N NMR relaxation. Blue represents greater rigidity with pThr peptide bound, while orange-red represents mobilization by pThr peptide binding. Dark blue indicated greater rigidification than lighter blue. The diameter of the backbone tube is proportional to greater amplitude of subnanosecond motion in the free state.
Some residues of the 3-4, 4-5, and 8-9 recognition loops are comparatively flexible in the free state 26. The unusual extension, exposure and nanosecond mobility of the 8-9 loop of KI-FHA might help it adjust to a broader range of receptor-like kinase partners. Flexibility of recognition surfaces of other proteins broadens their specificity while limiting affinity and aiding dissociation 33, 35. The pThr peptide-increased flexibility on the periphery of the binding site 26 could provide part of the favorable entropy of pThr peptide binding to KI-FHA 18. The solvation entropy gain may overcome entropic costs of the phosphopeptide becoming rigid upon binding 18.
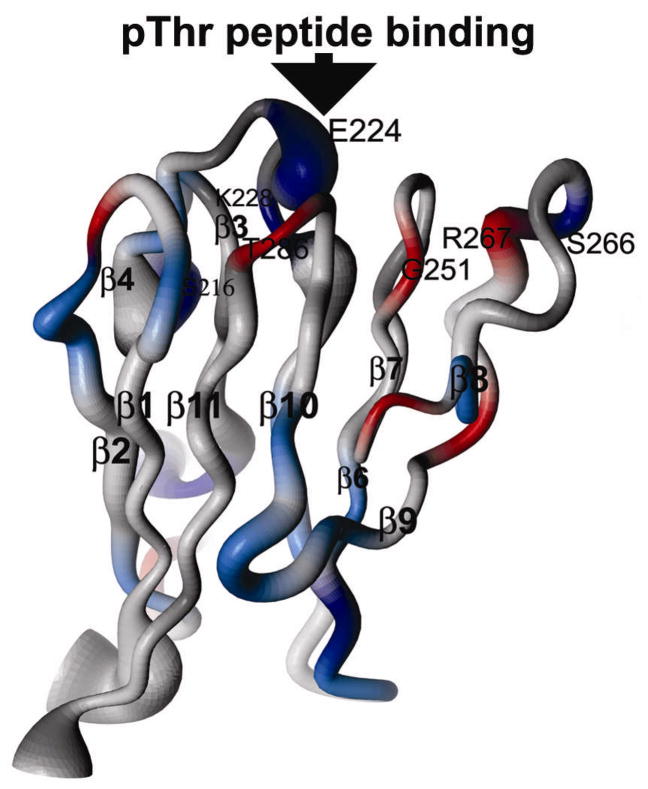
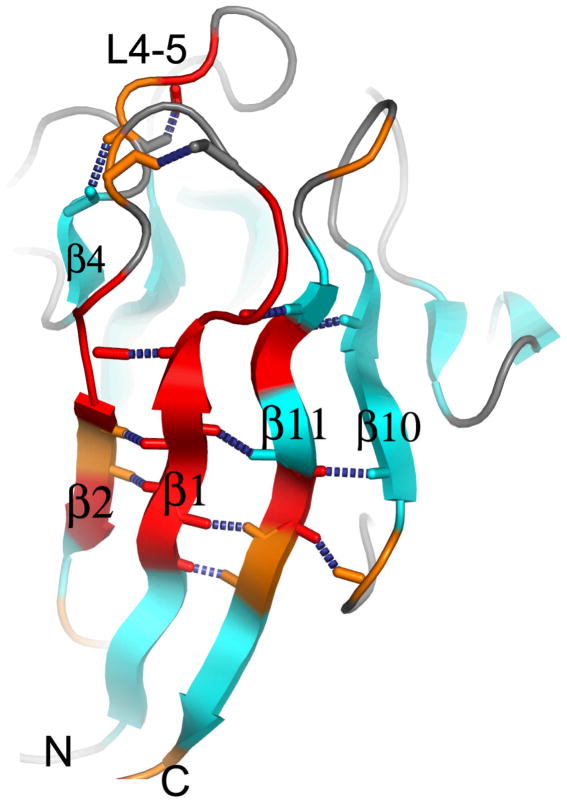
The dynamics of KI-FHA may also be linked to its stability. At pH 6.3, KI-FHA possesses a network of residues that undergoing backbone motions in millisec (Figure 6) 26. This network traverses β2, β1, β11, and β10 of the six-stranded sheet, plus β4 of the other sheet, where side chains are arrayed in two packed rows in the interior of the β-sandwich 26. At pH 7.3 where KI-FHA is at least 3 kcal/mol more stable 32, the NMR line broadening evidence for the slow motions of this network (Figure 6) vanishes. The stabilization and absence of slow correlated motions at pH 7.3 might reflect improved Van der Waals contacts among buried side chains.
