Abstract
We have developed a novel system for coupling reverse-phase (RP) and hydrophilic interaction liquid chromatography (HILIC) online in a micro-flow scheme. In this approach, the inherent solvent incompatibility between RP and HILIC is overcome through the use of constant-pressure online solvent mixing, which allows our system to perform efficient separations of both hydrophilic and hydrophobic compounds for mass spectrometry-based proteomics applications. When analyzing the tryptic digests of bovine serum albumin, ribonuclease B, and horseradish peroxidase, we observed near-identical coverage of peptides and glycopeptides when using online RP-HILIC—with only a single sample injection event—as we did from two separate RP and HILIC analyses. The coupled system was also capable of concurrently characterizing the peptide and glycan portions of deglycosylated glycoproteins from one injection event, as confirmed, for example, through our detection of 23 novel glycans from turkey ovalbumin. Finally, we validated the applicability of using RP-HILIC for the analysis of highly complex biological samples (mouse chondrocyte lysate, deglycosylated human serum). The enhanced coverage and efficiency of online RP-HILIC makes it a viable technique for the comprehensive separation of components displaying dramatically different hydrophobicities, such as peptides, glycopeptides, and glycans.
Electronic supplementary material
The online version of this article (doi:10.1007/s00216-010-3991-2) contains supplementary material, which is available to authorized users.
Keywords: RP-HILIC, UPLC, Hydrophilic analytes, Glycan separation, Full-component analysis
Introduction
Reverse-phase (RP) liquid chromatography (LC) is among the most frequently employed methods for sample separation prior to mass spectrometry (MS)-based proteomics analyses [1–5]. Hydrophobic interaction with the hydrocarbon stationary phase is the primary principle by which RP separates compounds; therefore, most relatively hydrophobic molecules, such as peptides and proteins, can be separated efficiently on RP columns according to their different hydrophobicities. Complex proteomics samples, however, often comprise species of vastly differing hydrophobicities, including relatively polar analytes, such as short peptides, glycopeptides, or phosphopeptides featuring short peptide moieties. These polar analytes tend to adsorb weakly on the RP stationary phase and often elute directly with the dead volume. The poor separation of these compounds leads to decreased effective sensitivity in electrospray MS, due to mutual suppression, and, as a consequence, potential loss of valuable biological information.
In recent years, there has been an increasing trend toward enhancing the separation and analysis of relatively hydrophilic analytes—for example, in non-targeted analysis involving whole-proteome or -glycome profiling of cells or tissues. Ion-pairing chromatography is one strategy that has been adopted to improve the retention of polar analytes in RP columns; in this approach, ion-pairing reagents are added to the mobile phase to serve as a bridge between the stationary phase and the polar molecules and, as such, modify their retention behavior [6–8]. The addition of an ion-pairing reagent, however, could lead to ion suppression at the electrospray during MS analysis. A popular alternative for the separation of these analytes is hydrophilic interaction liquid chromatography (HILIC) [9, 10], which operates on the basis of hydrophilic interactions between the analytes and the stationary phase. Here, retention of analytes is based on hydrophilic interactions with the stationary phase (e.g., polysulfoethyl A, TSKgel Amide 80), with highly polar or hydrophilic compounds interacting most strongly. The analytes are then eluted upon increasing the polarity of the mobile phase (e.g., water). The efficacy of this technique for the separation of polar compounds has been demonstrated for carbohydrates [11–15], polar peptides [16, 17], glycopeptides [18, 19], and phosphopeptides [20]. Despite its prospects for separating relatively polar compounds, HILIC tends to be less useful for separating hydrophobic peptides. To overcome this obstacle, two separate injections of the sample could, in theory, be performed using standalone RP and HILIC, but such analyses would require double the amount of sample.
Ideally, the combination of RP and HILIC in tandem would combine their complementary selectivities for the separation of both hydrophobic and hydrophilic species and enable “full component” analysis [21–24]. In particular, RP-HILIC is an attractive prospect as a hyphenated system for proteomics because common solvents would be used: for their solvent gradients, both RP and HILIC employ water and acetonitrile, which are readily amenable to electrospray ionization for MS analysis. In theory, offline coupling of RP and HILIC can be performed simply through collection of the flow-through from the RP extraction and then reinjection into the HILIC system following lyophilization. In practice, however, offline collection is labor intensive and often results in significant sample loss; as such, online coupling is generally more desirable.
The primary technical challenge of an automatic RP-HILIC system is, therefore, overcoming the solvent strength incompatibility between RP and HILIC online. Because RP requires a low-organic solvent for extraction, whereas HILIC extraction is performed using a high-organic medium, the RP flow-through must be converted to a high-organic solvent prior to its diversion into the HILIC column. Column-switching LC set-ups are frequently used in multidimensional LC to maximize analytical output [25]; recently, they have allowed online mixing of solvent for RP-HILIC in experiments using small molecules and chromatographic profiling [26, 27]. Because column switching involves using a miniaturized first column and a relatively-high-flow-rate second column to achieve efficient solvent exchange, this approach would, however, render a system flow outside the micro-flow scheme commonly employed in MS-based proteomics.
In this paper, we propose a novel system for performing micro-flow RP-HILIC with efficient extraction of both relatively hydrophobic and hydrophilic analytes in two separate capillary columns. We have used an ultrahigh-performance LC (UPLC) system to generate specific pressures and flow rates in the RP and HILIC channels to achieve effective online solvent mixing. The high-pressure output of UPLC also allows long columns (ca. 800 mm) to be used to increase the peak capacities for both RP and HILIC separation [28–30]. We have verified the applicability of this system though analyses of a number of protein and glycoprotein standards. In addition, we report the proteomics analyses of complex biological samples using RP-HILIC with mouse chondrogenic cell protein extracts and deglycosylated human serum.
Experimental section
Chemicals and materials
Polyimide-coated fused-silica capillaries were purchased from Polymicro Technologies (Phoenix, AZ). PEEK tubing was purchased from Upchurch Scientific (Oak Harbor, WA). Four- and six-port switching valves, stainless 2-μm screens, two-way tees, and zero-dead-volume unions were purchased from Valco Instruments (Houston, TX). Jupiter C18 packing materials (3-μm particles, 300-Å pores) were acquired from Phenomenex (Torrance, CA); TSKgel Amide 80 packing materials (5-μm particles, 300-Å pores) were obtained from Tosoh Bioscience (Montgomeryville, PA). Dithiothreitol (DTT), iodoacetamide (IAA), human serum, and all protein standards were purchased from Sigma-Aldrich (St. Louis, MO). Modified trypsin was acquired from Promega (Madison, WI); formic acid and trifluoroacetic acid were obtained from Fluka (St. Louis, MO). The high-performance liquid chromatography (HPLC)-grade water and acetonitrile used as solvents and for sample preparation were purchased from Tedia (Fairfield, OH).
Sample preparation
In the protein standards study, 100 μg of the protein standards were digested. The proteins were dissolved in 100 mM ammonium bicarbonate and reduced by 50 mM DTT at 60 °C for 60 min. The proteins were cooled to room temperature prior to alkylation in 100 mM IAA for 60 min at room temperature in the dark. Trypsin digestion was performed using modified trypsin (protein-to-enzyme ratio, 33:1) and incubation at 37 °C overnight for ca. 17 h. Digestion was terminated by heating to 90 °C for 10 min; samples were stored at −20 °C until required for use. In the peptide-glycan study, the tested glycoproteins were first deglycosylated with 1 unit of peptide-N 4-(acetyl-β-glucosaminyl)asparagine amidase (PNGaseF) at 37 °C overnight prior to trypsin digestion following the procedure described above. The resulting standard protein digest (0.5 μg/μL) was diluted 25 times; 5 μL (100 ng) was injected for the experiments. The human serum, which contained proteins at concentrations in the range 4.0–9.0 g/dL, was used without further purification. A solution (600 μL) of 100 mM ammonium bicarbonate containing 8 M urea was added to human serum (400 μL). The serum was then deglycosylated with PNGaseF (5 equiv.) and trypsinized as described above; 25 μg of the resulting peptide-glycan mixture was used for the experiment. Mouse chondrogenic cells (ATDC5) were grown in Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium supplemented with 5% fetal bovine serum (FBS), 1% penicillin/streptomycin, and fungizone (1 μg/mL) at 37 °C. The cells were harvested by direct scraping, spun down, and lysed with 0.5 M triethylammonium bicarbonate (TEAB) at pH 8.5, and then subjected to sonication on ice using 25%-amplitude 0.5-s/3.5-s pauses for 30 min. The lysed cells were centrifuged at 13,400 rpm at 4 °C for 1 h to remove insoluble pellets; the collected supernatant containing the soluble proteins was quantified through a Bradford assay, following the manufacturer’s protocol, and then digested with trypsin, as described above, to yield peptides at a concentration of 0.5 μg/μL. A portion (10 μL) of the digest was used in the complex biological sample analysis.
