SUMMARY
The hair-cell tip link, a fine filament directly conveying force to mechanosensitive transduction channels, is composed of two proteins, protocadherin-15 and cadherin-23, whose mutation causes deafness. However, their molecular structure, elasticity, and deafness-related structural defects are unknown. We present crystal structures of the first and second extracellular cadherin repeats of cadherin-23. Overall, structures show typical cadherin folds, but reveal an elongated N-terminus that precludes classical cadherin interactions and contributes to an N-terminal Ca2+-binding site. The deafness mutation D101G, in the linker region between the repeats, causes a slight bend between repeats and decreases Ca2+ affinity. Molecular dynamics simulations suggest that cadherin-23 repeats are stiff, and that either removing Ca2+ or mutating Ca2+-binding residues reduces rigidity and unfolding strength. The structures define an uncharacterized cadherin family and, with simulations, suggest mechanisms underlying inherited deafness and how cadherin-23 may bind with itself and with protocadherin-15 to form the tip link.
INTRODUCTION
Hair cells are extremely sensitive mechanoreceptors that transform mechanical stimuli into electrical signals, providing vertebrates with the senses of hearing and balance. At a hair cell's apical end, a bundle of stereocilia arranged in a staircase of increasing heights can be deflected by forces produced by sound or head movement (Gillespie and Müller, 2009). Positive deflection of this hair bundle causes shearing of adjacent stereocilia and tensioning of a fine filament, termed the tip link, which connects the tip of each stereocilium to the side of its tallest neighbor (Pickles et al., 1984). Mechanosensitive ion channels located at the lower end of each tip link are opened by tension, subsequently depolarizing hair cells (Assad et al., 1991; Beurg et al., 2009; Denk et al., 1995). In vertebrates, these are the first steps underlying auditory and vestibular perception.
Channel gating in hair cells occurs in <10 μs and is mediated by a gating spring, a biophysically defined element which may be the tip link itself or another elastic element connected in series (Corey and Hudspeth, 1983; Howard and Hudspeth, 1988). Removal of extracellular Ca2+ abolishes transduction currents and simultaneously eliminates the tip links (Assad et al., 1991; Vollrath et al., 2007). Tip links reappear along with transduction currents several hours after extracellular Ca2+ is restored (Zhao et al., 1996). Thus, tip links are essential for mechanotransduction and apparently convey tension directly to hair-cell transduction channels.
The tip link is formed by cadherin-23 and protocadherin-15, non-classical cadherins with unusually long extracellular domains, single transmembrane domains, and C-terminal cytoplasmic domains (Ahmed et al., 2006; Kazmierczak et al., 2007; Siemens et al., 2004; Söllner et al., 2004). Sequence alignments suggest that they have 27 and 11 extracellular cadherin (EC) repeats, respectively, with inter-repeat Ca2+-binding sites resembling those of classical cadherins. Cadherin-23 is proposed to form the upper two-thirds of the tip link as a parallel dimer while protocadherin-15 forms the lower part; the combined length matches observed tip-link lengths. Cadherin-23 and protocadherin-15 interact in vitro at their distal tips (Kazmierczak et al., 2007), but neither has the key residues that mediate trans classical cadherin binding interactions through N-terminal EC1 repeats (Chen et al., 2005; Katsamba et al., 2009; Leckband and Prakasam, 2006; Patel et al., 2003, 2006; Takeichi, 1990). Thus the mechanism of tip-link assembly is not known, nor is the reason why tip-link integrity requires Ca2+. Both electron micrographs of tip links and molecular dynamics (MD) simulations of classical cadherin repeats suggested that the tip link is too stiff to be the gating spring (Kachar et al., 2000; Sotomayor et al., 2005; Sotomayor and Schulten, 2008). Yet the elasticity of cadherin-23 and protocadherin-15 has not been directly determined, so the question remains open.
Cadherin-23 and protocadherin-15 are both mutated in Usher Syndrome, a hereditary deafness and blindness disease in humans, and also in the nonsyndromic deafnesses DFNB12 and DFNB23 (Ahmed et al., 2003; Alagramam et al., 2001; Astuto et al., 2002; Di Palma et al., 2001). Structural models of classical cadherins have been used to map the location of some mutations causing hereditary deafness (de Brouwer et al., 2003; Schwander et al., 2009). Yet neither the corresponding molecular defects caused by cadherin-23 and protocadherin-15 mutations, nor how these defects cause deafness, are known. How tip link proteins withstand large mechanical stimuli from loud sound, or how they break in excessive noise, is also unclear.
We determined structures of the two most distal extracellular repeats (EC1 and EC1+2) of wild-type cadherin-23, and of cadherin-23 with a mutation associated with DFNB12. Cadherin-23 EC1 features an elongated N-terminus and a novel Ca2+-binding site absent in all other EC1 repeats of known structure, making cadherin-23 EC1 incompatible with classical cadherin binding interfaces. In addition, MD simulations suggest that cadherin-23 EC1+2 repeats are stiff, with their molecular strength and inter-repeat dynamics mediated by Ca2+ binding to highly conserved acidic residues. These same residues are targeted by deafness mutations, which simulations suggest can render tip links prone to mechanical failure.
RESULTS
Structures of the cadherin-23 EC1 and EC1+2 repeats
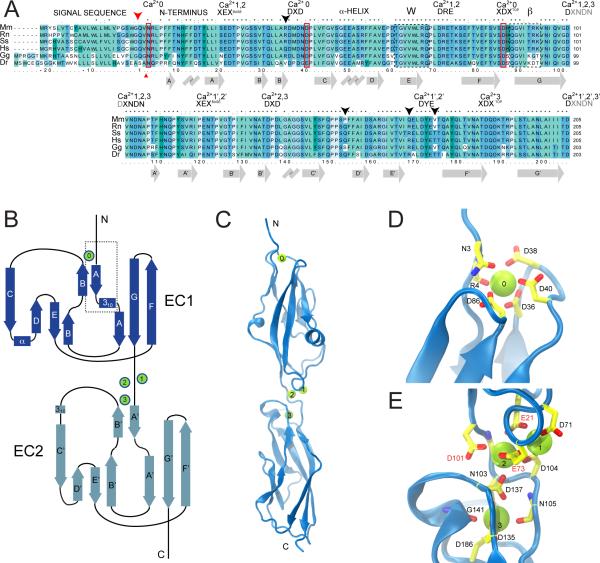
The EC1 repeat and combined EC1+2 repeats of mouse cadherin-23 were refolded from inclusion bodies produced in E. coli (Figure S1A) and used for crystallization and structure determination. The native signal sequence was replaced by a methionine, which is not expected to significantly alter cadherin-23 properties, as the length of the processed N-terminus is not conserved (Figure 1A). Residue numbering throughout the text corresponds to the processed protein (EC1: Q1 to D101; EC1+2: Q1 to D205). Structures of wild-type cadherin-23 repeat EC1 and of repeats EC1+2 at high (1.1 M) Ca2+ concentration (“EC1” and “EC1+2 Ca2+” in Table 1) exhibit an overall fold closely matching that of classical cadherins: a Greek key motif with seven ß-strands forming a ß-sandwich fold (Figures 1B and 1C). EC1+2 also features a conserved Ca2+-binding motif at the linker region with Ca2+ ions at sites 1, 2, and 3 bridging acidic residues in canonical sequence motifs (20XEXBASE22 and 71DRE73 of EC1; 101DXNDN105, 135DXD137, and 185XDXTOP187 of EC2, Figure 1E; Boggon et al. (2002); Nagar et al. (1996)).
Figure 1.
Structure of cadherin-23 extracellular repeats 1 and 2. (A) Alignment of cadherin 23 sequences corresponding to EC1 (top) and EC2 (bottom) for Mus musculus, Rattus norvegicus, Sus scrofa, Homo sapiens, Gallus gallus, and Danio rerio (NCBI reference sequences: NP_075859.2, NP_446096.1, XP_001925718.1, NP_071407.4, XP_421595.2, NP_999974.1). Signal sequence cleavage site is indicated by a large red arrowhead (small red arrowhead for Gallus gallus and Danio rerio). Conserved Ca2+-binding motifs are labeled XEXBASE, DXD, DRE, XDXTOP, and DXNDN. Crystallographic W and ß interfaces are enclosed in dashed rectangles. Mus musculus and Homo sapiens sequences differ at four sites indicated by black arrowheads. Secondary structure is indicated below the alignment. (B) Topology diagram of cadherin-23 EC1+2. A typical cadherin fold with seven ß-strands (labeled A to G) is observed for both repeats. An elongated N-terminus in EC1 features a 310 helix and helps to form a Ca2+-binding site (labeled 0). Standard Ca2+-binding sites at the linker region between cadherin repeats are labeled 1, 2, & 3. (C) Ribbon diagram of cadherin-23 EC1+2 with Ca2+ ions as green spheres. (D,E) Detail of Ca2+-binding site 0 at the N-terminus and Ca2+-binding sites 1, 2, & 3 at the EC1+2 linker region, respectively. Protein back-bone and side-chains are in cartoon and stick representations, respectively. Numbering corresponds to Mm-cadherin-23 without its signal sequence. See also Figure S1.
Table 1.
Statistics for Cadherin-23 Repeat Structures
| Data Collection and Refinement | EC1 | EC1+2 Na+ | EC1+2 Ca2+ | EC1+2 D101G Na+ |
|---|---|---|---|---|
| Space group | C2 | R32 | R32 | P42212 |
| Unit cell parameters: | ||||
| a, b, c (Å) | 45.89, 49.54, 45.88 | 151.46, 151.46, 133.46 | 151.29, 151.29, 136.88 | 179.69, 179.69, 63.98 |
| α, β, γ (°) | 90, 99, 90 | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 |
| Molecules per asymmetric unit | 1 | 1 | 1 | 2 |
| Beam source | APS 24-ID-E | APS 24-ID-E | MicroMax007 | NSLS-X25H |
| Wavelength (Å) | 0.97918 | 0.97918 | 1.5418 | 0.9795 |
| Resolution limit (Å) | 1.50 | 1.98 | 2.36 | 2.74 |
| Unique reflections | 16,290 (819) | 40,718 (2,014) | 24,916 (1,198) | 27,463 (1,361) |
| Redundancy | 3.4 (3.3) | 6.3 (6.1) | 6.0 (4.4) | 7.4 (5.2) |
| Completeness (%) | 99.7 (98.5) | 99.7 (98.3) | 99.7 (97.1) | 97.9 (97.4) |
| Average I/σ(I) | 18.7 (2.5) | 13.2 (3.2) | 17.7 (2.9) | 13.8 (4.5) |
| Rmerge | 0.05 (0.49) | 0.07 (0.50) | 0.09 (0.45) | 0.09 (0.49) |
|
Refinement | ||||
| Resolution range (Å) | 25.54 - 1.50 (1.53 - 1.50) |
30.0 - 1.98 (2.03 - 1.98) |
22.25 - 2.36 (2.40-2.36) |
29.77 - 2.74 (2.81 - 2.74) |
| Residues (atoms) | 102 (815) | 208 (1,626) | 208 (1,626) | 416 (3,248) |
| Water molecules | 105 | 262 | 187 | 52 |
| Rwork (%) | 17.2 (21.9) | 17.3 (22.7) | 20.0 (32.6) | 21.9 (35.8) |
| Rfree (%) | 19.9 (25.6) | 18.8 (21.7) | 22.1 (33.7) | 24.9 (41.4) |
| RMS deviations | ||||
| Bond lengths (Å) | 0.012 | 0.012 | 0.010 | 0.011 |
| Bond angles (°) | 1.368 | 1.363 | 1.143 | 1.099 |
| B-factor average | ||||
| Protein | 24.02 | 45.01 | 43.39 | 44.86 |
| Ligand/ion | 14.07 | 49.17 | 53.54 | 40.69 |
| Water | 27.02 | 31.20 | 30.26 | 18.63 |
| B-factor | ||||
| Site 0 | Ca2+ 14.07 | Ca2+ 32.11 | Ca2+ 32.01 | Ca2+ 39.89; 60.84 |
| Site 1 | - | Na+ 34.93 | Ca2+ 30.93 | Na+ 39.43; 38.48 |
| Site 2 | - | Ca2+ 31.87 | Ca2+ 31.08 | Ca2+ 31.10; 37.60 |
| Site 3 | - | Ca2+ 35.74 | Ca2+ 34.59 | Ca2+ 36.35; 35.84 |
|
Ramachandran Plot Regionsa | ||||
| Most favored (%) | 93.3 | 91.7 | 91.7 | 88.3 |
| Additionally allowed (%) | 5.6 | 7.8 | 7.8 | 11.5 |
| Generously allowed (%) | 1.1 | 0.6 | 0.6 | 0.3 |
| Disallowed (%) | 0.0 | 0.0 | 0.0 | 0.0 |
| PDB ID code | 2wbx | 2wcp | 2whv | 2wd0 |
Computed with PROCHECK
Despite similarities with classical cadherins, several features make cadherin-23 unique. An α-helix between EC1 ß-strands C and D (Figures 1B and 1C) has not been observed in published cadherin EC1 repeat structures. More importantly, cadherin-23's elongated N-terminus includes a 310 helix within strand A and contributes residues N3 and R4 to an additional Ca2+-binding site at the very tip of cadherin-23 EC1, referred to here as site 0 (Figures 1D and S1B). Site 0 is homologous to site 3 in the linker region between EC1+2 repeats (see below) and is further lined by a modified DXD Ca2+-binding motif in the loop between ß-strands B and C (36DXDXD40), and an XDXTOP motif at position 85-87 found in EC1s of classical type-II cadherins, but not type I.
