Abstract
Collagen is an extracellular matrix structural component that can regulate cellular processes through its interaction with the integrins, α1β1, α2β1, α10β1, and α11β1. Collagen-like proteins have been identified in a number of bacterial species. Here, we used Scl2 from Streptococcus pyogenes serotype M28 strain MGAS6274 as a backbone for the introduction of discrete integrin-binding sequences. The introduced sequences GLPGER, GFPGER, or GFPGEN did not affect triple helix stability of the Scl (Streptococcal collagen-like) protein. Using ELISA and surface plasmon resonance, we determined that Scl2GLPGER and Scl2GFPGER bound to recombinant human α1 and α2 I-domains in a metal ion-dependent manner and without a requirement for hydroxyproline. We predicted a novel and selective integrin-binding sequence, GFPGEN, through the use of computer modeling and demonstrated that Scl2GFPGEN shows specificity toward the α1 I-domain and does not bind the α2 I-domain. Using C2C12 cells, we determined that intact integrins interact with the modified Scl2 proteins with the same selectivity as recombinant I-domains. These modified Scl2 proteins also acted as cell attachment substrates for fibroblast, endothelial, and smooth muscle cells. However, the modified Scl2 proteins were unable to aggregate platelets. These results indicate that Scl2 is a suitable backbone for the introduction of mammalian integrin-binding sequences, and these sequences may be manipulated to individually target α1β1 and α2β1.
Keywords: Cell Adhesion, Collagen, Fibroblast, Integrin, Platelet, Scl, Collagen-like
Introduction
Collagen is a major constituent of the extracellular matrix where it functions as a structural component. In addition, collagen can directly or indirectly interact with cellular receptors and regulate a variety of cellular processes (1). There are at least 28 identified mammalian collagens each consisting of three polypeptides that can be genetically identical or distinct (2). A defining feature of collagens is the tightly packed left-handed triple helix made of polypeptide segments with repeating GXY triplets. The small Gly residue fits in the interior of the triple helix, and the X and Y positions are often occupied by proline and hydroxyproline residues. Hydroxylation of proline residues stabilizes the triple helical structure and inhibition of post-translational hydroxylation decreases the melting temperature of the mammalian collagen triple helix by ∼15 °C (3).
Surface proteins with collagen-like domains recently have been found on a number of prokaryotic organisms, including Streptococcus pyogenes, Streptococcus equi, and Bacillus anthracis (4–6). These collagen-like domains contain conserved GXY repeats but lack the hydroxyproline found in mammalian collagens. S. pyogenes often contain two collagen-like proteins, Scl1 and Scl2 (4, 7–13). The primary sequences and length of different domains, including the GXY repeat segments, vary considerably in Scl1 and Scl2 proteins from different strains (4). In electron micrographs, both proteins form a lollipop-like structure with an N-terminal variable globular domain connected to an extended strand. The strands are composed of the GXY repeats and behave in biochemical analysis as triple helical collagens (4). The collagen-like domains of both Scl proteins have melting temperatures near 37 °C, similar to those of mammalian collagens (4). Because bacterial collagens lack hydroxyproline, the triple helix is likely stabilized by a higher proportion of charged residues at the X and Y positions in comparison with mammalian collagens (12).
Several ligands have been identified for the globular domains of various Scl1 proteins (7, 13). In addition, the collagen-like domain of Scl1 from S. pyogenes serotype M41 is recognized by the collagen-binding α2β1 integrin in an interaction that can lead to bacterial host cell invasion (8, 11). Prokaryotic collagens also form higher order structures (14), as often observed with mammalian collagens (15). Because of these similarities to mammalian collagens, prokaryotic collagen-like proteins are a potential replacement of animal-derived collagens in medical and bioengineering applications (9, 14, 16, 17).
Integrins are cell surface heterodimeric receptors that participate in a variety of cellular processes through activation of different signaling pathways. There are four collagen-binding integrins, α1, α2, α10, and α11, which each complex with a β1 subunit to form a functioning receptor. The α1β1 integrin has been implicated in activation of the Ras-MAPK signaling cascade, which directs cellular functions such as proliferation and survival (18). The α2β1 integrin on endothelial cells is involved in angiogenesis (19, 20), and, on platelets, it participates in platelet activation and aggregation (21, 22).
Several integrin recognition sequences have been identified (23–26). The sequences GFOGER and GLOGER (where O is hydroxyproline) represent binding sites in collagen for the integrins α1β1 and α2β1. To interact with the integrins, these sequences must reside in a triple helical structure, and both sequences are recognized by each integrin (24–25).
Each α-chain of integrins contains an I-domain responsible for collagen binding. The crystal structures of the apo-α1 and apo-α2 I-domains and of α2 in complex with a collagen-like peptide containing the GFOGER sequence revealed details and possible differences of ligand-binding mechanisms by the two integrins. In the α2 I-domain, a divalent cation coordinates the glutamate in the collagen peptide to the integrin metal ion-dependent adhesion site. Surface packing of a phenylalanine or possibly other hydrophobic groups in collagen and an arginine salt bridge to Asp219 in the I-domain also play important roles in the molecular interaction (27). The structure of the α1 I-domain indicated a similar overall structure, but the ligand-binding trench in α1 is predicted to be longer and more flexible than in α2 (28). These observations suggest that α1 and α2 integrin-specific sequences may exist in collagen and may contribute to the observed differences in α1 and α2 integrin binding to various collagens (23, 25, 29).
Here, we have used the frame of S. pyogenes collagen-like protein, Scl2, for analyzing integrin interactions with specific engineered collagen sequences. We demonstrate that the introduction of integrin-binding sequences does not disrupt triple helix formation or alter thermal stability of the modified Scl2 protein and that, in general, the presence of hydroxyproline is not required for integrin binding or cell adherence to substrates composed of the modified Scl2 proteins. We generated a Scl2 protein with a novel integrin-binding site (GFPGEN) that specifically recognizes α1 but not α2 integrins. We also showed that Scl2 proteins containing engineered integrin-binding sequences can support the adherence of fibroblast, endothelial, and smooth muscle cells. Additionally, we determined that Scl2-modified proteins did not aggregate platelets. Thus, we describe a novel recombinant system for the study of collagen-integrin interactions with potential bioengineering applications.