Figure 6.
Sites of slow internal motions of KI-FHA at pH 6.3. Residues with μs to ms motion at pH 6.3 are colored red where NMR line broadening is larger or orange where it is smaller. This line broadening and motion is quenched at pH 7.3 where KI-FHA is more stable.
Folding of an FHA Domain and its Recognition Loops
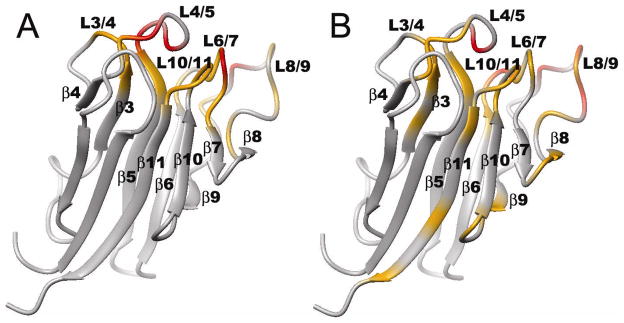
Compact, near-native intermediates were identified in the folding of KI-FHA 32. The most stable core of KI-FHA contains β-strands 1, 5, 6, 9, 10 and 11, as well as the 1-2 and 9-10 loops, each minutely populated in the partially unfolded form labeled PUF2 32. Flanking the most stable core, β-strands 2, 3 and 7 as well as the 6-7 and 10-11 recognition loops display an intermediate level of stability (green in Figure 7). β-strands 2 and 3 appear to form a subglobal folding motif. Neighboring recognition loops form another subglobal motif of similar stability in PUF1 32. The least stable subglobal motifs from the 4-5 and 8-9 recognition loops and β-strand 7 appear only in the native state (Figure 7) 32. The near-native intermediate states of KI-FHA suggest that it may fold progressively. An initial hydrophobic collapse may form the stable 6-stranded core (PUF2; blue in Figure 7). The 6-7 and 10-11 recognition loops may then organize (PUF1; green in Figure 7). Subsequently, less stable β-strands and loops might add on the flanks of the β-sandwich 32 (Figure 7). Loops of the phosphoprotein binding surface (at top in Figure 7) exhibit striking stability absent from the loops on the opposite end of the β-sandwich. The stable formation of the 6-7 and 10-11 recognition loops in PUF1 (green in Fig. 7) is significant because (i) these two loops are central to the phosphoprotein binding surface and (ii) they assume considerable stability of 5 to 6 kcal/mol 32 that may contribute to the aforementioned rigidity of the recognition surface that seems favorable for phosphopeptide binding.
Figure 7.

Hierarchy of folding states of KI-FHA observed under equilibrium conditions. PUF1 and PUF2 are near-native, partially unfolded forms. U is unfolded. U* has residual structure melted. N is the native state. Stabilities are represented by blue for high global stability, green for intermediate subglobal stability, red for lower subglobal stability, and black for higher-than-global stability.
Conclusions
In representative FHA, SH2, and PTB domains, the subsites important for affinity are rigid. While FHA domains seem to avoid substantial conformational change upon binding, more subtle dynamics changes might accompany their associations with pThr-containing partners, judging from changes propagating to the periphery of KI-FHA’s binding surface and across its β-sandwich. The rigidity expected of pre-formed phosphoThr-binding sites of FHA domains could be conducive to favorable enthalpy from van der Waals contacts. The folding of one FHA domain features a near-native intermediate in which the central 6-7 and 10-11 recognition loops are already very stable; this may promote rigidity of the center of the nascent phosphoprotein recognition surface.
