Abstract
In eukaryotic cells, degradation of many mRNAs is initiated by removal of the poly(A) tail followed by decapping and 5′-3′ exonucleolytic decay. Although the order of these events is well established, we are still lacking a mechanistic understanding of how deadenylation and decapping are linked. In this report we identify human Pat1b as a protein that is tightly associated with the Ccr4-Caf1-Not deadenylation complex as well as with the Dcp1-Dcp2 decapping complex. In addition, the RNA helicase Rck and Lsm1 proteins interact with human Pat1b. These interactions are mediated via at least three independent domains within Pat1b, suggesting that Pat1b serves as a scaffold protein. By tethering Pat1b to a reporter mRNA, we further provide evidence that Pat1b is also functionally linked to both deadenylation and decapping. Finally, we report that Pat1b strongly induces the formation of processing (P) bodies, cytoplasmic foci that contain most enzymes of the RNA decay machinery. An amino-terminal region within Pat1b serves as an aggregation-prone domain that nucleates P bodies, whereas an acidic domain controls the size of P bodies. Taken together, these findings provide evidence that human Pat1b is a central component of the RNA decay machinery by physically connecting deadenylation with decapping.
By controlling gene expression at the posttranscriptional level, cells can rapidly induce, suppress, or fine-tune the production of specific proteins. A well-studied example is a class of mRNAs that contain AU-rich elements (AREs) in their 3′ untranslated region (UTR), which causes rapid mRNA decay. The degradation of ARE mRNAs can be inhibited by extracellular signals, a mechanism that contributes to the efficient production of cytokines in activated cells of the immune system (37). Another prominent example are mRNAs targeted by microRNAs (miRNAs), which generally cause translational inhibition and/or accelerated mRNA decay (7). The reversible transit of an actively translating mRNA to a state of translational silencing and the irreversible step that elicits mRNA degradation involve crucial rearrangements of the ribonucleoprotein (RNP) composition. In some cases, the transit of an mRNA to a repressed state is associated with its recruitment to processing (P) bodies (3, 14, 35). P bodies are cytoplasmic foci that contain many enzymes of the general mRNA decay pathway, such as the Ccr4-Caf1-Not deadenylase complex and the decapping enzyme Dcp2 together with its activators Dcp1, Edc3, and Hedls, as well as the 5′-3′ exoribonuclease Xrn1 that is tightly associated with the heptameric ring of the Lsm1-7 proteins (2, 10, 13, 21, 24, 35, 41, 43, 47).
For most eukaryotic mRNAs, deadenylation is the first step in the decay pathway. Once the poly(A) tail is removed, the mRNA is either degraded in the 3′-5′ direction by the exosome or decapped at the 5′ end and subsequently degraded in the 5′-3′ direction by Xrn1 (15). Both ARE-mediated mRNA decay and miRNA-mediated mRNA decay make use of this general decay pathway by delivering mRNAs at an accelerated rate to the decay machinery.
An unsolved question is how deadenylation at the 3′ end of an mRNA is linked to decapping at the 5′ end. Since deadenylated but capped mRNAs are virtually undetectable in wild-type (wt) cells, the two events seem to be coupled very tightly. However, it is not clear which factors physically connect the deadenylation and decapping complexes.
Studies of Saccharomyces cerevisiae suggest that Pat1 (protein associated with topoisomerase II, also termed MRT1) might play a role in coupling deadenylation with decapping. On the one hand, yeast mutants lacking Pat1 show reduced rates of mRNA degradation and accumulate deadenylated but capped mRNA (18). Thus, Pat1 was proposed to enhance mRNA decay by activating the decapping reaction. On the other hand, Pat1 was found to interact with the cytoplasmic Lsm1-7 proteins, which form a heptameric ring structure (5, 6). The Lsm1-7-Pat1 complex was shown to preferentially bind to the 3′ end of mRNAs that contain short poly(A) tails (8, 42). Since strains lacking Lsm1 or Pat1 generate transcripts that are truncated at the 3′ end, the Lsm1-7-Pat1 complex also appears to protect mRNA from further trimming after deadenylation (19). Thus, the Lsm1-7-Pat1 complex was proposed to serve as a linker that recognizes oligo- or deadenylated mRNAs at the 3′ end and activates the subsequent step of decapping at the 5′ end.