EXPERIMENTAL PROCEDURES
Recombinant Proteins
The recombinant P163 protein, derived from Scl2.28, was generated using the Strep-Tag II expression and purification system as described previously and referred here as Scl2 (4). Single amino acid mutations were introduced in Scl2 at position 127 Gln → Pro generating Scl2GLPGER; using Scl2GLPGER as a template, at position 126 Leu → Phe generating Scl2GFPGER; and using Scl2GFPGER at position 130 Arg → Asn generating Scl2GFPGEN using synthesized PCR primers (Integrated DNA Technologies) and the QuikChange site-directed mutagenesis kit (Stratagene) per the manufacturer's instructions. The mutations were verified by sequencing (SeqWright, Houston, TX). Recombinant proteins were expressed in Escherichia coli BL21 (Novagen) and purified by affinity chromatography on a StrepTrap HP column (GE Healthcare). Protein purity was determined by SDS-PAGE followed by Coomassie Blue staining and Western blot analysis. The α2 and α1 I-domains were expressed in E. coli BL21 and purified as described previously (25). Recombinant proteins were dialyzed against PBS, pH 7.4, and the protein concentration was determined using an extinction coefficient for each protein.
Gel Migration Analysis
SDS-PAGE analysis was used to determine multimer formation of collagen-like proteins as described (4). Briefly, proteins were denatured by incubation at 95 °C for 5 min in the presence of 0.1% SDS and 2% β-mercaptoethanol. Nondenatured samples were incubated in 5% glycerol and kept on ice prior to electrophoresis on 12% SDS gels. Gels were stained with Coomassie Blue, and protein migration as it corresponds to size was determined using protein standards. Experiments were replicated at least three times.
Circular Dichroism Spectroscopy
Circular dichroism spectra of protein samples in water were recorded on a Jasco J720 spectropolarimeter in a thermostatically controlled cuvette with a 0.5-mm path length. Data were collected at ambient temperature in a wavelength range from 250 to 190 nm and integrated for 1 s at 0.2-nm intervals with a bandwidth of 1 nm. For each spectrum, 10 scans were averaged, and the contribution from the buffer was subtracted. For thermal transition experiments, the ellipticity at 220 nm was monitored as the sample temperature was increased from 25 to 45 °C, with an average temperature slope of 10 °C/h. Each independently prepared batch of protein was analyzed.
Solid Phase Binding Assays
Microtiter wells were coated with 1 μg per well of Scl2, Scl2GLPGER, Scl2GFPGER, Scl2GFPGEN, or rat tail-derived collagen type I (Cultrex R&D) in PBS containing 1 mm MgCl2 or 1 mm EDTA overnight at 4 °C. The samples were blocked with PBS containing 1% BSA (w/v) for 1 h. 5 μm α1 or α2 I-domains were added to the wells and incubated for 2 h at room temperature. A mouse monoclonal anti-His-HRP conjugate (Alpha Diagnostics) followed by SigmaFast OPD (Sigma) was used to detect bound I-domains. The absorbance at 450 nm was measured using a Thermomax plate reader (Molecular Devices Corp, Menlo Park, CA). Experiments were replicated in triplicate.
Surface Plasmon Resonance (SPR)-based Binding Assays
Experiments were performed at 25 °C on a Biacore 3000 (GE Healthcare Biacore Life Sciences, Uppsala, Sweden). A ligand-capturing surface was prepared by immobilizing ∼8000 resonance units of StrepMAB-Immo (IBA GmbH, Germany), a monoclonal antibody against the Strep-tag, on each flow cell of a CM5 chip (GE Healthcare/Biacore). Briefly, 35 μl of antibody (10 μg/ml in 10 mm sodium acetate, pH 5.0) was injected onto a 7-min activated surface at a flow rate of 5 μl/min. The recombinant Scl Strep-tag II fusion proteins were then captured to the antibody surface by injecting 10 μg/ml of protein in a binding buffer (10 mm HEPES, pH 7.3, 150 mm NaCl, 5 mm β-mercaptoethanol, 1 mm MgCl2, 0.005% Tween 20) until a density of ∼600 resonance units was reached. The control protein, Scl2, was captured to the first flow cell and served as a reference surface. All binding experiments were carried out in the running buffer at a flow rate of 20 μl/min. After dissociation, bound integrin was removed by injection of 10 mm EDTA in running buffer without MgCl2. All measurements were baseline-corrected by subtracting the response obtained with the reference surface. For comparing purposes, SPR2 response curves from different density surfaces were rescaled to the response equivalent to that generated by every 1000 resonance units of ligand captured.
Cell Culture
Human lung fibroblast cells, MRC5 and HT1080, were maintained in DMEM with 10% FBS. Human umbilical vein endothelial cells (HUVECs) and mouse vascular smooth muscle cells were maintained in EGM2 medium with 2% FBS and in SMGM-2 medium with 5% FBS, respectively (Clonetics). Mouse myoblast C2C12 cells, C2C12-α1 cells stably expressing the human α1 integrin subunit, and C2C12-α2 cells stably expressing the human α2 integrin subunit were kindly provided by Dr. Donald Gullberg (University of Bergen, Bergen, Norway) and maintained in DMEM with 10% FBS in the absence or presence of 1 mg/ml geneticin (Invitrogen) for C2C12-α1 cells and 10 μg/ml of puromycin (InvivoGen) for C2C12-α2 cells.
Cell Adherence Analysis
Microtiter wells were coated with 1 μg per well or 0.0–10 μg per well of Scl2, Scl2GLPGER, Scl2GFPGER, Scl2GFPGEN, or collagen type I overnight at 4 °C. The wells were washed with PBS and incubated with blocking buffer (PBS containing 1% (w/v) BSA) for 1 h at room temperature. Cells were kept overnight in serum-free medium containing 1 mm CaCl2 and 1 mm MgCl2 and then detached using 0.005% trypsin and 0.1 mm EDTA at 37 °C for 2 min. The cells were washed with PBS containing 1 mm CaCl2 and 1 mm MgCl2 and suspended in serum-free medium containing 0.2% BSA supplemented with 1 mm CaCl2 and 1 mm MgCl2. 1 × 104 cells per well were added and allowed to adhere for 60 min in atmospheric conditions of 37 °C with 5% CO2. Cells were washed three times with PBS. The attached cells were fixed with 96% ethanol for 10 min and stained with 0.1% crystal violet for 30 min at room temperature. 0.1% Triton X-100 was added to the wells to dissolve the dye, and the absorbance at 590 nm was measured using a Thermomax plate reader (Molecular Devices Corp.). Experiments were performed in triplicate and repeated at least three times.