Cadherin-23 EC1 defines the Cr-2 family of cadherin proteins
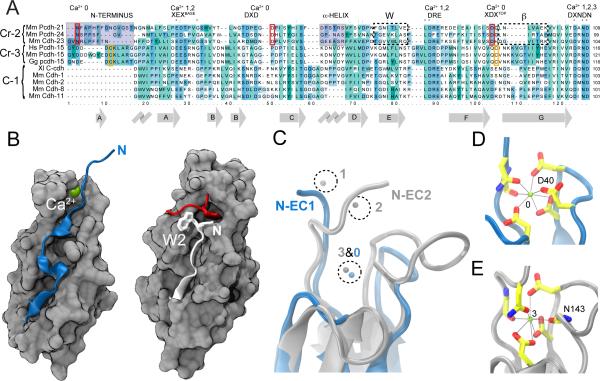
Placing cadherins into a phylogenetic relationship has been difficult due to the variable number of EC repeats and widely divergent C-termini. Nevertheless, a reasonable phylogeny has been proposed using EC1 sequences to segregate the superfamily into six branches: two branches of true cadherins (C-1 and C-2), a protocadherin branch (Cr-1a), and three other “cadherin-related” branches (Cr-1b, Cr-2, and Cr-3; Hulpiau and van Roy (2009)). In this scheme, cadherin-23 is in branch Cr-2 along with protocadherin-24 and protocadherin-21. Cr-2 and Cr-3 family members sport an N-terminus that is at least six residues longer than classical C-1 cadherins (Figure 2A). All three mammalian Cr-2 family members have all sequence motifs involved in Ca2+ binding at site 0. Protocadherin-15, in the Cr-3 group, has an even longer N-terminus, yet it lacks the DXDXD and XDXTOP motifs of site 0. Instead, protocadherin-15 features two cysteine residues that likely form a disulfide bond at the distal tip in regions aligning closely with cadherin-23 site 0 (Figure 2A). Thus, site 0 is likely a hallmark of Cr-2 family members. Notably, the elongated cadherin-23 N-terminus lacks not only the tryptophan residues at positions 2 and 4, but also the corresponding deep binding pockets that mediate antiparallel binding through strand-exchange in classical type-I and type-II cadherins (Figure 2B). Cr-2 family members must therefore use a different dimerization mechanism.
Figure 2.
The first extracellular repeat of cadherin-23 structurally defines a family of cadherin proteins. (A) Sequence alignment of EC1 from the cadherin-related families 2 and 3 (Cr-2 and Cr-3, respectively) and classical cadherins (C-1). Alignment notations are as in Figure 1A. Cr-2 members protocadherin-21 and protocadherin-24 feature elongated N-termini and share critical conserved cadherin-23 residues involved in Ca2+-binding site 0 (red boxes). Protocadherin-15's extended N-terminus is likely involved in a disulfide bridge instead (yellow boxes), and has three acidic residues encoded by exon 2, and another two by exon 3 (sometimes spliced out as shown for the human sequence). NCBI reference sequences: NP_570948.1, NP_001028536.1, NP_075859.2, NP_149045.3, NP_075604.2, NP_001038119.1, NP_001080946.1, NP_033994.1, BAA23549.1, NP_001034243.1, NP_033996.4. (B) Molecular surface representation of EC1 repeats from cadherin-23 (left) and classical type I C-cadherin (right, pdb code 1L3W, Boggon et al. (2002)), with the N-terminal ß-strands shown in blue (cadherin-23) or white (C-cadherin) cartoon. Cadherin-23 EC1 displays an elongated N-terminus and lacks the characteristic tryptophan residues at position 2 (W2) of classical type I cadherins (or positions 2 & 4 of classical type II cadherins). A piece of the N-terminus and W2 of the interacting C-cadherin protomer is shown in red. (C) Superposition of cadherin-23 EC1 (blue) with C-cadherin EC2 (grey). The location of the cadherin-23 EC1 Ca2+-binding site 0 matches that of classical cadherin site 3. (D,E) Detail of the cadherin-23 site 0 and C-cadherin Ca2+-binding site 3, respectively, with residues coordinating Ca2+ ions in stick. The overall architecture of both Ca2+-binding sites is similar, but C-cadherins's N143 carbonyl oxygen is replaced by cadherin-23's D40 side-chain.
Cadherin-23's site 0 closely resembles Ca2+-binding site 3 of classical cadherins and cadherin-23 itself (Figure 2C), which might suggest that cadherin-23 and Cr-2 family members simply lack a “true” or classical EC1. However, several structural features of cadherin-23 EC1 distinguish it from classical EC2s, namely the subtly different Ca2+ coordination (Figures 2D and 2E), the 310 and α-helices, and the shorter loops between ß-strands B-C and F-G. Thus, cadherin-23 EC1 seems to represent a unique repeat structure, perhaps adapted to perform specific tasks like those of cadherin-23 in the tip link.
Cadherin-23 repeats are stiff and Ca2+ determines their mechanical strength
Tip links are constantly subjected to small and large forces (1 to > 100 pN) and have been proposed to function as gating springs (Pickles et al., 1984). Mechanical measurements of hair bundles indicate that the gating spring stiffness is about 1 mN/m (Cheung and Corey, 2005; Howard and Hudspeth, 1988). To test cadherin-23 elasticity and its compatibility with the gating spring model, steered molecular dynamics (SMD) simulations of EC1 and EC1+2 repeats were carried out using explicit water and 150 mM KCl (Grubmüller, 2005; Isralewitz et al., 2001; Sotomayor and Schulten, 2007). Simulations are summarized in Table 2.
Table 2.
Overview of Simulations.
| Label | # | PDB | Binding Site | tsim (ns) | Slowest SMD Speed (nm/ns) | Size (# atoms) | Size (nm3) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |||||||
| C23 WT EC1 | S1a-g | 2wbx | Ca2+ | - | - | - | 102.72 | 0.1 | 65,592 | 18.0 × 6.0 × 6.5 |
| C23 WT EC1 | S2a-f | 2wbx | - | - | - | - | 85.49 | 0.1 | 65,815 | 18.0 × 6.0 × 6.5 |
| C23 WT EC1 | S3a-g | 2wbx | - | - | - | - | 92.62 | 0.1 | 65,591 | 18.0 × 6.0 × 6.5 |
| C23 WT EC1 β | S4a-g | 2wbx | Ca2+ at 2 available sites | 56.30 | 0.1 | 84,904 | 18.0 × 6.9 × 7.3 | |||
| C23 WT EC1 β | S5a-g | 2wbx | - | - | - | - | 61.32 | 0.1 | 84,902 | 18.0 × 6.9 × 7.3 |
| C23 WT EC1+2 | S6a-l | 2wcp | Ca2+ | Na+ | Ca2+ | Ca2+ | 191.49 | 0.1 | 130,919 | 24.8 × 7.6 × 7.4 |
| C23 WT EC1+2 | S7a-b | 2wcp | Ca2+ | Na+ | Ca2+ | Ca2+ | 49.49 | - | 125,350 | 14.8 × 9.6 × 9.4 |
| C23 WT EC1+2 | S8a-b | 2wcp | Ca2+ | Na+ | Ca2+ | Ca2+ | 43.63 | - | 172,807 | 13.8 × 11.6 × 11.4 |
| C23 WT EC1+2 | S9a-k | 2wcp | - | - | - | - | 165.86 | 0.1 | 130,909 | 24.8 × 7.6 × 7.4 |
| C23 WT EC1+2 | S10a-b | 2wcp | - | - | - | - | 32.95 | 0.1 | 125,343 | 14.8 × 9.6 × 9.4 |
| C23 WT EC1+2 | S11a-j | 2wcp | Ca2+ | Na+ | Na+ | Ca2+ | 103.67 | 0.1 | 130,918 | 24.8 × 7.6 × 7.4 |
| C23 WT EC1+2 | S12a-k | 2whv | Ca2+ | Ca2+ | Ca2+ | Ca2+ | 175.30 | 0.1 | 132,325 | 24.8 × 7.6 × 7.4 |
| C23 WT EC1+2 | S13a-b | 2whv | Ca2+ | Ca2+ | Ca2+ | Ca2+ | 33.00 | - | 126,924 | 14.8 × 9.6 × 9.4 |
| C23 WT EC1+2 | S14a-b | 2whv | Ca2+ | Ca2+ | Ca2+ | Ca2+ | 25.00 | - | 174,615 | 13.8 × 11.6 × 11.4 |
| C23 WT EC1+2 CUT | S15a-j | 2whv | Ca2+ | Ca2+ | Ca2+ | Ca2+ | 121.86 | 0.1 | 132,307 | 24.8 × 7.6 × 7.4 |
| C23 WT EC1+2 W | S16a-g | 2whv | Ca2+ at 8 available sites | 65.77 | 0.1 | 231,800 | 23.6 × 9.9 × 10.4 | |||
| C23 D101G EC1+2 | S17a-l | 2wd0 | Ca2+ | Na+ | Ca2+ | Ca2+ | 180.64 | 0.1 | 115,303 | 24.6 × 7.4 × 6.8 |
| C23 D101G EC1+2 | S18a-b | 2wd0 | Ca2+ | Na+ | Ca2+ | Ca2+ | 45.48 | - | 112,882 | 14.6 × 9.4 × 8.8 |
| C23 D101G EC1+2 | S19a-b | 2wd0 | Ca2+ | Na+ | Ca2+ | Ca2+ | 37.95 | - | 182,226 | 13.6 × 12.0 × 11.8 |
| C23 D101G EC1+2 | S20a-f | 2wd0 | - | - | - | - | 77.33 | 0.1 | 115,299 | 24.6 × 7.4 × 6.8 |
| C23 D101G EC1+2 | S21a-b | 2wd0 | - | - | - | - | 26.21 | - | 112,880 | 14.6 × 9.4 × 8.8 |
| C23 D101G EC1+2 | S22a-j | 2wd0 | Ca2+ | Na+ | Na+ | Ca2+ | 97.56 | 0.1 | 115,302 | 24.6 × 7.4 × 6.8 |
| C23 E21A IS EC1+2 | S23a-c | 2whv | Ca2+ | Ca2+ | Ca2+ | Ca2+ | 46.47 | - | 155,690 | 14.8 × 10.6 × 10.4 |
| C23 D101G IS EC1+2 | S24a-c | 2whv | Ca2+ | Ca2+ | Ca2+ | Ca2+ | 46.54 | - | 155,690 | 14.8 × 10.6 × 10.4 |
| C23 E73V IS EC1+2 | S25a-c | 2whv | Ca2+ | Ca2+ | Ca2+ | Ca2+ | 46.63 | - | 155,696 | 14.8 × 10.6 × 10.4 |
| C23 E21A IS EC1+2 | S26a-k | 2whv | Ca2+ | Ca2+ | Ca2+ | Ca2+ | 151.53 | 0.1 | 121,228 | 24.6 × 7.6 × 6.9 |
| C23 E73V IS EC1+2 | S27a-k | 2whv | Ca2+ | Ca2+ | Ca2+ | Ca2+ | 166.10 | 0.1 | 129,165 | 24.3 × 7.6 × 7.5 |
| C WT | S28a-e | 1l3w | Ca2+ at 12 available sites | 50.31 | 1 | 355,208 | 37.4 × 10.5 × 9.4 | |||
| C E69A IS | S29a-i | 1l3w | Ca2+ at 12 available sites | 78.68 | 1 | 355,208 | 37.4 × 10.5 × 9.4 | |||
| C E11A IS | S30a-c | 1l3w | Ca2+ at 12 available sites | 13.10 | 10 | 355,205 | 37.4 × 10.5 × 9.4 | |||
| C WT DIM | S31a-m | 1l3w | Ca2+ at 12 available sites | 154.39 | 0.1 | 203,769 | 27.8 × 9.4 × 8.2 | |||
| C8 WT DIM | S32a-f | 2a62 | Ca2+ at 12 available sites | 35.10 | 1 | 155,379 | 25.9 × 8.5 × 7.6 | |||
| C8 WT DIM | S33a-i | 2a62 | Ca2+ at 12 available sites | 139.67 | 0.1 | 203,376 | 27.9 × 9.5 × 8.2 | |||
| C11 WT DIM | S34a-c | 2a4e | Ca2+ at 6 available sites | 9.88 | 1 | 150,108 | 24.2 × 9.2 × 7.2 | |||
Overview of cadherin simulations. Labels indicate the system and protein variant (C23: cadherin-23; WT: wild-type; EC1: extracellular repeat 1; β: dimer interface mediated by β-strands; EC1+2: extracellular repeats 1 & 2; CUT: peptide bond N103-C-D104-N deleted; W: dimer interface mediated by W65; IS: in silico mutation; DIM: strand-exchanged dimer interface). Unique simulation identifiers are denoted by Snnx. Initial size of the system (in nm3) is indicated in the last column for each system. A detailed list of all simulations is in Tables S1-S6. The overall computational effort involved a cumulative total of 2,810.04 nanoseconds of simulation for systems ranging in size from 65K to 355K atoms (equivalent to a single simulation of a 25K-atom system lasting 16 μs using a uniform time-step of 2 fs).
