SUMMARY
Most metazoan microRNA target sites have perfect pairing to the seed region, located near the miRNA 5’ end. Although pairing to the 3’ region sometimes supplements seed matches or compensates for mismatches, pairing to the central region has been known to function only at rare sites that impart Agronaute-catalyzed mRNA cleavage. Here we present “centered sites,” a class of microRNA target sites that lacks both perfect seed pairing and 3’-compensatory pairing and instead has 11–12 contiguous Watson–Crick pairs to the center of the microRNA. Although centered sites can impart mRNA cleavage in vitro (in elevated Mg2+), in cells they repress protein output without consequential Agronaute-catalyzed cleavage. Our study also identified extensively paired sites that are cleavage substrates in cultured cells and human brain. This expanded repertoire of cleavage targets and the identification of the centered site type help explain why central regions of many microRNAs are evolutionarily conserved.
INTRODUCTION
MicroRNAs (miRNAs) are a class of ~23 nucleotide (nt) RNAs that direct the post-transcriptional repression of protein-coding genes (Bartel, 2004). After processing from hairpin precursors, miRNAs are loaded into Argonaute-containing silencing complexes, which down-regulate mRNA targets through two distinct modes, either Argonaute-catalyzed cleavage or a second mode that involves mRNA destabilization and translational repression, at least in part through poly(A) shortening (Filipowicz et al., 2008).
Argonaute-catalyzed cleavage of the target strand occurs in the context of extensive base pairing, at the linkage joining the mRNA nucleotides that pair to miRNA positions 10 and 11 (Elbashir et al., 2001b; Hutvagner and Zamore, 2002; Yekta et al., 2004). In mammals, this slicing activity is catalyzed by Argonaute2 (AGO2), which leaves a 3’ hydroxyl on the 5’ cleavage fragment and a 5’ monophosphate on the other (Liu et al., 2004; Meister et al., 2004; Schwarz et al., 2004). Unlike miRNAs in plants, very few examples of miRNA-dependent cleavage targets have been reported in mammals (Yekta et al., 2004; Davis et al., 2005; Jones-Rhoades et al., 2006). Nonetheless, artificially designed small interfering RNAs (siRNAs) that silence target genes through this mechanism are widely used reagents, illustrating that in principle the cleavage mode of repression can function in many contexts and for many targets (Elbashir et al., 2001a).
Sites that confer slicing-independent destabilization and translational repression typically pair to the 5’ region of the miRNA, centering on miRNA nucleotides 2–7, known as the miRNA seed (Lewis et al., 2005; Bartel, 2009). Introducing an siRNA/miRNA or deleting an endogenous miRNA leads to modest yet detectable changes in the output of hundreds of genes containing seed sites in their 3’UTRs (Krutzfeldt et al., 2005; Lim et al., 2005; Giraldez et al., 2006; Grimson et al., 2007; Rodriguez et al., 2007; Baek et al., 2008; Selbach et al., 2008). Moreover, most mammalian protein-coding genes are under selection to maintain pairing to the seed of one or more miRNAs, and thousands of genes have also evolved to specifically avoid pairing to the seeds of preferentially co-expressed miRNAs (Farh et al., 2005; Lewis et al., 2005; Stark et al., 2005; Friedman et al., 2009). These observations illustrate both the broad scope of seed-type regulation and the widespread influence of this targeting mode on mRNA evolution.
Pairing to the 3’ region of the miRNA can supplement seed pairing to enhance target recognition, or it can even compensate for a mismatch to the seed; such sites are known as “3’-supplementary sites” and “3’-compensatory sites”, respectively (Bartel, 2009). However, pairing to the 3’ region appears to be consequential for relatively few (<10%) of sites (Bartel, 2009). In principle, pairing to the central region of the miRNA could also supplement pairing to the other regions of the miRNA, but a role for such pairing has been demonstrated only for sites that mediate Ago-catalyzed cleavage and not for sites that mediate destabilization and translation repression.
Here we describe “centered sites”, a unique class of microRNA target sites that lacks both perfect seed pairing and 3’-compensatory pairing and instead has 11–12 contiguous Watson–Crick pairs to miRNA nucleotides 4–15. In the process of characterizing these sites, we found that Mg2+ concentration profoundly influences both the specificity and efficacy of miRNA-directed cleavage, and we performed whole-transcriptome analyses that substantially add to the number of known instances in which metazoan miRNAs direct mRNA cleavage.
RESULTS
A Unique Class of MicroRNA Target Site
The most highly conserved region of metazoan miRNAs is the 5’ region containing the seed (Lewis et al., 2003; Lim et al., 2003), which is the region most important for recognizing most targets. The next most highly conserved region spans nucleotides 13–16, which is the region most important for 3’-supplementary and 3’-compensatory pairing (Grimson et al., 2007). Despite being less conserved than other miRNA regions, we noted that the central region of vertebrate miRNAs is significantly more conserved than is the opposite arm of the pre-miRNA hairpin (Figure 1A and S1A). Because both arms participate equally in the pairing required to form the pre-miRNA hairpin, preferential conservation of the miRNA observed in this region suggested that these central nucleotides play a role beyond that of miRNA biogenesis. One such role would be to aid in target recognition, but among the previously characterized targeting modes, the central region is known to function only for cleavage sites, which seemed too rare to provide the additional selective pressure for conserving nucleotide identity at the miRNA central regions. Therefore, we searched for another type of site that might explain this preferential conservation.
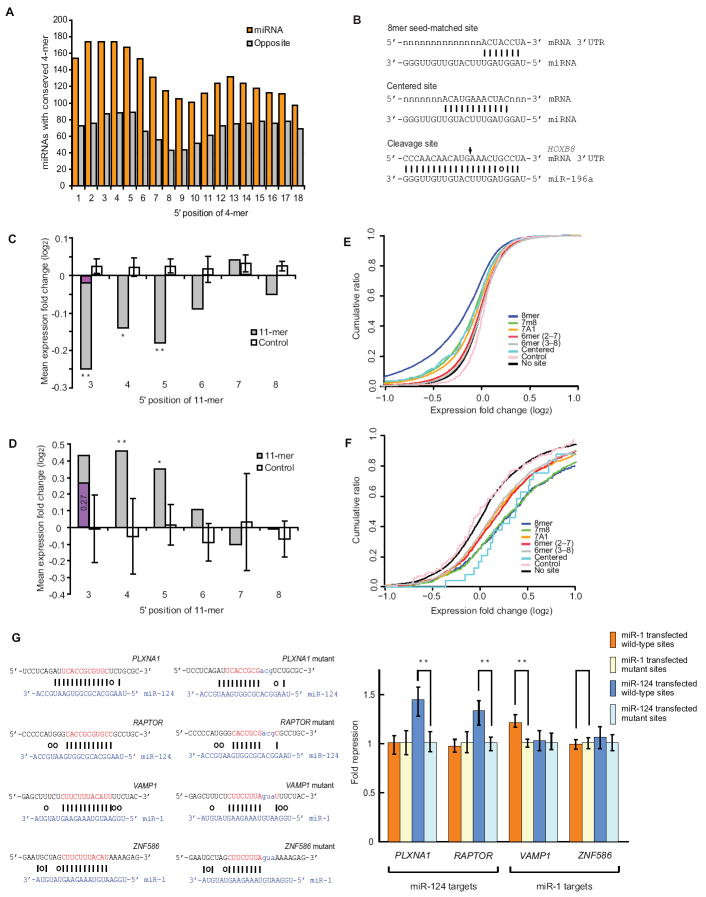
Figure 1. Centered Sites Regulate both mRNA Accumulation and Protein Output.
(A) Conservation of 4-nt segments of mammalian miRNAs. As schematized in Figure S1A, segments from the mature miRNA (orange) were compared with opposing segments from the other arm of the hairpin (grey). At each position of the mouse miRNAs, the number of segments perfectly conserved in the whole-genome alignments of the other 29 species (Blanchette et al., 2004) is plotted.