Fluorescence Microscopy
Glass chamber slides (BD Biosciences) were coated with 1 μg per well of Scl2, Scl2GLPGER, Scl2GFPGER, Scl2GFPGEN, or collagen type I overnight at 4 °C. 1 × 104 C2C12, C2C12-α1, or C2C12-α2 cells per well were added to chamber slides and allowed to adhere for 60 min in atmospheric conditions of 37 °C with 5% CO2. Cells were washed with PBS, fixed in 3% paraformaldehyde for 10 min at room temperature, washed, blocked with 2.5% BSA, and stained with Alexa Fluor 488 conjugated phalloidin (1:250) and DAPI (1:500) for 1 h at room temperature (Invitrogen). After washing with PBS, the slides were mounted with cover slips using ProLong Gold antifade reagent (Invitrogen). Images were captured using an AxioCam MRc5 digital camera and were superimposed using the AxioVision software (Zeiss). Experiments were performed in duplicate and repeated at least three times. Representative images were selected by an unbiased observer.
Platelet Aggregation Analysis
Human blood was drawn from healthy, medication-free, donors into one-ninth of the volume of 3.8% sodium citrate. Platelet-rich plasma was obtained by centrifugation at 160 g for 20 min at 25 °C. 2 × 108/ml of platelet-rich plasma were stimulated by collagen-like proteins (2 μm) or collagen type I (0.2 μm) in an aggregometer (platelet aggregation profiler model PAP-8E, Bio/Data Corp., Horsham, PA) at 37 °C, under continuous stirring at 1000 rpm. Aggregation was monitored by optical turbidometry. To ensure that observations were not restricted to an individual platelet donor, all experiments were repeated several times using blood from different donors.
RESULTS
Engineered Integrin-binding Sequences in Collagen-like Proteins Did Not Alter the Triple Helical Structure
We used P163, a well characterized recombinant Scl2 protein, as the backbone triple helical protein for the introduction of known and putative integrin-binding sequences. Scl2 contains a globular N-terminal variable region, a collagen-like region containing 79 GXY repeats, and a short linker region followed by an engineered C-terminal Strep-tag (4).
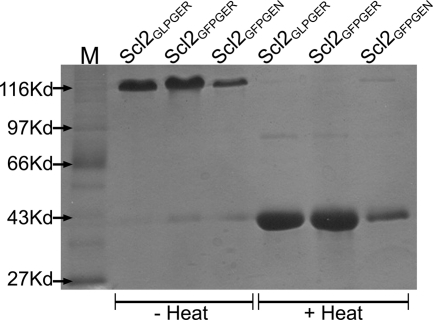
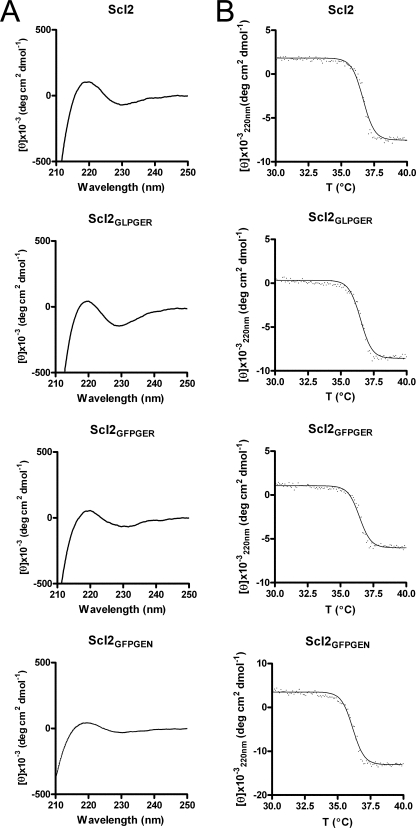
Initially, two previously identified integrin-binding sequences, GLOGER and GFOGER (24, 25), were introduced in Scl2 by site-directed mutatgenesis, except that hydroxyproline was replaced by proline. Each integrin sequence was introduced in the N-terminal 1/3 region of the collagen-like domain, and the proteins were named after their binding sequences (Scl2GLPGER and Scl2GFPGER, respectively). Scl2GLPGER and Scl2GFPGER recombinant proteins were expressed in E. coli, purified, and analyzed by SDS-PAGE under denaturing and nondenaturing conditions. In a previous report, Scl2 appeared as a trimer on SDS-PAGE gels, and the trimer was denatured to monomers after boiling (4). The engineered Scl2GLPGER and Scl2GFPGER proteins also appeared as trimers on SDS-PAGE gels and as monomers after boiling (Fig. 1). As observed with Scl2, the monomer proteins migrated slightly slower in SDS-PAGE gels than expected from their molecular weights (35 kDa calculated molecular weight migrated as a 45 kDa protein) (4). The triple helical structure of collagens and Scl proteins has a characteristic CD spectrum with a positive peak at 220 nm. CD analysis of both Scl2GLPGER and Scl2GFPGER also revealed similar spectra with the characteristic 220 nm peak, indicating that a typical collagen triple helical structure is retained in the modified Scl2 constructs (Fig. 2A). The introduction of the GLPGER or the GFPGER sequences did not alter the triple helix thermal stability of the modified Scl proteins. A thermal transition at ∼ 37 °C for both Scl2GLPGER and Scl2GFPGER was determined by monitoring the 220 nm ellipticity as a function of increasing temperature (Fig. 2B). Thus, the introduction of the integrin-binding motifs into Scl2 did not affect the triple helix structure of the proteins. These data highlight the suitability of Scl2 as a template for the introduction of biologically active collagen sequences.
FIGURE 1.
SDS-PAGE analysis reveals the presence of trimers by collagen-like proteins containing integrin-binding sequences. Coomassie-stained SDS-PAGE analysis of purified recombinant Scl2GLPGER, Scl2GFPGER, and Scl2GFPGEN. Proteins were kept at 4 °C (− Heat) or heat-treated at 95 °C for 5 min in the presence of 0.1% SDS (+ Heat) prior to electrophoresis. Trimers demonstrate decreased electrophoretic migration in comparison with monomers as determined by molecular mass standards (M).
FIGURE 2.