A ligand’s phosphoThr is consistently bound by the conserved Arg, Ser, and another residue of the 4-5 loop of the FHA domain. Otherwise, FHA domains employ diverse strategies and sequences in recognizing pThr-containing sequences. Dun1 FHA from yeast is distinctive in selectively binding two phosphoThr residues 36. Three groups of FHA domains select their partners somewhat by the pT+3 position. Negatively charged aspartate is preferred by yeast Rad53p FHA1 and Cds1 13. A branched hydrophobic Ile or Leu is preferred by Rad53p FHA2 and FHA1, human Chk2, and human Mdc1 9, 13, 14. An uncharged polar or hydrophobic residue is preferred by plant KAPP and Y127 from M. tuberculosis 13. Promiscuity by FHA domains is suggested by alternative preferences for D or I at pT+3 by FHA1 of Rad53p 14, acceptance of polar or hydrophobic residue at pT+3 by KI-FHA and Y127 13, and PNK’s residual preferences 27. Two important exceptions to pT+3 recognition have appeared. Mammalian PNK primarily selects for pT−3, pT−2, and pT+1 27. When these positions are negatively charged, they interact with three positively charged side chains of PNK 17. Human Ki67FHA instead recognizes in NIFK a phenylalanine from a helix and the addition of a β-strand to β-strand 4 16 (Figure 3E).
Structural contacts that FHA domains use to recognize the key residues listed have been identified but are difficult to categorize since they emanate from variable 4-5, 6-7, and 10-11 loops. Systematic explanation of structural determinants of specificities will require further research. For a greater cross-section of FHA domains, quantitative affinities for phosphorylated partners are needed for describing and differentiating specificities. Since an FHA domain can bind a range of phosphoThr-containing sequences, additional factors may target it to its intended partners. Explorations of the mechanisms by which more complex, physiological ligands present multiple recognition elements to the FHA domain will be desirable. Additional surfaces of FHA domains may participate in protein interactions. Another frontier is to investigate how FHA domains interact with neighboring modules and how the tandem assemblies jointly recognize their targets in signaling.
Acknowledgments
The authors gratefully acknowledge the grant support of NSF MCB0111589 and NIH R01GM57289. We thank Dr. Zhaofeng Ding for unpublished NMR relaxation data.
Biographies
Xiangyang Liang received his Ph.D. in bioinorganic chemistry from the University of Edinburgh in 2002. He is Research Scientist at the University of Missouri-Columbia where he has investigated protein folding, dynamics, protein–protein interactions, drug binding, and structure-based drug design. He is interested in the role of metal ions in biology and medicine.
Steven Van Doren earned his Ph.D. in molecular biophysics at the University of Illinois at Urbana-Champaign in 1991. He is Associate Professor of Biochemistry at the University of Missouri-Columbia. He is interested in physical chemical mechanisms of protein-protein interactions, especially those of inflammatory diseases. His favorite approach is NMR spectroscopy.