To further pursue the mechanism by which deadenylation is coupled to decapping, we sought to study the function of human Pat1 homologs. For many years, a metazoan counterpart of yeast Pat1 has not been identified since routine BLAST analysis does not reveal homologous proteins. In 2007, Scheller et al. identified two human homologs which they termed PatL1 and PatL2 (34). This report showed that human PatL1 localizes to P bodies and that its depletion by small interfering RNA (siRNA) causes loss of P bodies. We have independently identified the same two proteins as possible homologs of yeast Pat1. In accordance with the two orthologs in Xenopus laevis, we termed the two proteins Pat1a and Pat1b (Nancy Standard, personal communication). In the present study we provide evidence that human Pat1b (PatL1) physically and functionally connects the deadenylation and decapping machinery. Whereas no function could be attributed to Pat1a (PatL2), we found that Pat1b also controls the assembly of P bodies.
MATERIALS AND METHODS
Plasmids.
The following plasmids have been described previously: pCMV-FLAG-NOT1 (45), pcDNA3-Myc-Ccr4, pcDNA3-Flag-Dcp1a, pcDNA3-Flag-Dcp2 (25), and pCIneo-RL (31).
To generate pcDNA3-HA (p2003), oligonucleotides G85/G86 (for primer sequences, see Table S1 in the supplemental material) were annealed and cloned into the HindIII-EcoRI sites of pcDNA3 (Invitrogen). For pcDNA3-YFP (p2168), yellow fluorescent protein (YFP) was amplified with oligonucleotides G243/G244 and then ligated into the HindIII-KpnI sites of pcDNA3-HA (p2003), thereby replacing the hemagglutinin (HA) tag.
For plasmid pEYFP-Pat1a (p2639), the human Pat1a coding sequence was amplified in three portions from the cDNA of HeLa cells and ligated sequentially into the BglII-XbaI sites of pEYFP-C1 (Clontech/BD Biosciences). The following primer pairs were used: G1332/G1222 (1st PCR) and G1332/G1198 (2nd PCR) for the Pat1a C-terminal portion, ligated as a KpnI-XbaI fragment; G1315/G1221 for the middle portion, ligated as a BglII-KpnI fragment; and G1219/G1340 (1st PCR) and G1353/G1340 (2nd PCR) for the Pat1a N-terminal portion, ligated as a BamHI-BglII fragment and thereby introducing an EcoRI site upstream of the start codon. For pcDNA3-HA-Pat1a (p2638), the human Pat1a coding sequence was excised from pEYFP-Pat1a (p2639) as an EcoRI-XbaI fragment and cloned into the same sites of pcDNA3-HA (p2003).
A human Pat1b cDNA clone (DKFZp451I053) fused at the C terminus to YFP was kindly provided by Stefan Wiemann (German Cancer Research Center, Heidelberg, Germany). In order to generate HA-tagged Pat1b constructs, fragments were amplified by PCR from human Pat1b cDNA and inserted into the KpnI-XhoI sites of pcDNA3-HA (p2003). The following primers were used: G1225/G1230 for pcDNA3-HA-Pat1b (p2516), G1225/G1226 for pcDNA3-HA-AN (p2511), G1225/G1228 for pcDNA3-HA-ANH (p2514), G1227/G1228 for pcDNA3-HA-H (p2512), G1227/G1230 for pcDNA3-HA-HC (p2515), G1229/G1230 for pcDNA3-HA-C (p2513), G1225/G1425 for pcDNA3-HA-A (p2591), G1408/G1226 for pcDNA3-HA-N (p2592), and G1408/G1230 for pcDNA3-HA-dA (p2675). For pcDNA3-HA-dH (p2542), the N- and C-terminal regions of Pat1b were first amplified as separate fragments with primers G1225/G1295 and G1296/G1230, respectively. The PCR products were then aligned, Pat1b lacking the H domain was amplified with primers G1225/G1230, and the PCR product was cloned into the KpnI-XhoI sites of pcDNA3-HA (p2003).
pcDNA3-YFP-Pat1b full length (p2521), pcDNA3-YFP-A (p2630), pcDNA3-YFP-AN (p2626), pcDNA3-YFP-N (p2627), pcDNA3-YFP-ANH (p2522), pcDNA3-YFP-H (p2632), pcDNA3-YFP-C (p2628), pcDNA3-YFP-HC (p2629), pcDNA3-YFP-dA (p2631), and pcDNA3-YFP-dH (p2633) were generated by replacing the HA tag in the corresponding HA-tagged Pat1b constructs with YFP excised as a HindIII-KpnI fragment from pcDNA3-YFP (p2168). For pcDNA3-HA-Pat1b-YFP (p2520), Pat1b-YFP was amplified by PCR with primers G1225/G1253, using clone DKFZp451I053 as a template, and inserted into the KpnI-XhoI sites of pcDNA3-HA-Pat1b (p2516).