CD analysis demonstrates triple helical structures within Scl2, Scl2GLPGER, Scl2GFPGER, and Scl2GFPGEN. A, CD wavelength scans revealed the presence of a peak at 220 nm indicative of a triple helical structure. B, CD thermal transition monitored at 220 nm with a temperature slope of 10 °C/h revealed a transition at ∼37 °C.
Hydroxyproline in Mammalian Collagen Integrin-binding Sequences Is Not Required for Integrin Binding
GLOGER is a high affinity integrin-binding sequence that is found in the α1 chain of collagens I, II, III, and VII, and in the α2 chain of collagens I and VIII (23). Recent reports show that the GLPGER sequence found within a Scl1 protein or introduced into a Scl protein is biologically active and recognized by α2β1 and α11β1 (11, 30). Thus, the hydroxyproline residue in the GLOGER sequence is not an absolute requirement for binding to at least the α2β1 and α11β1 integrins and in these interactions can be replaced with proline.
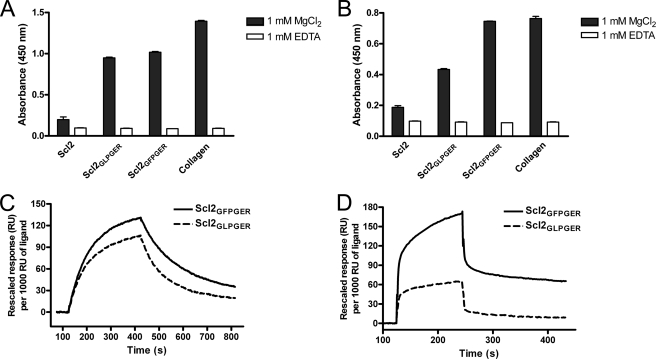
The GFOGER sequence also has been described as a high affinity binding site for α1β1 and α2β2 and is present in the α1 chain of collagens I, II, V, VII, and XI, the α2 chain of collagen XI, and in α3, α4, and α5 of collagen IV (25). We introduced the GFPGER sequence into Scl2 to determine whether hydroxyproline was required for the interaction of GFOGER with the two integrins. In solid phase binding assays, unmodified Scl2 did not bind human α1 or α2 recombinant I-domains compared with collagen type I, which binds both I-domains (Fig. 3, A and B). Both Scl2GLPGER and Scl2GFPGER were recognized by both α1 and α2 I-domains in the presence of MgCl2. As expected, the presence of EDTA abolished α1 and α2 I-domain binding to collagen type I, Scl2GLPGER, and Scl2GFPGER. These interactions were further characterized in surface plasmon resonance experiments where the modified Scl proteins were coupled to a Biacore chip. The results showed that both α1 and α2 I-domains could bind to Scl2GLPGER and Scl2GFPGER (Fig. 3, C and D), though there was a decreased amount of α2 binding to Scl2GLPGER compared with Scl2GFPGER (as also observed in ELISA-type assays). These data indicate that, similar to mammalian collagens, specific sequences within the bacterial triple helix of the Scl proteins mediate metal ion-dependent binding to the α1 and α2 I-domains. These data also show that the GLPGER and GFPGER sequences do not require hydroxyproline for binding to the α1 and α2 I-domains.
FIGURE 3.
Integrin α1 and α2 I-domains bind to Scl2GLPGER and Scl2GFPGER. Recombinant human α1 (A) and α2 (B) I-domains (5 μm) were incubated with immobilized Scl2, Scl2GLPGER, Scl2GFPGER, and collagen type I (Collagen) (1 μg/well) with 1 mm Mg2+ (black bars) or 1 mm EDTA (white bars). Integrin binding was detected with Scl2GLPGER, Scl2GFPGER, and collagen type I in the absence of EDTA indicating a metal ion-dependent binding mechanism. C and D, Biacore analysis indicated that both I-domains interacted with Scl2GFPGER and Scl2GLPGER. In an SPR-based binding assay, 0.8 μm of α1 (C) or 25 μm of α2 (D) I-domains in binding buffer (10 mm HEPES, pH 7.3, 150 mm NaCl, 5 mm β-mercaptoethanol, 1 mm MgCl2, 0.005% Tween 20) was injected over the captured Scl2GFPGER and Scl2GLPGER surface. The captured Scl2 surface was used as a control (which did not have detectable binding by either I-domain; data not shown), and SPR response was obtained by subtraction of that from a reference surface. For comparison, each response curve was rescaled to the response equivalent to that generated by every 1000 resonance units (RU) of ligand captured.
A Novel Integrin-binding Sequence Results in α1-Specific Integrin Binding
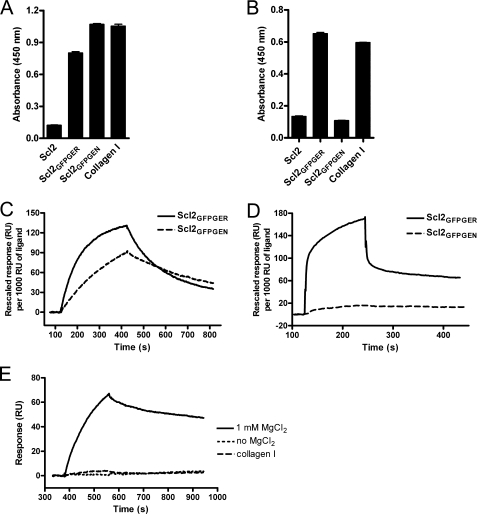
The α1 and α2 I-domains are predicted to have similar structures but differ in trench dimensions (28). This variable trench dimension may explain the difference in binding affinities between α1 and α2 for collagen types I and IV (29, 31). Therefore, we hypothesized that certain sequences may show selectivity in binding to the I-domains. We investigated this possibility using computer modeling to predict residues in the binding region that would preferentially bind α1 but not α2 (23). The sequences identified were introduced into Scl2 in the same position as the GLPGER and GFPGER sequences, and the recombinant proteins were examined for I-domain binding. One of the sequences, GFPGEN, showed selective binding to the α1 I-domain in a preliminary screen and was further characterized. The Scl2GFPGEN recombinant protein formed trimers as indicated by SDS-PAGE analysis (Fig. 1), had a CD spectrum with a positive peak at 220 nm and a melting temperature around 37 °C, suggesting that the introduction of GFPGEN into Scl2 did not affect the stability of the triple helix structure (Fig. 2). Scl2GFPGEN bound the α1 I-domain but not the α2 I-domain in solid phase binding assays (Fig. 4, A and B). Biacore analysis also indicated that Scl2GFPGEN could be recognized by α1 (Fig. 4C) but not by α2 (Fig. 4D). Furthermore, α1 I-domain binding to Scl2GFPGEN was Mg2+-dependent and could be inhibited by collagen type I (Fig. 4E). These data show that GFPGEN is a novel integrin-binding sequence that selectively binds the α1 I-domain.