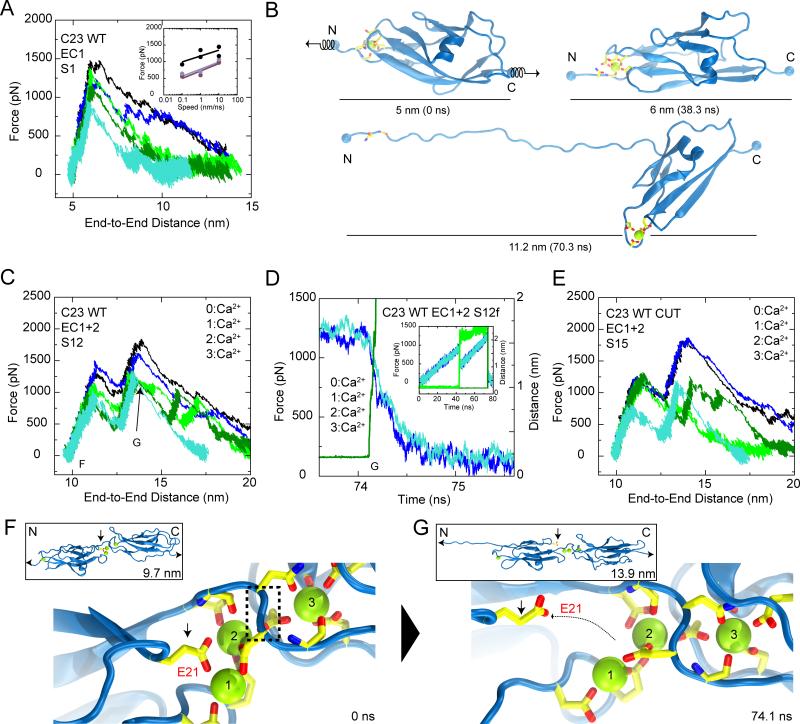
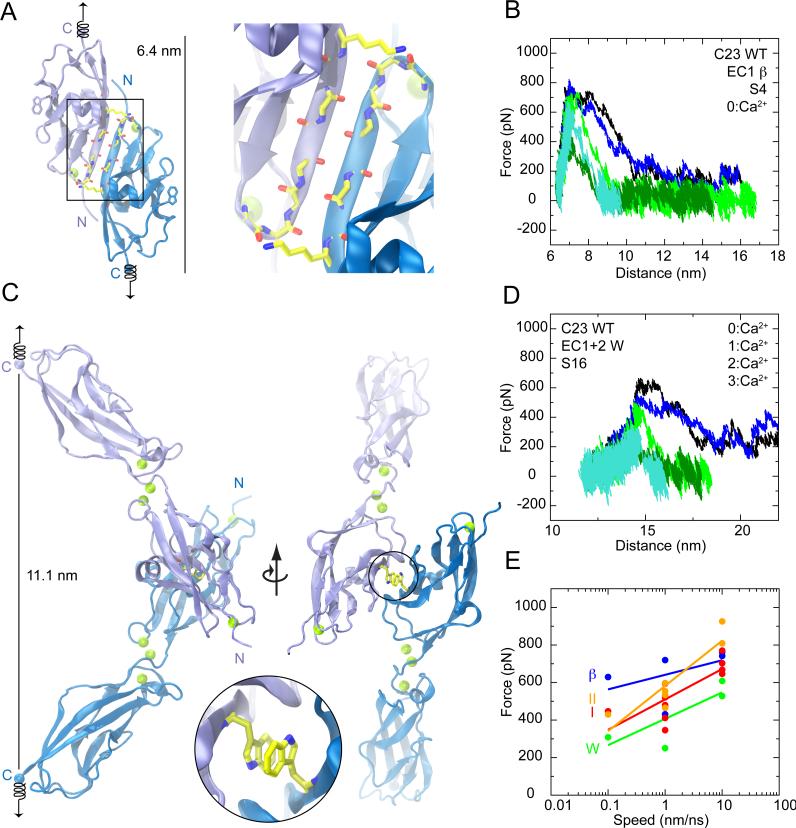
Cadherin-23 EC1, including Ca2+ at site 0, was equilibrated and stretched from both ends in constant velocity SMD simulations (simulations S1a-g in Table 2). Stretching speeds ranged from 0.1 to 10 nm/ns; in all cases the monitored force increased to several hundred pN with little protein extension until unfolding occurred by rupture of non-covalent interactions at site 0 accompanied by a sudden drop in applied force (Figures 3A, 3B, and S2A; Movie I). The slope for the force versus end-to-end distance during initial stretching stages indicated a stiffness of 710 mN/m per repeat for the slowest SMD simulation. Maximum unfolding-force peak values follow the well-known dependency on stretching speed (Evans and Ritchie, 1997; Izrailev et al., 1997), increasing with faster stretching (Figure 3A inset). Simulations in the absence of Ca2+ at site 0 show reduced maximum force peaks across all stretching speeds (simulations S2a-f and S3a-g; Figure 3A inset; Figures S2B and S2C). The dependence of force-peak magnitude on the presence of Ca2+ is consistent with Ca2+-dependent stabilization of classical cadherins (Prasad and Pedigo, 2005; Sotomayor et al., 2005; Sotomayor and Schulten, 2008), indicating that Ca2+ at site 0 may further protect cadherin-23 EC1 from large mechanical stimuli.
Figure 3.
SMD simulations show stiff cadherin-23 repeats with mechanical responses controlled by Ca2+ ions. (A) Force applied to N-terminus versus end-to-end distance for constant velocity stretching of cadherin-23 EC1 at 10 (simulation S1c: black; S1d: blue), 1 (S1e: green; S1f dark green), and 0.1 nm/ns (S1g: turquoise). Inset: force peak values for cadherin-23 EC1 versus stretching speed fitted to logarithmic regressions in the presence (S1c-g: black) or absence of Ca2+ (S2b-f: brown; S3c-g: gray, see Figures S2A-C). (B) Snapshots of initial conformation and mechanically induced unfolding states (S1g). Springs indicate position and direction of applied forces. (C) Force applied to N-terminus versus end-to-end distance for simulations of cadherin-23 EC1+2 with Ca2+ at all sites (S12b-f). Colors denote independent simulations using different stretching speeds and thermodynamic ensembles (black, 10 nm/ns & NVE; blue, 10 nm/ns & NpT; green, 1 nm/ns & NVE; dark green, 1 nm/ns & NpT; turquoise, 0.1 nm/ns & NpT). (D) Force applied to cadherin-23 EC1+2 N-terminus (blue) and C-terminus (cyan) along with distance between Ca2+ at site 1 and E21-Oε (dark green) versus time for part of simulation S12f. Inset shows entire trajectory, and includes the distance between Ca2+ at site 0 and R4-O (green). (E) Force applied to N-terminus versus end-to-end distance for cadherin-23 EC1+2 with Ca2+ at all sites and with the peptide bond between N103 and D104 deleted (S15b-f, colors as in C). (F,G) Snapshots of linker region conformations (entire protein in inset) during simulation S12f at time points indicated in (C). Vertical arrows indicate residue E21 and dashed box, peptide bond N103-C-D104-N. See also Figure S2.
The overall elastic response of cadherin-23 is likely to be dominated by the elasticity of single cadherin repeats rather than tertiary structure elasticity, since ultrastructural studies of tip links and TEM images of cadherin-23, within their limitations and possible artifacts, show rather straight filaments (Kachar et al., 2000; Kazmierczak et al., 2007). Nevertheless, some elasticity might arise from mechanical failure of linker regions between repeats. Thus, SMD simulations were performed on cadherin-23 EC1+2 structures (simulations S12a-k in Table 2). Stretching simulations of EC1+2 again indicated a stiff protein, with force increasing to several hundred pN with little protein extension (Figure 3C). The overall stiffness was estimated to be 570 mN/m for the slowest SMD simulation (S12f).
After initial stretching, however, a more complex response was observed, with at least two well-defined peaks at several hundred pN at all stretching speeds (Figure 3C). The first peak correlates with rupture of site 0 as residues N3 and R4 detach, followed by unfolding of the 5-residue 310 helix within strand A and leading to a 2.5 nm extension that corresponds to the distance between both force peaks. The second force peak involves rupture of the EC1+2 linker region, with either residue E21 and ß-strand A detaching from Ca2+ at sites 1 and 2 (simulations S12b,c,f; Figures 3C, 3D, 3F, 3G; Movie II) or ß-strands of repeat EC2 detaching from Ca2+ at site 3 (simulations S12d,e). In both scenarios, the synchronized rupture of Ca2+-protein interactions, rapid extension of the protein's end-to-end distance, and drop in stretching forces suggest that Ca2+ is essential for holding cadherin-23 EC1 and EC2 repeats together. The Ca2+ bond provides stiffness, but at the same time confers “break points” when excessive force is applied. This notion is supported by simulations of wild type and mutant EC1+2 stripped of Ca2+ presented below.
To determine whether Ca2+ alone can keep EC repeats together, we performed additional SMD simulations (S15b-f) in which the peptide backbone was cut between N103 and D104, disrupting the covalent EC1-to-EC2 linker, but which retained all Ca2+ ions bridging acidic residues from both repeats. Forces required to unfold this complex were comparable to those observed for the intact protein (Figure 3E). Furthermore, force peaks originated from the same unfolding and detachment of ß-strands observed for the EC1+2 structure with an intact linker, and separation of EC1 from EC2 was never observed in the simulations (Figures S2D and S2E). Ca2+-protein interactions alone are therefore sufficient to keep EC repeats together even under tension, and Ca2+ binding must be crucial for cadherin strength.