In order to generate pcDNA3-HA-PP7cp-Pat1b (p2634), pcDNA3-HA-PP7cp-AN (p2658), pcDNA3-HA-PP7cp-HC (p2659), pcDNA3-HA-PP7cp-dA (p2663), pcDNA3-HA-PP7cp-dH (p2660), and pcDNA3-HA-PP7cp-Pat1a (2661), PP7cp (46) was amplified by PCR using primers G1501/G1502 and inserted into the KpnI site of corresponding pcDNA3-HA-Pat1b constructs and pcDNA3-HA-Pat1a (p2638). Orientation of the insert was checked by sequencing.
For pcDNA3-Bgl (p2001), the BglII site in pcDNA3 (Invitrogen) was removed by digestion with BglII and blunt-end religation, thereby introducing a novel ClaI site. pcDNA3-7B (p2308) expressing a T7-tagged rabbit β-globin gene was subsequently generated by amplifying β-globin by PCR with primers G18/G19 from plasmid puroMXbglobin (39). The fragment was digested with EcoRV and SalI and ligated into the BamHI/blunt-XhoI sites of pcDNA3-Blg. For pcDNA3-7B-PP7bs (p2314), six repeats of the PP7 binding site were excised as a BamHI-BglII fragment from pSP73-6xPP7bs (46) and introduced into the BglII site of pcDNA3-7B (p2308). For pFLB (p2524), firefly luciferase (FL) cDNA was amplified by PCR using primers G1256/G1257 from pGL3-Control (Promega) and cloned into the HindIII-KpnI sites of pcDNA3-7B (2308). From there, the 6× PP7bs repeats were cloned as an EcoRI-BglII fragment from pcDNA3-7B-PP7bs (p2314) into the corresponding sites of pFLB (p2524) to generate pFLB-PP7bs (p2646).
For TOpuro-Caf1a-mycSG (p2485), the human Caf1a cDNA was amplified by reverse transcription (RT)-PCR using primers G1122 and G1123 and cloned into the BamHI-XhoI sites of TOpuro-mycSG (p2484). For pTOpuro-Caf1a-AA-mycSG (p2737) containing the D40A/E42A mutation, a fragment amplified by PCR with primers G1730 and G1731 was inserted into the KpnI-EcoRI sites of TOpuro-Caf1a-mycSG (p2485). For pEGFP-TEV-Caf1a (p2793), Caf1a cDNA was cloned as a BamHI-XhoI fragment into the BglII-XhoI sites of pEGFP-N1 containing a tobacco etch virus (TEV) cleavage site. To generate pcDNA3-Flag-Dcp2-AA (p2798), the E147A/E148A mutation was introduced into the KpnI-EcoRV sites of pcDNA3-Flag-Dcp2 (25) using a PCR fragment amplified with primers G1883 and G1884.
Cell culture and transfection.
HeLa, U2OS, HEK293, and COS7 cells were cultured in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (PAA Laboratories), 2 mM l-glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin (all PAN Biotech) at 37°C/5% CO2. Plasmids were transfected with Lipofectamine 2000 (Invitrogen) for protein interaction studies or with Fugene HD (Roche) for immunofluorescence analysis according to the manufacturer's recommendations. Alternatively, polyethyleneimine (Polysciences Europe; 1 mg/ml, pH 7.0) was also used as a transfection reagent for interaction and functional studies. siRNAs were transfected at a concentration of 100 nM with Lipofectamine 2000 twice over a time period of 4 days. Where indicated, cells were treated for 2 h with 5 μg/ml cycloheximide (Roth).
siRNAs.
siRNAs used in this study were synthesized by Ambion against the following sequences (sense strand): D0 (control), 5′-GCAUUCACUUGGAUAGUAAdTdT-3′; T2 (human Pat1b), 5′-GCAGAAAGGCUCAGUAAGAdTdT-3′; and T3 (human Pat1b), 5′-GGACGGAGGUGAUGUUCAUdTdT-3′.
Immunofluorescence microscopy.