(B) Spectrum of functional miRNA target sites.
(C) Response of HeLa mRNAs with contiguous perfect pairing to 11-mer sites starting at the indicated positions of 78 transfected mi/siRNAs. Plotted are mean fold changes for mRNAs with 3’UTR sites to the cognate mi/siRNAs (gray) and the cohorts of chimeric miRNA-like controls sequences (white, error bars indicate standard deviation for 10 cohorts). Also plotted is the mean expression change for mRNAs with a shifted 6-mer 3’UTR site (purple), which includes all mRNAs with 11-mer sites starting at position 3. The 11-mer sites with repression distributions significantly different from that the no-site distribution are indicated (*, P < 0.05; ** P < 0.01; K–S test).
(D) In vivo response of mRNAs with 11-mer sites to endogenous miRNAs after loss of all miRNAs. Plotted is the fold change of zebrafish mRNAs with 11-mer sites to 21 endogenous miRNAs depleted in the dicer mutant. Otherwise as in panel (C).
(E) Reduced levels of HeLa messages with either centered sites or canonical sites to 78 transfected mi/siRNAs. Shown is analysis of microarray data, plotting cumulative changes of mRNAs with single 3’UTR sites of the indicated types. Canonical sites (8mer, 7m8, 7A1, and 6mer) had either 8, 7 or 6 nt matches centered on the miRNA seed (Bartel, 2009); centered sites had 11 contiguous Watson–Crick pairs to miRNA positions 4–14 or 5–15; control sites were centered sites to the chimeric miRNA-like control sequences (combining results for all 10 cohorts).
(F) Efficacy of endogenous centered sites in vivo. Shown is analysis of microarray data, plotting cumulative changes of zebrafish mRNAs with single 3’UTR sites to 21 miRNAs depleted in the dicer mutant. Otherwise as in panel (E).
(G) miRNA-mediated repression at centered sites. Shown is the fold repression of luciferase reporter genes fused to 3’UTR fragments of the indiciated genes with the indicated sites or mutant sites. Plotted are the geometric means, normalized to the geometric means observed for reporters with mutant sites. Error bars represent the second largest and the second smallest values among 12 replicates (from 4 independent experiments). Statistical significance is indicted (**, P value < 0.001, Wilcoxon rank-sum test). See also Figure S1 and Table S1.
Examination of array data investigating the response of mRNAs after transfecting 11 miRNAs into HeLa cells (Lim et al., 2005; Grimson et al., 2007) revealed a unique type of site that was associated with mRNA down-regulation (Figure S1B). This site type, which we call the “centered site”, was characterized by at least 11 nucleotides of contiguous Watson–Crick base pairing to the center region of the miRNA at either nucleotides 4–14 or 5–15, without substantial pairing to either the 5’ or the 3’ ends of the miRNA. Because of the location and extent of their base pairing, these sites occupy a unique position intermediate between seed sites and the extensively complementary cleavage sites (Figure 1B).
Because these sites are relatively rare, pooling of data from multiple miRNA transfections was initially required to achieve statistical significance for determining their efficacy. In order to systematically analyze these sites, we therefore compiled additional array data from HeLa experiments with similarly transfected miRNAs and siRNAs (Birmingham et al., 2006; Jackson et al., 2006a; Jackson et al., 2006b; Schwarz et al., 2006; Anderson et al., 2008). To ensure that the transfected mi/siRNAs were loaded and active within the silencing complex, the pooled datasets were restricted to those of the 78 HeLa experiments for which the canonical 8mer 3’UTR site to the transfected mi/siRNA was associated with downregulated mRNAs with high statistical significance (P < 0.0001, K–S test, Table S1A). Testing matches that did not include the canonical seed match to miRNA positions 2–7 showed that perfect 11-mer matches starting at miRNA positions 3, 4, and 5 were each significantly associated with repression (Figure 1C), whereas perfect 10-mer matches, perfect 9-mer matches, and near-perfect 11-mer matches (those with single mismatches or wobbles) were not significantly associated with repression (Figure S1C and D).
The efficacy of centered sites matching ectopically introduced miRNAs and siRNAs raised the possibility that such sites might also mediate endogenous miRNA targeting. Array results examining the effects of miRNA loss in zebrafish embryos lacking Dicer provided data on a sufficient number of messages with centered sites to enable a systematic analysis of targeting interactions in vivo. MicroRNAs present at 24 hours post fertilization (hpf), the developmental stage used for mRNA analysis, were identified by high-throughput sequencing (Table S1B). Sites were considered for 21 of these miRNAs for which the canonical 8mer 3’UTR sites were significantly associated with mRNAs derepressed in the dicer mutant (P < 0.01, Table S1B). As observed for the ectopic interactions, perfect 11-mer matches starting at miRNA positions 3, 4, and 5 were each associated with significant repression, although efficacy of sites starting at position 3 was mostly attributable to overlap with the “shifted 6mer” seed match (Figure 1D), which comprises pairing to nucleotides 3–8 (Friedman et al., 2009). Perfect 10-mer matches, perfect 9-mer matches, and near-perfect 11-mer matches generally were not associated with significant repression, although for a few of the numerous possibilities examined, marginal significance was observed (Figure S1E and F).
When considering both ectopic and endogenous interactions, contiguous Watson–Crick 3’UTR pairing to the central region of the miRNA, at either nucleotides 4–14 or 5–15, was unique among the tested possibilities in that it was both consistently associated with mRNA repression and not attributable to overlap with previously described site types. A previous array study had reported a handful of siRNA off-targets with similarly long stretches of contiguous Watson–Crick base pairing, but these sites were offset further towards the 3’ end of the miRNA, at nucleotides 6–16 or 7–17 (Jackson et al., 2003), a region not significantly associated with targeting when examined using our larger datasets. Effective miRNA-target prediction algorithms rely heavily on perfect pairing to the seed region, and thus miss this additional class of targets (Bartel, 2009).
The transfected mi/siRNAs had an average of 11 and a median of eight centered sites in 3’UTRs of human mRNAs. About a quarter of the mRNAs with a centered site lacked conventional seed sites to the transfected RNA and were sufficiently expressed in HeLa such that changes could be accurately measured on the arrays. Analysis of cumulative distributions of log-fold changes indicated that >20% of these mRNAs responded to the transfected mi/siRNAs in a manner attributable to the site, with a lower bound for site efficacy resembling that of canonical 7-mer sites (Figure 1E). Likewise, >30% of the endogenous centered sites analyzed appeared to mediate repression in zebrafish embryos (Figure 1F).
To examine whether centered sites also function in other animals, we analyzed mRNA array datasets monitoring the impact of knocking down proteins required for Drosophila miRNA biogenesis (Kadener et al., 2009). Following either Drosha or Dicer1 knockdown in Drosophila S2 cells, messages with 3’UTR centered sites matching the endogenous miRNAs had a significant propensity to be derepressed (Figure S1G and H, P = 0.00045 and 0.027, respectively, for Drosha and Dicer1 knockdown datasets, Table S1C and D).
To confirm that centered sites can be directly targeted by miRNAs, luciferase reporter constructs and their mutant counterparts with disrupted pairing were prepared and tested in both HeLa cells and S2 cells (Figure 1G and S1I). For three of the four UTR fragments tested, the sites reduced protein output in a manner that depended on the presence of both the wild-type site and the cognate miRNA. Taken together, the reporter and microarray results suggested that the centered site is a miRNA target site capable of downregulation comparable with that observed for single 7-nt seed sites. Although they are much less abundant than both seed-matched sites and sites with 3’-supplementary pairing, centered sites are present in similar numbers as 3’-compensatory sites and could help explain the preferential conservation observed in the central region of most miRNAs.
