Abstract
Background
Merkel cell carcinoma (MCC) is a rare skin cancer associated with immunosuppression and the integration of Merkel cell polyomavirus (MCPyV) DNA into the tumor cell genome. Little is known about the natural history of MCPyV infection.
Objectives
To investigate the presence of MCPyV, BK and JC polyomaviruses in serum and urine from immunosuppressed kidney transplant patients (KTx) and a control group of normal volunteers.
Study design
Quantitative real time PCR (qPCR) was used to assess MCPyV, BKV and JCV viral load in urine and serum samples collected from normal donors (Group A), prospectively enrolled KTx patients (Group B) and from KTx with documented BK reactivation and/or nephropathy (Group C).
Results
Low levels of MCPyV viruria was seen in 15% of the subjects in Group A, 30% of Group B, and was not detected in Group C. No individuals in the study developed MCPyV viremia. BK viruria was seen in 5% of Group A, 30% of Group B, and 100% of Group C. Consistent with previous reports, the mean BKV urinary load was significantly higher in immunosuppressed patients compared to non-immunosuppressed controls and also higher in urine compared to serum samples.
Conclusions
Like BKV and JCV, MCPyV is likely a common infection in adult humans. Low level shedding of MCPyV in urine was similar in immunosuppressed organ transplant recipients to non-immunosuppressed subjects. However, MCPyV was not detected and JCV was infrequent in samples from KTx patients with clinical BKV reactivation.
Keywords: Merkel cell polyomavirus, BK virus, JC virus, kidney transplantation
1. Background
Merkel cell carcinoma (MCC) is an uncommon but aggressive malignant skin cancer of the elderly and immunocompromised. While rare, the incidence of MCC is increasing(1) and it frequently has a poor prognosis(2). MCC occurs more frequently in organ transplant recipients and the ratio of MCC to melanoma is higher in this group than in the general population(3,4). A novel human polyomavirus, Merkel cell polyomavirus (MCPyV), was identified recently in samples of MCC by digital transcriptome subtraction(5). MCPyV was detected in 8 of 10 human MCC and in addition, clonal integration of the viral DNA in was seen in 6 of 8 cases of MCPyV-positive MCC(5). Several reports from the United States, Europe, and Australia found MCPyV in MCC cases but only rarely in other cancers(6–13). Based on this evidence, MCPyV has been suggested to be a contributing factor in the pathogenesis of Merkel cell carcinoma(5). Subsequent PCR studies have reported that MCPyV is widespread at low levels in various human tissues from individuals without MCC(14) and was detected in forehead swabs from 62% of healthy subjects(15). Currently the natural history of MCPyV infection in humans is not well understood. Other polyomaviruses, including BK (BKV), JC (JCV), and simian virus 40 (SV40) viruses, have been implicated as etiologic agents of human cancer, but definitive links to oncogenesis remain controversial(16).
Kidney transplant recipients (KTx) are at risk of polyomavirus reactivation and polyomavirus-induced renal dysfunction. Nephropathy associated with polyomaviruses has been reported in 1%–10% of renal-transplant recipients, and it frequently leads to loss of the allograft(17). BKV is the infectious agent commonly implicated in nephropathy, although renal dysfunction related to JCV or SV40 has also been described(18,19). BKV establishes viral latency in the urogenital tract after initial infection occurs in childhood via respiratory route(20). Impaired BKV-specific immune control due to therapeutic immunosuppression appears to lead to a high titer of BK viral DNA in the urine which may be followed by viremia and is associated with BK nephropathy(21–23).
2. Objectives
The aim of this study was to simultaneously quantify and compare MCPyV, BKV and JCV viral loads in urine and blood samples obtained from immunosuppressed KTx, KTx with documented BKV reactivation, and a control group of normal donors.
3. Study design
3.1. Study participants and sample collection
Urine samples were obtained at a single timepoint from 20 healthy anonymous donors (Group A), and serum (82 samples) and urine (70 samples) at monthly intervals from 20 KTx prospectively enrolled at the time of transplant (Group B) and from 5 KTx (15 urine and 15 serum samples) enrolled at the time of documented BKV reactivation in PCR screening assays performed as part of standard patient care at UCLA (Group C). All KTx gave informed consent and samples were collected in accordance with clinical protocols approved by the UCLA and COH IRBs. Blood samples were collected in Vacutainer tubes, and the serum fraction separated out by low speed centrifugation within 24 h.