Twenty-four hours after transient transfection, cells grown on glass coverslips were fixed in 4% paraformaldehyde for 10 min at room temperature. Cell membranes were then permeabilized in −20°C cold methanol for 10 min. Phosphate-buffered saline (PBS) containing 0.1% sodium azide and 5% horse serum was used for blocking and antibody dilution. DNA was visualized using Hoechst dye (no. 33342, 1 μg/ml; Sigma). After washing in PBS, cells were mounted onto glass slides using a solution of 14% polyvinyl alcohol (P8136; Sigma) and 30% glycerol in PBS. P bodies were counted on an upright epifluorescence microscope (BX60; Olympus). Images were acquired at the Nikon Imaging Center, Heidelberg, Germany, either on an upright epifluorescence microscope (90i; Nikon) or by spinning-disc confocal microscopy (Ultraview ERS [Perkin Elmer] on a TE2000 inverted microscope [Nikon]) using an electron-multiplying charge-coupled-device (EM-CCD) camera (Hamamastu).
Coimmunoprecipitation (co-IP) and Western blot analysis.
Twenty-four hours after transient transfection, HEK293 cells from a confluent 10-cm dish were collected and lysed in 400 μl ice-cold hypotonic lysis buffer (10 mM Tris [pH 7.5], 10 mM NaCl, 10 mM M EDTA, 0.5% Triton X-100 with freshly added protease inhibitors [Complete; Roche]). Nuclei were removed by centrifugation at 500 × g for 5 min at 4°C. The cytoplasmic lysate was precleared by the addition of 30 μl protein A/G-agarose beads (Pierce or Santa Cruz) for 1 h at 4°C and incubated with 1 μg of antibody for 2 h. A total of 30 μl of protein A/G beads was added for an additional 2 h and washed six times in NET2 buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.5% Triton X-100). Protein complexes were eluted with 25 μl SDS sample buffer with or without 100 mM dithiothreitol (DTT). Where indicated, RNase A (Sigma; 0.1 mg/ml) was added to the lysates after preclearing. Proteins were resolved on 10% polyacrylamide gels and transferred onto a 0.2-μm-pore-size nitrocellulose membrane (Peqlab) for Western blotting. Horseradish peroxidase-coupled secondary antibodies (Jackson Immunoresearch) in combination with Western Lightning enhanced chemiluminescence substrate (Perkin Elmer) were used for detection. The green fluorescent protein (GFP)-binder was used for immunoprecipitation (IP) of YFP-tagged proteins as described previously (33).
Antibodies.
Polyclonal antibody Ab387 against human Pat1b was produced at Eurogentec by immunizing rabbits with peptide QGPEDDRDLSERALPR. The following antibodies were used for IP and Western blotting, and immunofluorescence analysis: mouse monoclonal antibodies anti-HA (HA.11; Covance) and anti-Edc3 (Abcam; ab57780-100); a cross-reacting mouse monoclonal phospho-S6-kinase antibody (sc-8416; Santa Cruz) for detection of Hedls; polyclonal chicken antibodies against Lsm1 (15-288-22100F; Genway Biotech) and Lsm4 (19101; Abcam); polyclonal rabbit antibodies against Xrn1 (A300-461A; Bethyl Laboratories), p54/Rck (A300-443A; Bethyl Laboratories), eIF4G (sc-11373; Santa Cruz), 14-3-3 (sc-629; Santa Cruz), and HA (sc-805; Santa Cruz); and a polyclonal goat antibody against eIF3B (sc-16377; Santa Cruz). Polyclonal rabbit anti-Caf1a was kindly provided by Ann-Bin Shyu (University of Texas, Houston, TX).
Northern blot analysis.
For RNA decay experiments, actinomycin D (Applichem) was added to cell cultures at 5 μg/ml, and total RNA was extracted using the Genematrix RNA purification kit (Eurx, Roboklon). A total of 7 to 15 μg of RNA was resolved by 1.1 or 1.6% agarose-2% formaldehyde-MOPS (morpholinepropanesulfonic acid) gel electrophoresis and blotted overnight with 8× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) onto Hybond-N+ Nylon membranes (Amersham, GE). Membranes were hybridized overnight with digoxigenin-labeled RNA probes at 55°C and washed twice with 2× SSC-0.1% SDS for 5 min and twice with 0.5× SSC-0.1% SDS for 20 min at 65°C. Alkaline phosphatase-labeled anti-DIG Fab fragments and CDP-Star substrate (both Roche) were used for detection according to the manufacturer's instructions. The following primers were used to generate templates for Sp6 probes by PCR: G1000/G1001 for the probe against exon 1 and 2 of rabbit β-globin, G78/G1008 for the probe against human RPS7, and G083/G1009 for the probe against nucleolin. Primer sequences are listed in Table S1 in the supplemental material.