MicroRNA-directed Cleavage Detected at Centered Sites
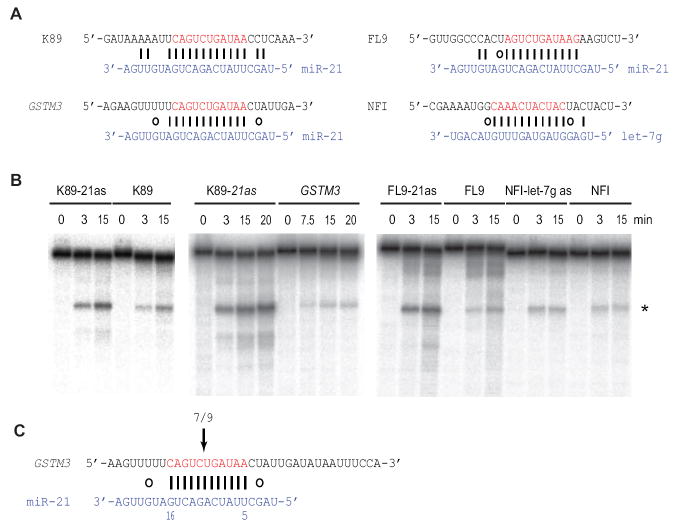
Because of pairing to the central region of the miRNA, centered sites might be subject to AGO2-dependent cleavage similar to that occurring for known cleavage sites of plants and animals, which are more extensively paired (Yekta et al., 2004; Davis et al., 2005; Jones-Rhoades et al., 2006). To test this possibility, we employed an in vitro cleavage assay using S100 extract prepared from HeLa cells (Martinez and Tuschl, 2004; Shin, 2008), focusing on mRNA fragments containing centered sites for miR-21 or let-7g miRNA, which are abundant in HeLa cells (Figure 2A, Table S4A). Cleavage was observed at the position expected for AGO2-catalyzed cleavage of the centered sites (Figure 2B).
Figure 2. MicroRNA-directed Cleavage at Centered Sites.
(A) Centered sites for miR-21 or let-7g within 3’UTR fragments of the indicated mRNAs tested in cleavage assays. K89, KIAA1189; FL9, FLJ40919; NFI, NFIA (sequences provided in Supplementary Information).
(B) Cleavage of target fragments directed by endogenous miRNAs in vitro. 5’-Cap-radiolabeled mRNA fragments were incubated in HeLa cytoplasmic S100 extract for the indicated time and analyzed on denaturing gels. As a control, fragments modified to be fully complementary to the cognate miRNA, designated as antisense (as) substrates, were tested and analyzed in parallel. Whereas most fragments were cleaved predominantly at the expected site, NFIA was cleaved at two positions (*, Figure S2), as is sometimes observed in vitro (Martinez and Tuschl, 2004).
(C) miR-21–directed cleavage of GSTM3 mRNA in HeLa cells. 5’-RACE using primers specific for GSTM3 mRNA was performed on mRNA isolated from HeLa cells treated with two siRNAs targeting XRN-1. Seven of nine sequenced clones mapped to the position expected for miR-21–directed cleavage. The other two clones mapped 52 nt downstream.
To examine whether cleavage was also occurring in the cells, we tested for miR-21-directed cleavage of GSTM3 mRNA (moderately expressed in HeLa cells), using RNA ligase–mediated rapid amplification of cDNA 5’ ends (5’-RACE). By directly cloning and sequencing the 5’ end of the 3’ cleavage product, this assay can be used to validate miRNA-directed cleavage (Llave et al., 2002; Yekta et al., 2004). To increase the sensitivity of the assay, XRN1, the 5’→3’ exonuclease responsible for degrading the 3’ cleavage product (Souret et al., 2004; Orban and Izaurralde, 2005), was knocked down (Aleman et al., 2007). 5’-RACE fragments within ~50 bp of the expected cleavage site were cloned for sequencing. The 5’ ends for seven of nine sequenced clones precisely matched that expected for cleavage at the centered site in the cell (Figure 2C). These results indicated that for an endogenous mRNA targeted at a centered site by an endogenous miRNA, at least some transcripts underwent AGO2-catalyzed cleavage in the cell.
Pairing Requirements for Cleavage are Sensitive to Mg2+ Concentration
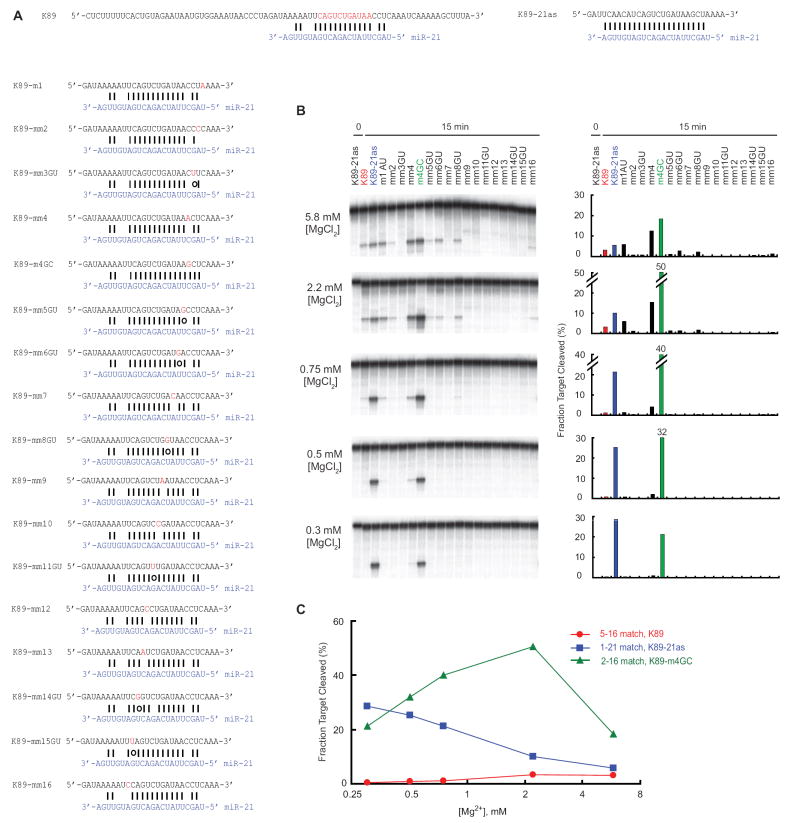
To understand the specificity of cleavage at centered sites, miR-21 recognition of the K89 mRNA fragment (Figure 2) was examined further. The K89 RNA sequence, which was perfectly complementary to positions 5–16 of miR-21, was systematically mutated at each nucleotide corresponding to miR-21 positions 1–16, substituting an A:C mismatch or a G:U wobble for each Watson–Crick match, and substituting a Watson–Crick match for each of the two mismatches (Figure 3A). When using 5.8 mM Mg2+, as in Figure 2B, or 2.2 mM Mg2+, both of which were within the ~2–6 mM range used previously to study in vitro cleavage (Martinez and Tuschl, 2004; Gregory et al., 2005; Maniataki and Mourelatos, 2005; Miyoshi et al., 2005; Rand et al., 2005; Ameres et al., 2007; Wang et al., 2009a; Wang et al., 2009b), cleavage was retained after changing positions outside of the centered site and was reduced after changing most positions within the centered site, although wobble pairs were tolerated at positions 6 and 8 (Figure 3B, top two panels).
Figure 3. Pairing Requirements for Cleavage at a Centered Site and the Influence of Mg2+ Concentration.
(A) Sequences used to examine pairing requirements for cleavage. Seqeunces were derivatives of the K89 3’UTR fragment, a miR-21 target. K89-21as, fully complementary version of K89; K89-m1, matched to position 1 of miR-21; k89-mm2, A-C mismatch at position 2; K89-mm3GU, G:U wobble pairing position 3.
(B) The influence of Mg2+ on cleavage specificity and efficacy in vitro. Reactions were performed as in Figure 2B, using the substrates depicted in (A), with the indicated Mg2+concentrations. Quantification of the fraction cleaved is plotted on the right.