RP-HILIC chromatographic equipment and conditions
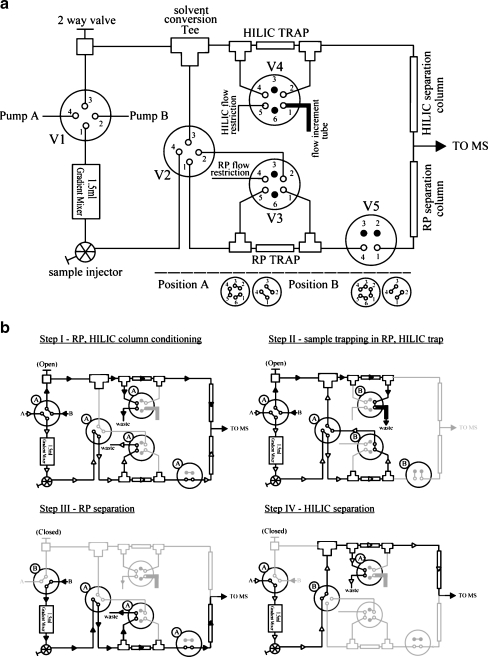
The simplified diagram in Fig. 1a illustrates the system assembly. The micro-flow RP-HILIC system consisted of a UPLC system featuring two 65D isocratic syringe pumps and a system controller (Isco; Lincoln, NE), a gradient mixer, three four-port/two-position mechanical switch valves (valves 1, 2, 5), two six-port/two-position switch valves (valves 3, 4), and a sample injector valve (Valco Instruments; Houston, TX). Two trap columns and two analytical columns were used. The RP and HILIC trap [150-μm i.d. × 20-mm length] and analytical (150-μm i.d. × 800-mm length) columns were packed in-house using C18 and Amide80 materials, as described previously [29]. All connecting capillaries used were 75-μm i.d. × 200-mm long, unless otherwise stated; the flow-increment tube was 1.0-mm (0.04-in.) i.d. × 200-mm long; the flow restriction capillary was 20-μm i.d × 1-m long. The i.d. and length can be fine-tuned if a different gradient speed is desired. The separated analytes were detected and analyzed using MS. The switching valve positions and the solvent flow path at each step of the experiment are provided in Fig. 1b. Detailed descriptions are given below:
-
Step I
(RP, HILIC column conditioning): all valves were set to position A during the column conditioning. Pumps A and B were operated at constant pressure (7,000 and 6,000 PSI, respectively). The aqueous solvent (97.5% water, 2% acetonitrile, 0.5% formic acid; v/v/v) was directed to the gradient mixer through the sample injection valve to valve 2, where it was diverted to the RP trap and analytical column after flow-splitting; meanwhile, the organic solvent (2% water, 97.5% acetonitrile, 0.5% formic acid; v/v/v) was delivered through a two-way valve to the HILIC trap and analytical column after flow-splitting. The conditioning time for the RP and HILIC columns was 20 min.
-
Step II
(sample trapping in RP, HILIC trap): valves 3–5 were set to position B; the sample was injected via the injector and the hydrophobic analytes were extracted in the RP trap. Any unretained, hydrophilic analytes in the flow-through were then diverted through valves 2 and 3 to the solvent conversion tee, where they were mixed with the organic solvent. At this point, the flow rate, as indicated by the residual pump volume readings on the Isco UPLC, of the organic solvent was considerably higher (ca. 120 μL/min) than that of the incoming aqueous solvent (ca. 3 μL/min). The hydrophilic analytes in the then-organic medium were subsequently extracted by the HILIC trap column. The sample trapping time was 20 min.
-
Step III
(RP separation): valves 3 and 4 were set back to position A, the two-way valve was closed, and pump B was set to 8,000 psi (constant pressure). Valve 1 was then set to position B and pump B delivered organic solvent that slowly changed the aqueous composition inside the mixing chamber and generated the solvent gradient used for separation. The hydrophobic analytes trapped in the RP trap column at this point were eluted into the RP analytical column for gradient separation. The %B gradient progression over time follows the shape of “1 minus exponential decay” and is a function of the restriction flow rate [29]. Most peptides eluted from the RP column within 4 h, whereas the HILIC run was shorter at ca. 2.5 h. Longer experiments can be conducted for complex samples to take advantage of the greater plate number of the long column by modifying the gradient mixing rate. The post-column flow rate was measured using a volumetric capillary; it was ca. 1 μL/min throughout most of the gradient.
-
Step IV
(HILIC separation): The gradient mixer was prefilled with organic solvent prior to the HILIC run. Pump A was then set to deliver aqueous solvent at 8,000 PSI to generate the HILIC solvent gradient. The solvent gradient egressing from the mixer was diverted at valve 2 directly to the HILIC channel. The analytes trapped in the HILIC trap column were eluted into the HILIC analytical column for separation.
Fig. 1.
Simplified diagram illustrating the (a) assembly of the micro-flow RP-HILIC system and (b) the flow path of solvent and analytes during conditioning, sample trapping, RP separation and HILIC separation
Mass spectrometry
The eluents from the RP and HILIC analytical columns were coupled online continuously to a MDS Sciex QSTAR XL quadrupole time-of-flight hybrid mass spectrometer (Applied Biosystems; Framingham, MA) through a nano-ionspray interface. Typical mass spectrometer parameters were set as follows: DP, 20 V; FP, 125 V; CUR, 20 arbitrary units (a.u.); GSI, 0 a.u.; GS2, 0 a.u.; nanospray voltage, 3000 V. Each MS/MS experiment was set to 8 h in length; the mass spectrometer was left to run in excess of the LC gradient length in information-dependent acquisition (IDA) mode to ensure all eluents were analyzed. In IDA settings, every 1-s survey scan in the range m/z 300–2000 was followed by up to three 1-s product ion scans in the range m/z 100–2,000. Selection criteria for product ion scans were the three most-intense peaks in each precursor ion scan having more than 50 counts and charge states between +2 and +4. Enhanced ion settings were used to improve sensitivity; 60-s dynamic exclusion and rolling collision energy were used to acquire non-redundant, high-quality product ion spectra. The calibration curve for the collision energy for the +2-to-+4 ions was determined through direct infusion of BSA tryptic peptides. The acquired MS/MS data were analyzed directly using the Paragon algorithm in the ProteinPilot 2.0.1 software (Applied Biosystems, Framingham, MA). The MS/MS spectra were matched against theoretical spectra generated from sequences in the Uniprot SwissProt (release 14.8) mouse (16,116 entries) and human (20,336 entries) protein databases (http://www.uniprot.org). Trypsin was set as the enzyme used; IAA was chosen as the cysteine alkylation agent; a thorough search with biological modifications settings, which contain a number of built-in modifications, was performed. The precursor mass accuracy and product ion mass accuracy were pre-set by the software. The identified peptides from the Paragon algorithm were grouped into minimal non-redundant protein sets by the ProGroup algorithm of the software. For protein identifications to be considered, a minimal unused ProtScore of 2.0 was needed. The unused ProtScore is the sum of individual peptide scores, based on identification confidence, that are not already assigned to higher-ranked protein sets; it served to minimize redundancy of protein identification based on common peptide sequences. The peptide score was based on statistical confidence of sequence matching according to the software; 99% confidence translates into a score of 2.0 and 95% confidence scores 1.3. In the complex sample analysis experiment, protein sets containing two or more peptides passing the 95% confidence threshold were automatically accepted, whereas other protein sets were manually confirmed. Each spectrum in the protein standard and peptide-glycan standard experiments was inspected manually. Possible N-glycan compositions were determined with the aid of free software [Glycomod [31] and GlycoWorkbench [32]]. The hydrophobicities of the detected peptides were calculated using Sequence Specific Retention Calculator (v. 3.0) [33].