Luciferase assay.
One third of a 6-cm dish of transiently transfected HeLa cells was lysed in 150 μl of passive lysis buffer (dual-luciferase reporter assay system; Promega) and frozen at −20°C. After thawing, nuclei were removed by centrifugation for 2 min at 13,000 rpm at 4°C. A total of 20 μl of the supernatant was mixed with 50 μl of substrates from the dual-luciferase reporter assay system, diluted 1:3. Firefly and Renilla luciferase activities were measured on a Fluostar Optima (BMG Labtech) plate reader.
RESULTS
Identification of two human Pat1 homologs.
In order to identify a human homolog of yeast Pat1, we compared Pat1 sequences of different fungi and observed a highly conserved central region of about 40 amino acids (aa), hereinafter termed the homology (H) domain. Using this domain, BLAST analysis was able to identify homologous sequences in various metazoa, including Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, and Xenopus laevis (see Fig. S1 in the supplemental material). Two proteins sharing the H domain were found in mice and humans, indicative of a gene duplication in mammals. Outside the H domain, the only detectable sequence conservation between the fungal and metazoan proteins lies in a highly acidic stretch of 50 to 70 aa at the very amino terminus, hereinafter referred to as the acidic domain (A). We termed the two human proteins Pat1a and Pat1b. Pat1b is identical to PatL1 previously identified by Scheller et al. (34). Pat1a matches the partial sequence that was previously suggested as the human Pat1 ortholog (9) and differs from the predicted PatL2 sequence at the N terminus (34). An alignment between the human Pat1a and Pat1b sequence (see Fig. S2 in the supplemental material) shows that Pat1a lacks a region of about 150 aa in the amino-terminal part following the A domain.
Human Pat1b, but not Pat1a, localizes to P bodies.
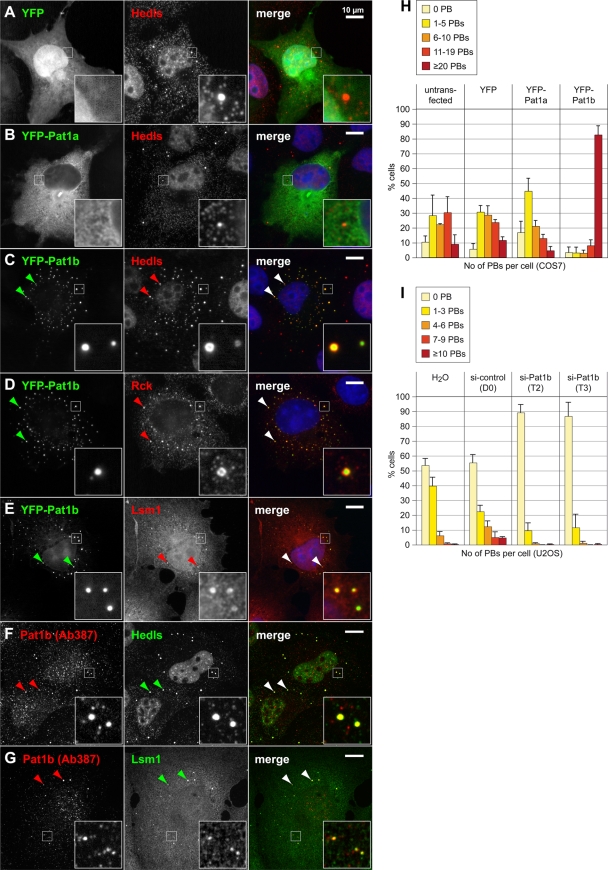
We first examined the localization of human Pat1a and Pat1b by expressing yellow fluorescent protein (YFP)-tagged fusion proteins in COS7 cells. Epifluorescence microscopy showed that YFP-Pat1a is expressed throughout the cytoplasm, where it has a reticular distribution, but does not accumulate in P bodies (Fig. 1B). YFP-Pat1b, however, strongly accumulates in small cytoplasmic foci (Fig. 1C to E). These foci correspond to P bodies as determined by colocalization with different markers, including the enhancer of decapping Hedls (Fig. 1C), the helicase Rck (Fig. 1D), and Lsm1 (Fig. 1E). The localization of Pat1b in P bodies was also observed with a C-terminal fusion of YFP and with N-terminal HA and Flag tags (data not shown). We confirmed the localization of Pat1b in P bodies of human HeLa (data not shown) and U2OS cells (see Fig. S3 in the supplemental material). Thus, human Pat1b, but not Pat1a, localizes in P bodies. This is in line with the previously observed localization of Pat1b (PatL1) in HeLa cells (34).