(C) Plot of the effect of Mg2+ on cleavage efficacy for the model centered site (K89), a more extensively paired site (K89-m4GC), and a fully paired site (K89-21as).
Mg2+ is essential for the in vitro cleavage reaction (Schwarz et al., 2004) but also has a striking effect on the relative stabilities of matched and mismatched RNA duplexes (Serra et al., 2002). Indeed, lowering the Mg2+ concentration increases the fidelity of RNA 2’-O-methylation, another reaction specified by Watson-Crick pairing between small guide RNAs and their targets (Appel and Maxwell, 2007). We found that lowering Mg2+ gave maximal target RNA cleavage specificity and efficacy for substrates that were extensively paired to miR-21, whereas higher Mg2+ was optimal for more weakly pairing substrates (Figure 3B). For example, the cleavage of K89-21as RNA, which is fully paired to the miRNA, was the most efficient at 0.3 mM Mg2+, whereas cleavage of the wild-type K89 substrate containing the centered site with only 12 contiguous pairs was undetectable at 0.3 mM Mg2+ and most efficient at 5.8 mM Mg2+, and K89-m4GC, which had an intermediate number of contiguous pairs, had an intermediate Mg2+ optimum (Figure 3B and C).
The free-Mg2+ levels in the cytoplasm of various cells and tissues is less than 1 mM (Gunther, 2006), a concentration at which we found that efficient cleavage required pairing more extensive than that of typical centered sites (Figure 3B). Nonetheless, some cleavage at the centered site was detected at physiological Mg2+ concentrations (Figure 3B, 0.75 mM Mg2+), which explained why the 5’-RACE assay yielded fragments diagnostic of miR-21-directed cleavage in the cell (Figure 2C).
Whole-transcriptome Analysis of miRNA-directed Cleavage
The poor efficacy of cleavage at the centered site at physiological Mg2+ concentration called into question whether miRNA-directed cleavage plays a consequential role during repression mediated by centered sites and suggested that most repression at centered sites might resemble the destabilization and translational repression observed for most seed-matched targets. To better characterize the scope of miRNA and siRNA-directed cleavage in mammals, and to examine the extent to which cleavage at centered sites is relevant to target gene regulation in vivo, we applied degradome sequencing to mammalian cells. Degradome sequencing generates short sequence tags representing the 5’ ends of uncapped mRNA fragments found in the cell (Addo-Quaye et al., 2008; German et al., 2008). Although these fragments are predominantly 5’→3’ exonuclease degradation intermediates, they also include 3’ fragments of Argonaute-catalyzed mRNA cleavage in sufficient numbers to enable empirical detection of endogenous cleavage targets of plant miRNAs and siRNAs (Addo-Quaye et al., 2008; German et al., 2008). Inspired by this success in plants and the ability to detect miR-21-directed cleavage by 5’-RACE, we applied the method to HeLa cells following XRN1 knockdown by RNAi (Figure S3A).
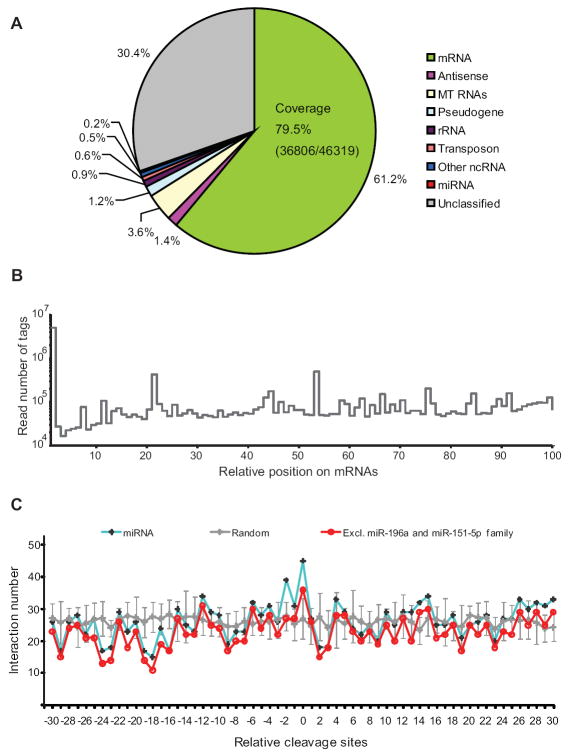
Sequencing yielded 14,323,668 tags mapping to the human genome, with a diversity of 2,069,190 unique tag sequences. Of the total tags, 61.2% came from protein-coding genes and represented 36,806 out of 46,319 ENSEMBL mRNAs (Figure 4A). The tags showed a relatively uniform distribution across the mRNAs, with a very strong peak at the 5’ terminus (Figure 4B). About 30% of tags were not classified because they did not map to mature annotated RNAs (Figure 4A). Many of these were from introns and processing fragments from pri-miRNAs, mitochondrial tRNAs, ribosomal RNAs, and snRNAs, illustrating how unstable 3’ products of endonucleases can be detected in mammalian cells by using degradome sequencing (Table S2A and B).
Figure 4. Rare MicroRNA-directed Cleavage Detected by Degradome Sequencing.
(A) Mapping of HeLa degradome sequencing tags to the transcriptome. Antisense corresponds to tags mapping antisense to annotated mRNAs, noncoding RNAs (ncRNAs), pseudogenes, and mitochondrial (MT) RNAs. Coverage indicates the fraction of the 46,319 annotated mRNAs that were represented by at least one tag. Unclassified tags mapped primarily to introns and 3’ flanking regions of ncRNAs (Table S2A).
(B) The distribution of degradome tags along the length of the mature mRNAs. mRNAs were split along their length into 100 bins, and tag 5’ ends were tallied for each bin. Shown are the aggregate tallies for all mRNAs.
(C) Search for evidence of cleavage at centered sites. Plotted are the numbers of interactions with evidence for cleavage at the expected position (0) or at positions either upstream (negative values) or downstream (positive values). Interactions were counted if a centered site matching a conserved miRNA expressed in HeLa had a tag supporting cleavage at the indicated position (blue). Analysis excluding miR-196, miR-28, miR-151-5p is also shown (red). As a control, the analysis was repeated using 10 cohorts of artificial tags, generated by randomly positioning tags on mRNAs (gray; error bars, standard deviation). See also Figure S3 and Table S2.
To determine if miRNA centered sites were associated with cleavage at the expected position within the mRNA 3’UTR, we searched for centered matches to 50 distinct, conserved miRNAs most highly expressed in HeLa cells and tabulated the frequency of degradome tags corresponding to mRNA cleavage at the tenth position of these sites. Tags corresponding to cleavage at the expected position were found much more frequently for authentic miRNA:site pairs than for negative-control pairs (Figure 4C). However, when we excluded miR-196a, miR-151, and miR-28, which target several extensively paired sites, the signal above background was greatly reduced, suggesting that most centered sites lacked the complementarity required for robust miRNA-directed cleavage (Figure 4C). The abundance of degradome tags mapping to the expected cleavage sites of the siRNAs targeting XRN1 illustrated that the method can identify tags diagnostic of AGO2-catalyzed cleavage in human cells (Figure S3). These results supported those from the in vitro cleavage assays (Figure 3B) in suggesting that under physiological Mg2+ conditions the mRNA downregulation mediated by centered sites is usually accompanied by very little AGO2-catalzyed cleavage.
Genome-wide Search for miRNA:site Duplexes with High Complementarity
Our observation of significant cleavage at the small subset of centered sites with unusually extensive complementarity to the miRNA indicated that miRNA-directed cleavage at extensively paired sites was more frequent in animals than had been appreciated. This insight prompted a systematic examination of mammalian sites with extensive miRNA complementarity of the type that would mediate cleavage in plants, but might not have fulfilled our criteria for classification as centered sites because they either had perfect seed pairing or lacked 11 contiguous pairs within positions 4–15.