Structure of cadherin-23 EC1+2 with a Na+ bound to its linker region
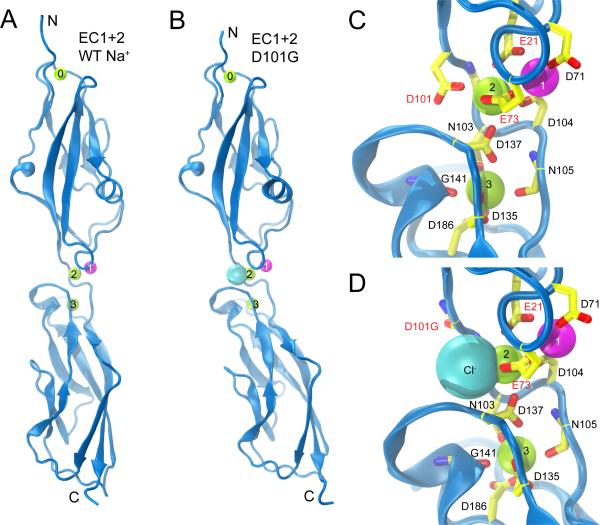
Structures and simulations highlight the importance of the three Ca2+ ions at cadherin-23's linker for its mechanical response. Ca2+ is required for tip-link integrity (Assad et al., 1991), but cochlear endolymph surrounding hair cells has an unusually low Ca2+ concentration of 20-40 μM (or 100-300 μM in vestibular endolymph; Bosher and Warren (1978); Salt et al. (1989)). Consequently cadherin-23 must have relatively high affinity for Ca2+. The E-cadherin EC1+2 linker region has a KD for Ca2+ of 20 μM (Courjean et al., 2008); if the same is true for cadherin-23 (see below), it suggests that its Ca2+-binding sites are only partially occupied in vivo. We crystallized wild-type cadherin-23 EC1+2 in conditions containing >2 M NaCl and ~1 mM CaCl2, and in this second structure observed replacement of Ca2+ with Na+ at site 1 (Figures 4A and 4C). Assignment of Na+ to the electron density at site 1 was supported by comparison of B-factors of the Na+ ion and surrounding residues (see “EC1+2 Na+” in Table 1; Supplemental Text; Figures S1C and S1D). Overall, the structure closely resembles the first cadherin-23 EC1+2 structure, obtained in >1 M CaCl2 (RMSD of 0.45 Å for all atoms, Figure 4A). The substitution of Ca2+ by a cation of similar ionic radius (Na+) at site 1, although an obvious consequence of the crystallization conditions, indicates a likely hierarchy of Ca2+-binding affinities with the outermost site 1 as the weakest. This structure led us to test the elasticity and strength of cadherin-23 when Ca2+ is not occupying all sites.
Figure 4.
Structures of cadherin-23 EC1+2 with modified linkers. (A) Wild-type cadherin-23 crystallized in high NaCl conditions features a linker where Na+ replaced the Ca2+ normally present at site 1. (B) Cadherin-23 EC1+2 D101G crystallized in high (1M) NaCl conditions as well. Proteins are shown in cartoon representation; ions as spheres (Ca2+: green; Cl-: cyan; Na+: magenta). (C,D) Detail of wild-type and D101G linker regions, respectively. The latter shows a Cl- ion near Ca2+-binding site 2. See also Figures S1 and S4.
Ca2+ ions bound to sites 2 & 3 are sufficient to prevent cadherin unfolding
Low Ca2+ concentrations (such as those of the endolymph) can be effectively mimicked in simulations by progressively eliminating Ca2+ from different binding sites. Thus, we used SMD simulations to test the elasticity of cadherin-23 EC1+2 Na+, a model with Ca2+ at both sites 1 and 2 replaced by Na+, and a model with all binding sites empty.
Simulations with Na+ replacing Ca2+ at site 1 (S6a-g) show a two-peak force response similar to that of cadherin-23 with Ca2+ at all sites (Figure 5A; c.f. Figure 3C). While the first force peak is again associated with rupture of site 0, the second force peak correlated to detachment of ß-strands in EC2 (Figures S2F and S2G), as opposed to detachment of strand A in EC1 in simulations with all Ca2+ sites loaded.
Figure 5.
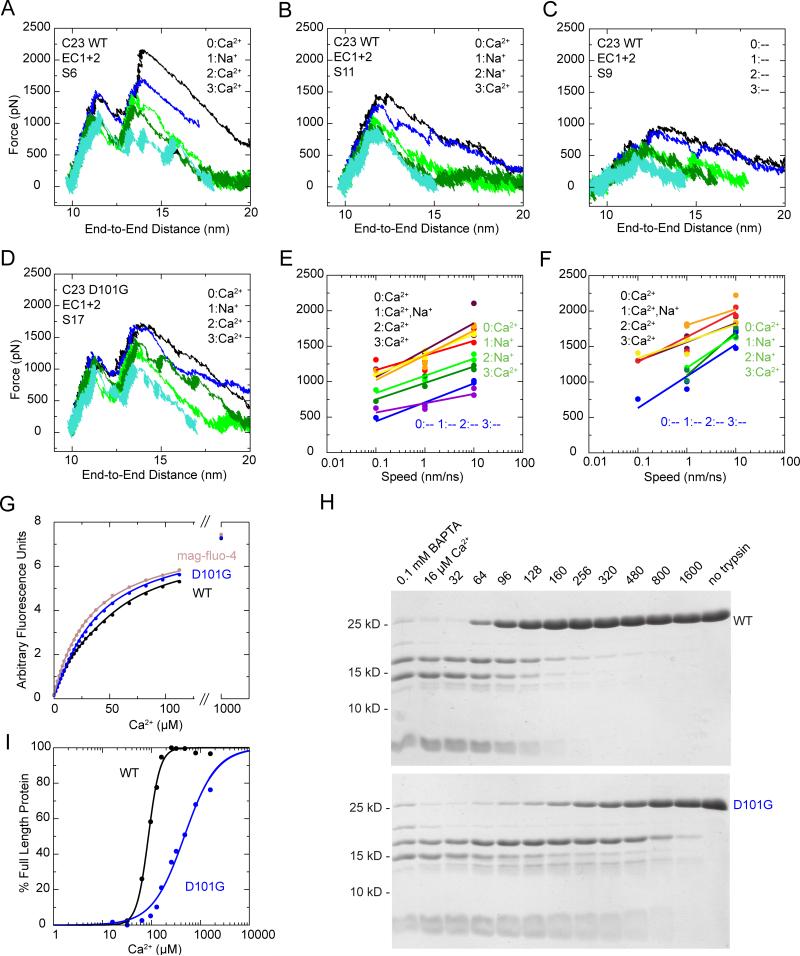
SMD simulations of cadherin-23 with modified linkers and Ca2+-binding assays. (A-C) Force applied to N-terminus versus end-to-end distance for stretching simulations of wild-type cadherin-23 EC1+2 with Ca2+ at sites 0, 2, & 3 and Na+ at site 1 (A, S6c-g); Ca2+ at sites 0 & 3 and Na+ at sites 1 & 2 (B, S11b-f), and all binding sites empty (C, S9b-f). Colors denote independent simulations using different stretching speeds and thermodynamic ensembles as in Figure 3C. See also Figure S2. (D) Force applied to N-terminus versus end-to-end distance for simulations of EC1+2 D101G with Ca2+ at sites 0, 2, & 3 and Na+ at site 1. (E) Force peak maxima for cadherin-23 EC1+2 simulations versus stretching speed. Force peaks of simulations with Ca2+ at least at sites 0, 2, & 3 are red (S12b-f), maroon (S6c-g), yellow (S17c-g; D101G), and orange (S15b-f; CUT); with Ca2+ at sites 0 & 3 and Na+ at sites 1 & 2 are green (S11b-f) and dark green (S22b-f; D101G); with all Ca2+-binding sites empty are blue (S9b-f) and violet (S20b-f; D101G). (F) Force peak maxima for cadherin-23 EC1+2 when force is applied to center of mass of Cα1-5;36-41;86-89 atoms at the N-terminus and Cα118-121;171-173;176;203-205 at the C-terminus. Peak values color-coded as in (E), corresponding to simulations with Ca2+ at least at sites 0, 2, & 3 (red, S12g-k; maroon, S6h-l; yellow, S17h-l; orange, S15g-j); with Ca2+ at sites 0 & 3 and Na+ at sites 1 & 2 (green, S11g-j; dark green, S22g-j); and with all Ca2+-binding sites empty (blue, S9g-k). Logarithmic regression fits are shown for each simulation set. (G) Fluorescence vs added Ca2+ in competition assays between the Ca2+ chelator mag-fluo-4 and the wild-type cadherin-23 EC1+2 (black) or the D101G EC1+2 mutant (blue). Data were fitted using a four-binding-site model (Table S7). (H) Trypsin digests of wild-type and D101G mutant cadherin-23 EC1+2 analyzed by SDS-PAGE. Incubation in a range of Ca2+ concentrations shows different Ca2+-dependence of proteolysis protection. The intact proteins migrate at 25 kDa. (I) Quantification of the intact protein in the gels presented in H. Fits using the Hill equation indicate an effective KD of 86.8 μM for wild-type and 470.4 μM for the D101G mutant (Hill coefficients: 3.7 and 1.3, respectively). See also Figures S4, S5, and S6.
In contrast, simulations of EC1+2 with one more Ca2+ ion replaced (Ca2+ at sites 0 & 3 and Na+ at sites 1 & 2; S11), show a single unfolding-force peak that is significantly smaller than those observed for Ca2+-loaded cadherin-23. It coincides with rupture of the EC1+2 linker region and ß-strand detachment (simulations S11a-f, Figure 5B; Figures S2H and S2I). Simulations of cadherin EC1+2 with all binding sites empty (S9) show a further decrease in mechanical strength with unfolding at even lower forces. Force peaks correspond to rupture of hydrogen bonds between ß-strands (S9a-f, Figure 5C). Simulations therefore suggest that Ca2+ ions at sites 2 and 3 are sufficient to maintain cadherin-23's mechanical strength, which may gradually decrease as Ca2+ concentration is decreased.
All these stretching simulations applied force to Cα atoms at the protein's N- and C-termini. This approach may favor unfolding of the terminal ends or the structures that are in line with the ß-strands to which force is applied. To address this concern, a second set of simulations was performed in which force was applied to groups of ~12 Cα atoms at each end of the protein, effectively distributing the applied force over various ß-strands (simulations S12g-k, S6h-l, S11g-j, S9g-k; Figure S3). Such simulations showed that unfolding of the EC1-EC2 linker was similar to previous simulations, and thus that Ca2+ stabilization of cadherin-23 under force is robust and independent of the stretching protocol.
Structure of the deafness-related mutant D101G of cadherin-23 EC1+2
Multiple missense mutations in cadherin-23's extracellular domain cause the nonsyndromic deafness DFNB12, and most of these are located at its Ca2+-binding motifs (Astuto et al., 2002; Baux et al., 2008; Bolz et al., 2001; Bork et al., 2001; de Brouwer et al., 2003; Ouyang et al., 2005; Roux et al., 2006; Schultz et al., 2005; Schwander et al., 2009; Wagatsuma et al., 2007) (Figure S4). Of these, D101G is the only one located in cadherin-23 EC1+2 (E120Q is located at the EC2+3 linker, unresolved in our structures). In compound heterozygous individuals the D101G mutation together with R2442W causes recessive non-syndromic deafness. The nonsyndromic and progressive characteristics of DFNB12 associated with D101G/R2442W suggest that these mutations cause neither protein misfolding nor mislocalization. Thus, the EC1+2 D101G mutant was generated to gain insight into molecular mechanisms underlying deafness.
The mutant protein exhibited a slightly smaller apparent molecular weight than wild type (Figure S1A) and crystallized in a different space group with two EC1+2 molecules in the asymmetric unit. The individual repeats adopt conformations nearly identical to their wild-type counterparts (Figure 4B; Table 1). However, the arrangement of EC2 with respect to EC1 was different for the two molecules of the asymmetric unit, with one similar to wild type, and the other displaying a different inter-repeat arrangement discussed further below. Remarkably, the mutant protein retains a Ca2+ ion at site 2 despite the absence of the Ca2+-coordinating D101 side-chain (Figure 4D). Density was observed at the previous location of the aspartate's carboxylate group (Figures S1E and S1F) and was assigned to a Cl- ion located 2.6 Å from Ca2+ at site 2. As in the EC1+2 Na+ structure, Na+ was modeled at site 1. The D101G structure demonstrates that the mutant protein retains the ability to fold and bind Ca2+. The mutation may therefore affect the function of cadherin-23 by one or both of two mechanisms: (i) the mutation may directly affect the unfolding strength and flexibility of the linker region or (ii) the mutation may shift the Ca2+ affinity.
Mechanical strength of the deafness-related D101G mutant is altered by decreased Ca2+-binding affinity
To understand how the D101G mutation affects cadherin stability, SMD simulations of the D101G mutant structure were performed following the same two stretching protocols used for wild type. For all simulations done with the same ions at sites 1-3, observed forces in the mutant EC1+2 were comparable to those in wild-type (simulations S17a-l, S22a-j, S20a-f; Figures 5D-F, S5A-E, S3E, and S3F). Thus, when Ca2+ is bound, the D101G mutant is as strong as wild-type.