Results and discussion
Online system for micro-flow RP-HILIC
Previous attempts to combine RP and HILIC have invariably employed the so-called column-switching technique, with miniaturization of the first column to solve the problem of solvent incompatibility. To develop an RP-HILIC system in which both separations would be performed in a micro-flow scheme suitable for MS-based proteomics, we had to solve the problem of solvent incompatibility between the RP and HILIC columns through effective online mixing. Our system converts the aqueous solvent used in RP trapping to the organic solvent used for HILIC trapping through the application of various flow restrictions in the RP and HILIC channels. The aqueous UPLC channel, operating at constant pressure, delivered solvent at a lower flow rate, albeit with higher pressure, because of the higher restriction from the flow path. In comparison, the organic solvent channel UPLC pump operated at a lower pressure, but with a considerably higher flow rate, through the use of a much wider flow-splitting capillary (flow-increment tube in Fig. 1). Thus, the aqueous flow-through from the RP column, upon entering the organic channel, was readily mixed with a high volume of organic solvent, rendering it amenable for HILIC extraction (see Fig. 1 and the Experimental section for details).
Retention of hydrophobic and hydrophilic peptides in RP-HILIC
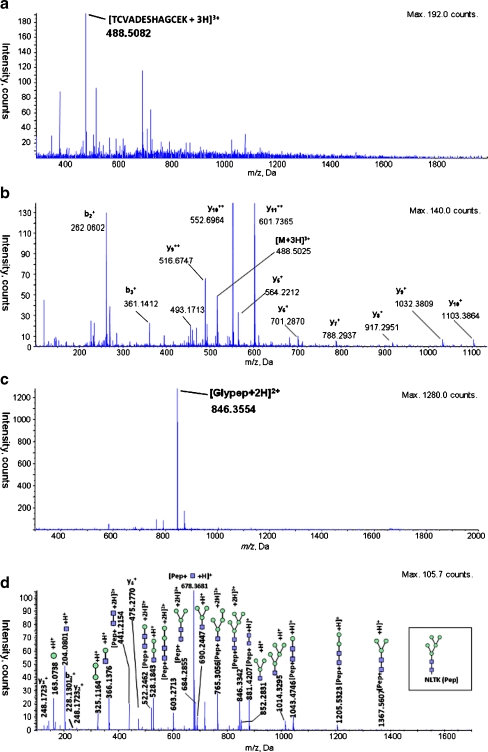
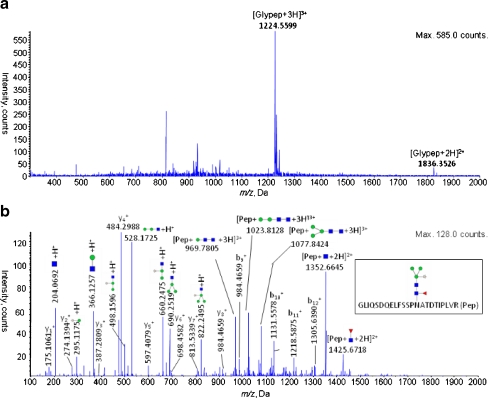
The coverage of peptides using hyphenated RP-HILIC was increased over that of either RP or HILIC alone, as readily demonstrated from the analyses of three protein standards: ribonuclease B, bovine serum albumin, and horseradish peroxidase. We analyzed the mixture of peptides and glycopeptides resulting from trypsin digestion of the proteins through tandem mass spectrometry (MS/MS) after separation using either RP or HILIC alone or the online RP-HILIC system. Figure 2 presents the mass spectral identification of a relatively hydrophilic peptide and a glycopeptide that eluted solely from the HILIC system; i.e., they were not detected during RP separation. Similarly, we detected a number of relatively hydrophobic peptides solely from the RP system that were unretained on the HILIC column (Table 1). Based on mass spectral evidence, the individual RP and HILIC separations resulted in the identification of a total of 58 unique native (non-glycosylated) peptides and 11 glycopeptides. The online RP-HILIC system recovered all of the native peptides and all but one glycopeptide from a single injection event; 53 native peptides were revealed in the RP part of the system and the remaining five, presumably unretained by the RP trap column, were captured by the coupled HILIC system. The HILIC system separated only six of the 10 recovered glycopeptides. Table 1 summarizes the identified peptides and glycopeptides, from the standalone RP, standalone HILIC, and RP-HILIC separations. As expected, the peptides and glycopeptides retained by the RP column generally contained longer, more-hydrophobic peptide sequences. In contrast, those retained by the HILIC system were either short and contained many polar residues (e.g., serine, threonine, glutamic acid) or they were rendered more hydrophilic as a result of extensive glycosylation. Figure 3a and b display example full-scan and product ion spectra of a detected glycopeptide from horseradish peroxidase, analyzed using RP-HILIC MS/MS. The ions of the peptide fragments as well as the characteristic N-glycan products are readily apparent, allowing characterization of the peptide sequence, the position of the glycosylation site, and the glycan structure.
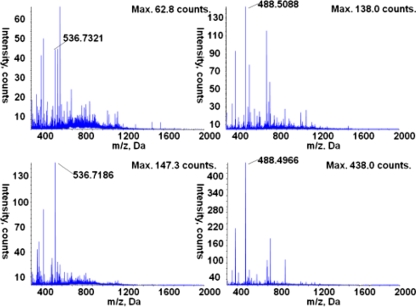
Fig. 2.
Mass spectra of a short peptide and a glycopeptides typically retained only by HILIC and undetected in RP; (a) full-scan spectrum of the protonated peptide [TCVADESHAGCEK]3+ (m/z 488.51), eluted from standalone HILIC at 73.297 min and (b), the corresponding product ion scan showing characteristic fragment ions; (c) full-scan spectrum and (d) product ion spectrum of the protonated glycopeptide [NLTK-GlcNAc2Man5 + 2H]2+ (m/z 846.36), eluted from standalone HILIC at 93.497. The proposed composition of the glycopeptides was first determined based on the mass-to-charge ratio of the precursor ion, through a database search using the Glycomod website, and subsequently confirmed using the software ProteinPilot and GlycoWorkbench
Table 1.