References
- 1.Westheimer FH. Why nature chose phosphates. Science. 1987;235(4793):1173–8. doi: 10.1126/science.2434996. [DOI] [PubMed] [Google Scholar]
- 2.Yaffe MB, Cantley LC. Grabbing phosphoproteins. Science. 1999;402:30–31. doi: 10.1038/46925. [DOI] [PubMed] [Google Scholar]
- 3.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7(7):473–83. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 4.Yaffe MB, Smerdon SJ. The use of in vitro peptide-library screens in the analysis of phosphoserine/threonine-binding domain structure and function. Annu Rev Biophys Biomol Struct. 2004;33:225–44. doi: 10.1146/annurev.biophys.33.110502.133346. [DOI] [PubMed] [Google Scholar]
- 5.Durocher D, Jackson SP. The FHA domain. FEBS Lett. 2002;513(1):58–66. doi: 10.1016/s0014-5793(01)03294-x. [DOI] [PubMed] [Google Scholar]
- 6.Hofman K, Bucher P. The FHA Domain: A Putative Nuclear Signalling Domain Found in Protein Kinases and Transcription Factors. Trends Biochem Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- 7.Pallen M, Chaudhuri R, Khan A. Bacterial FHA domains: neglected players in the phospho-threonine signalling game? Trends Microbiol. 2002;10(12):556–63. doi: 10.1016/s0966-842x(02)02476-9. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Smith GP, Walker JC. Kinase interation domain of kinase-associated protein phosphatase, a phosphoptotein-binding domain. Proc Natl Acad Sci USA. 1999;96:7821–7826. doi: 10.1073/pnas.96.14.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, Yaffe MB, Jackson SP, Smerdon SJ. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell. 2002;9(5):1045–54. doi: 10.1016/s1097-2765(02)00527-0. [DOI] [PubMed] [Google Scholar]
- 10.Liao H, Yuan C, Su MI, Yongkiettrakul S, Qin D, Li H, Byeon IJ, Pei D, Tsai MD. Structure of the FHA1 domain of yeast Rad53 and identification of binding sites for both FHA1 and its target protein Rad9. J Mol Biol. 2000;304:941–951. doi: 10.1006/jmbi.2000.4291. [DOI] [PubMed] [Google Scholar]
- 11.Yuan C, Yongkiettrakul S, Byeon IJ, Zhou S, Tsai MD. Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53. J Mol Biol. 2001;314(3):563–575. doi: 10.1006/jmbi.2001.5140. [DOI] [PubMed] [Google Scholar]
- 12.Byeon IJ, Yongkiettrakul S, Tsai MD. Solution structure of the yeast Rad53 FHA2 complexed with a phosphothreonine peptide pTXXL: comparison with the structures of FHA2-pYXL and FHA1-pTXXD complexes. J Mol Biol. 2001;314(3):577–588. doi: 10.1006/jmbi.2001.5141. [DOI] [PubMed] [Google Scholar]
- 13.Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell. 2000;6(5):1169–82. doi: 10.1016/s1097-2765(00)00114-3. [DOI] [PubMed] [Google Scholar]
- 14.Mahajan A, Yuan C, Pike BL, Heierhorst J, Chang CF, Tsai MD. FHA domain-ligand interactions: importance of integrating chemical and biological approaches. J Am Chem Soc. 2005;127(42):14572–3. doi: 10.1021/ja054538m. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Byeon IJ, Ju Y, Tsai MD. Structure of human Ki67 FHA domain and its binding to a phosphoprotein fragment from hNIFK reveal unique recognition sites and new views to the structural basis of FHA domain functions. J Mol Biol. 2004;335(1):371–81. doi: 10.1016/j.jmb.2003.10.032. [DOI] [PubMed] [Google Scholar]
- 16.Byeon IJ, Li H, Song H, Gronenborn AM, Tsai MD. Sequential phosphorylation and multisite interactions characterize specific target recognition by the FHA domain of Ki67. Nat Struct Mol Biol. 2005;12(11):987–93. doi: 10.1038/nsmb1008. [DOI] [PubMed] [Google Scholar]
- 17.Bernstein NK, Williams RS, Rakovszky ML, Cui D, Green R, Karimi-Busheri F, Mani RS, Galicia S, Koch CA, Cass CE, Durocher D, Weinfeld M, Glover JN. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol Cell. 2005;17(5):657–70. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Ding Z, Wang H, Liang X, Morris ER, Gallazzi F, Pandit S, Skolnick J, Walker JC, Van Doren SR. Phosphoprotein and phosphopeptide interactions with the FHA domain from Arabidopsis kinase-associated protein phosphatase. Biochemistry. 2007;46(10):2684–96. doi: 10.1021/bi061763n. [DOI] [PubMed] [Google Scholar]