FIG. 1.
Intracellular localization of human Pat1a and Pat1b in COS7 cells. COS7 cells were transiently transfected with YFP (A), YFP-Pat1a (B), or YFP-Pat1b (C to E) and processed for immunofluorescence microscopy. P bodies in the cytoplasm were counterstained in red using an antibody against Hedls (A to C), Rck (D), or Lsm1 (E). In addition, cells were stained with Hoechst 33342 in blue to visualize nuclei. Images in panels A to E were acquired by epifluorescence microscopy. Merged images are shown on the right, and the size bar represents 10 μm. Arrowheads point toward P bodies. (F and G) Untransfected COS7 cells were stained in red for endogenous Pat1b with antibody 387 and in green for P bodies using Hedls (F) or Lsm1 (G) as a marker. Images in panels F and G were acquired by spinning-disc confocal microscopy; maximum projections of z-stacks are depicted. (H) COS7 cells transiently transfected with either YFP, YFP-Pat1a, or YFP-Pat1b were fixed and stained for P bodies using an antibody against Hedls. The number of P bodies (PBs) in transfected cells was counted, and the average distribution is represented in the graph. Error bars show the standard deviations (SD) based on three independent repeats. Untransfected COS7 cells serve as an additional control. (I) The expression of Pat1b was knocked down in U2OS cells by transfection of two different siRNAs (T2 and T3) at a concentration of 100 nM. An unspecific siRNA (D0) was transfected as a negative control. The number of PBs per cell was counted after staining for Helds, and the average distribution is represented in the graph. Error bars show the SD based on three independent repeats.
In order to determine whether endogenous Pat1b would also localize in P bodies, we raised a peptide antibody in rabbit (Ab387). This antibody was able to detect overexpressed Pat1b by Western blot analysis (see Fig. S4A in the supplemental material). By immunofluorescence microscopy, Ab387 gave a punctate staining in both the nucleus and the cytoplasm of untransfected COS7 (Fig. 1F and G) and U2OS cells (see Fig. S3F and G in the supplemental material). The larger foci in the cytoplasm perfectly overlap with Hedls and Lsm1, indicating that endogenous Pat1b also localizes to P bodies.
Human Pat1b induces P-body formation.
We noticed that cells transfected with YFP-Pat1b showed strongly increased numbers of P bodies compared to control cells transfected with YFP or YFP-Pat1a (compare Hedls staining in Fig. 1A to C). By counting P bodies using Hedls as a marker, we found that most COS7 cells overexpressing YFP-Pat1b have >20 P bodies per cell compared to an average of about 9 P bodies per cell in untransfected or YFP-expressing cells (Fig. 1H). Overexpression of YFP-Pat1a, however, did not increase the number of P bodies. The strong increase in P-body numbers by Pat1b overexpression was confirmed with an HA-tagged version, and the same was also observed with other cell lines, such as HeLa and U2OS (data not shown). Since Pat1b overexpression causes a dramatic amplification of P bodies, many of which are very small, Pat1b may play an active role in nucleating the assembly of P bodies.
To further address the role of Pat1b in P-body assembly, we knocked down endogenous Pat1b in U2OS cells. Two different siRNAs, T2 and T3, reduced Pat1b mRNA levels to 5% and 6%, respectively, compared to the expression level in cells transfected with control siRNA (see Fig. S5A in the supplemental material). Indeed, knockdown of Pat1b led to a nearly complete loss of P bodies (Fig. 1I; see also micrographs shown in Fig. S5B in the supplemental material). This result supports the notion that Pat1b nucleates the assembly of P bodies.
Pat1b domains control P-body assembly.