Peptides from mixture of ribonuclease B, bovine serum albumin, and horseradish peroxidase detected using standalone RP, standalone HILIC, and RP-HILIC
| Expt. m/z | z | Identified Sequence | Standalone RP | Standalone HILIC | RP-HILIC RP | RP-HILIC HILIC | Hydrophobicity | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Relative Retention | Peak width (s) | Intensity (counts) | Relative Retention | Peak width (s) | Intensity (counts) | Relative Retention | Peak width (s) | Intensity (counts) | Relative Retention | Peak width (s) | Intensity (counts) | ||||
| (Native peptides of bovine serum albumin) | |||||||||||||||
| 305.15 | 2 | AFDEK | – | – | – | 0.13 | 16.5 | 9.6E + 3 | – | – | – | 0.00 | 10.5 | 1.2E + 3 | 4.92 |
| 409.69 | 2 | ATEEQLK | – | – | – | 0.31 | 54.7 | 3.3E + 4 | – | – | – | 0.08 | 10.5 | 4.0E + 3 | 6.4 |
| 488.51 | 3 | TCVADESHAGCEK | – | – | – | 0.75 | 45.1 | 6.9E + 4 | – | – | – | 0.66 | 48.6 | 2.2E + 5 | 6.81 |
| 569.73 | 2 | CCTESLVNR | 0.03 | 13.3 | 5.1E + 3 | – | – | – | 0.01 | 11.1 | 1.2E + 4 | – | – | – | 12.73 |
| 536.73 | 2 | SHCIAEVEK | – | – | – | 0.65 | 49.5 | 2.7E + 4 | – | – | – | 0.51 | 52,4 | 1.2E + 5 | 12.83 |
| 739.76 | 2 | ETYGDMADCCEK | 0.01 | 24.9 | 5.5E + 4 | – | – | – | 0.01 | 10.6 | 1.7E + 5 | – | – | – | 13.32 |
| 722.32 | 2 | YICDNQDTISSK | 0.01 | 30.9 | 1.1E + 5 | 0.49 | 72.0 | 1.0E + 5 | 0.01 | 13.4 | 3.7E + 5 | – | – | – | 14.82 |
| 449.73 | 2 | LCVLHEK | 0.03 | 37.0 | 1.2E + 5 | 0.35 | 71.2 | 9.3E + 4 | 0.01 | 15.8 | 1.4E + 5 | – | – | – | 15.76 |
| 325.48 | 3 | DLGEEHFK | 0.02 | 10.6 | 6.6E + 3 | – | – | – | 0.01 | 13.6 | 4.2E + 3 | – | – | – | 16.23 |
| 517.75 | 2 | NECFLSHK | 0.02 | 14.3 | 8.1E + 4 | 0.53 | 37.8 | 1.4E + 4 | 0.02 | 9.4 | 2.4E + 4 | – | – | – | 16.6 |
| 395.23 | 2 | LVTDLTK | 0.04 | 68.1 | 4.5E + 5 | 0.07 | 50.6 | 8.3E + 4 | 0.03 | 16.5 | 1.8E + 5 | – | – | – | 17.02 |
| 379.70 | 2 | GACLLPK | 0.08 | 80.8 | 4.3E + 5 | 0.11 | 43.3 | 2.4E + 4 | 0.05 | 23.2 | 1.9E + 5 | – | – | – | 17.65 |
| 419.21 | 4 | QEPERNECFLSHK | 0.02 | 11.9 | 3.1E + 4 | – | – | – | 0.01 | 12.9 | 9.1E + 3 | – | – | – | 17.93 |
| 751.81 | 2 | EYEATLEECCAK | 0.01 | 12.7 | 2.9E + 5 | 0.43 | 58.5 | 4.2E + 4 | 0.02 | 14.0 | 2.6E + 5 | – | – | – | 18.23 |
| 554.25 | 2 | EACFAVEGPK | 0.03 | 12.6 | 5.1E + 4 | 0.22 | 42.3 | 1.9E + 4 | 0.04 | 16.2 | 5.2E + 4 | – | – | – | 19.69 |
| 874.40 | 2 | YNGVFQECCQAEDK | 0.03 | 11.4 | 1.1E + 5 | – | – | – | 0.04 | 12.1 | 2.2E + 4 | – | – | – | 19.98 |
| 461.74 | 2 | AEFVEVTK | 0.09 | 18.1 | 2.6E + 4 | 0.12 | 27.3 | 8.3E + 4 | 0.04 | 20.0 | 2.2E + 5 | – | – | – | 20.05 |
| 583.89 | 3 | ECCHGDLLECADDR | 0.03 | 16.5 | 1.9E + 5 | 0.66 | 64.2 | 7.7E + 4 | 0.04 | 16.6 | 1.2E + 5 | – | – | – | 20.09 |
| 511.65 | 3 | LKECCDKPLLEK | 0.02 | 20.4 | 8.1E + 4 | – | – | – | 0.02 | 12.4 | 9.0E + 4 | – | – | – | 20.83 |
| 643.28 | 3 | CCAADDKEACFAVEGPK | 0.03 | 13.7 | 3.6E + 4 | – | – | – | 0.03 | 14.1 | 5.0E + 3 | – | – | – | 20.98 |
| 518.88 | 3 | DDPHACYSTVFDK | 0.04 | 13.1 | 6.7E + 3 | – | – | – | 0.06 | 12.6 S | 4.6E + 3 | – | – | – | 21.87 |
| 417.20 | 3 | FKDLGEEHFK | 0.01 | 13.9 | 1.6E + 5 | 0.66 | 64.8 | 6.8E + 4 | 0.02 | 13.3 | 1.2E + 5 | – | – | – | 22.54 |
| 464.25 | 2 | YLYEIAR | 0.06 | 14.2 | 1.6E + 5 | 0.01 | 47.4 | 3.1E + 4 | 0.09 | 22.5 | 2.4E + 5 | – | – | – | 24.44 |
| 526.26 | 3 | LKPDPNTLCDEFK | 0.06 | 29.8 | 2.1E + 5 | – | – | – | 0.10 | 20.4 | 1.9E + 5 | – | – | – | 25.03 |
| 674.01 | 3 | LKPDPNTLCDEFKADEK | 0.05 | 30.2 | 1.3E + 5 | – | – | – | 0.09 | 39.8 | 1.9E + 5 | – | – | – | 25.36 |
| 653.35 | 2 | HLVDEPQNLIK | 0.05 | 19.3 | 2.2E + 5 | 0.41 | 71.3 | 8.9E + 4 | 0.07 | 20.4 | 2.9E + 5 | – | – | – | 25.69 |
| 636.06 | 4 | QEPERNECFLSHKDDSPDLPK | 0.03 | 12.8 | 9.6E + 4 | – | – | – | 0.05 | 23.5 | 1.1E + 5 | – | – | – | 26.22 |
| 634.62 | 3 | NECFLSHKDDSPDLPK | 0.02 | 14.3 | 8.6E + 4 | 0.74 | 53.2 | 2.3E + 4 | 0.04 | 13.2 | 5.8E + 4 | – | – | – | 26.24 |
| 749.99 | 3 | ECCHGDLLECADDRADLAK | 0.04 | 9.4 | 3.5E + 4 | 0.35 | 49.8 | 2.9E + 4 | 0.07 | 12.9 | 1.0E + 5 | – | – | – | 26.73 |
| 547.31 | 3 | KVPQVSTPTLVEVSR | 0.07 | 23.9 | 2.5E + 5 | 0.54 | 75.3 | 8.6E + 4 | 0.11 | 40.0 | 3.6E + 5 | – | – | – | 26.99 |
| 582.31 | 2 | LVNELTEFAK | 0.13 | 35.1 | 5.1E + 5 | 0.06 | 54.0 | 1.4E + 5 | 0.21 | 39.5 | 5.4E + 5 | – | – | – | 28.48 |
| 710.35 | 2 | SLHTLFGDELCK | 0.16 | 23.7 | 4.0E + 5 | 0.32 | 61.1 | 8.2E + 4 | 0.25 | 38.0 | 4.9E + 5 | – | – | – | 30.18 |
| 480.60 | 3 | RHPEYAVSVLLR | 0.11 | 27.3 | 2.1E + 5 | 0.57 | 70.6 | 3.6E + 4 | 0.18 | 38.7 | 3.0E + 5 | – | – | – | 32.16 |
| 507.81 | 2 | QTALVELLK | 0.26 | 25.5 | 1.6E + 5 | 0.05 | 43.2 | 2.6E + 4 | 0.38 | 26.5 | 1.5E + 5 | – | – | – | 32.23 |