3.3. PCR cloning of VP1 fragment of MCPyV
To generate a reference clone of MCPyV to be used as a standard for quantitative PCR, a standard nested PCR was performed to amplify a fragment of the VP1 region (140 bp) of MCPyV using primers designed by reference to the MCPyV 339 sequence (NCBI gene accession number EU375804). The first (outside) PCR amplification employed primers MCV VP1AF and MCV VP1BR (Table 1) and as template 5ul of total DNA extracted from the urine of KTx patients. The second (inside) amplification was performed using 5ul of the outside amplification reaction mixture as template and primers MCV VP1BF and MCV VP1BR. Cycling scheme was 5 min at 95°C followed by 40 cycles of 30s at 95°C and 30s at 60°C, and 30s at 72°C, followed by 7 min at 72°C. PCR amplicons were cloned into the vector pCR2.1 (Invitrogen), and sequenced. The resulting sequences were homologous to the MCPyV 339 genome sequence except for a single nucleotide substitution causing an isoleucine to valine change at amino acid residue 228 of the VP1 ORF.
Table 1.
Primers used for cloning and for q-PCR
| Primer | Sequence |
|---|---|
| MCV | |
| MCV VP1AF | TGTTCATTATTGGGACATGAAAAGA |
| MCV VP1BF | CAGGGCCTAGATCCACAAGCT |
| MCV VP1 BR | AACTGTAGGAGTCTGAGAGCCTGTC |
| BKV Dunlop | |
| BKV F | GCAGCTCCCAAAAAGCCAAA |
| BKV R | CTGGGTTTAGGAAGCATTCTA |
| JCV | |
| JCV F | AGAAAAGGAGAAAGGAAGGACCC |
| JCV R | TCTGTAATTGAGTCAACCCCAGTTT |
3.4. Quantitative real-time PCR assays
q-PCR was performed with a 7300 Real time PCR instrument (Applied Biosystems, Foster City, CA). Each reaction contained 5μl of purified DNA isolated from urine or serum using QIAamp Blood mini kit (Qiagen, Valencia, CA), 12.5μl Power SYBR Green Master Mix (Applied BioSystems), and 500 nM (each) forward and reverse primers in a total volume of 25 μl. Primers used were MCV VP1BF and MCV VP1BR for MCPyV, BKV F and BKV R for BKV, and JCV F and JCV R for JCV q-PCR (Table 1). Thermal cycling was initiated with a first denaturation step of 10 min at 95°C, followed by 40 cycles of 95°C for 15s, 60°C for 60s. Standard curves for the quantification of BKV and JCV were constructed using serial dilutions of plasmids containing the linearized genomes of the BKV Dunlop and JCV Mad-1 strains (ATCC 45025 and 45027 respectively). Standard curves for the quantification of MCPyV were constructed using serial dilutions of the pCR2.1 MCPyV VP1 plasmid. The MCPyV primers did not amplify BKV or JCV sequences, and the BKV and JCV primers were also specific. All patient samples were tested in duplicate. Data were expressed as copies of viral DNA per milliliter of urine or serum. A lower limit of detection of 25 gc/ml in these q-PCR assays was defined for all three polyomaviruses. Standard precautions designed to prevent contamination during q-PCR were followed and multiple no-template controls were included in each run.
4. Results
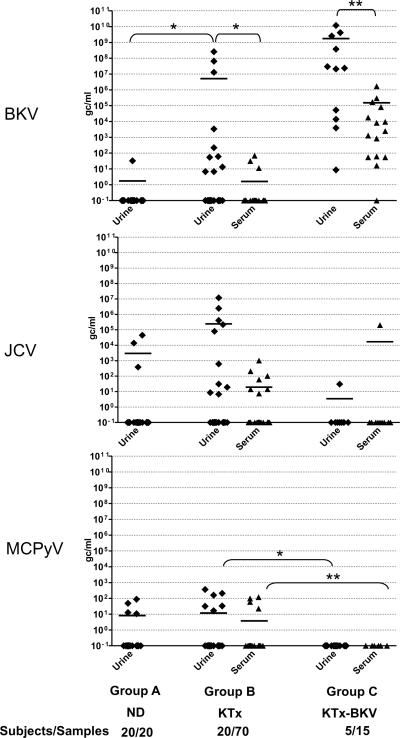
In an analysis of urine samples from 20 normal donors (Group A), MCPyV viruria was observed at low levels in 3 subjects (15%), with viral loads varying from 10 to 90 gc/ml. Low-level BK viruria was documented in 1 individual (5%), while JC viruria was seen in 3 donors (15%) with viral loads varying from 10 to 4.5 × 104gc/ml (Table 2 and Figure 1).