The D101G mutation may instead change cadherin-23 strength indirectly, by reducing its affinity for Ca2+. A competition assay based on a fluorescent Ca2+ indicator was thus used to determine Ca2+-binding affinities for cadherin-23 EC1+2 and for EC1+2 D101G (Figure 5G). Fluorescence of a Ca2+ indicator mixed with EC1+2, at Ca2+ concentrations ranging between 20 and 200 μM, was consistently smaller than fluorescence obtained with the D101G EC1+2 mutant, indicating a stronger Ca2+-binding affinity for the wild-type protein. Fitting these data using models with three or four Ca2+-binding sites, we found that Ca2+ dissociation constants (KD) are larger for the D101G EC1+2 protein. Dissociation constants for the wild-type protein were (in increasing order) 1.9, 5.0, 44.3 and 71.4 μM, whereas for the D101G mutant the best fit was obtained using a three-site model with KD values of 3.9, 40.6, >100 μM. Interestingly, the same experiment for the EC1 fragment indicates a KD of 1.0 μM for site 0 (Figures S5F and S5G). Thus, a tentative assignment of KD values to Ca2+-binding sites is possible: The two lowest KD values in EC1+2 proteins are likely to be site 0 and the structurally similar site 3; site 1 probably has the highest KD, as Ca2+ at this site is replaced by Na+ in the crystals prepared in high Na+ (Figure 4C and 4D); and site 2 corresponds to the intermediate KD values (Table S7).
The lower affinity for Ca2+ in the D101G mutant was confirmed using a trypsin sensitivity assay. Significantly higher Ca2+ concentrations were required to protect the EC1+2 D101G protein from trypsin digestion than for the wild-type protein (Figures 5H and 5I). Quantification of the trypsin digestion data revealed half-maximal proteolysis rates at 86 μM Ca2+ for the wild-type EC1+2 protein, consistent with the KD values estimated from fluorescence. A Hill coefficient of 3.7 for the fitted curve indicates strong cooperativity in Ca2+ binding, which was reduced in the D101G mutant (Hill coefficient 1.3). These results are in agreement with Courjean et al. (2008), where the D103A mutation in E-cadherin EC1+2 (a different aspartate residue in the same DXNDN motif) increased the KD for Ca2+ (from 20 to 240 μM) and eliminated cooperativity. Our data and simulations suggest that the reduced Ca2+ affinity of the D101G mutant leads to reduced mechanical strength at the physiological Ca2+ concentrations of cochlear endolymph (20-40 μM).
Mechanical strength can be directly altered in other cadherin-23 mutants
Do all deafness mutations in these Ca2+-binding sites cause the same structural and functional defects? While structural information on other cadherin-23 EC repeats is not available, sequence alignments can be used to map the mutations either onto our cadherin-23 EC1+2 structure or onto classical cadherin structures, to test their effect on mechanical strength. Among all known cadherin-23 mutations (Figure S4), two classes were selected for in silico studies.
The first modifies the DRE site. The salsa mouse with progressive hearing loss, one of the animal models for human nonsyndromic deafness DFNB12 (Schwander et al., 2009), has the mutation E714V at the EC7+8 linker. A human deafness mutation, E1572K at the EC15+16 linker, modifies the same motif residue. The salsa E-to-V substitution was modeled into the cadherin-23 EC1+2 structure, mapping to residue E73. The resulting model was equilibrated for 10 ns (S25a-b) and subsequently stretched in SMD simulations (S27a-k). This mutant showed a behavior similar to wild-type when Ca2+ was loaded at all binding sites (Figures S6A, S6C, S6D, and S6F). Like that of D101G, the phenotype is more likely to arise from changes in Ca2+ affinity leading indirectly to altered elasticity and strength.
The second class alters the glutamate of the XEXBASE motif, e.g., mutations E120Q at the EC2+3 linker and E224K at the EC3+4 linker in humans (Baux et al., 2008; Roux et al., 2006). These correspond to position E21 in cadherin-23 EC1+2, coordinating Ca2+ at sites 1 and 2 (Figure 1E). Since either charge neutralization (Q) or reversal (K) causes a phenotype, we chose to evaluate an alanine substitution (E21A), which simply removes the side chain. The E21A mutant shows only one peak in stretching simulations, as opposed to the two peaks observed for wild type, indicating that the mutation directly alters the elastic properties of the protein, reducing its mechanical strength when stretched from Cα atoms (S26a-f, Figures S6B and S6C). The mutation likely affects affinities for Ca2+ at sites 1 & 2 as well, which could further impair protein function.
The cadherin-23 EC1+2 background did not affect the behavior of modeled mutations: Equivalent mutations were created in the five-repeat C-cadherin structure and similar behavior was found in simulations (S28 for WT, S29 for E69A, and S30 for E11A; Figures S6G-J).
Thus, deafness mutations targeting Ca2+ motifs can affect cadherin-23 in at least two ways: they can reduce Ca2+ binding and indirectly affect mechanical strength, or they can modify favorable Ca2+-protein interactions to directly reduce the mechanical strength of the protein.
Deafness mutations modulate inter-repeat motion
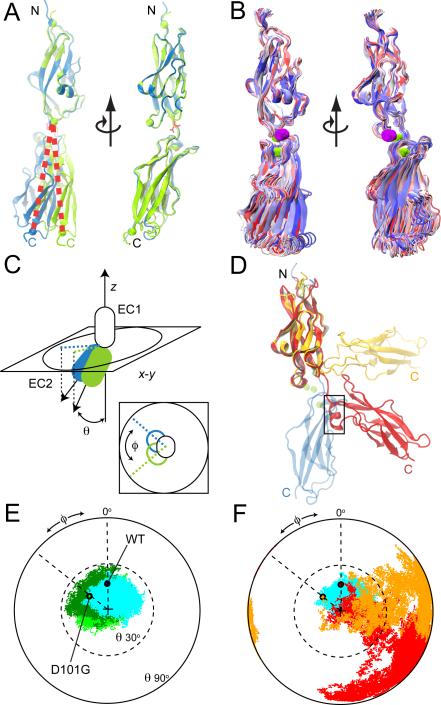
The two conformations observed in the crystal structure of the D101G mutant (Figure 6A) suggest a third possible effect of deafness mutations: altered inter-repeat orientation and dynamics, that may impair cis and trans interactions. The conformation of classical and tip-link cadherin extracellular domains indeed depends on availability of Ca2+, being extended and rigid in its presence and collapsed and flexible in its absence (Cailliez and Lavery, 2005; Kazmierczak et al., 2007; Pokutta et al., 1994; Sotomayor and Schulten, 2008). Equilibrium MD simulations were thus performed to compare inter-repeat motion of wild-type and mutant cadherin-23 EC1+2 repeats in the presence and absence of Ca2+.
Figure 6.
Inter-repeat arrangement and dynamics of cadherin-23 EC1+2. (A) Two views of wild-type (blue) and D101G (green) EC1+2, with EC1 repeats superimposed. Spheres denote Cα atoms at positions 3, 74, and 205. Residue D101 is in red sticks. (B) Analogous views of the superposition of wild-type cadherin-23 conformations taken every 100 ps during simulation S8a-b (9.2-12.3 ns). Color indicates time (red-white-blue). (C) To quantify the conformational freedom of EC2 with respect to EC1, the principal axis of EC1 was aligned to the z-axis, and the EC2 principal axis projection in the x-y plane plotted (perspective and top views). Vector length relates to the tilt angle (sin θ), while the phase angle corresponds to the azimuthal angle φ. (D) Initial (transparent blue cartoon) and final (opaque cartoon) conformations of cadherin-23 EC1+2 without Ca2+ are shown for the wild-type protein (red) and the D101G mutant (orange). An α-helix between ß-strands B and C at EC2 spontaneously formed during simulation S10a-b (box). (E) Plotted projections (as in C) shown for every picosecond of equilibrium simulation of the wild-type protein (S8a-b in cyan) and two independent simulations of the D101G mutant protein (S18a-b and S19a-b in light and dark green, respectively). Initial projections for wild-type (WT) and mutant (D101G) are highlighted by black circles. (F) Projections of EC2's principal axis along its longest dimension on the x-y plane for every picosecond of equilibrium simulations of the WT (red) and D101G (orange) proteins with all Ca2+-binding sites empty.
Simulations of cadherin-23 EC1+2 with Ca2+ at all binding sites or with Na+ substituting at site 1 show considerable inter-repeat motion (simulations S7a-b, S8a-b, S13a-b, & S14a-b; Figure 6B). Domain motions can be characterized by the tilt angle (θ) between the principal axes of EC1 and EC2 repeats, and the azimuthal angle (φ) of EC2 with respect to EC1 (Figure 6C). Two independent simulations of wild-type EC1+2 with Ca2+ at site 1 and two additional simulations with Na+ at site 1 display overlapping and similar distributions of tilt and azimuthal angles. Two independent simulations of the D101G mutant show that it explores an overlapping and wider range of values for both θ and φ (S18a-b and S19a-b, Figure 6E), i.e., it both bends and twists more than wild-type.
To rule out the possibility that the Cl- ion near site 2 in the D101G mutant was responsible for the observed differences, or that the simulations were biased by the initial conditions used, the structure of wild-type EC1+2 with Ca2+ at all sites was mutated (D101G) in silico and equilibrated (S24a-c). Inter-repeat motion was similar to that observed for simulations of the D101G cadherin-23 EC1+2 crystal structure (data not shown). Thus, simulated dynamics and inter-repeat motion of the D101G mutant seem robust and distinct from the wild-type. Similar equilibrium MD simulations were performed on the in silico E21A and E73V mutants of cadherin-23 EC1+2 (S23a-c and S25a-c). E21A shows an overall shift in tilt and azimuthal angles, opposite to the shift observed for D101G, whereas the E73V trajectories show a moderately broadened tilt angle distribution but little azimuthal angle preference (data not shown).
If these mutations also compromise Ca2+ binding, as shown for the D101G mutant above, an even more pronounced effect in inter-repeat dynamics is expected in low Ca2+ environments: In the absence of ions at all binding sites, and regardless of the presence of the D101G mutation, repeats EC1 and EC2 move dramatically with respect to each other (S10a-b & S21a-b; Figures 6D and 6F). Consistently, the extracellular domain of cadherin-23 loses its filamentous shape in the absence of Ca2+ to become a chain of randomly oriented repeats (Kazmierczak et al., 2007). The same simulated behavior has been observed for E-cadherin EC1+2 (Cailliez and Lavery, 2005), C-cadherin EC1-5 (Sotomayor and Schulten, 2008), and the type-II classical cadherin cadherin-8 (data not shown). Overall, simulations suggest that mutations and Ca2+ binding modulate cadherin inter-repeat orientation and compromise rigidity, which in turn may affect cis and trans dimerization.
Possible cadherin-23 interfaces differ from the strand-exchanged classical cadherin interface
Although cadherin-23 in the tip link is thought to bind to protocadherin-15 in an antiparallel configuration and bind to the partner-strand cadherin-23 in parallel, it may also engage in antiparallel homophilic binding. Cadherin-23 alone has been reported to enable aggregation of cultured cells (Siemens et al., 2004). In addition, hair-cell bundles are splayed in cadherin-23 mutants, indicating a developmental role perhaps independent of tip links (Söllner et al., 2004). The novel structure of cadherin-23 EC1 indicates that interaction with itself or with other proteins like protocadherin-15 must involve a non-classical binding interface. Although EC1 or EC1+2 dimerization was not observed in size exclusion chromatography (Figure S1A), the cadherin-23 crystal packing hints at possible dimer interfaces.
The crystal lattices include symmetric antiparallel dimeric EC1 interfaces, with two different observed arrangements that we labeled ß and W interfaces. The ß interface observed in the EC1 crystal lattice buries 599 Å2 of surface area by forming an intermolecular ß-sheet through residues 88-94 of ß-strands G (Figure 7A). In the EC1+2 structures, including the D101G mutant, the opposite faces pack together to form the W interface, burying 480 Å2 with contact between conserved tryptophan residues (W65, Figure 7C). Neither of these interfaces is targeted by known deafness mutations and classical cadherin interfaces bury larger surface areas (850 Å2 and 1270 Å2 for type I and II, respectively). Consistently, SMD simulations that forced unbinding in either the ß or W interface showed an unbinding strength similar than that observed for classical cadherins. However, a single force peak upon unbinding without unfolding was observed in all cases (Figure 7B, 7D and 7E; Figure S7; Movies III, IV, and V; see also Bayas et al. (2004)). While suggestions from crystal packing interactions must be considered cautiously (Bahadur et al., 2004; Kobe et al., 2008), possible cadherin-23 homophilic interfaces identified here are probably weaker than and likely differ from the mechanically strong heterophilic interaction expected with protocadherin-15.
Figure 7.