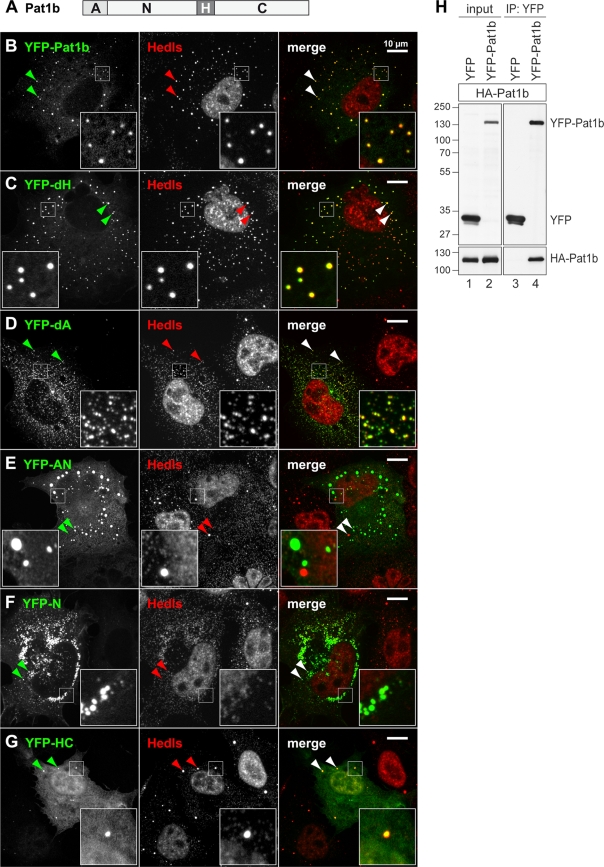
Next, we wanted to examine the contribution of different Pat1b domains, schematically depicted in Fig. 2A, to its intracellular localization. Fragments and deletions of Pat1b were expressed as YFP fusion proteins in COS7 cells and examined by spinning-disc confocal microscopy. Localization data are summarized in Table S2 in the supplemental material, and examples are shown in Fig. 2. As stated above, YFP-Pat1b forms many small- or medium-size foci, reminiscent of a “salt”-like appearance, and these foci colocalize with all P-body markers tested, including Hedls, Rck, Lsm1, and Lsm4 (Fig. 1C to E and 2B; see also Fig. S6E in the supplemental material). Thus, overexpressed Pat1b localizes to and strongly amplifies the number of P bodies. Deletion of the homology domain (YFP-dH) did not change this localization pattern (Fig. 2C). Deletion of the acidic region (YFP-dA), however, caused the protein to localize to much smaller and more numerous foci, reminiscent of a “pepper”-like appearance. These foci colocalize with Hedls (Fig. 2D) and with Rck (data not shown). Thus, YFP-dA triggers the formation of small P bodies, yet deletion of the acidic domain seems to prevent small aggregates from growing or merging into larger P bodies.
FIG. 2.
Localization of Pat1b subdomains. (A) Schematic representation of Pat1b domains. A, acidic domain; N, amino-terminal region; H, homology domain; C, carboxy-terminal region. COS7 cells were transiently transfected with YFP-Pat1b (B), YFP-dH (C), YFP-dA (D), YFP-AN (E), YFP-N (F), or YFP-HC (G) and processed for immunofluorescence microscopy. P bodies in the cytoplasm were counterstained in red using an antibody against Hedls. Images were acquired by spinning-disc confocal microscopy; maximum projections of z-stacks are depicted. Merged images are shown on the right; arrowheads point toward P bodies. Size bar, 10 μm. Localization of Pat1b subdomains is also summarized in Table S2 in the supplemental material. (H) HEK293 cells were transiently transfected with HA-Pat1b together with either YFP or YFP-Pat1b. Cytoplasmic lysates (input) were prepared for IP with GFP-binder, and Western blot analysis was carried out to visualize interaction between HA-Pat1b and YFP-Pat1b. The sizes of the molecular weight markers (in thousands) are indicated on the left.
Localization of the YFP-AN fragment was very heterogeneous: in about half of the cells, YFP-AN showed a diffuse localization in the cytoplasm, and in these cells endogenous P bodies were strongly suppressed (see Table S2 in the supplemental material). In other cells, YFP-AN formed aggregates apparent as large round foci (Fig. 2E) or irregular patches (not shown). Many of these round foci and patches did not contain other P-body markers, suggesting that the AN fragment had the intrinsic ability to form aggregates. This aggregation-prone behavior was also observed with the YFP-N fragment lacking the A region (Fig. 2F). In contrast, the YFP-A fragment had a diffuse localization indistinguishable from YFP alone (see Table S2). The YFP-HC fragment primarily showed a diffuse localization in the nucleus and the cytoplasm. In some cells, YFP-HC also accumulated weakly in P bodies (Fig. 2G) yet did not affect the size or number of P bodies. Taken together, these results suggested that the HC fragment is passively recruited to P bodies, whereas the N fragment contains an active aggregation-prone domain. In the context of full-length Pat1b, the N region can strongly promote the formation of P bodies. The idea that Pat1b actively promotes P-body assembly is supported by our observation that Pat1b interacts with itself in a coimmunoprecipitation (co-IP) assay (Fig. 2H).