Table 2. Distribution of Viral Loads in Patients Studied.
Incidence indicates patients with one or more samples positive for BKV, JCV or MCPyV. Viral loads are expressed in copies per milliliter for urine and serum.
| Viruria | Viremia | ||||||
|---|---|---|---|---|---|---|---|
| Incidence | Range (viral load) | Mean | Incidence | Range (viral load) | Mean | ||
| Group A Normal donors | BKV | 1/20 (5%) | 0– 32.8 | 1.7 | ND | ||
| JCV | 3/20 (15%) | 0– 4.5×104 | 3×103 | ND | |||
| MCV | 3/20 (15%) | 0– 88.8 | 8.1 | ND | |||
| Group B KTx recipients | BKV | 6/20 (30%) | 0– 2.6×108 | 5.1×106 | 3/20 (15%) | 0– 69.5 | 1.6 |
| JCV | 5/20 (25%) | 0– 1.2×107 | 2.5×105 | 5/20 (25%) | 0– 1.02×103 | 19.42 | |
| MCV | 6/20 (30%) | 0– 3.7×102 | 11.8 | 3/20 (15%) | 0– 1.2×102 | 3.8 | |
| Group C KTx with BKV Reactivation | BKV | 5/5 (100%) | 8.7– 4.2×109 | 1.7×109 | 5/5 (100%) | 0– 1.7×106 | 1.5×105 |
| JCV | 1/5 (20%) | 0– 30.5 | 3.5 | 1/5 (20%) | 0– 2.03×105 | 1.7×104 | |
| MCV | 0/5 | 0 | 0 | 0/5 | 0 | 0 | |
Fig. 1. BK, JC and MCPy viral loads in urine and serum of normal donors (Group A n=20) prospectively enrolled KTx (Group B n=20), and KTx enrolled on clinical BKV reactivation (Group C, n=5).
Shown are the viral loads in each patient group, with each point representing one measurement. The data include multiple samples at different time points for each patient in the two transplant groups for a total of 70 urine and 82 serum samples in Group B and 15 urine and 15 serum samples in Group C. One urine sample was examined from each of the donors in Group A. Viral loads in serum are shown as triangles, viral loads in urine are shown as diamond symbols. Black bars indicate the mean viral load in each group. Data are shown as viral copies per milliliter serum or urine. To avoid bias due to repeated measurements on the same subjects, mean gc/ml were calculated for the set of samples from each study subject and these values used for statistical comparison between groups. Mann-Whitney test was used for tests of significance. * indicates a P value of <0.05, ** P< 0.01.
The study of immunosuppressed patients included both prospectively enrolled kidney transplant patients (Group B) and kidney transplant patients enrolled at the time of documented BK viremia (Group C) (see Table 2 and Figure 1). Six (30%) of the patients in Group B experienced MCPyV viruria, with viral loads in six samples from this group varying from 10 to 3.7×102 gc/ml. BK viruria was also documented in 8 samples from 6 subjects from Group B (30%), with viral loads in these 6 patients varying from 10 to 2.6×108 gc/ml. while JC viruria was found in 8 samples from 5 patients (25%), with JC viral loads varying from 10 to 1.2×107 gc/ml.
Low level MCPyV viremia (3 subjects, 15%), BKV viremia (3 subjects, 15%) and JCV viremia (5 subjects, 25%) was observed in a minority of samples from Group B. In agreement with previous reports(17,22), the BKV and JCV viral loads in serum were generally several logs lower than the urine loads in the corresponding patients. In the case of BKV, this difference was statistically significant (P<0.05).
MCPyV was not detected in the urine or serum samples of any of the five KTx enrolled due to documented BK viremia (Group C), whereas, as expected, BK viremia (up to1.7×106 copies/ml with a mean of 1.5×105) as well as viruria (8.7 to 4.2×109 copies/ml, mean 1.7×109) was observed in almost all samples from all five subjects. In this group of patients, JC viremia was only seen in 1 subject at a single timepoint, (2.03×105 copies/ml), and at a low level in urine. This absence of MCPyV in the samples from Group C compared to groups A and B was statistically significant (P<0.05) but the basis for this is unclear.