SMD simulations probing the strength of possible cadherin-23 binding interfaces. (A) The ß-strand interface; detailed view at right panel. Backbone of residues 87 to 95 and side chain of K94 are in sticks. C-terminal Cα and Ca2+ atoms are blue and green spheres, respectively. (B) Force versus distance between C-termini ends of two molecules forming the ß interface with Ca2+ in binding sites 0. Traces correspond to independent simulations at stretching speeds of 10 (S4c, black; S4d, blue), 1 (S4e, green; S4f, dark green), and 0.1 nm/ns (S4g, turquoise). (C) The W interface; overall structure shown in two perpendicular views, with W65 in sticks. Inset: detailed view of W65 residues. (D) Force versus distance between C-termini ends of two protomers forming a cadherin-23 W interface. Traces correspond to independent simulations at stretching speeds of 10 (S16c, black; S16d, blue), 1 (S16e, green; S16f, dark green), and 0.1 nm/ns (S16g, turquoise). (E) Maximum force-peak values for unbinding simulations of cadherin-23 ß and W interfaces (blue, S4c-g; green, S16c-g). Unbinding force-peak values for simulations of C-cadherin and Cadherin-8 dimers are shown in red (S31d-m) and orange (S33c-i, see Figure S7). Logarithmic regression fits are shown for each simulation set.
DISCUSSION
The structures and simulations of cadherin-23 EC1+2 repeats presented here shed light on at least three aspects of its function in hair-cell mechanotransduction: its role as a stiff cable rather than a soft spring, its weakening by deafness mutations, and its possible binding mechanisms.
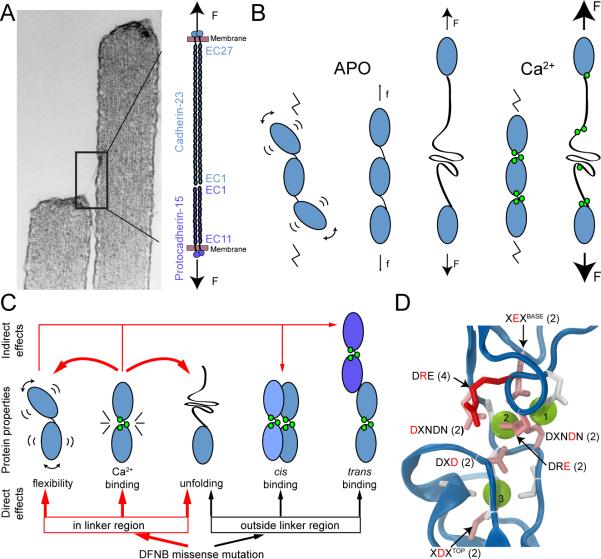
The elasticity of repeats EC1+2 computed using SMD simulations is strongly dependent on the presence of Ca2+ at sites 0, 2, & 3, but not site 1. Simulations corroborate our previous studies on classical cadherins (Sotomayor et al., 2005; Sotomayor and Schulten, 2008) which suggested that the tip link may not be the molecular correlate of the long-sought “gating spring”. SMD simulations, although using stretching speeds somewhat faster than physiological hair bundle motions, allowed us to calculate the approximate stiffness of the cadherin-23 structures (see also Supplemental Text). Stretching simulations of one or two repeats predict a stiffness ranging from ~710 to ~1140 mN/m per repeat at pulling speeds of 0.1 nm/ns (in the presence of Ca2+). Extrapolated to 38 repeats in each strand of a parallel dimer (27 contributed by cadherin-23 and 11 by protocadherin-15; Figure 8A; Sarkar et al. (2007)), the stiffness of the tip link is predicted to be ~40-60 mN/m, much stiffer than the ~1 mN/m measured for the gating spring (Cheung and Corey, 2005; Howard and Hudspeth, 1988). Even in the absence of Ca2+, the effective stiffness of the whole tip link is predicted to be ~16 mN/m. It is possible that another unknown component of the transduction apparatus or tip-link (mechanically in series with the transduction channel) provides the required elasticity, or even that structures of the other 36 repeats are different enough to render the tip link more compliant. However, sequence analyses suggest that most of all cadherin-23 and protocadherin-15 repeats are closely related to canonical repeats with Ca2+-binding sites in the linker regions. It is possible that domains of ~100 residues proximal to the membrane in each tip-link cadherin, not identified as cadherin repeats in sequence motifs searches but perhaps forming a 28th and a 12th repeat, have significantly different mechanical properties. But even if compliant, they could have only a limited range of extension. Overall, structures and simulations both suggest that the extracellular repeats of cadherin-23 are rather stiff elements conveying force (Figure 8B), instead of being an elastic gating spring.
Figure 8.
Structural determinants of cadherin-23 function in hearing and deafness. (A) Hair-cell tip links made of cadherin-23 and protocadherin-15. (B) Tip link dynamics and elasticity are controlled by Ca2+. In the absence of Ca2+, cadherin repeats move independently of each other; small stretching forces can straighten the chain of repeats and cause unfolding. In the presence of Ca2+ cadherin repeats are stiff and unfold at large and likely unphysiological stretching forces. (C) Missense mutations can alter protein behavior in multiple ways. Mutations at linker regions are more likely to directly affect Ca2+ binding, inter-repeat flexibility, and mechanical strength, whereas mutations outside of the linker regions could also directly affect trans and cis binding, as well as unfolding strength. Indirect effects of the mutations are also indicated by the arrows at the top. Bold arrows indicate effects observed here in structures and simulations. (D) Sixteen of the 35 DFNB12 mutations in human cadherin-23's extracellular domain target residues in Ca2+-binding motifs: four target the arginine (red) at a DRE motif, and the other 12 are equally distributed among six other residues coordinating Ca2+ (pink). Ca2+-coordinating residues with no known mutation are white.
These studies also address the effect of deafness mutations on tip-link function (Figure 8C). The crystal structure of the cadherin-23 EC1+2 D101G mutant, the first of a mutant cadherin linker region, demonstrates that mutations targeting Ca2+-binding sites do not necessarily compromise the overall architecture of the protein. Furthermore, Ca2+ ions can still bind at the altered linker region, although the measured binding affinities are lower. Simulations suggest that the D101G mutation does not directly affect the mechanical strength of cadherin-23 EC1+2, as long as Ca2+ is bound, but could render the protein weaker through a shift in Ca2+ affinity. The D101G structure and equilibrium simulations also suggest that inter-repeat motion is directly, although moderately, increased by this mutation. Results are consistent with binding assays showing altered protocadherin-15/cadherin-23 interactions for the cadherin-23 EC1-27 D101G mutant (Schwander et al., 2009), in that a change in inter-repeat motion and dynamics caused by a shift in Ca2+ affinity would likely compromise trans dimerization. Simulations also suggest that the cadherin-23 D101G and salsa mutants are mechanically weaker at low Ca2+ concentrations. Because vestibular endolymph has more Ca2+ than cochlear endolymph (Nakaya et al., 2007; Salt et al., 1989), this may explain the lack of vestibular phenotypes in salsa mice and some human DFNB12. In contrast, we found that other mutations, such as E21A studied here, can directly affect both the mechanical strength of the protein and its dynamics, regardless of whether Ca2+ ions are bound. Further studies will be needed to address the effects of all classes of missense deafness mutations (Figure 8D) and if wild-type and mutated tip-links are affected by local variations in Ca2+ concentration around hair-cell bundles (Yamoah et al., 1998).
Structures and simulations also provide insight into cadherin-23 binding. The architecture of cadherin-23 EC1 is incompatible with classical cadherin interactions (Patel et al., 2006; Posy et al., 2008). Two possible antiparallel interfaces, ß and W, observed in crystal packing interactions, provide suggestions of how cadherin-23 may interact with itself (see also Supplemental Text). Simulations suggest that these cadherin-23 homophilic interfaces exhibit a mechanical strength comparable to that of classical cadherins, with the W interface somewhat weaker than classical cadherins type I and II. Considering that classical cadherins unbind at forces of ~50 pN and stretching speeds of 4 μm/s (Baumgartner et al., 2000), but the tip link must withstand larger forces, these results suggest that cadherin-23's heterophilic binding with protocadherin-15 to form the tip link must use a different, mechanically stronger, interface.
A trans-binding interface between cadherins mediated by Ca2+, like that between EC repeats within cadherin-23, might provide the strength required for tip-link function. Our SMD simulations show that the strength of the linker region between EC repeats is provided by the bound Ca2+ ions rather than the peptide bond, and that EC repeats interacting through Ca2+ (without a peptide bond linking them) unfold before they unbind (Figures S2D and S2E). Protocadherin-15, with its long N-terminus beginning with a conserved QYDDD sequence, could form an analogous Ca2+-mediated bridge to the cadherin-23 N-terminus and its newly discovered Ca2+-binding site 0. Although further structural studies are needed to test this hypothesis, it would provide a natural explanation for why extracellular Ca2+ chelation cleaves tip links (Assad et al., 1991).
The results presented have implications beyond inner-ear mechanotransduction, providing the first cadherin-repeat structure of Cr-2 family proteins including protocadherin-24, protocadherin-21, and cadherin-23. Protocadherin-24 is involved in contact inhibition of cell proliferation and cancer (Okazaki et al., 2002). Protocadherin-21 is found at the base of the outer segment of both rod and cone photoreceptors (Rattner et al., 2001) and in the olfactory bulb (Nakajima et al., 2001). It is essential for photoreceptor survival, but its function is otherwise unclear. Our work strongly suggests that Cr-2 proteins feature the same EC1 Ca2+-binding site 0 observed for cadherin-23 and may share properties such as homologous partners, interface configurations, and physiological activities. Finally, the possible mechanisms underlying hearing loss discussed here might be applicable to other diseases caused by missense mutations at cadherin linker regions, such as progressive myocardial dystrophy (Pilichou et al., 2009).
EXPERIMENTAL PROCEDURES
Cloning, expression, and purification of cadherin-23 repeats
Mouse cadherin-23 repeats EC1 and EC1+2 comprising residues Q1 to D101 (Q24 to D124 in NP_075859.2) and Q1 to D205 respectively were sub-cloned into the NdeI and XhoI sites of the vector pET21a. The D101G mutation in EC1+2 was generated using the QuikChange Lightning mutagenesis kit (Stratagene). Cadherin-23 repeats were expressed in BL21CodonPlus(DE3)-RIPL (Stratagene) cultured in LB and induced at OD600=0.6 with 100 μM IPTG at room temperature for ~16 hrs. Cells were lysed by sonication in denaturing buffer (20 mM HEPES at pH 7.5, 6 M guanidine hydrochloride, 2 mM CaCl2, 20 mM imidazole at pH 7.0). The cleared lysates were loaded onto Ni-sepharose (GE Healthcare), eluted with denaturing buffer supplemented with 500 mM imidazole, and refolded by overnight dialysis against 20 mM HEPES at pH 7.5, 150 mM NaCl, 2 mM CaCl2, and 50 mM KCl using MWCO 2000 membranes. Refolded proteins were further purified by size-exclusion chromatography on a Superdex75 column (GE Healthcare) in 20 mM TrisHCl pH 7.5, 2 mM CaCl2, and 200 mM NaCl (EC1 and EC1+2 Na+) or 150 mM KCl & 50 mM NaCl (EC1+2 Ca2+ and EC1+2 D101G) and concentrated by ultrafiltration to 10 mg/ml for crystallization (Vivaspin 10 kDa).
Crystallization, data collection, and structure determination
Crystals were grown by vapor diffusion at 4°C by mixing equal volumes of protein and reservoir solution of (0.1 M sodium cacodylate pH 6.0, 40 % MPD) for cadherin-23 EC1, (0.1 M MES pH 6.5, 1.1 M CaCl2) for EC1+2 Ca2+, (0.1 M Tris HCl pH 8.0, 2.7 M NaCl, 10 % glycerol) for EC1+2 Na+, and (0.1 M sodium cacodylate pH 7.1, 1 M NaCl) for cadherin-23 EC1+2 D101G. Cadherin-23 EC1+2 crystals were cryoprotected in reservoir solution plus 25% glycerol. All crystals were cryo-cooled in N2. X-ray diffraction data were collected as indicated in Table 1 and processed with HKL2000 (Otwinowski and Minor, 1997). The cadherin-23 EC1 and EC1+2 Na+ structures were determined by molecular replacement using a cadherin-23 EC1+2 homology model based on E-cadherin EC1+2 (pdb code 1FF5, Pertz et al. (1999)) as a search model with Phaser (McCoy et al., 2007), while cadherin-23 EC1+2 Na+ was used for the EC1+2 Ca2+ and D101G EC1+2 structures. Model building was done using COOT (Emsley and Cowtan, 2004) and restrained TLS refinement using REFMAC5 (Murshudov et al., 1997). The final models include residues M0 to D101 (EC1), M0 to E207 (EC1+2 Ca2+ and EC1+2 Na+), and Q1 to H208 (EC1+2 D101G). Data collection and refinement statistics are provided in Table 1. Coordinates have been deposited in the Protein Data Bank with entry codes 2wbx (EC1), 2wcp (EC1+2 Na+), 2whv (EC1+2 Ca2+), and 2wd0 (EC1+2 D101G).