FIGURE 4.
Scl2GFPGEN binds the recombinant integrin α1 I-domain. Recombinant human α1 (A) and α2 (B) I-domains were incubated with immobilized Scl2, Scl2GFPGER, Scl2GFPGEN, and collagen type I (Collagen I) (1 μg/well) with 1 mm Mg2+. α1 Binding was detected with Scl2GFPGER, Scl2GFPGEN, and collagen type I and α2 binding was detected with Scl2GFPGER and collagen type I only. C–E, Biacore analysis revealed that unlike Scl2GFPGER, Scl2GFPGEN is only recognized by the α1 I-domain, and this recognition is Mg2+-dependent and can be inhibited by collagen type I. In an SPR-based binding assay, 0.8 μm of α1 (C) or 25 μm of α2 (D) I-domains in the binding buffer were injected over the captured Scl2GFPGER and Scl2GFPGEN surfaces. E, 2 μm of α1 in the binding buffer, binding buffer without MgCl2, or in the presence of 0.5 μm collagen type I was injected over the Scl2GFPGEN surface. All SPR responses were obtained by subtraction of that from a reference surface.
Both Previously Identified and Novel Integrin-binding Sequences in Collagen-like Proteins Support Cell Attachment and Spreading
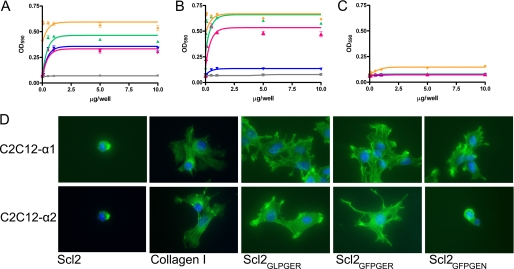
To investigate whether the identified integrin-binding sequences incorporated in Scl proteins can support the different stages of cell adhesion, we first focused on the ability of Scl2GLPGER, Scl2GFPGER, or Scl2GFPGEN to serve as substrates for cell attachment. We used mouse myoblast C2C12 cells, which in their native state lack the α-subunits of collagen-binding integrins but express the β1 subunit, and the stably transfected cells lines, C2C12-α1, expressing the human α1 subunit, and C2C12-α2, expressing the human α2 subunit, to evaluate the interactions of α1β1 and α2β1, individually. Collagen type 1, Scl2, Scl2GLPGER, Scl2GFPGER, and Scl2GFPGEN were used in increasing concentrations to coat microtiter wells and cell attachment to the collagenous surfaces was measured after 1 h incubation at 37 °C. The C2C12 cells lacking both α1 and α2 did not attach to any of the substrates (Fig. 5C). With the exception of Scl2, the C2C12-α1 cells attached to all substrates (Fig. 5A). Scl2GFPGEN served as a substrate for attachment of α1 but not α2-expressing cells (Fig. 5B). On the other hand, C2C12-α2 cells readily attached to Scl2GFPGER and Scl2GLPGER. The cell attachment profiles mimicked the protein-protein binding results with cells showing greater attachment to Scl2GFPGER when compared with Scl2GLPGER, and the attachment was Scl concentration-dependent. These data demonstrate that collagen-like proteins containing integrin-binding sequences are recognized by α1β1 and α2β1 on cell surfaces, leading to cell attachment. As a result, α1 and α2, which are usually expressed in the same cell types, may be selectively targeted by specific collagen sequences for signaling activation.
FIGURE 5.
Collagen-like proteins act as substrates for cell adherence and spreading. Increasing concentrations of Scl2 (gray square), Scl2GLPGER (pink triangle), Scl2GFPGER (green triangle), Scl2GFPGEN (purple inverted triangle), and collagen type I (yellow diamond) were coated on microtiter wells (0–10 μg/well) in duplicate. C2C12-α1 (A), C2C12-α2 (B), and C2C12 (C) cells were added to wells for 1 h and washed to remove unbound cells. Bound cells were stained with crystal violet and quantified by optical density measurement at 590 nm (A590). D, glass chamber slides were coated with Scl2, collagen type I (Collagen I), Scl2GLPGER, Scl2GFPGER, or Scl2GFPGEN (1 μg/well). C2C12-α1 (top row) or C2C12-α2 (bottom row) cells were added to wells, and unbound cells were washed after 1 h of incubation. Bound cells were stained with fluorescently tagged phalloidin (green) and DAPI (blue). Bound cells demonstrated a spread morphology as apposed to a rounded morphology. Details are as described under “Experimental Procedures.” The representative experiments and images are shown.
To further investigate adherence of cells to the prokaryotic collagen-like proteins, we analyzed the spreading and morphology of adhered cells on different substrates using fluorescence microscopy. Nuclei and actin were both visualized. (Nuclei were stained with DAPI (blue) and actin with Alexa Fluor 488-tagged phalloidin (green).) The results were in line with the cell attachment assays (shown in representative merged images, Fig. 5D). The few cells that remained on the Scl2 substrate and were not washed away had a round appearance. Collagen type I, Scl2GLPGER, and Scl2GFPGER supported the spreading of both C2C12-α1 and C2C12-α2 cells. Scl2GFPGEN was a substrate for cell spreading of α1-expressing cells but not α2-expressing cells. Adhered cells demonstrated a spread morphology in comparison with a rounded shape.
Interactions of Collagen-like Proteins Containing Integrin-binding Sequences with Different Cell Types
We next investigated the ability of modified Scl proteins to support the adherence of a collection of commonly used cell lines, including the human fibroblast cell lines, HT10180 and MRC5, HUVECs, and mouse vascular smooth muscle cells. Fibroblasts and endothelial cells express both α1β1 and α2β1; however, the expression profile of α1β1 and α2β1 on smooth muscle cells is dependent on cell phenotype and therefore is not clearly defined (11, 32–34).