5. Discussion
Our study quantifies and compares three human polyomaviruses, MCPyV, BKV and JCV in samples from normal donors and KTx patients receiving immunosuppressive therapy. We also examined a small group of KTx who were previously identified by serum PCR as failing to control BKV. Our results indicate very low-level MCPyV and JCV shedding in the urine of normal subjects (15%) and the prospectively enrolled renal transplant recipients (30%). This indicates that despite the rarity of MCC, the associated virus, MCPyV is a common human infection, and is consistent with a recent serology study using MCPyV VP1 capsids as antigen that indicated approximately 80% of adults over 50 years of age have MCPyV antibodies(24). The presence of MCPyV DNA in urine suggests that like BKV and JCV, the virus is likely present in renal tubule or bladder epithelial cells. It is also formally possible that the low levels of MCPyV detected in the urine and serum samples reflects contamination of the skin or urethral openings with virus shed from another location. The primary site of JCV persistence is the kidney even though the site of JCV-associated clinical disease (progressive multifocal leukoencephalopathy, PML) is the central nervous system where it infects oligodendrocytes and astrocytes. In an analogous manner, MCPyV may persist in the kidney and upon reactivation in elderly or immunocompromised individuals may infect skin cells with subsequent integration and transformation. It has been shown that MCC patients with MCPyV present in their tumors have higher levels of MCPyV antibodies compared to infected healthy controls, suggesting higher levels of antigen(24).
Acknowledgements
The authors thank Eileen Kim for technical assistance.
Funding was from R21 DK077374 NIDDK to SFL.
Abbreviations
- (MCPyV)
Merkel cell polyomavirus
- (MCC)
Merkel cell carcinoma
- (JCV)
JC virus
- (BKV)
BK virus
- (SV40)
simian virus 40
- RT-PCR
real time polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest The authors have no conflicts of interest to declare.
References
- 1.Hodgson NC. Merkel cell carcinoma: changing incidence trends. J Surg. Oncol. 2005;89:1–4. doi: 10.1002/jso.20167. [DOI] [PubMed] [Google Scholar]
- 2.Swann MH, Yoon J. Merkel cell carcinoma. Semin. Oncol. 2007;34:51–56. doi: 10.1053/j.seminoncol.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 3.Penn I, First MR. Merkel's cell carcinoma in organ recipients: report of 41 cases. Transplantation. 1999;68:1717–1721. doi: 10.1097/00007890-199912150-00015. [DOI] [PubMed] [Google Scholar]
- 4.Harwood CA, McGregor JM, Swale VJ, Proby CM, Leigh IM, Newton R, Khorshid SM, Cerio R. High frequency and diversity of cutaneous appendageal tumors in organ transplant recipients. J Am. Acad. Dermatol. 2003;48:401–408. doi: 10.1067/mjd.2003.97. [DOI] [PubMed] [Google Scholar]
- 5.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foulongne V, Kluger N, Dereure O, Brieu N, Guillot B, Segondy M. Merkel cell polyomavirus and Merkel cell carcinoma. France. Emerg. Infect. Dis. 2008;14:1491–1493. doi: 10.3201/eid1409.080651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharp CP, Norja P, Anthony I, Bell JE, Simmonds P. Reactivation and mutation of newly discovered WU, KI, and Merkel cell carcinoma polyomaviruses in immunosuppressed individuals. J Infect. Dis. 2009;199:398–404. doi: 10.1086/596062. [DOI] [PubMed] [Google Scholar]
- 8.Kassem A, Schopflin A, Diaz C, Weyers W, Stickeler E, Werner M, Zur HA. Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer Res. 2008;68:5009–5013. doi: 10.1158/0008-5472.CAN-08-0949. [DOI] [PubMed] [Google Scholar]
- 9.Becker JC, Houben R, Ugurel S, Trefzer U, Pfohler C, Schrama D. MC polyomavirus is frequently present in Merkel cell carcinoma of European patients. J Invest Dermatol. 2009;129:248–250. doi: 10.1038/jid.2008.198. [DOI] [PubMed] [Google Scholar]