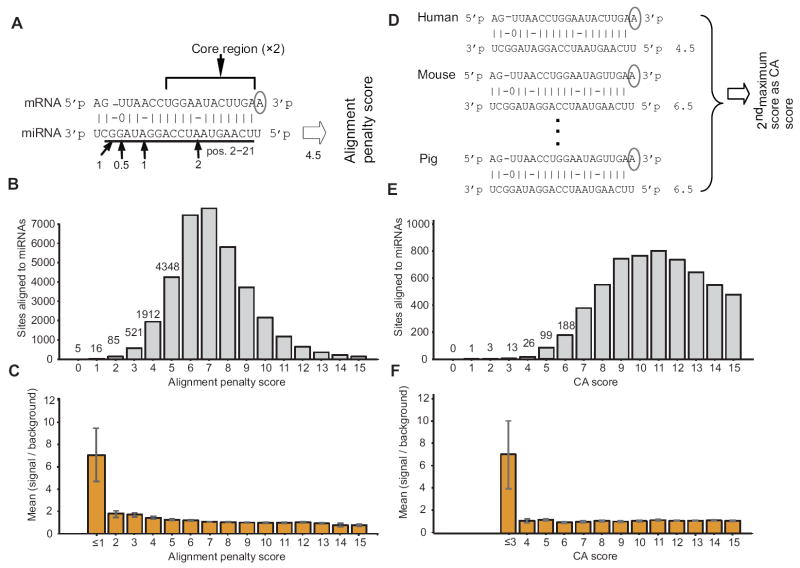
To search for potential cleavage sites in mammals, we used a scoring rubric similar to those that successfully identify miRNA target sites in plants (Figure 5A) (Jones-Rhoades and Bartel, 2004; Allen et al., 2005). The search yielded 106 predicted miRNA:site duplexes scoring ≤2.0 (Figure 5B), including 47 in annotated ORFs, 16 in 5’UTRs, and 43 in 3’UTRs (Table S3A). At the mid-to-higher penalty scores, sites were no more abundant than expected by chance, but at scores ≤3.0, sites were at least 1.5-fold enriched compared to the control sets of chimeric miRNAs constructed so as to preserve the seeds as well as the overall dinucleotide and trinucleotide compositions of authentic miRNAs (Figure 5C). Repeating the analyses with annotated murine miRNAs yielded analogous results (Figure S4C–E, Table S3B).
Figure 5. Enrichment and Conservation of miRNA:site Duplexes with Extensive Complementarity.
(A) Illustration of the alignment penalty score, used to judge the quality of pairing to miRNAs. Pairing to miRNA nucleotides 2–21 was considered, assigning a 2-point penalty for each mismatch or alignment insertion/deletion (indel) within the miRNA core (nucleotides 2–13) (Mallory et al., 2004), a 1-point penalty for each mismatch or insertion/deletion outside the core, or each G:U wobble within the core, a 0.5-point penalty for each G:U wobble outside the core. An additional 1-point penalty was assigned to sites lacking an A across from miRNA nucleotide 1 (Lewis et al., 2005).
(B) Distribution of scores for potential miRNA:site duplexes with at least seven consecutive base pairs and 13 base pairs in total. Sites were considered for the 620 distinct human miRNA/miRNA*s annotated in miRBase, version 11.0, excluding four miRNAs that paired to multiple repeat loci, skewing the distribution to the left (Figure S4B).
(C) Analysis of site enrichment. To estimate the signal-to-background ratio for each score bin, the number of miRNA:site duplexes was compared with the number of miRNA:site duplexes found when using chimeric control miRNAs (error bars, standard deviation for 10 chimeric miRNA cohort sets).
(D) Illustration of the conservation alignment (CA) score, used to identify conserved miRNA:site pairs. Alignment penalty scores were considered for human sites aligned in orthologous genomic regions of mouse, rat, dog, horse, and pig, with the second highest (second worst) among the six assigned as the CA score.
(E) Distribution of CA scores for miRNA:site duplexes. Sites were considered for 165 distinct miRNAs conserved in mammals.
(F) Analysis of preferential conservation of extensively paired sites. To estimate the signal-to-background ratio for each score bin, the fraction of miRNA:site duplexes that were conserved was compared with the fraction of analogous duplexes that were conserved when using chimeric control miRNAs, accounting for the lower abundance of matches to control sequences (error bars, standard deviation for 10 chimeric miRNA cohort sets). See also Figure S4 and Table S3.
The higher abundance of extensive matches to miRNAs compared to that of controls might indicate biological function. However, eukaryotic genomes, complex tapestries containing remnants of innumerable duplications and repetitive elements, are far from random, and thus this abundance might simply be a consequence of the miRNAs and sites sharing common ancestry. To distinguish between these possibilities, we examined the conservation of orthologous sites in five mammalian species, as assessed using a conservation-alignment (CA) score (Figure 5D). When applied to sites for distinct miRNAs conserved throughout mammals, 17 miRNA:site duplexes had CA scores ≤3.0 (Figure 5E), most of which were unlikely to be conserved by chance (Figure 5F). Four of the 17 top-scoring sites were miR-151-5p targets (Table S3C).
Cleavage at Highly Complementary Predicted Duplexes
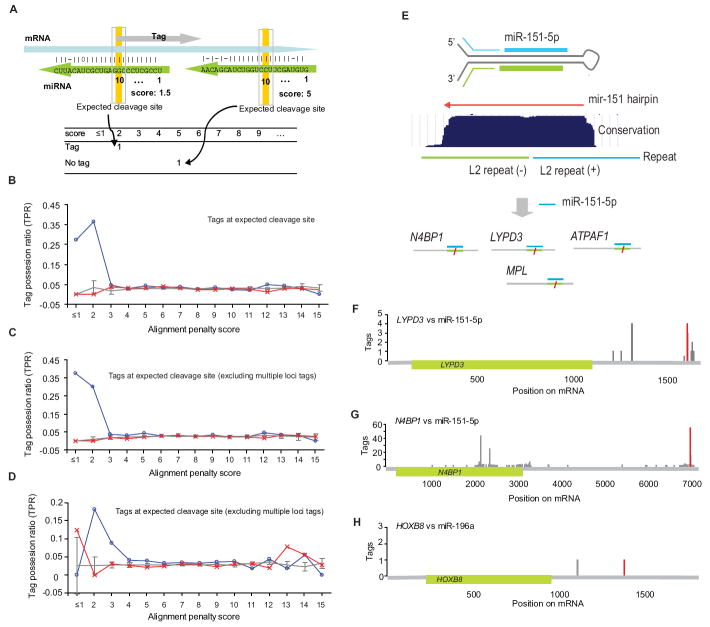
Having found evidence that the most extensively paired sites were more abundant and more conserved than expected by chance, we returned to the degradome sequencing data to search for evidence that these sites were cleaved in the cell. Because the degradome sequencing data included intermediates of normal mRNA decay, steps were taken to distinguish AGO2 cleavage products from other decay intermediates. To do this, we considered the tag possession ratio (TPR), which represented the proportion of predicted miRNA:site duplexes that were represented by tags at the expected cleavage site (Figure 6A). When focusing on the miRNAs and mRNAs expressed in HeLa, miRNA:site duplexes with alignment penalty scores ≤2.5 possessed significantly more cleavage tags at the expected cleavage site than did control duplexes (Figure 6B and Table S4B, Fisher’s exact test, P = 1.1 × 10 −04). Even after excluding tags mapping to multiple loci, this TPR difference remained both substantial and significant (Figure 6C and Table S4B, P = 2.6 × 10 −04). MicroRNA-directed cleavage in Arabidopsis sometimes occurs at ±1 nt from the expected cleavage site (Addo-Quaye et al., 2008). When applying a window of ±1 nt, there was no improvement in the TPR of expressed miRNA:site pairs (Figure S5A and Table S4B). As an added control, we repeated the analysis for miRNAs that were not expressed in HeLa cells and found that these miRNAs performed similarly to the chimeric miRNA controls (P = 1.0) and significantly worse than the miRNAs expressed in HeLa cells (P = 5.3 × 10 −05). These results strongly indicated that for miRNA:site pairs with favorable alignment scores (≤2.5), most tags at the expected cleavage site did not arise from background 5’→3’ degradation but instead were the consequence of miRNA-directed mRNA cleavage.