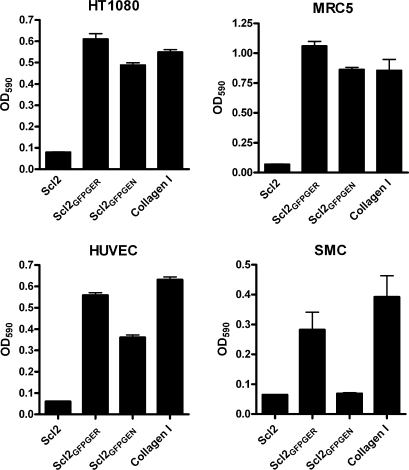
The cell types tested did not attach to an unmodified Scl2 substrate in significant numbers (Fig. 6). Both HT1080 and MRC5 cells attached to modified Scl proteins at levels similar to collagen type I. Endothelial cells attached to Scl2GFPGER at levels similar to collagen type I but attached less efficiently to Scl2GFPGEN (65% of GFPGER adherence). The observed reduced attachment may be due to Scl2GFPGEN acting as a ligand for only α1β1 and may indicate that the α1-dependent interaction is not enough for maximal adherence of HUVECs. Smooth muscle cells attached to Scl2GFPGER and collagen type 1; however, attachment to Scl2GFPGER was reduced by ∼25% in comparison with collagen type 1. Attachment of smooth muscle cells to Scl2GFPGEN was not observed. Adhered cells demonstrated a spread morphology as observed with the C2C12-transfected cell lines. These results demonstrated that integrin-binding sequences introduced into collagen-like proteins are recognized by the receptors on a variety of cell lines. Furthermore, sequences selectively recognized by specific integrins could potentially be used in biomaterials supporting the adherence of specific cell types.
FIGURE 6.
Human fibroblast, endothelial, and smooth muscle cells adhere to collagen-like protein substrates. Recombinant proteins Scl2, Scl2GFPGER, and Scl2GFPGEN and collagen type I (Collagen I) were coated on microtiter wells (1 μg/well) in triplicate. Human fibroblast cells (HT10180), human fibroblast cells (MRC5), HUVECs, and mouse vascular smooth muscle cells (SMC), were allowed to adhere to substrates for 1 h, and unbound cells were washed away. Bound cells were stained with crystal violet and quantified by measurement at 590 nm. Details are as described under “Experimental Procedures.” A representative experiment is shown.
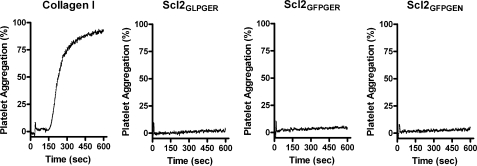
Collagen type I induces platelet activation and aggregation. The binding of collagen type I to α2β1 contributes to activation, but binding is not sufficient to initiate activation (21, 35). In view of these observations, we investigated whether the modified Scl proteins could induce platelet aggregation. Human platelet-rich plasma was used to assess aggregation by measuring optical density during a 10-min time frame. As expected, collage type I aggregated platelets to >80% after 5 min of incubation and aggregation rose to >90% aggregation at the completion of the assay. Collagen-like proteins, regardless of integrin-binding sequence, did not aggregate platelets (Fig. 7). Scl2GLPGER, Scl2GFPGER, and Scl2GFPGEN demonstrated <5% aggregation, regardless of a 10-fold increase in protein concentration compared with collagen type I. Despite the ability of α2β1 to bind to Scl2GLPGER and Scl2GFPGER, platelets were not aggregated. This suggests that these collagen-like proteins are not sufficient to induce platelet aggregation.
FIGURE 7.
Collagen-like proteins do not aggregate platelets. Collagen type I (Collagen I), Scl2GLPGER, Scl2GFPGER, and Scl2GFPGEN were tested as agonists for platelet aggregation. Briefly, platelet-rich plasma was obtained and stimulated by collagen-like proteins (2 μm) or collagen type I (0.2 μm) in an aggregometer under stirring conditions for 10 min. Platelet aggregation (y axis) was measured by optical turbidometry over time (x axis). A representative experiment is shown.
DISCUSSION
In this report, we examined the potential use of bacteria-derived, collagen-like proteins as hosts for bio-specific collagen guest sequences. We found that introducing integrin-binding sequences, which are two GXY triplets in length, into Scl2 does not disrupt triple helix formation or stability (24–26). Each integrin-binding sequence retains the consensus GXY repeats and likely contributes to their acceptance within the triple helix. The collagen-like protein used in these studies, Scl2, contains a noncollagenous N-terminal globular domain that is required for refolding after denaturation (4, 14). The globular domain did not affect integrin interactions with the collagen-like domain.
Identified mammalian collagen-integrin recognition sequences contain hydroxyproline. It has been reported that less hydroxyproline within recombinant collagen type I resulted in decreased α1β1 binding (36). The ability of bacteria-derived proteins containing GLPGER to interact with integrin α2 and α11 I-domains has been reported (30). We report here that the α1 I-domain recognizes GLPGER introduced into a collagen-like protein, as well as two additional sequences, GFPGER and GFPGEN. These data indicate that hydroxyproline is not an absolute requirement for collagen-integrin interactions.
Structural studies of the α2 I-domain and a synthetic collagen-like peptide carrying the integrin-binding sequence, GFOGER, indicated the glutamate residue coordinates the metal ion and the arginine residue interacts with the I-domain residue Asp219 (27, 37, 38). These studies also suggest that phenylalanine facilitates surface packing but may be replaced by other hydrophobic residues. It is therefore not surprising that the sequence GFPGER, which previously was shown to be an integrin ligand when presented as a synthetic triple helix peptide, is recognized by α2 (as well as α1) when present in Scl2.
We created an α1-specific binding protein, Scl2GFPGEN. A BLAST search of GFPGEN sequence within mammalian proteins did not result in identification of the sequence in human, mouse, pig, or bovine proteins (39). A single residue change Arg → Asn significantly reduces binding of GFPGER to α2. The change in binding may be related to the different trench sizes and hydrophobicity of the α1 and α2 I-domains (28). Further characterization of this protein may provide a clean background for the elucidation of α1β1-specific cell responses and the intracellular signaling pathways that mediate these functions.