Ca2+-binding affinity measurements and trypsin digestion
F-buffer (100 mM KCl, 10 mM MOPS, pH 7.5) was incubated with 2% Chelex-100 (Bio-Rad) resin in a dialysis bag and stirred for 3 days before use. Cadherin-23 EC1, EC1+2, EC1+2 D101G, and calmodulin (CaM) at 5-10 mg/mL were stripped of Ca2+ by three consecutive batch incubations of 100 μl Chelex-100 resin per ml of protein solution for 1 h. This procedure yielded buffer and protein solutions that contained <0.2 μM Ca2+ as assessed by Ca2+-dependent Fura-2 fluorescence. Protein concentrations were determined by averaging absorbance reads at 280 nm and data from amino-acid analyses. Ca2+-competition assays used for the determination of Ca2+ dissociation constants (KD) were modified from André and Linse (2002). Assays at 25°C were performed in cuvettes containing 2 mL of 2.5 μM mag-fluo-4 (Invitrogen) and 3-4.2 μM protein in F-buffer. CaCl2 solutions were titrated in 5 μl aliquots; after a 10 min incubation the change in fluorescence was recorded on a fluorescence spectrometer (Fluorolog-3, Instruments S. A., Inc.) set at excitation and emission wavelengths of 430 nm and 530 nm, respectively. Experiments were conducted in duplicate and repeated twice. Titration of the chelator in absence of protein was used to determine the apparent KD of the chelator as 33.88 μM as described in (http://probes.invitrogen.com/media/pis/mp03008.pdf). Fluorescence signals were scaled according to sample volume and fitted with models assuming one, three, or four Ca2+-binding sites using the CaLigator software (André and Linse, 2002). For trypsin proteolysis protection assays, 15 ng of trypsin was mixed with 15 μg of decalcified EC1+2 or EC1+2 D101G in 50 μL F-buffer supplemented with Ca2+ concentrations ranging from 0 to 1.6 mM and incubated for 1 h at 37°C. Digestions were stopped by addition of PMSF to a final concentration of 1 mM. Samples were analyzed on Coomassie-stained 20% SDS-PAGE gels quantified with ImageJ.
Simulated systems
The psfgen, solvate, autoionize, and mutator VMD (Humphrey et al., 1996) plugins were used to build all systems (Table 2 and Figure S8). Hydrogens were automatically added to protein structures and crystallographic water molecules. Structures with non-native N- and C-terminal tails were modified back to native sequences. Disulfide bonds in C-cadherin were explicitly modeled. Residues D, E, K, and R were assumed to be charged. Histidine residues were assumed neutral and protonation states chosen to favor the formation of evident hydrogen bonds. Additional water molecules and randomly placed ions were used to solvate the systems at the desired KCl concentration (150 mM for cadherin-23 systems). For SMD simulations, molecules were aligned such that the vector joining the terminal residue Cα atoms was oriented along the x-axis.
Molecular dynamics
MD simulations (Karplus and Petsko, 1990) were performed using NAMD 2.6/2.7 (Phillips et al., 2005), the CHARMM27 force field for proteins with CMAP correction (MacKerell et al., 1998, 2004), and the TIP3P model for water (Jorgensen et al., 1983). A 12-Å cutoff (switching function starting at 10 Å) for van der Waals interactions was used along with periodic boundary conditions. The Particle Mesh Ewald method was used to compute long-range electrostatic forces without cut-off and with a grid point density of >1 Å-3. A uniform 2-fs integration time step was used along with SHAKE. Langevin dynamics was utilized to enforce constant temperature (T = 300 K) when indicated, with a damping coefficient of 0.1 ps-1 unless otherwise stated. Constant pressure simulations (NpT) at 1 atm were conducted using the hybrid Nosé-Hoover Langevin piston method with a 200 fs decay period and a 50 fs damping time constant.
Simulations and analysis tools
Each system was energy-minimized, then equilibrated in the constant number, pressure, and temperature ensemble (NpT), and the resulting state used to perform subsequent equilibrium and SMD simulations (Tables S1 to S6). Coordinates of all atoms were saved for analysis every picosecond of simulation. Constant velocity stretching simulations used the SMD method and NAMD Tcl Forces interface (Isralewitz et al., 2001; Sotomayor and Schulten, 2008). The stretching direction was set along the x-axis matching the vector connecting terminal regions of the protein. SMD simulations were performed by attaching Cα atoms of N- and C-terminal residues to virtual (independent) springs of stiffness ks = 1 (kcal/mol)/Å2, or where indicated, by attaching the center of mass of groups of Cα atoms to the same type of virtual springs. In unbinding simulations, virtual springs were attached to Cα atoms of C-terminal residues of independent molecules. The stretching direction was set along the x-axis matching the vector connecting terminal regions of the protein, the free ends of springs were moved away from the protein in opposite directions at a constant velocity, and applied forces were computed using the extension of the virtual springs. Plotted forces correspond to those applied to N-terminal atoms unless otherwise stated. Stiffness was computed through linear regression fits of force-distance plots. Maximum force peaks were computed from 50 ps running averages used to eliminate local fluctuations. End-to-end and CM-to-CM distances were computed as the distances between individual SMD atoms or the center-of-mass of SMD atoms at opposite protein ends, respectively. Regression fits to data points of maximum force peaks versus stretching speeds were performed using a logarithmic expression of the form y = a + b log x. Principal axes of EC repeats were computed using the VMD Orient plugin. Sequence alignments were performed using ClustalX. Structural alignments were performed using VMD/STAMP (Roberts et al., 2006; Russell and Barton, 1992). Plots and curve fits were prepared using xmgrace. Molecular images in this paper were created with the molecular graphics program VMD (Humphrey et al., 1996).
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Ricci and members of the Corey and Gaudet laboratories for helpful discussions; Joy Sircar, Seppo Sahrakorpi, David Hart, John R. Boisseau, Chris Hempel, and John Estabrook for assistance with supercomputer usage; and the RapiData2009 course team at BNL/NSLS for training. This work was supported by the National Institutes of Health (R01 DC02281 to D.P.C.), by a Klingenstein Award to R.G, and by the National Science Foundation through TeraGrid resources provided by NCSA and TACC (TRAC MCB080015 to M.S. and D.P.C.). Simulations were performed at the NCSA-Abe and TRAC-Ranger supercomputers, and exploratory simulations at the Harvard SEAS Blue Gene supercomputer. Use of APS beamlines was supported by National Institutes of Health award RR-15301 and Department of Energy contract No. DE-AC02-06CH11357. Use of the NSLS beamline X25 through the Mail in program is supported by the US Department of Energy and the National Institutes of Health. M.S. is a Howard Hughes Medical Institute Fellow of the Helen Hay Whitney Foundation and D.P.C. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- Cadherin-23 N-terminus defines a cadherin family and suggests new binding mechanisms
- Cadherin-23 mechanical strength is dominated by Ca2+ ions bound between EC repeats
- Cadherin-23 is stiffer than the biophysically defined hair-cell gating spring
- Cadherin-23 deafness mutations reduce Ca2+ affinity and unfolding strength
SUPPLEMENTAL DATA
The Supplemental Data include Supplemental Text, 8 Supplemental Figures, and 7 Supplemental Tables.
REFERENCES
- Ahmed ZM, Goodyear R, Riazuddin S, Lagziel A, Legan PK, Behra M, Burgess SM, Lilley KS, Wilcox ER, Riazuddin S, et al. The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. Journal of Neuroscience. 2006;26:7022–7034. doi: 10.1523/JNEUROSCI.1163-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Aye S, Ali RA, Venselaar H, Anwar S, Belyantseva PP, Qasim M, Riazuddin S, Friedman TB. Gene structure and mutant alleles of PCDH15: nonsyndromic deafness DFNB23 and type 1 Usher syndrome. Hum. Genet. 2003;124:215–223. doi: 10.1007/s00439-008-0543-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagramam KN, Murcia CL, Kwon H, Pawlowski KS, Wright CG, Woychik RP. The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat. Genet. 2001;27:99–102. doi: 10.1038/83837. [DOI] [PubMed] [Google Scholar]
- André I, Linse S. Measurement of Ca2+-binding constants of proteins and presentation of the CaLigator software. Analytical Biochemistry. 2002;305:195–205. doi: 10.1006/abio.2002.5661. [DOI] [PubMed] [Google Scholar]
- Assad JA, Shepherd G, Corey DP. Tip-link integrity and mechanical transduction in vertebrate hair cells. Neuron. 1991;7:985–994. doi: 10.1016/0896-6273(91)90343-x. [DOI] [PubMed] [Google Scholar]
- Astuto LM, Bork JM, Weston MD, Askew JW, Fields RR, Orten DJ, J. OS, Riazuddin S, Morell RJ, Khan S, et al. CDH23 mutation and phenotype heterogeneity: a profile of 107 diverse families with Usher syndrome and nonsyndromic deafness. Am. J. Hum. Genet. 2002;71:262–275. doi: 10.1086/341558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahadur RP, Chakrabarti P, Rodier F, Janin J. A dissection of specific and non-specific protein-protein interfaces. J. Mol. Biol. 2004;336:943–955. doi: 10.1016/j.jmb.2003.12.073. [DOI] [PubMed] [Google Scholar]
- Baumgartner W, Hinterdorfer P, Ness W, Raab A, Vestweber D, Schindler H, Drenckhahn D. Cadherin interaction probed by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 2000;97:4005–4010. doi: 10.1073/pnas.070052697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baux D, Faugère V, Larrieu L, Le Guédard S, Hamroun D, Béroud C, Malcolm S, Claustres M, Roux AF. UMD-USHbases: A comprehensive set of databases to record and analyse pathogenic mutations and unclassified variants in seven usher syndrome causing genes. Hum. Mutat. 2008;29:E76–E87. doi: 10.1002/humu.20780. [DOI] [PubMed] [Google Scholar]
- Bayas MV, Schulten K, Leckband D. Forced dissociation of the strand dimmer interface between C-cadherin ectodomains. Mechanics and Chemistry of Biosystems. 2004;1:101–111. [PubMed] [Google Scholar]
- Beurg M, Fettiplace R, Nam JH, Ricci AJ. Localization of inner hair cell mechanotransducer channels using high-speed calcium imaging. Nature Neuroscience. 2009;12:553–558. doi: 10.1038/nn.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- Bolz H, von Brederlow B, Ramírez A, Bryda EC, Kutsche K, Nothwang HG, Seeliger M, del C-Salcedó Cabrero M, Vila MC, Molina OP, et al. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat. Genet. 2001;27:108–112. doi: 10.1038/83667. [DOI] [PubMed] [Google Scholar]
- Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, Polomeno R, Ramesh A, Schloss M, Srisailpathy CRS, et al. Usher syndrome 1D and non-syndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am. J. Hum. Genet. 2001;68:26–37. doi: 10.1086/316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosher SK, Warren RL. Very low calcium content of cochlear endolymph, an extracellular fluid. Nature. 1978;273:377–378. doi: 10.1038/273377a0. [DOI] [PubMed] [Google Scholar]
- Cailliez F, Lavery R. Cadherin mechanics and complexation: the importance of calcium binding. Biophys. J. 2005;89:3895–3903. doi: 10.1529/biophysj.105.067322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CP, Posy S, Ben-Shaul A, Shapiro L, Honig BH. Specificity of cell-cell adhesion by classical cadherins: Critical role for low-affinity dimerization through ß-strand swapping. Proc. Natl. Acad. Sci. USA. 2005;102:8531–8536. doi: 10.1073/pnas.0503319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung EL, Corey DP. Ca2+ changes the force sensitivity of the hair-cell transduction channel. Biophys. J. 2005;90:124–139. doi: 10.1529/biophysj.105.061226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey DP, Hudspeth AJ. Kinetics of the receptor current in bullfrog saccular hair cells. J. Neurosci. 1983;3:962–976. doi: 10.1523/JNEUROSCI.03-05-00962.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courjean O, Chevreux G, Perret E, Morel A, Sanglier S, Potier N, Engel J, van Dorsselaer A, Feracci H. Modulation of E-cadherin monomer folding by cooperative binding of calcium ions. Biochemistry. 2008;47:2339–2349. doi: 10.1021/bi701340d. [DOI] [PubMed] [Google Scholar]