Figure 6. Enrichment of Degradome Tags at Sites of Expected miRNA-directed mRNA Cleavage.
(A) The tag possession ratio (TPR), used to search for evidence of miRNA-directed cleavage. At each alignment penalty score, the number of miRNA:site duplexes that had at least one tag with its 5’ terminal nucleotide mapping to the expected site of cleavage (across from miRNA position 10) was tallied, as was the number duplexes that lacked a tag indicative of cleavage. The TPR was calculated as the number of duplexes with a tag divided by the sum of all duplexes.
(B) Enrichment of tags at the expected sites. TPR values for miRNAs expressed in HeLa (blue; Table S4A) were compared to values for miRNAs not expressed in HeLa (red) and values for 10 cohorts of chimeric control miRNAs (gray; error bar, the standard deviation).
(C) Enrichment of tags at expected site after excluding tags mapping to multiple loci.
(D) Enrichment of tags at the expected sites, after omitting tags mapping to multiple loci, in human brain. TPR values for miRNAs expressed in human brain (blue line; Table S4C) were compared to values for miRNAs not expressed in human brain (red) and values for 10 cohorts of chimeric control miRNAs (gray; error bar, the standard deviation).
(E) The mir-151 hairpin and its cleavage targets. Schematic depicts the mir-151 hairpin, the positions of the two ancestral L2 LINE repeats that gave rise to the hairpin, the region of high conservation in sequenced mammals. Once processed from the hairpin the mature miR-151-5p pairs to and directs cleavage of mRNAs in HeLa cells.
(F–H) The distribution of degradome tags along the length of the indicated mRNAs (omitting tags mapping to >10 genomic loci). The red peaks are cleavage tags corresponding to cleavage at the site expected for the indicated miRNA. Shown are results from HeLa cells. Similar graphs are provided for the other mRNAs with evidence of miRNA-directed cleavage in HeLa cells (Figure S5B) and in human brain (Figure S5E). See also Figure S5 and Table S4.
MicroRNA-directed Cleavage in HeLa Cells and Human brain
Using an alignment penalty score of 2.5, a threshold at which the cumulative TPR difference between signal and background was most significant in HeLa data (Table S4B), we found eight miRNA-directed cleavage targets with tags precisely at the expected cleavage site (Table 1 and Figure S5B). All eight cleavage sites were in 3’UTRs, and half were conserved in other mammals (Table 1 and Figure S5B). Four of the pairs involved miR-151-5p (Figure 6E–G and Table 1). miR-196a and its cleavage target HOXB8 are both known to be moderately expressed in HeLa cells (Lim et al., 2005), and as expected HOXB8 was among the eight (Figure 6H).
Table 1. mRNAs with Degradome Tags at the Expected Cleavage Site.
Listed are mRNA:site pairs with alignment penalty scores (Score) for which the TPR (counting only tags mapping precisely to the expected cleavage site) significantly exceeded background (≤2.5 in Hela cells and ≤3.0 in brain; Table S4B). The expression of each miRNA is indicated by its miRNA reads in a high-throughput sequencing experiment (Table S4A and S4C). Cleavage tags were those tags mapping precisely to the expected site of cleavage and were normalized by the number of times they mapped to the genome. For each mRNA the fraction of degradome tags that were cleavage tags is indicated (Degradome fraction). Sites with CA ≤3.0 are categorized as conserved (Figure S5B and S5E)
| Tissue | miRNA | miRNA reads | mRNA | Location of site | Score | Cleavage tags | Degradome fraction (%) | Conserved |
|---|---|---|---|---|---|---|---|---|
| HeLa cell | miR-196a | 3682 | HOXB8 | 3’UTR | 1 | 1 | 50.0 | * |
| miR-545* | 34 | IGFBP4 | 3’UTR, LINE L2 | 2 | 1 | 0.7 | ||
| miR-28-5p | 1829 | LYPD3 | 3’UTR, LINE L2 | 2.5 | 0.75 | 3.4 | ||
| miR-151-5p | 2526 | N4BP1 | 3’UTR, LINE L2 | 2 | 55 | 14.1 | Yes | |
| miR-151-5p | 2526 | MPL | 3’UTR, LINE L2 | 1.5 | 0.5 | 6.9 | ||
| miR-151-5p | 2526 | LYPD3 | 3’UTR, LINE L2 | 0 | 4 | 17.9 | ||
| miR-151-5p | 2526 | ATPAF1 | 3’UTR, LINE L2 | 1 | 1 | 1.4 | Yes | |
| miR-182 | 624 | FRS2 | 3’UTR | 2.5 | 1 | 0.7 | Yes | |
| Human brain | miR-28-5p | 2544 | MDGA1 | 3’UTR, LINE L2 | 3 | 1 | 0.1 | |
| miR-151-5p | 33,007 | MDGA1 | 3’UTR, LINE L2 | 3 | 9 | 0.9 | ||
| miR-151-5p | 33.007 | N4BP1 | 3’UTR, LINE L2 | 2 | 6 | 2.1 | Yes | |
| miR-873 | 2033 | MAN2C1 | ORF | 2.5 | 1 | 0.2 | Yes | |
| miR-330-5p | 744 | FAM62C | ORF | 3 | 2 | 1.3 | ||
| miR-95 | 523 | EGLN3 | 3’UTR | 2.5 | 1 | 1.2 | ||
| miR-182 | 115 | FRS2 | 3’UTR | 2.5 | 2 | 1.4 | Yes | |
| miR-877 | 41 | SPTBN1 | ORF | 3 | 2 | 0.1 | ||
| miR-185 | 1014 | PMVK | 5’UTR | 3 | 2 | 0.8 | ||
| miR-383 | 593 | DCTN4 | ORF | 2 | 1 | 0.4 | Yes | |
| miR-598 | 8131 | EFTUD2 | ORF | 2.5 | 1 | 0.3 |
Although miR-196a:HOXB8 was not classified as conserved using our CA score because the site is missing in pig and horse, this pairing is known to be conserved in more distant lineages, including frog and fish (Yekta et al., 2004).
To extend our results beyond cells in culture, we performed degradome sequencing using poly(A)-selected RNA from whole human brain. Sequencing yielded 9,240,114 reads mapping to the human genome, with a diversity of 2,360,502 unique tag sequences. MicroRNAs expressed in human brain tissues were found by small-RNA sequencing (Table S4C). As in HeLa cells, we found a statistical association between the miRNA:site pairs and cleavage tags for miRNAs and mRNAs expressed in brain (Figures 6D and S5D). For pairs with alignment score ≤3.0, the TPR was significantly higher than for that of the controls (Table S4B, P = 0.008 and P = 0.030, nonexpressed and chimeric controls, respectively). Statistical significance was retained when also including tags mapping 1 nt downstream of the expected cleavage site as diagnostic of cleavage (Table S4B, P = 0.011 and P = 0.013, nonexpressed and chimeric controls, respectively), perhaps because some 5’→3’ trimming occured in the animal, where we could not knock down XRN1 activity. Eleven sites with scores ≤3.0 had tags suggestive of miRNA-directed cleavage (Table S4D) at the expected position (Table 1) and two had tags suggestive of cleavage at position −1 (Figure S5E). Three of the 13 matched miR-151-5p and included N4BP1, which was also identified in HeLa cells. FRS2, a proposed target of miR-182, was also identified in HeLa cells. Four of the miRNA:site pairs newly identified in brain appeared conserved in other mammals (CA ≤3.0; Table 1 and Figure S5E).