We assayed for the ability of prokaryotic collagen-like proteins containing mammalian integrin-binding sequences to act as substrates for cell adherence. We used C2C12-α1 and C2C12-α2 cells to determine whether collagen-like proteins containing integrin-binding sequences would serve as substrates for cell attachment. The cell attachment profiles closely mimicked the results from the protein-protein assays, where C2C12-α2 cells attached selectively to Scl2GFPGER and C2C12-α1 cells attached to both Scl2GFPGER and Scl2GFPGEN. The utilization of C2C12 myoblasts allows us to attribute cell attachment to a single integrin subunit. Cell attachment gives rise to several points of observation. The first is that the collagen-like proteins can interact with intact integrins on cell surfaces as opposed to recombinant I-domains resulting in cell attachment to the modified Scl substrate. Secondly, initial cell attachment is followed by induced cellular responses, including cell spreading, indicating that the integrin-Scl interaction induces appropriate cell signaling that results in cytoskeletal rearrangements. Though this is a relatively crude measurement of integrin activation and signaling, it provides precedence for further investigation of these processes. Humstoe et al. (11) addressed whether Scl1, which contains GLPGER, induced integrin-mediated signaling and found that fibroblasts showed increased phosphorylation levels of focal adhesion kinase, paxillin, Crk-associated substrate, and JNK. Though the conclusions were drawn to indicate α2 involvement, the fibroblasts used in the study likely expressed both α1 and α2 (11). Our results suggest that these signaling levels could have resulted from a combination of α1- and α2-mediated signaling. It will be interesting to evaluate Scl2GFPGER- and Scl2GFPGEN-induced signaling pathways in different cell types.
The presence of GFPGER or GFPGEN in collagen-like proteins also was sufficient to support adherence of other cultured mammalian cells. Fibroblasts showed similar adherence levels to all modified Scl2 proteins and collagen type I. Endothelial cells adhered to Scl2GFPGER and Scl2GFPGEN, although at a reduced number compared with collagen type I. Smooth muscle cells adhered to both Scl2GFPGER and collagen type I but not to Scl2GFPGEN. Perhaps the decrease in adherence to Scl2GFPGER (∼25% reduction in cell number) and the lack of adherence observed with Scl2GFPGEN is due to minimal or no expression of active α1β1. The integrin expression profiles of smooth muscle cells are thought to depend on cell phenotype as well as environmental factors (11, 32–34).
Platelet activation and aggregation make up a multistep process. α2β Integrin and GPIV are the major collagen receptors on platelet surfaces (21, 35). Collagen peptides containing GFOGER are capable of binding to platelet surfaces (40). Collagen peptides containing GFOGER also bound and induced signaling events involved in cell spreading, but this pathway is not detectable in suspended cells (41). The bacterial collagen-like proteins with α2β1 recognition sequences are not capable of inducing platelet aggregation. It remains to be tested whether α2β1 on platelet surfaces is engaged by bacterial collagens with GFPGER and GLPGER.
The idea of utilizing bacteria-derived collagen-like proteins for biomaterials has been proposed previously (9). The development of the modified Scl2 proteins described in this study may provide novel materials suitable for manipulation of cell adherence, proliferation, and differentiation, which can be employed in the field of tissue engineering (42). The characteristics of Scl2GFPGEN are ideal for blood contacting vascular materials (17, 43). Medical devices within vascular tissue need to overcome several hurdles, including efficient endothelialization, limiting intimal layer thickening, decreasing thrombosis, and appropriate burst strength. The utilization of Scl2GFPGEN as a blood-contacting surface coating in combination with other biomaterials may achieve this by limiting smooth muscle cell adherence, limiting platelet activation, and promoting efficient endothelial cell adherence and spreading (17).
In conclusion, we present a novel system for the study of collagen-integrin interactions. These modified Scl2 proteins may be used as substrates for cell adherence and demonstrate cell specificity. The utilization of these modified Scl2 proteins could lead to advances in biomaterial development and may provide an insight into the individual contribution of α1β1 and α2β1 with regard to cellular behavior.
Footnotes
- SPR
- surface plasmon resonance
- HUVEC
- human umbilical vein endothelial cell.
REFERENCES
- 1.Leitinger B., Hohenester E. (2007) Matrix Biol. 26, 146–155 [DOI] [PubMed] [Google Scholar]
- 2.Hulmes D. J. (1992) Essays Biochem. 27, 49–67 [PubMed] [Google Scholar]
- 3.Jimenez S. A., Dehm P., Olsen B. R., Prokop D. J. (1973) J. Biol. Chem. 248, 720–729 [PubMed] [Google Scholar]
- 4.Xu Y., Keene D. R., Bujnicki J. M., Höök M., Lukomski S. (2002) J. Biol. Chem. 277, 27312–27318 [DOI] [PubMed] [Google Scholar]
- 5.Sylvestre P., Couture-Tosi E., Mock M. (2002) Mol. Microbiol. 45, 169–178 [DOI] [PubMed] [Google Scholar]
- 6.Karlström A., Jacobsson K., Flock M., Flock J. I., Guss B. (2004) Vet. Microbiol. 104, 179–188 [DOI] [PubMed] [Google Scholar]
- 7.Caswell C. C., Han R., Hovis K. M., Ciborowski P., Keene D. R., Marconi R. T., Lukomski S. (2008) Mol. Microbiol. 67, 584–596 [DOI] [PubMed] [Google Scholar]