| 642.33 | 2 | HPEYAVSVLLR | 0.16 | 24.8 | 3.0E + 4 | 0.85 | 18.5 | 4.3E + 3 | 0.25 | 14.6 | 2.1E + 4 | – | – | – | 32.31 |
| 627.64 | 3 | RPCFSALTPDETYVPK | 0.09 | 25.8 | 2.4E + 5 | 0.47 | 56.2 | 5.9E + 4 | 0.15 | 42.7 | 3.3E + 5 | – | – | – | 32.51 |
| 571.86 | 2 | KQTALVELLK | 0.15 | 22.2 | 1.5E + 5 | 0.37 | 58.4 | 6.0E + 4 | 0.24 | 22.7 | 2.4E + 5 | – | – | – | 33.2 |
| 740.41 | 2 | LGEYGFQNALIVR | 0.27 | 24.2 | 3.4E + 5 | 0.04 | 51.9 | 5.5E + 4 | 0.40 | 27.8 | 3.9E + 5 | – | – | – | 35.04 |
| 636.64 | 3 | LFTFHADICTLPDTEK | 0.24 | 34.6 | 4.0E + 5 | 0.33 | 74.8 | 1.4E + 5 | 0.38 | 45.9 | 4.8E + 5 | – | – | – | 35.93 |
| 820.07 | 3 | DAIPENLPPLTADFAEDKDVCK | 0.28 | 46.4 | 5.5E + 5 | 0.71 | 67.8 | 9.2E + 4 | 0.42 | 41.3 | 4.5E + 5 | – | – | – | 35.96 |
| 630.32 | 3 | HPYFYAPELLYYANK | 0.68 | 36.2 | 4.7E + 4 | – | – | – | 0.48 | 43.4 | 9.3E + 4 | – | – | – | 36.86 |
| 700.35 | 2 | TVMENFVAFVDK | 0.56 | 45.2 | 8.6E + 5 | 0.03 | 48.5 | 1.5E + 5 | 0.69 | 46.5 | 9.9E + 5 | – | – | – | 37.53 |
| 878.68 | 4 | SHCIAEVEKDAIPENLPPLTADFAEDKDVCK | 0.28 | 45.0 | 7.5E + 5 | 0.77 | 53.2 | 2.1E + 5 | 0.43 | 43.8 | 9.0E + 5 | – | – | – | 38.36 |
| 784.36 | 2 | DAFLGSFLYEYSR | 0.81 | 54.0 | 4.6E + 3 | 0.12 | 33.3 | 7.3E + 3 | 0.52 | 27.3 | 1.4E + 4 | – | – | – | 39.68 |
| 862.90 | 2 | MPCTEDYLSLILNR | 0.67 | 25.2 | 1.1E + 5 | 0.08 | 45.2 | 1.9E + 4 | 0.80 | 26.1 | 1.1E + 5 | – | – | – | 43.23 |
| 831.45 | 3 | GLVLIAFSQYLQQCPFDEHVK | 0.87 | 20.7 | 3.0E + 4 | – | – | – | 1.00 | 19.5 | 3.4E + 4 | – | – | – | 49.35 |
| (Native peptides and glycopeptides of horseradish peroxidase) | |||||||||||||||
| 511.72 | 2 | DAFGNANSAR | 0.01 | 22.6 | 1.1E + 5 | 0.22 | 69.4 | 3.9E + 4 | 0.01 | 10.0 | 2.0E + 4 | – | – | – | 10.22 |
| 468.23 | 2 | TPTIFDNK | 0.05 | 11.4 | 4.0E + 3 | 0.14 | 26.2 | 1.5E + 5 | 0.07 | 30.2 | 4.1E + 5 | – | – | – | 18.67 |
| 593.28 | 2 | YYVNLEEQK | 0.04 | 16.6 | 2.6E + 5 | 0.18 | 68.0 | 4.9E + 4 | 0.06 | 19.2 | 2.9E + 5 | – | – | – | 19.78 |
| 372.23 | 2 | IAASILR | 0.06 | 10.5 | 1.8E + 4 | 0.00 | 39.4 | 2.3E + 4 | 0.06 | 20.8 | 2.7E + 5 | – | – | – | 19.85 |
| 480.25 | 2 | DTIVNELR | 0.06 | 18.4 | 2.5E + 5 | 0.12 | 23.0 | 1.9E + 5 | 0.08 | 24.5 | 3.7E + 5 | – | – | – | 20.71 |
| 402.21 | 2 | GFPVIDR | 0.07 | 26.4 | 3.1E + 5 | 0.04 | 50.1 | 6.1E + 4 | 0.10 | 38.4 | 3.4E + 5 | – | – | – | 22.12 |
| 793.90 | 2 | MGNITPLTGTQGQIR | 0.06 | 11.1 | 9.5E + 4 | – | – | – | 0.10 | 32.7 | 1.1E + 5 | – | – | – | 26.72 |
| 738.36 | 2 | SSDLVALSGGHTFGK | 0.07 | 14.5 | 1.4E + 5 | 0.47 | 37.5 | 2.2E + 4 | 0.11 | 16.6 | 3.0E + 5 | – | – | – | 26.92 |
| 921.89 | 2 | NVGLNR[GlcNAc2Man3XylFuc] | – | – | – | 0.86 | 46.5 | 4.2E + 5 | – | – | – | 0.82 | 44.3 | 8.5E + 5 | – |
| 1118.45 | 3 | SFANSTQTFFNAFVEAMDR[GlcNAc2Man3XylFuc] | 1.00 | 23.2 | 1.6E + 4 | 0.69 | 16.7 | 3.5E + 3 | – | – | – | – | – | – | – |
| 1224.55 | 3 | GLIQSDQELFSSPNATDTIPLVR[GlcNAc2Man3XylFuc] | 0.38 | 35.8 | 4.1E + 5 | 0.71 | 51.7 | 1.3E + 5 | 0.53 | 41.6 | 4.3E + 5 | – | – | – | – |
| 1425.68 | 2 | GLIQSDQELFSSPNATDTIPLVR[GlcNAcFuc] | 0.38 | 31.9 | 6.0E + 3 | – | – | – | 0.57 | 21.7 | 7.1E + 3 | – | – | – | – |
| 1661.70 | 3 | LYNFSNTGLPDPTLNTTYLQTLR[GlcNAc2Man3XylFuc]2 | 0.43 | 15.9 | 5.6E + 4 | 0.87 | 49.2 | 3.5E + 4 | 0.57 | 28.5 | 6.9E + 4 | – | – | – | – |
| (Native peptides and glycopeptides of ribonuclease B) | |||||||||||||||
| 331.67 | 2 | TTQANK | – | – | – | 0.50 | 17.9 | 8.9E + 4 | – | – | – | 0.38 | 15.4 | 1.1E + 4 | 2.17 |
| 458.19 | 2 | YPNCAYK | 0.00 | 11.9 | 2.3E + 3 | 0.14 | 30.9 | 2.8E + 4 | 0.00 | 12.2 | 7.9E + 4 | – | – | – | 7.78 |
| 839.71 | 3 | CKPVNTFVHESLADVQAVCSQK | 0.21 | 17.8 | 2.7E + 4 | – | – | – | 0.35 | 26.0 | 2.1E + 4 | – | – | – | 34.83 |
| 742.03 | 3 | HIIVACEGNPYVPVHFDASV | 0.25 | 69.9 | 1.6E + 6 | 0.38 | 78.2 | 2.8E + 5 | 0.39 | 81.0 | 1.5E + 6 | – | – | – | 37.91 |
| 846.34 | 2 | NLTK [GlcNAc2Man5] | – | – | – | 0.89 | 45.9 | 5.1E + 5 | – | – | – | 0.86 | 48.4 | 9.5E + 5 | – |
| 927.37 | 2 | NLTK [GlcNAc2Man6] | – | – | – | 0.93 | 48.7 | 3.7E + 5 | – | – | – | 0.90 | 40.7 | 6.5E + 5 | – |
| 1008.40 | 2 | NLTK [GlcNAc2Man7] | – | – | – | 0.95 | 48.9 | 8.7E + 4 | – | – | – | 0.94 | 39.7 | 1.4E + 5 | – |
| 1089.43 | 2 | NLTK [GlcNAc2Man8] | – | – | – | 0.98 | 49.5 | 8.1E + 4 | – | – | – | 0.97 | 38.1 | 1.3E + 5 | – |
| 1170.44 | 2 | NLTK [GlcNAc2Man9] | – | – | – | 1.00 | 46.2 | 2.3E + 4 | – | – | – | 1.00 | 37.0 | 2.9E + 4 | – |
The glycopeptides were revealed by the XIC of the diagnostic ions [HexNAc + H] + (m/z 204.10), [HexNacHex + H] + (m/z 366.10), and [HexNAcHex2 + H] + (m/z 528.10). Hydrophobicity values were calculated using Sequence Specific Retention Calculator v. 3.0 [33]
Fig. 3.