- 19.Foiani M, Pellicioli A, Lopes M, Lucca C, Ferrari M, Liberi G, Muzi Falconi M, Plevani P. DNA damage checkpoints and DNA replication controls in Saccharomyces cerevisiae. Mutat Res. 2000;451(1–2):187–96. doi: 10.1016/s0027-5107(00)00049-x. [DOI] [PubMed] [Google Scholar]
- 20.Pike BL, Yongkiettrakul S, Tsai MD, Heierhorst J. Mdt1, a novel Rad53 FHA1 domain-interacting protein, modulates DNA damage tolerance and G(2)/M cell cycle progression in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24(7):2779–88. doi: 10.1128/MCB.24.7.2779-2788.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Z, Hsiao J, Fay DS, Stern DF. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 22.Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW. DNA damage-induced activation of p53 by the checkpoint kinase Chk2.[comment] Science. 2000;287(5459):1824–7. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 23.Braun DM, Stone JM, Walker JC. Interaction of the maize and Arabidopsis kinase interaction domains with a subset of receptor-like protein kinases: implication for transmembrane signaling in plants. Plant J. 1997;12(1):83–95. doi: 10.1046/j.1365-313x.1997.12010083.x. [DOI] [PubMed] [Google Scholar]
- 24.Stone JM, Trotochaud AE, Walker JC, Clark SE. Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interaction. Plant Physiol. 1998;117:1217–1225. doi: 10.1104/pp.117.4.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee GI, Ding Z, Walker JC, Van Doren SR. NMR structure of the forkhead-associated domain from the Arabidopsis receptor kinase-associated protein phosphatase. Proc Natl Acad Sci U S A. 2003;100(20):11261–6. doi: 10.1073/pnas.2031918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding Z, Lee GI, Liang X, Gallazzi F, Arunima A, Van Doren SR. PhosphoThr peptide binding globally rigidifies much of the FHA domain from Arabidopsis receptor kinase-associated protein phosphatase. Biochemistry. 2005;44(30):10119–34. doi: 10.1021/bi050414a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koch CA, Agyei R, Galicia S, Metalnikov P, O’Donnell P, Starostine A, Weinfeld M, Durocher D. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. Embo J. 2004;23(19):3874–85. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410(6826):380–3. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 29.Takagi M, Sueishi M, Saiwaki T, Kametaka A, Yoneda Y. A novel nucleolar protein, NIFK, interacts with the forkhead associated domain of Ki-67 antigen in mitosis. J Biol Chem. 2001;276(27):25386–91. doi: 10.1074/jbc.M102227200. [DOI] [PubMed] [Google Scholar]
- 30.Lichtarge O, Sowa ME. Evolutionary predictions of binding surfaces and interactions. Curr Opin Struct Biol. 2002;12(1):21–7. doi: 10.1016/s0959-440x(02)00284-1. [DOI] [PubMed] [Google Scholar]
- 31.Stavridi ES, Huyen Y, Loreto IR, Scolnick DM, Halazonetis TD, Pavletich NP, Jeffrey PD. Crystal Structure of the FHA Domain of the Chfr Mitotic Checkpoint Protein and Its Complex with Tungstate. Structure. 2002;10:891–899. doi: 10.1016/s0969-2126(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 32.Liang X, Lee GI, Van Doren SR. Partially unfolded forms and non-two-state folding of a beta-sandwich: FHA domain from Arabidopsis receptor kinase-associated protein phosphatase. J Mol Biol. 2006;364(2):225–40. doi: 10.1016/j.jmb.2006.08.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kay LE, Muhandiram DR, Wolf G, Shoelson SE, Forman-Kay JD. Correlation between binding and dynamics at SH2 domain interfaces. Nat Struct Biol. 1998;5(2):156–63. doi: 10.1038/nsb0298-156. [DOI] [PubMed] [Google Scholar]
- 34.Olejniczak ET, Zhou MM, Fesik SW. Changes in the NMR-derived motional parameters of the insulin receptor substrate 1 phosphotyrosine binding domain upon binding to an interleukin 4 receptor phosphopeptide. Biochemistry. 1997;36(14):4118–24. doi: 10.1021/bi963050i. [DOI] [PubMed] [Google Scholar]
- 35.Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci U S A. 1996;93(21):11504–9. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee H, Yuan C, Hammet A, Mahajan A, Wu M-R, Chen ES-W, Su MI, Heierhorst J, Tsai MD. Di-phosphothreonine-specific interaction between an SQ/TQ cluster and an FHA domain in the Rad53-Dun1 kinase cascade. Mol Cell. doi: 10.1016/j.molcel.2008.05.013. Submitted. [DOI] [PubMed] [Google Scholar]