- 8.Caswell C. C., Lukomska E., Seo N. S., Höök M., Lukomski S. (2007) Mol. Microbiol. 64, 1319–1331 [DOI] [PubMed] [Google Scholar]
- 9.Han R., Zwiefka A., Caswell C. C., Xu Y., Keene D. R., Lukomska E., Zhao Z., Höök M., Lukomski S. (2006) Appl. Microbiol. Biotechnol. 72, 109–115 [DOI] [PubMed] [Google Scholar]
- 10.Hoe N. P., Lukomska E., Musser J. M., Lukomski S. (2007) FEMS Microbiol. Lett. 277, 142–149 [DOI] [PubMed] [Google Scholar]
- 11.Humtsoe J. O., Kim J. K., Xu Y., Keene D. R., Höök M., Lukomski S., Wary K. K. (2005) J. Biol. Chem. 280, 13848–13857 [DOI] [PubMed] [Google Scholar]
- 12.Mohs A., Silva T., Yoshida T., Amin R., Lukomski S., Inouye M., Brodsky B. (2007) J. Biol. Chem. 282, 29757–29765 [DOI] [PubMed] [Google Scholar]
- 13.Han R., Caswell C. C., Lukomska E., Keene D. R., Pawlowski M., Bujnicki J. M., Kim J. K., Lukomski S. (2006) Mol. Microbiol. 61, 351–367 [DOI] [PubMed] [Google Scholar]
- 14.Yoshizumi A., Yu Z., Silva T., Thiagarajan G., Ramshaw J. A., Inouye M., Brodsky B. (2009) Protein Sci. 18, 1241–12451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadler K. E., Holmes D. F., Trotter J. A., Chapman J. A. (1996) Biochem. J. 316, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng Y. Y., Yoshizumi A., Danon S. J., Glattauer V., Prokopenko O., Mirochnitchenko O., Yu Z., Inouye M., Werkmeister J. A., Brodsky B., Ramshaw J. A. (2010) Biomaterials 31, 2755–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosgriff-Hernandez E., Hahn M., Russell B., Wilems T., Munoz-Pinto D., Browning M. B., Rivera J., Höök M. (2010) Acta Biomater., in press [DOI] [PubMed] [Google Scholar]
- 18.Pozzi A., Wary K. K., Giancotti F. G., Gardner H. A. (1998) J. Cell Biol. 142, 587–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zweers M. C., Davidson J. M., Pozzi A., Hallinger R., Janz K., Quondamatteo F., Leutgeb B., Krieg T., Eckes B. (2007) J. Invest. Dermatol. 127, 467–478 [DOI] [PubMed] [Google Scholar]
- 20.San Antonio J. D., Zoeller J. J., Habursky K., Turner K., Pimtong W., Burrows M., Choi S., Basra S., Bennett J. S., DeGrado W. F., Iozzo R. V. (2009) Am. J. Pathol. 175, 1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munnix I. C., Gilio K., Siljander P. R., Raynal N., Feijge M. A., Hackeng T. M., Deckmyn H., Smethurst P. A., Farndale R. W., Heemskerk J. W. (2008) J. Thromb. Haemost. 6, 2132–2142 [DOI] [PubMed] [Google Scholar]
- 22.Cosemans J. M., Iserbyt B. F., Deckmyn H., Heemskerk J. W. (2008) J. Thromb. Haemost. 6, 1253–1261 [DOI] [PubMed] [Google Scholar]
- 23.Kim J. K., Xu Y., Xu X., Keene D. R., Gurusiddappa S., Liang X., Wary K. K., Höök M. (2005) J. Biol. Chem. 280, 32512–32520 [DOI] [PubMed] [Google Scholar]
- 24.Knight C. G., Morton L. F., Peachey A. R., Tuckwell D. S., Farndale R. W., Barnes M. J. (2000) J. Biol. Chem. 275, 35–40 [DOI] [PubMed] [Google Scholar]
- 25.Xu Y., Gurusiddappa S., Rich R. L., Owens R. T., Keene D. R., Mayne R., Höök A., Höök M. (2000) J. Biol. Chem. 275, 38981–38989 [DOI] [PubMed] [Google Scholar]
- 26.Raynal N., Hamaia S. W., Siljander P. R., Maddox B., Peachey A. R., Fernandez R., Foley L. J., Slatter D. A., Jarvis G. E., Farndale R. W. (2006) J. Biol. Chem. 281, 3821–3831 [DOI] [PubMed] [Google Scholar]
- 27.Emsley J., Knight C. G., Farndale R. W., Barnes M. J. (2004) J. Mol. Biol. 335, 1019–1028 [DOI] [PubMed] [Google Scholar]
- 28.Rich R. L., Deivanayagam C. C., Owens R. T., Carson M., Höök A., Moore D., Symersky J., Yang V. W., Narayana S. V., Höök M. (1999) J. Biol. Chem. 274, 24906–24913 [DOI] [PubMed] [Google Scholar]
- 29.Calderwood D. A., Tuckwell D. S., Eble J., Kühn K., Humphries M. J. (1997) J. Biol. Chem. 272, 12311–12317 [DOI] [PubMed] [Google Scholar]
- 30.Caswell C. C., Barczyk M., Keene D. R., Lukomska E., Gullberg D. E., Lukomski S. (2008) J. Biol. Chem. 283, 36168–36175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Käpylä J., Ivaska J., Riikonen R., Nykvist P., Pentikäinen O., Johnson M., Heino J. (2000) J. Biol. Chem. 275, 3348–3354 [DOI] [PubMed] [Google Scholar]
- 32.Deb A., Skelding K. A., Wang S., Reeder M., Simper D., Caplice N. M. (2004) Circulation 110, 2673–2677 [DOI] [PubMed] [Google Scholar]
- 33.Graf K., Kappert K., Stawowy P., Bokemeyer J., Blaschke F., Schmidt G., Kintscher U., Goetze S., Fleck E. (2003) J. Cardiovasc. Pharmacol. 41, 89–96 [DOI] [PubMed] [Google Scholar]
- 34.Ogle B. M., Mooradian D. L. (1999) Tissue Eng. 5, 387–402 [DOI] [PubMed] [Google Scholar]
- 35.Farndale R. W., Slatter D. A., Siljander P. R., Jarvis G. E. (2007) J. Thromb. Haemost. 5, 220–229 [DOI] [PubMed] [Google Scholar]
- 36.Perret S., Eble J. A., Siljander P. R., Merle C., Farndale R. W., Theisen M., Ruggiero F. (2003) J. Biol. Chem. 278, 29873–29879 [DOI] [PubMed] [Google Scholar]
- 37.Bella J., Berman H. M. (2000) Structure 8, R121–126 [DOI] [PubMed] [Google Scholar]
- 38.Emsley J., Knight C. G., Farndale R. W., Barnes M. J., Liddington R. C. (2000) Cell 101, 47–56 [DOI] [PubMed] [Google Scholar]
- 39.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siljander P. R., Hamaia S., Peachey A. R., Slatter D. A., Smethurst P. A., Ouwehand W. H., Knight C. G., Farndale R. W. (2004) J. Biol. Chem. 279, 47763–47772 [DOI] [PubMed] [Google Scholar]
- 41.Inoue O., Suzuki-Inoue K., Dean W. L., Frampton J., Watson S. P. (2003) J. Cell Biol. 160, 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lutolf M. P., Hubbell J. A. (2005) Nat. Biotechnol. 23, 47–55 [DOI] [PubMed] [Google Scholar]
- 43.Burkel W. E. (1988) Med. Prog. Technol. 14, 165–175 [PubMed] [Google Scholar]