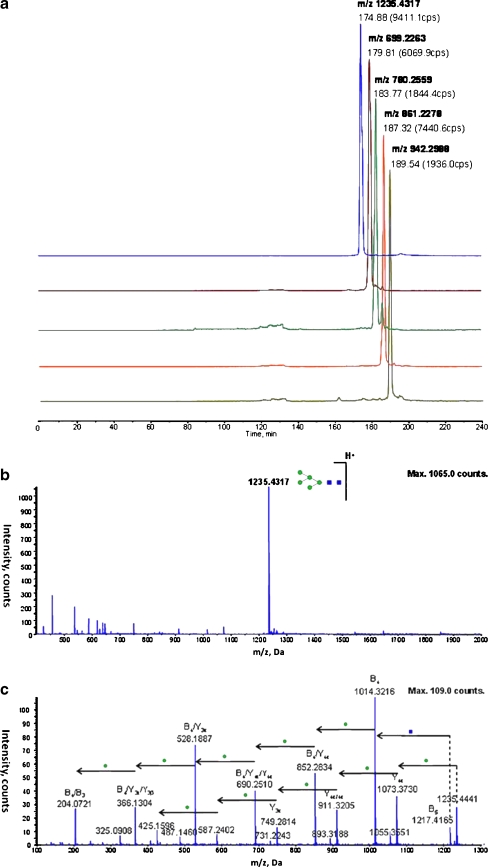
The (a) full-scan and (b) product ion spectra of a typical glycopeptide [GLIQSDQELFSSPNATDTIPLVR-GlcNAc2Man3XylFuc + 3H]3+ from horseradish peroxidase as identified using RP-HILIC, showing characteristic fragment ions. The peptide sequence was elucidated from the b- and y- ion series, whereas the N-glycan product ions were identified using the software GlcyoWorkbench. The characteristic signals of protonated Man (m/z 163.07), GlcNAc (m/z 204.08), ManXyl (m/z 295.10), GlcNAcMan (m/z 366.15), GlcNAcManXyl (498.17), GlcNAcMan2 (m/z 528.18), GlcNAcMan2Xyl (m/z 660.24), GlcNAcMan3 (m/z 690.25) and GlcNAcMan3Xyl (m/z 822.26) can be discerned. Manual examination identified further glycopeptide signals such as protonated y11-Man (m/z 1399.74), y12-Man (m/z 1486.76), y13-Man (m/z 1573.75), y14-Man (m/z 1720.78) as well as GLIQSDQELFSSPNATDTIPLVR -Man (m/z 1356.65) and GLIQSDQELFSSPNATDTIPLVR -ManFuc (m/z 1425.64)
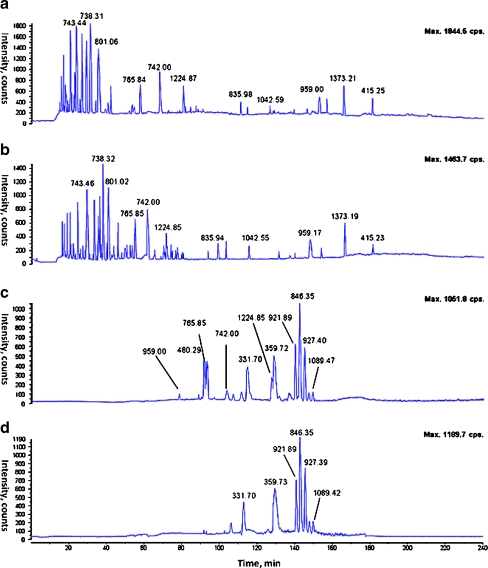
Figure 4 displays mass spectrometric base peak chromatographs (BPCs) of the protein standards, recorded after performing standalone RP, standalone HILIC, and hyphenated RP-HILIC separations. From a comparison of the BPCs for the standalone RP separation and the RP separation of the RP-HILIC process, it appears that the hyphenated system did not deteriorate the performance of the RP peptide separation; we observe only minimal shifts in the retention times (within 1–2 s). Furthermore, relative to the BPC of the standalone HILIC, the HILIC chromatograph from the RP-HILIC system is considerably simpler because the RP trap column removed the signals of the relatively hydrophobic peptides. For example, chromatographic peaks corresponding to the peptide ion at m/z 742.00 are present in the BPCs of the standalone RP separation, the standalone HILIC separation, and the RP separation of the RP-HILIC system, but is absent in the BPC from the coupled HILIC system. In addition, the increased sensitivity in the coupled HILIC system, arising from the reduced sample complexity, is evident when comparing the intensities of the two peptide ions in the full-scan mass spectrum of the RP-HILIC system (Fig. 5) with those in the corresponding spectra after standalone HILIC; the latter are populated by co-eluting peptides that mutually suppress their signal strengths through competition during electrospray ionization. We suspect that such improved ionization efficiency will benefit complex sample analyses when some of the eluting peptides are present in only marginally detectable concentrations. In addition, because only one sample injection event is required for RP-HILIC, the coupled system is essentially capable of harnessing the combined separation power of the standalone RP and HILIC systems while consuming only half the amount of sample.
Fig. 4.
The base peak chromatograph (BPC) of all peptide peaks from the mixture of ribonuclease B, bovine serum albumin and horseradish peroxidase in (a) standalone RP; (b) RP in RP-HILIC; (c) standalone HILIC and (d) HILIC in RP-HILIC. The elution profile of standalone RP and RP in RP-HILIC show high degree of reproducibility, with highly similar profile and only minor shifts in retention. The HILIC chromatograph in RP-HILIC shows a simpler chromatographic profile especially in analytes with shorter retention time, i.e., relatively hydrophobic compounds. The chromatographic peak corresponding to m/z 742.00, observed in both standalone RP and HILIC, is missing in coupled HILIC and shows up instead in the RP part of RP-HILIC. The peak is from the relatively hydrophobic peptide [HIIVACEGNPYVPVHFDASV + 3H]3+ with a calculated relative hydrophobicity of approximately 38
Fig. 5.
The full-scan spectra of two peptide ions [TCVADESHAGCEK +3H]3+ at m/z 488.5 at 54.3 min, and [SHCIAEVEK + 2H]2+ at m/z 536.72 at 58.9 min in (a) and (b) standalone HILIC, and (c) and (d) RP-HILIC, showing the simplified elution profile in RP-HILIC. Several co-eluting ions such as m/z 583.87 and m/z 699.99 found in standalone HILIC are extracted instead in the RP part of RP-HILIC
Retention of released glycans in deglycosylated glycoproteins
Next, we demonstrate the applicability of RP-HILIC to studies of a wide range of compounds in non-targeted analyses through the simultaneous separation of peptides and PNGaseF-released glycan chains from deglycosylated glycoproteins. We analyzed turkey ovalbumin, a previously uncharacterized glycoprotein featuring highly complex glycoforms, as well as the glycoprotein standard ribonuclease B after PNGaseF-mediated deglycosylation and trypsin-mediated digestion. For turkey ovalbumin, the mixture of peptides and highly hydrophilic glycan chains was successfully extracted and separated by the RP-HILIC system, enabling us to report 23 novel N-glycan compositions (Table 2). From ribonuclease B, all five known major N-linked glycans (GlcNAc2Man5, GlcNAc2Man6, GlcNAc2Man7, GlcNAc2Man8, GlcNAc2Man9) were separated and detected through unequivocal spectral evidence (Fig. 6); the peptide coverage was similar to that obtained using standalone RP (data not shown).
Table 2.
Glycoforms in turkey ovalbumin detected using RP-HILIC MS/MS, revealing 23 previously uncharacterized N-glycans
| Exp. m/z | Charge (+) | Experimental Mass | Theoretical Mass | Δm/z (ppm) | Glycan composition | |
|---|---|---|---|---|---|---|
| 1 | 659.2420 | 2 | 1316.4680 | 1316.4762 | −6.23 | (HexNAc)2 + (Man)3(GlcNAc)2 |
| 2 | 537.1875 | 2 | 1072.3590 | 1072.3690 | −9.33 | (Hex)1 + (Man)3(GlcNAc)2 |
| 3 | 638.7215 | 2 | 1275.4270 | 1275.4480 | −16.46 | (Hex)1(HexNAc)1 + (Man)3(GlcNAc)2 |
| 4 | 760.7841 | 2 | 1519.5522 | 1519.5547 | −1.65 | (HexNAc)3 + (Man)3(GlcNAc)2 |
| 5 | 862.3154 | 2 | 1722.6148 | 1722.6340 | −11.15 | (HexNAc)4 + (Man)3(GlcNAc)2 |
| 6 | 1235.4082 | 1 | 1234.4002 | 1234.4227 | −18.23 | (Hex)2 + (Man)3(GlcNAc)2 |
| 7 | 740.2648 | 2 | 1478.5136 | 1478.5292 | −10.55 | (Hex)1(HexNAc)2 + (Man)3(GlcNAc)2 |
| 8 | 841.8002 | 2 | 1681.5844 | 1681.6077 | −13.86 | (Hex)1(HexNAc)3 + (Man)3(GlcNAc)2 |
| 9 | 1397.4729 | 1 | 1396.4649 | 1396.4762 | −8.09 | (Hex)3 + (Man)3(GlcNAc)2 |
| 10 | 719.7494 | 2 | 1437.4828 | 1437.5017 | −13.15 | (Hex)2(HexNAc)1 + (Man)3(GlcNAc)2 |
| 11 | 821.2895 | 2 | 1640.5630 | 1640.5817 | −11.40 | (Hex)2(HexNAc)2 + (Man)3(GlcNAc)2 |
| 12 | 922.8255 | 2 | 1843.6350 | 1843.6612 | −14.21 | (Hex)2(HexNAc)3 + (Man)3(GlcNAc)2 |
| 13 | 943.3355 | 2 | 1884.6550 | 1884.6878 | −17.40 | (Hex)1(HexNAc)4 + (Man)3(GlcNAc)2 |
| 14 | 1024.3636 | 2 | 2046.7112 | 2046.7402 | −14.17 | (Hex)2(HexNAc)4 + (Man)3(GlcNAc)2 |
| 15 | 1044.8703 | 2 | 2087.7246 | 2087.7672 | −20.40 | (Hex)1(HexNAc)5 + (Man)3(GlcNAc)2 |
| 16 | 800.7698 | 2 | 1599.5236 | 1599.5552 | −19.76 | (Hex)3(HexNAc)1 + (Man)3(GlcNAc)2 |
| 17 | 902.3268 | 2 | 1802.6376 | 1802.6342 | 1.89 | (Hex)3(HexNAc)2 + (Man)3(GlcNAc)2 |
| 18 | 1003.8547 | 2 | 2005.6934 | 2005.7142 | −10.37 | (Hex)3(HexNAc)3 + (Man)3(GlcNAc)2 |
| 19 | 1105.3951 | 2 | 2208.7742 | 2208.7932 | −8.60 | (Hex)3(HexNAc)4 + (Man)3(GlcNAc)2 |
| 20 | 1125.9025 | 2 | 2249.7890 | 2249.8202 | −13.87 | (Hex)2(HexNAc)5 + (Man)3(GlcNAc)2 |
| 21 | 804.9570 | 3 | 2411.8470 | 2411.8732 | −10.86 | (Hex)3(HexNAc)5 + (Man)3(GlcNAc)2 |
| 22 | 834.2967 | 3 | 2499.8661 | 2499.9142 | −19.24 | (HexNAc)3(Deoxyhexose)4(Pent)3 + (Man)3(GlcNAc)2 |
| 23 | 858.9686 | 3 | 2573.8818 | 2573.9252 | −16.86 | (Hex)4(HexNAc)5 + (Man)3(GlcNAc)2 |
Fig. 6.