DISCUSSION
Centered Sites
We present centered sites as a type of miRNA target site. Centered sites contain at least 11 contiguous nucleotides that pair to a miRNA at positions 4–14 or 5–15, a pairing pattern distinct from that of most 3’-compensatory sites and seed sites. However, because a centered site might include additional nucleotide pairing on either side and a 3’-compensatory site might have additional pairing extending into the miRNA central region, there is potential overlap between a few extended centered sites and a few 3’-compensatory sites. Similarly, a seed site might include 3’-supplementary pairing extending into the miRNA central region, which creates potential overlap between a few extended centered sites and a few 3’-supplementary sites. However, such overlap with previously known site types is very rare. For example, a search of annotated human 3’UTRs revealed that for most human miRNAs, no seed-matched sites extend into centered sites; i.e., most human miRNAs have no 3’UTR match with contiguous Watson–Crick pairing to nucleotides 2–14. Furthermore, conservation analysis and array data show that seed-type targets prefer to acquire supplemental pairing at positions 13–16 rather than extending pairing through nucleotides 9–12 (Grimson et al., 2007).
The reason that centered sites had not been described previously can be explained by their relatively low abundance, which resembles that of 3’-compensatory sites and is far lower than that of seed-matched sites. Although no more effective than 7-nt seed-matched sites, centered sites are 4 nt longer, leading to an informational complexity ~250-fold (~44-fold) greater than that of 7-nt sites and a correspondingly increased difficulty for their emergence and retention during evolution. The rarity of centered sites hampers statistical assessment of whether they are subject to evolutionary conservation. Nonetheless, the conserved miRNAs of mammals each match an average of 13 centered sites in human 3’UTRs (Figure S1J), and based on our zebrafish analyses we estimate that on average about two sites per miRNA both reside in messages coexpressed with the miRNA and mediate repression. The presence of even a few beneficial interactions (species-specific or more broadly conserved) for a subset of the miRNAs could impart at least intermittent pressure to preserve the miRNA sequence, thereby explaining the preferential conservation observed in the central region of vertebrate miRNAs (Figure 1A). Moreover, centered sites resemble 3’-compensatory sites in providing a mechanism by which different members of the same miRNA seed family can repress distinct targets (Bartel, 2009).
Why would centered sites require so much more contiguous pairing than that required by seed sites? When bound by the Argonaute protein within the silencing complex, the seed is thought to be pre-organized to favor Watson-Crick pairing to the mRNA (Bartel, 2004). In the current version of this seed-nucleation model, pairing cannot propagate to the center of a miRNA without a substantial conformational change in which the original contacts between Argonaute and the miRNA central and 3’ regions are disrupted (Bartel, 2009). Disrupting these contacts offsets some binding energy gained in forming the central pairs, causing contiguous pairing adjacent to the seed to contribute less affinity than might have otherwise been expected. This lower contribution of pairing to the central region, combined with the higher contribution achieved by the pre-organized seed, would explain why so much more pairing is needed for centered sites to achieve the same outcome as 7-nt seed sites.
Mg2+ Effect on Cleavage Specificity and Efficiency
Our results shed a new light on the biochemistry of RNAi. We suggest that at 37°C in the low Mg2+ concentrations present in the cell, only the extensively paired sites can be bound with the stability and conformation that favors mRNA cleavage, and that after cleavage the products are not so tightly bound so as to slow multiple turnover. In higher Mg2+, however, less extensively paired sites achieve the stability and conformation needed for cleavage, and product release is more apt to slow turnover. This model explains the reduction of both specificity and efficiency at extensively paired sites observed in high Mg2+ concentrations. Under these conditions, less extensively paired sites are more readily cleaved—hence, the reduced specificity. The more extensively paired sites, on the other hand, undergo slower product release and gain little benefit from this more permissive binding-and-cleavage regime. Indeed, any benefit gained is more than offset by the tighter binding of the miRNA to less extensively paired sites, which causes the total cellular RNA present in extracts used for cleavage reactions to more effectively inhibit utilization of the labeled substrates—hence, the reduced efficiency. The free cytoplasmic Mg2+ concentration in most cells and tissues is < 1 mM (Gunther, 2006), suggesting that cleavage specificity is very high in vivo.
Our results explain previous observations regarding the effects of adding phosphate-containing compounds to in vitro cleavage reactions. Many diverse phosphate compounds, including inorganic monophosphate, stimulate the multiple-turnover cleavage by the mammalian silencing complex (Gregory et al., 2005). We suggest that these phosphate compounds titrate the free Mg2+, which in turn increases product turnover through decreased RNA duplex stability.
Endogenous miRNA-directed mRNA Cleavage in Human
We find that miRNA-directed cleavage of mammalian mRNAs, although even more rare than repression at centered sites, occurs more frequently than previously appreciated. Two endogenous cleavage targets had been reported in mammals, HOXB8 and RTL1 (Yekta et al., 2004)(Davis et al., 2005). We substantially add to this list, with evidence for cleavage of seven additional targets in HeLa cells and cleavage of thirteen in human brain, two of which overlapped with HeLa targets. This small overlap, largely attributed to differential expression of the miRNAs or mRNAs in the two samples (Tables 1, S4A, S4C and S4D), suggests that as more tissues are examined, more cleavage targets will be found.
The fraction of degradome sequencing tags that provided evidence of miRNA-directed cleavage was generally higher in the HeLa analysis than in the brain analysis (Table 1; Figure S5B and S5E). In the brain, this fraction of cleavage tags was sufficiently low so as to suggest that some might represent degradation intermediates not indicative of miRNA-directed cleavage. Whether a smaller fraction of brain messages are cleaved, however, is unclear. The brain analysis lacked the benefit of the XRN1-endonuclease knockdown, designed to stabilize the transient 3’ cleavage product so that it could be more readily detected over the background of metastable mRNA-decay intermediates. Moreover, whole brain has many cell types, with the possibility that differential expression of a miRNA and its cleavage targets might decrease the signal relative to background. Nonetheless, for most cleavage targets in HeLa and for some in brain, degradome profiles resembled those of plant targets with validated biological relevance (Figures 6F–H, S5Bi and S5Ei) (Addo-Quaye et al., 2008; German et al., 2008), strongly supporting the hypothesis that the miRNA-directed cleavage pathway is an important degradation pathway for those mRNAs.
In both brain and HeLa cells, several cleavage targets identified were targets of miR-151-5p. This miRNA derives from a hairpin that has homology to the L2 subclass of repeat elements known as long interspersed nuclear elements (LINEs). L2 LINE elements are remnants of a non-LTR retrotransposon activity present in the common ancestor of mammals. They make up over 3% of the human genome (Kamal et al., 2006). Indeed, the miR-151 hairpin is derived from a tail-to-tail arrangement of two L2 fragments (Figure 6E). Hence, miR-151-5p derived from L2(+) is strongly complementary to several target sites derived from L2(−) repeats. Analogous tail-to-tail arrangements of short (S)INE fragments produce transcripts with longer hairpins that are processed in mouse ES cells into endogenous siRNAs (Babiarz et al., 2008). However, miR-151-5p and miR-151-3p are typical miRNAs, in that 1) their accumulation depends on both Drosha/DGCR8 and Dicer endonucleases (Babiarz et al., 2008), 2) they pair to each other with 2-nt 3’ overhangs, 3) they are the two dominant products accumulating from the hairpin (Figure S5C), and 4) their hairpin has a conservation pattern typical of other conserved miRNAs (Figure 6E).
Two other miRNAs that direct cleavage in HeLa, miR-28-5p and miR-545*, are also L2 repeat–derived miRNAs. The notion that these miRNAs and their targets ultimately derived from the same ancestral elements is reminiscent of the origin of some plant miRNAs, which derive from duplicated fragments of their cleavage targets (Allen et al., 2004; Rajagopalan et al., 2006). In mammals, however, the miRNAs and target sites evolved in parallel from the common ancestor, rather than one from the other. Moreover, in mammals, common ancestry between the miRNAs and their targets can be detected for older, conserved miRNAs, such as miR-151 and miR-28, whereas in plants common ancestry has been detected only for younger, nonconserved miRNAs.