- 10.Duncavage EJ, Zehnbauer BA, Pfeifer JD. Prevalence of Merkel cell polyomavirus in Merkel cell carcinoma. Mod. Pathol. 2009;22:516–521. doi: 10.1038/modpathol.2009.3. [DOI] [PubMed] [Google Scholar]
- 11.Garneski KM, Warcola AH, Feng Q, Kiviat NB, Leonard JH, Nghiem P. Merkel Cell Polyomavirus Is More Frequently Present in North American than Australian Merkel Cell Carcinoma Tumors. J. Invest Dermatol. 2008 doi: 10.1038/jid.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goh S, Lindau C, Tiveljung-Lindell A, Allander T. Merkel cell polyomavirus in respiratory tract secretions. Emerg. Infect. Dis. 2009;15:489–491. doi: 10.3201/eid1503.081206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ridd K, Yu S, Bastian BC. The presence of polyomavirus in non-melanoma skin cancer in organ transplant recipients is rare. J Invest Dermatol. 2009;129:250–252. doi: 10.1038/jid.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loyo M, Guerrero-Preston R, Brait M, Hoque MO, Chuang A, Kim MS, Sharma R, Liegeois NJ, Koch WM, Califano JA, Westra WH, Sidransky D. Quantitative detection of Merkel cell virus in human tissues and possible mode of transmission. Int. J Cancer. 2010;126:2991–2996. doi: 10.1002/ijc.24737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wieland U, Mauch C, Kreuter A, Krieg T, Pfister H. Merkel cell polyomavirus DNA in persons without merkel cell carcinoma. Emerg. Infect. Dis. 2009;15:1496–1498. doi: 10.3201/eid1509.081575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moens U, Johannessen M. Human polyomaviruses and cancer: expanding repertoire. J Dtsch. Dermatol. Ges. 2008;6:704–708. doi: 10.1111/j.1610-0387.2008.06810.x. [DOI] [PubMed] [Google Scholar]
- 17.Hirsch HH, Brennan DC, Drachenberg CB, Ginevri F, Gordon J, Limaye AP, Mihatsch MJ, Nickeleit V, Ramos E, Randhawa P, Shapiro R, Steiger J, Suthanthiran M, Trofe J. Polyomavirus-associated nephropathy in renal transplantation: interdisciplinary analyses and recommendations. Transplantation. 2005;79:1277–1286. doi: 10.1097/01.tp.0000156165.83160.09. [DOI] [PubMed] [Google Scholar]
- 18.Kazory A, Ducloux D, Chalopin JM, Angonin R, Fontaniere B, Moret H. The first case of JC virus allograft nephropathy. Transplantation. 2003;76:1653–1655. doi: 10.1097/01.TP.0000090749.42791.14. [DOI] [PubMed] [Google Scholar]
- 19.Milstone A, Vilchez RA, Geiger X, Fogo AB, Butel JS, Dummer S. Polyomavirus simian virus 40 infection associated with nephropathy in a lung-transplant recipient. Transplantation. 2004;77:1019–1024. doi: 10.1097/01.tp.0000119156.52197.ca. [DOI] [PubMed] [Google Scholar]
- 20.Nickeleit V, Hirsch HH, Binet IF, Gudat F, Prince O, Dalquen P, Thiel G, Mihatsch MJ. Polyomavirus infection of renal allograft recipients: from latent infection to manifest disease. J. Am. Soc. Nephrol. 1999;10:1080–1089. doi: 10.1681/ASN.V1051080. [DOI] [PubMed] [Google Scholar]
- 21.Hirsch HH, Knowles W, Dickenmann M, Passweg J, Klimkait T, Mihatsch MJ, Steiger J. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N. Engl. J. Med. 2002;347:488–496. doi: 10.1056/NEJMoa020439. [DOI] [PubMed] [Google Scholar]
- 22.Nickeleit V, Klimkait T, Binet IF, Dalquen P, Del Z, V, Thiel G, Mihatsch MJ, Hirsch HH. Testing for polyomavirus type BK DNA in plasma to identify renal- allograft recipients with viral nephropathy. N. Engl. J. Med. 2000;342:1309–1315. doi: 10.1056/NEJM200005043421802. [DOI] [PubMed] [Google Scholar]
- 23.Binggeli S, Egli A, Schaub S, Binet I, Mayr M, Steiger J, Hirsch H. Polyomavirus BK-Specific Cellular Immune Response to VP1 and Large T-Antigen in Kidney Transplant Recipients. American Journal of Transplantation. 2007 doi: 10.1111/j.1600-6143.2007.01754.x. [DOI] [PubMed] [Google Scholar]
- 24.Tolstov YL, Pastrana DV, Feng H, Becker JC, Jenkins FJ, Moschos S, Chang Y, Buck CB, Moore PS. Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. Int. J Cancer. 2009;125:1250–1256. doi: 10.1002/ijc.24509. [DOI] [PMC free article] [PubMed] [Google Scholar]