(a) The extracted ion chromatograph (XIC) of the five major N-linked glycans in ribonuclease B analyzed by RP-HILIC after PNGaseF deglycosylation: GlcNAc2Man5 (m/z 1235.43, 1+), GlcNAc2Man6 (m/z 699.23, 2+), GlcNAc2Man7 (m/z 780.26, 2+), GlcNAc2Man8 (m/z 861.23, 2+), and GlcNAc2Man9 (m/z 942.30, 2+). Typical (b) full-scan and (c) product ion scan spectra of [GlcNAc2Man9 + H]+ (m/z 1235.43), showing characteristic glycan fragment ions; the glycan composition was identified and labeled according to GlycoWorkbench
Because the released glycans are considerably more hydrophilic than typical glycopeptides, we obtained a larger discrepancy in their analyses when using the RP and HILIC systems alone for the efficient separation of the different components in the glycoprotein samples. The great number of similar glycoforms present in complex glycoproteins, such as turkey ovalbumin, makes them difficult to characterize without efficient separation, suggesting that RP-HILIC has great potential for allowing sophisticated analyses of highly heterogeneous mixtures.
Increased component coverage in complex biological samples
To test the performance of RP-HILIC in the analysis of highly complex proteomics samples, we analyzed the total protein extract of mouse chondrogenic cells without further purification. From a single injection of a 10-μg sample, we identified a total of 6,812 distinct peptides using the RP component of the RP-HILIC system, translating into the identification of 493 unique proteins (95% confidence). The coupled HILIC system contributed a further 985 distinct peptides and allowed the overall identification of an additional 11 proteins (see Electronic Supplementary Material Figure S1). We attribute the increased coverage mainly to the separation and detection of hydrophilic peptides in the coupled HILIC. The calculated average hydrophobicity of the detected peptides in the RP component was 30.5 (ranging from 2.48 to 63.4); in contrast, the coupled HILIC allowed the detection of considerably-more-hydrophilic peptides, with a calculated average hydrophobicity of 12.3 (from 3.17 to 20.24; see Electronic Supplementary Material Figure S2). Our results obtained using the RP and RP-HILIC systems reveal that at least 10% of the detected proteins saw increases in sequence coverage when we included the coupled HILIC component; this improvement is likely to benefit peptide-based proteomics applications, such as bottom-up quantification and characterization of post-translational modifications.
Finally, when we evaluated the performance of the RP-HILIC system in the separation of deglycosylated and digested human serum, we identified more than 80 potential glycans without prior immunodepletion of abundant serum proteins. We foresee that these glycan structures might be resolved through the combined use of RP-HILIC and mass-analyzed laser desorption ionization (MALDI)–time-of-flight (TOF)/TOF MS/MS. After RP analysis of the relatively hydrophobic peptides, it might be worthwhile to directly deposit the HILIC eluent onto a MALDI plate for offline analysis. Because MALDI-TOF/TOF systems display higher glycan ionization efficiencies than electrospray ionization MS systems, they might potentially reveal cross-ring cleavage information of N-glycans when using high-energy collision-induced dissociation [34–36].
Conclusion
The online coupling of RP and HILIC systems in a micro-flow scheme allows the simultaneous analyses of relatively hydrophobic and hydrophilic compounds. We solved the problem of solvent incompatibility between RP and HILIC columns through the use of constant-pressure solvent delivery and flow-splitting capillaries of different internal diameters. Thus, the RP flow-through was converted from highly aqueous to highly organic by mixing it with a large volume of organic solvent prior to HILIC extraction. In contrast to common column-switching set-ups, our RP-HILIC system is simpler to use: it requires only two solvent pumps and is readily compatible with the flow-rate schemes required in proteomics applications. We demonstrated the power of RP-HILIC to separate peptides, glycopeptides, and glycans from a single injection event through the analyses of protein standards and complex proteomics samples. This approach has the potential to extend the scope of current proteomics and non-targeted “omics” analyses by aiding the detection of highly heterogeneous compounds.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 5448 kb)
Acknowledgments
This study was supported by the Hong Kong Research Grant Council (Project Nos. HKU7640/07M, HKU7650/09P, and HKU1/07C), Hong Kong Special Administrative Region, China, and the HKU seed funding program (200811159027). We thank Dr. W. L. Chan for providing the mouse chondrogenic cell protein extracts.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Maggie P. Y. Lam and S. O. Sin contributed equally to this work.