The observation that many of the cleaved mRNAs were the targets of repeat-derived mRNAs can be explained by the fact that repeat-derived miRNAs are more likely to encounter extensively complementary matches, since repeat-element remnants are found within many mRNAs. Over the course of evolution, repeat-derived miRNAs presumably had access to a wide variety of cleavage targets, providing the opportunity for some favorable regulatory interactions to emerge and be retained as conserved cleavage interactions. Thus, the repeat-derived miRNAs and their cleavage targets provide yet another avenue for repetitive elements to shape the regulation of cellular genes.
Concluding Remarks
The discovery of centered sites raises the question of how many additional site types remain to be found. On the one hand, transcriptome/proteome changes observed after introducing or deleting a miRNA can all be explained by direct interactions between the miRNA and messages with the five known site types (seed sites, 3’ supplementary seed sites, 3’ compensatory sites, centered sites, and cleavage sites), combined with indirect effects as changes in the primary targets influence expression of secondary targets. On the other hand, detailed experimental follow-up on mRNAs that respond to the miRNA despite lacking any of these established site types seems to indicate that some of them should not be dismissed as secondary targets but might instead be direct targets (Lal et al., 2009). However, the pairing schemes proposed thus far for these unusual interactions have not been defined sufficiently to provide predictive utility. That is, in contrast to centered sites and the previously known site types, these pairing schemes lack the specificity required to distinguish other responsive messages with similar pairing from background. Hence, experiments like that shown in Figure 1C–F cannot distinguish responsive messages that satisfy these unusual pairing schemes from nonresponsive messages that do not. Perhaps unknown factors binding to neighboring UTR elements help achieve interaction specificity differently for each individual mRNA in a manner too idiosyncratic to be generalized into site types. Alternatively, future insights into miRNA targeting might identify commonalities in these unusual interactions, which could form the basis of novel site types with predictive value.
EXPERIMENTAL PROCEDURES
A detailed description of all materials and methods used can be found in the Supplemental Text.
Microarray and Molecular Analyses
Array analyses were as in (Grimson et al., 2007). Luciferase reporter constructs were prepared as in (Grimson et al., 2007), and assays were performed as in (Farh et al., 2005). In vitro cleavage reactions were essentially as in (Haley and Zamore, 2004; Shin, 2008). Uncapped 5’ ends of GSTM3 mRNA degradation products were identified using the 5’-RACE kit (Invitrogen), as in (Jones-Rhoades and Bartel, 2004), starting with cells in which XRN-1 mRNA was knocked-down more than 90%, as confirmed by RT-PCR (Aleman et al., 2007). Degradome libraries were constructed essentially as in (Addo-Quaye et al., 2008). Small-RNA libraries were prepared for Illumina sequencing as described (Grimson et al., 2008).
Analysis of miRNA Conservation
Out of 223 miRNA genomic loci producing 197 mouse miRNAs conserved in other mammals (Friedman et al., 2009), 203 miRNA loci producing miRNAs with 5’ ends validated from a large scale profiling of mouse miRNAs (Chiang et al. 2010) were used in the analysis of Figure 1A.
Processing of Degradome Tags
After removing linker sequences and tags shorter than 20 nt, degradome tags were mapped to RNAs annotated in the ENSEMBL (http://www.ensembl.org/), requiring a perfect match. To find “multiple loci tags”, and tags that did not map to annotated RNAs, filtered tags were mapped to the human genome (hg18, http://genome.ucsc.edu/). When determining TPRs, filtered tags were mapped to a curated set of distinct mRNAs (Baek et al., 2008). Expressed mRNAs were those represented by at least one degradome tag.
miRNA:site Duplexes
When searching for miRNA:site duplexes, distinct mRNAs and miRNAs were selected to avoid over-counting predicted duplexes involving miRNA families or mRNA isoforms. To select distinct miRNAs, all human miRNAs and miRNA* sequences (miRBase 11.0) were aligned and classified into groups whose members differed from each other at ≤5 positions. The miRNA with the lowest miRBase annotation number was selected as the representative from each group. For distinct mRNAs, the mRNA isoform with the longest 3’UTR (or, if all 3’UTRs were of the same length, a randomly chosen isoform) was selected from a previously filtered set of RefFlat and H-INV annotations (Baek et al., 2008).
CA Score
To search for orthologous sites, we used 165 distinct miRNAs conserved among mammals and a 6-way genome alignment (human, mouse, rat, dog, horse, and pig) from the UCSC genome browser (hg18, http://genome.ucsc.edu/). Alignment penalty scores were determined and the second worst score rather than the worst score was selected as the CA score to accommodate some genome-alignment errors, incomplete genome sequences, and species-specific losses.
Generation of miRNA-like Control Sequences
To generate controls with the same seed composition and same trinucleotide composition as authentic miRNAs, chimeric miRNA sequences were created by reciprocally recombining, using the link between nucleotides 10 and 11 as the crossover breakpoint, two miRNAs randomly chosen (without replacement) from miRNA pairs with the same dinucletide at positions 10 and 11 considering only our set of distinct miRNAs. Ten chimeric miRNA cohorts were generated to estimate the signal-to-background ratios.
Supplementary Material
Acknowledgments
We thank Andrew Grimson, Daehyun Baek, and Alexander Subtelny for helpful discussions, Shujun Luo and Gary Schroth for Illumina sequencing of the small-RNA library from brain, and the Whitehead Genome Technology Core for the remaining Illumina sequencing. Supported by a Damon Runyon postdoctoral fellowship (C.S.) and a grant from the N.I.H. D.B. is a Howard Hughes Medical Institute Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleman LM, Doench J, Sharp PA. Comparison of siRNA-induced off-target RNA and protein effects. Rna. 2007;13:385–395. doi: 10.1261/rna.352507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Anderson EM, Birmingham A, Baskerville S, Reynolds A, Maksimova E, Leake D, Fedorov Y, Karpilow J, Khvorova A. Experimental validation of the importance of seed complement frequency to siRNA specificity. Rna. 2008;14:853–861. doi: 10.1261/rna.704708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel CD, Maxwell ES. Structural features of the guide:target RNA duplex required for archaeal box C/D sRNA-guided nucleotide 2’-O-methylation. Rna. 2007;13:899–911. doi: 10.1261/rna.517307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, et al. 3’ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E, Caiment F, Tordoir X, Cavaille J, Ferguson-Smith A, Cockett N, Georges M, Charlier C. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001a;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001b;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther T. Concentration, compartmentation and metabolic function of intracellular free Mg2+ Magnes Res. 2006;19:225–236. [PubMed] [Google Scholar]
- Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. Rna. 2006a;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. Rna. 2006b;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Kadener S, Rodriguez J, Abruzzi KC, Khodor YL, Sugino K, Marr MT, 2nd, Nelson S, Rosbash M. Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. Rna. 2009;15:537–545. doi: 10.1261/rna.1319309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal M, Xie X, Lander ES. A large family of ancient repeat elements in the human genome is under strong selection. Proc Natl Acad Sci U S A. 2006;103:2740–2745. doi: 10.1073/pnas.0511238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3’UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5’ region. Embo J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Tuschl T. RISC is a 5’ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. Rna. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J, Linsley PS, Aronin N, Xu Z, Zamore PD. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Serra MJ, Baird JD, Dale T, Fey BL, Retatagos K, Westhof E. Effects of magnesium ions on the stabilization of RNA oligomers of defined structures. Rna. 2002;8:307–323. doi: 10.1017/s1355838202024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C. Cleavage of the star strand facilitates assembly of some MicroRNAs into Ago2-containing silencing complexes in mammals. Mol Cells. 2008;26:308–313. [PubMed] [Google Scholar]
- Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell. 2004;15:173–183. doi: 10.1016/j.molcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3’UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol. 2009a doi: 10.1038/nsmb.1712. [DOI] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009b;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.