- de Brouwer APM, Pennings RJ, Roeters M, van Hauwe P, Astuto LM, Hoefsloot LH, Huygen PLM, van den helm B, Deutman AF, Bork JM, et al. Mutations in the calcium-binding motifs of cdh23 and the 35delg mutation in GJB2 cause hearing loss in one family. Hum. Genet. 2003;112:156–163. doi: 10.1007/s00439-002-0833-0. [DOI] [PubMed] [Google Scholar]
- Denk W, Holt JR, Shepherd GM, Corey DP. Calcium imaging of single stereocilia in hair cells: localization of transduction channels at both ends of tip links. Neuron. 1995;15:1311–1321. doi: 10.1016/0896-6273(95)90010-1. [DOI] [PubMed] [Google Scholar]
- Di Palma F, Pellegrino R, Noben-Trauth K. Genomic structure, alternative splice forms and normal and mutant alleles of cadherin 23 (Cdh23). Gene. 2001;281:31–41. doi: 10.1016/s0378-1119(01)00761-2. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PG, Müller U. Mechanotransduction by hair cells: Models, molecules, and mechanisms. Cell. 2009;139:33–44. doi: 10.1016/j.cell.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubmüller H. Force probe molecular dynamics simulations. Methods Mol Biol. 2005;305:493–515. doi: 10.1007/978-1-59259-912-7_23. [DOI] [PubMed] [Google Scholar]
- Howard J, Hudspeth AJ. Compliance of the hair bundle associated with gating of mechanoelectrical transduction channels in the bullfrog's saccular hair cell. Neuron. 1988;1:189–199. doi: 10.1016/0896-6273(88)90139-0. [DOI] [PubMed] [Google Scholar]
- Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. The International Journal of Biochemistry and Cell Biology. 2009;41:349–369. doi: 10.1016/j.biocel.2008.09.027. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD - Visual Molecular Dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Isralewitz B, Gao M, Schulten K. Steered molecular dynamics and mechanical functions of proteins. Curr. Opin. Struct. Biol. 2001;11:224–230. doi: 10.1016/s0959-440x(00)00194-9. [DOI] [PubMed] [Google Scholar]
- Izrailev S, Stepaniants S, Balsera M, Oono Y, Schulten K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys. J. 1997;72:1568–1581. doi: 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- Kachar B, Parakkal M, Kurc M, Zhao Y, Gillespie P. High-resolution structure of hair-cell tip links. Proc. Natl. Acad. Sci. USA. 2000;97:13336–13341. doi: 10.1073/pnas.97.24.13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M, Petsko GA. Molecular dynamics simulations in biology. Nature. 1990;347:631–639. doi: 10.1038/347631a0. [DOI] [PubMed] [Google Scholar]
- Katsamba P, Carroll K, Ahlsen G, Bahna F, Vendome J, Posy S, Rajebhosale M, Price S, Jessell TM, Ben-Shaul A, et al. Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proc. Natl. Acad. Sci. USA. 2009;106:11594–11599. doi: 10.1073/pnas.0905349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak P, Sakaguchi H, Tokita J, Wilson-Kubalek EM, Milligan RA, Müller U, Kachar B. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449:87–91. doi: 10.1038/nature06091. [DOI] [PubMed] [Google Scholar]
- Kobe B, Guncar G, Buchholz R, Huber T, Maco B, Cowieson N, Martin JL, Marfori M, Forwood JK. Crystallography and protein-protein interactions: biological interfaces and crystal contacts. Biochem. Soc. Trans. 2008;36:1438–1441. doi: 10.1042/BST0361438. [DOI] [PubMed] [Google Scholar]
- Leckband D, Prakasam A. Mechanism and dynamics of cadherin adhesion. Annu. Rev. Biomed. Eng. 2006;8:259–287. doi: 10.1146/annurev.bioeng.8.061505.095753. [DOI] [PubMed] [Google Scholar]
- MacKerell A, Jr., Bashford D, Bellott M, Dunbrack RL, Jr., Evanseck J, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- MacKerell AD, Jr., Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comp. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nagar B, Overdulin M, Ikura M, Rini J. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380:360–364. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- Nakajima D, Nakayam M, Kikuno R, Hirosawa M, Nagase T, Ohara O. Identification of three novel non-classical cadherin genes through comprehensive analysis of large cDNAs. Molecular Brain Research. 2001;94:85–95. doi: 10.1016/s0169-328x(01)00218-2. [DOI] [PubMed] [Google Scholar]
- Nakaya K, Harbidge DG, Wangemann P, Schultz BD, Greem ED, Wall SM, Marcus DC. Lack of pendrin HCO-3 transport elevates vestibular endolymphatic Ca2+ by inhibition of acid-sensitive TRPV5 and TRPV6 channels. Am. J. Physiol. Renal. Physiol. 2007;292:F1314–F1321. doi: 10.1152/ajprenal.00432.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki N, Takahashi N, Kojima S, Masuho Y, Koga H. Protocadherin LKC, a new candidate for a tumor suppressor of colon and liver cancers, its association with contact inhibition of cell proliferation. Carcinogenesis. 2002;23:1139–1148. doi: 10.1093/carcin/23.7.1139. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Ouyang XM, Yan D, Du LL, Fielding Hejtmancik J, Jacobson SG, Nance WE, Li AR, Angeli S, Kaiser M, Newton V, et al. Characterization of Usher syndrome type I gene mutations in an Usher syndrome patient population. Hum. Genet. 2005;116:292–299. doi: 10.1007/s00439-004-1227-2. [DOI] [PubMed] [Google Scholar]
- Patel SD, Chen CP, Bahna F, Honig B, Shapiro L. Cadherin-mediated cell-cell adhesion: sticking together as a family. Curr. Opin. Struct. Biol. 2003;13:690–698. doi: 10.1016/j.sbi.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale NA, Schieren I, Jessell TM, Honig B, Price SR, Shapiro L. Type II cadherin ectodomain structures: Implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Pertz O, Bozic D, Koch AW, Fauser C, Brancaccio A, Engel J. A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. EMBO J. 1999;18:1738–1747. doi: 10.1093/emboj/18.7.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles JO, Comis SD, Osborne MP. Cross-links between stereocilia in the guinea pig organ of corti, and their possible relation to sensory transduction. Hear. Res. 1984;15:103–112. doi: 10.1016/0378-5955(84)90041-8. [DOI] [PubMed] [Google Scholar]
- Pilichou K, Remme CA, Basso C, Campian ME, Rizzo S, Barnett P, Scicluna BP, Bauce B, van den Hoff MJB, de Bakker JMT, et al. Myocyte necrosis underlies progressive myocardial dystrophy in mouse dsg2-related arrhythmogenic right ventricular cardiomyopathy. JEM. 2009;206:1787–1802. doi: 10.1084/jem.20090641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta K, Herrenknecht S, Kemler R, Engel J. Conformational changes of the recombinant extracellular domain of E-cadherin upon calcium binding. Eur. J. Biochem. 1994;223:1019–1026. doi: 10.1111/j.1432-1033.1994.tb19080.x. [DOI] [PubMed] [Google Scholar]
- Posy S, Shapiro L, Honig B. Sequence and structural determinants of strand swapping in cadherin domains: do all cadherins bind through the same adhesive interface? J. Mol. Biol. 2008;378:954–968. doi: 10.1016/j.jmb.2008.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Pedigo S. Calcium-dependent stability studies of domains 1 and 2 of epithelial cadherin. Biochemistry. 2005;44:13692–13701. doi: 10.1021/bi0510274. [DOI] [PubMed] [Google Scholar]
- Rattner A, Smallwood PM, Williams J, Cooke C, Savchenko A, Lyubarsky A, Pugh EN, Nathans J. A photoreceptor-specific cadherin is essential for the structural integrity of the outer segment and for photoreceptor survival. Neuron. 2001;32:775–786. doi: 10.1016/s0896-6273(01)00531-1. [DOI] [PubMed] [Google Scholar]
- Roberts E, Eargle J, Wright D, Luthey-Schulten Z. MultiSeq: Unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux AF, Faugère V, Le Guédard S, Pallares-Ruiz N, Vielle A, Chambert S, Marlin S, Hamel C, Gilbert B, Malcolm S, Claustres M, the French Usher Syndrome Collaboration Survey of the frequency of USH1 gene mutations in a cohort of Usher patients shows the importance of cadherin 23 and protocadherin 15 genes and establishes a detection rate of above 90%. J. Med. Genet. 2006;43:763–768. doi: 10.1136/jmg.2006.041954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RB, Barton GJ. Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins: Struct., Func., Gen. 1992;14:309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- Salt AN, Inamura N, Thalmann R, Vora A. Calcium gradients in inner ear endolymph. Am. J. Otolaryngol. 1989;10:371–375. doi: 10.1016/0196-0709(89)90030-6. [DOI] [PubMed] [Google Scholar]
- Sarkar A, Caamano S, Fernandez JM. The mechanical fingerprint of a parallel polyprotein dimer. Biophys. J. 2007;92:L36–L38. doi: 10.1529/biophysj.106.097741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz JM, Yang Y, Caride AJ, Filoteo AG, Penheiter AR, Lagziel A, Morell RJ, Mohiddin SA, Fananapazir L, Madeo AC, Penniston JT, Griffith AJ. Modification of human hearing loss by plasma-membrane calcium pump PMCA2. N. Engl. J. Med. 2005;352:1557–1564. doi: 10.1056/NEJMoa043899. [DOI] [PubMed] [Google Scholar]
- Schwander M, Xiong W, Tokita J, Lelli A, Elledge HM, Kazmierczak A, Sczaniecka P, Kolatkar A, Wiltshire T, Kuhn P, Holt JR, et al. A mouse model for nonsyndromic deafness (DFNB12) links hearing loss to defects in tip links of mechanosensory hair cells. Proc. Natl. Acad. Sci. USA. 2009;106:5252–5257. doi: 10.1073/pnas.0900691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman JM, Choi MH, Mihalas S, Mayo SL, Kennedy MB. Ca2+/calmodulin-dependent protein kinase II (CaMKII) is activated by calmodulin with two bound calciums. Proc. Natl. Acad. Sci. USA. 2006;103:13968–13973. doi: 10.1073/pnas.0606433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens J, Lillo C, Dumont RA, Reynolds A, Williams DS, Gillespie PG, Müller U. Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature. 2004;428:950–955. doi: 10.1038/nature02483. [DOI] [PubMed] [Google Scholar]
- Söllner C, Rauch GJ, Siemens J, Geisler R, Schuster SC, the Tübingen 2000 Screen Consortium. Müller U, Nicolson T. Mutations in cadherin 23 affect tip links in zebrafish sensory hair cells. Nature. 2004;428:955–958. doi: 10.1038/nature02484. [DOI] [PubMed] [Google Scholar]
- Sotomayor M, Corey DP, Schulten K. In search of the hair-cell gating spring: Elastic properties of ankyrin and cadherin repeats. Structure. 2005;13:669–682. doi: 10.1016/j.str.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Sotomayor M, Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- Sotomayor M, Schulten K. The allosteric role of the Ca++ switch in adhesion and elasticity of C-cadherin. Biophys. J. 2008;94:4621–4633. doi: 10.1529/biophysj.107.125591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. Cadherins: A molecular family important in selective cell-cell adhesion. Annu. Rev. Biochem. 1990;59:237–252. doi: 10.1146/annurev.bi.59.070190.001321. [DOI] [PubMed] [Google Scholar]
- Vollrath MA, Kwan KY, Corey DP. The micromachinery of mechanotransduction in hair cells. Annu. Rev. Neurosci. 2007;30:339–365. doi: 10.1146/annurev.neuro.29.051605.112917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagatsuma M, Kitoh R, Suzuki H, Fukuoka H, Takumi Y, Usami S. Distribution and frequencies of CDH23 mutations in Japanese patients with non-syndromic hearing loss. Clin. Genet. 2007;72:339–344. doi: 10.1111/j.1399-0004.2007.00833.x. [DOI] [PubMed] [Google Scholar]
- Yamoah EN, Lumpkin EA, Dumont RA, Smith PJS, Hudspeth AH, Gillespie PG. Plasma membrane Ca2+-ATPase extrudes Ca2+ from hair cell stereocilia. J. Neurosci. 1998;18:610–624. doi: 10.1523/JNEUROSCI.18-02-00610.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yamoah EN, Gillespie PG. Regeneration of broken tip links and restoration of mechanical transduction in hair cells. Proc. Natl. Acad. Sci. USA. 1996;93:15469–15474. doi: 10.1073/pnas.93.26.15469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.