References
- 1.Sandra K, Moshir M, D’Hondt F, Verleysen K, Kas K, Sandra P. Highly efficient peptide separations in proteomics. Part 1. Unidimensional high performance liquid chromatography. J Chromatogr B. 2008;866:48–63. doi: 10.1016/j.jchromb.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 2.Saz JM, Marina ML. Application of micro- and nano-HPLC to the determination and characterization of bioactive and biomarker peptides. J Sep Sci. 2008;31:446–458. doi: 10.1002/jssc.200700589. [DOI] [PubMed] [Google Scholar]
- 3.Garcia MC. The effect of the mobile phase additives on sensitivity in the analysis of peptides and proteins by high-performance liquid chromatography-electrospray mass spectrometry. J Chromatogr B. 2005;825:111–123. doi: 10.1016/j.jchromb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 4.Garcia MC, Hogenboom AC, Zappey H, Irth H. Effect of the mobile phase composition on the separation and detection of intact proteins by reversed-phase liquid chromatography—electrospray mass spectrometry. J Chromatogr A. 2002;957:187–199. doi: 10.1016/S0021-9673(02)00345-X. [DOI] [PubMed] [Google Scholar]
- 5.Ren D, Pipes G, Xiao G, Kleernann GR, Bondarenko PV, Treuheit MJ, Gadgil HS. Reversed-phase liquid chromatography-mass spectrometry of site-specific chemical modifications in intact immunoglobulin molecules and their fragments. J Chromatogr A. 2008;1179:198–204. doi: 10.1016/j.chroma.2007.11.088. [DOI] [PubMed] [Google Scholar]
- 6.Horvath C, Melander W, Molnar I, Molnar P. Enhancement of retention by ion-pair formation in liquid-chromatography with nonpolar stationary phases. Anal Chem. 1977;49:2295–2305. doi: 10.1021/ac50022a048. [DOI] [Google Scholar]
- 7.Toll H, Oberacher H, Swart R, Huber CG. Separation, detection, and identification of peptides by ion-pair reversed-phase high-performance liquid chromatography-electrospray ionization mass spectrometry at high and low pH. J Chromatogr A. 2005;1079:274–286. doi: 10.1016/j.chroma.2005.03.121. [DOI] [PubMed] [Google Scholar]
- 8.Doneanu CE, Chen WB, Gebler JC. Analysis of oligosaccharides derived from heparin by ion-pair reversed-phase chromatography/mass spectrometry. Anal Chem. 2009;81:3485–3499. doi: 10.1021/ac802770r. [DOI] [PubMed] [Google Scholar]
- 9.Hemstrom P, Irgum K. Hydrophilic interaction chromatography. J Sep Sci. 2006;29:1784–1821. doi: 10.1002/jssc.200600199. [DOI] [PubMed] [Google Scholar]
- 10.Boersema PJ, Mohammed S, Heck AJR. Hydrophilic interaction liquid chromatography (HILIC) in proteomics. Anal Bioanal Chem. 2008;391:151–159. doi: 10.1007/s00216-008-1865-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu YQ, Gilar M, Kaska J, Gebler JC. A rapid sample preparation method for mass spectrometric characterization of N-linked glycans. Rapid Commun Mass Spectrom. 2005;19:2331–2336. doi: 10.1002/rcm.2067. [DOI] [PubMed] [Google Scholar]
- 12.Shimizu Y, Nakata M, Kuroda Y, Tsutsumi F, Kojima N, Mizuochi T. Rapid and simple preparation of N-linked oligosaccharides by cellulose-column chromatography. Carbohydr Res. 2001;332:381–388. doi: 10.1016/S0008-6215(01)00113-6. [DOI] [PubMed] [Google Scholar]
- 13.Charlwood J, Birrell H, Organ A, Camilleri P. A chromatographic and mass spectrometric strategy for the analysis of oligosaccharides: determination of the glycan structures in porcine thyroglobulin. Rapid Commun Mass Spectrom. 1999;13:716–723. doi: 10.1002/(SICI)1097-0231(19990430)13:8<716::AID-RCM547>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Urgaonkar S, Verkade JG, Armstrong DW. Separation and characterization of underivatized oligosaccharides using liquid chromatography and liquid chromatography-electrospray ionization mass spectrometry. J Chromatogr A. 2005;1079:146–152. doi: 10.1016/j.chroma.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Rudd PM, Colominas C, Royle L, Murphy N, Hart E, Merry AH, Hebestreit HF, Dwek RA. A high-performance liquid chromatography based strategy for rapid, sensitive sequencing of N-linked oligosaccharide modifications to proteins in sodium dodecyl sulphate polyacrylamide electrophoresis gel bands. Proteomics. 2001;1:285–294. doi: 10.1002/1615-9861(200102)1:2<285::AID-PROT285>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida T. Peptide separation in normal phase liquid chromatography. Anal Chem. 1997;69:3038–3043. doi: 10.1021/ac9702204. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida T. Peptide separation by hydrophilic-interaction chromatography: a review. J Biochem Biophys Methods. 2004;60:265–280. doi: 10.1016/j.jbbm.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Wuhrer M, Koeleman CAM, Hokke CH, Deelder AM. Protein glycosylation analyzed by normal-phase nano-liquid chromatography-mass spectrometry of glycopeptides. Anal Chem. 2005;77:886–894. doi: 10.1021/ac048619x. [DOI] [PubMed] [Google Scholar]
- 19.Yu YQ, Fournier J, Gilar M, Gebler JC. Identification of N-Linked glycosylation sites using glycoprotein digestion with pronase prior to MALDI tandem time-of-flight mass spectrometry. Anal Chem. 2007;79:1731–1738. doi: 10.1021/ac0616052. [DOI] [PubMed] [Google Scholar]
- 20.McNulty DE, Annan RS. Hydrophilic interaction chromatography reduces the complexity of the phosphoproteome and improves global phosphopeptide isolation and detection. Mol Cell Proteomics. 2008;7:971–980. doi: 10.1074/mcp.M700543-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Dugo P, Favoino O, Luppino R, Dugo G, Mondello L. Comprehensive two-dimensional normal-phase (adsorption)–reversed-phase liquid chromatography. Anal Chem. 2004;76:2525–2530. doi: 10.1021/ac0352981. [DOI] [PubMed] [Google Scholar]
- 22.Dugo P, Skerikova V, Kumm T, Trozzi A, Jandera P, Mondello L. Elucidation of carotenoid patterns in citrus products by means of comprehensive normal-phase x reversed-phase liquid chromatography. Anal Chem. 2006;78:7743–7750. doi: 10.1021/ac061290q. [DOI] [PubMed] [Google Scholar]
- 23.François I, de Villiers A, Sandra P. Considerations on the possibilities and limitations of comprehensive normal phase-reversed phase liquid chromatography (NPLC × RPLC) J Sep Sci. 2006;29:492–498. doi: 10.1002/jssc.200500451. [DOI] [PubMed] [Google Scholar]
- 24.Hawryl MA, Soczewinski E. Separation of some flavonoids using RP-HPLC-NP-TLC off-line coupled system. Chromatographia. 2000;52:175–178. doi: 10.1007/BF02490451. [DOI] [Google Scholar]
- 25.Giddings JC. Concepts and comparisons in multidimensional separation. J High Resolut Chrom. 1987;10:319–323. doi: 10.1002/jhrc.1240100517. [DOI] [Google Scholar]
- 26.Wang Y, Lehmann R, Lu X, Zhao XJ, Xu GW. Novel, fully automatic hydrophilic interaction/reversed-phase column-switching high-performance liquid chromatographic system for the complementary analysis of polar and apolar compounds in complex samples. J Chromatogr A. 2008;1204:28–34. doi: 10.1016/j.chroma.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Lu X, Xu GW. Simultaneous separation of hydrophilic and hydrophobic compounds by using an online HILIC-RPLC system with two detectors. J Sep Sci. 2008;31:1564–1572. doi: 10.1002/jssc.200700663. [DOI] [PubMed] [Google Scholar]
- 28.Shen Y, Tolic N, Zhao R, Pasa-Tolic L, Li L, Berger SJ, Harkewicz R, Anderson GA, Belov ME, Smith RD. High-throughput proteomics using high-efficiency multiple-capillary liquid chromatography with on-line high-performance ESI FTICR mass spectrometry. Anal Chem. 2001;73:3011–3021. doi: 10.1021/ac001393n. [DOI] [PubMed] [Google Scholar]
- 29.Shen Y, Zhao R, Belov ME, Conrads TP, Anderson GA, Tang K, Pasa-Tolic L, Veenstra TD, Lipton MS, Udseth HR, Smith RD. Packed capillary reversed-phase liquid chromatography with high-performance electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry for proteomics. Anal Chem. 2001;73:1766–1775. doi: 10.1021/ac0011336. [DOI] [PubMed] [Google Scholar]
- 30.Shen YF, Zhao R, Berger SJ, Anderson GA, Rodriguez N, Smith RD. High-efficiency nanoscale liquid chromatography coupled on-line with mass spectrometry using nanoelectrospray ionization for proteomics. Anal Chem. 2002;74:4235–4249. doi: 10.1021/ac0202280. [DOI] [PubMed] [Google Scholar]
- 31.Cooper CA, Gasteiger E, Packer NH. GlycoMod—a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics. 2001;1:340–349. doi: 10.1002/1615-9861(200102)1:2<340::AID-PROT340>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 32.Ceroni A, Maass K, Geyer H, Geyer R, Dell A, Haslam SM. GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J Proteome Res. 2008;7:1650–1659. doi: 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- 33.Krokhin OV, Craig R, Spicer V, Ens W, Standing KG, Beavis RC, Wilkins JA. An improved model for prediction of retention times of tryptic peptides in ion pair reversed-phase HPLC—its application to protein peptide mapping by off-line HPLC-MALDI MS. Mol Cellular Proteomics. 2004;3:908–919. doi: 10.1074/mcp.M400031-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Stephens E, Maslen SL, Green LG, Williams DH. Fragmentation characteristics of neutral N-linked glycans using a MALDI-TOF/TOF tandem mass spectrometer. Anal Chem. 2004;76:2343–2354. doi: 10.1021/ac030333p. [DOI] [PubMed] [Google Scholar]
- 35.Lattova E, Chen VC, Varma S, Bezabeh T, Perreault H. Matrix-assisted laser desorption/ionization on-target method for the investigation of oligosaccharides and glycosylation sites in glycopeptides and glycoproteins. Rapid Commun Mass Spectrom. 2007;21:1644–1650. doi: 10.1002/rcm.3007. [DOI] [PubMed] [Google Scholar]
- 36.Mechref Y, Novotny MV, Krishnan C. Structural characterization of oligosaccharides using MALDI-TOF/TOF tandem mass spectrometry. Anal Chem. 2003;75:4895–4903. doi: 10.1021/ac0341968. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 5448 kb)