We then tested whether P bodies induced by YFP-Pat1b would be resistant to treatment with the translation inhibitor cycloheximide (CHX). CHX was shown to inhibit the formation of P bodies (10), presumably because mRNAs are trapped in polysomes by CHX. After 2 h of CHX treatment, P bodies were nearly absent in untransfected cells, whereas large numbers of foci were still present in YFP-Pat1b-expressing cells (see Fig. S6A, D, and F in the supplemental material). Colocalization of Hedls, Rck, Xrn1, and Lsm4 indicated that these structures had the typical composition of P bodies. Interestingly, Pat1b-induced foci persisted even after 6 h of CHX treatment (data not shown). Although we cannot tell whether these P bodies are functional in the presence of CHX, this results indicates that Pat1b not only nucleates P bodies but also stabilizes P bodies once assembled. Aggregates induced by the expression of YFP-AN were also resistant to CHX treatment (see Fig. S6B in the supplemental material). These aggregates showed distinct colocalization with Rck but not with Hedls, suggesting that Rck may interact with the AN fragment.
Tethering of Pat1b accelerates mRNA degradation.
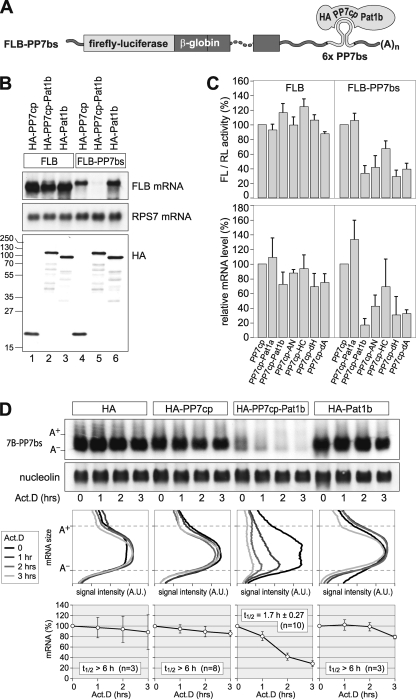
Since P bodies have been implicated in the control of both mRNA translation and decay, we went on to test if human Pat1b plays a role in the posttranscriptional control of gene expression. To this end, we tethered Pat1b to a reporter mRNA using a heterologous RNA-protein interaction of the Pseudomonas aeruginosa bacteriophage PP7 (23, 46). The PP7 coat protein (cp) binds as a dimer with high affinity to a stem loop in the bacteriophage RNA, the PP7 binding site (bs). We cloned six copies of the PP7bs into the 3′ UTR of a rabbit β-globin reporter gene containing firefly luciferase (FL) (FLB-PP7bs). A fusion protein containing an HA tag, PP7cp, and Pat1b, as shown schematically in Fig. 3A, brings Pat1b in close proximity to the reporter mRNA. When expressed together in HeLa cells, HA-PP7cp-Pat1b strongly suppressed the expression level of FLB-PP7bs mRNA compared to HA-PP7cp or HA-Pat1b alone (Fig. 3B, lanes 4 to 6). In contrast, HA-PP7cp-Pat1b had only a very weak effect on the FLB mRNA lacking PP7bs (Fig. 3B, lanes 1 to 3). Similar to HeLa cells used for Fig. 3, Pat1b was also found to suppress the expression of the tethered reporter mRNA in U2OS cells (data not shown).
FIG. 3.
Tethering of Pat1b accelerates mRNA decay. (A) Schematic representation of the reporter containing firefly luciferase fused to β-globin (FLB). Six copies of the PP7 binding site (bs) were inserted in the 3′ UTR for the FLB-PP7bs reporter. An HA-tagged PP7 coat protein (cp) fused to Pat1b specifically binds to the PP7bs RNA. (B) HeLa cells were transiently transfected with FLB or FLB-PP7bs reporter together with either HA-PP7cp, HA-PP7cp-Pat1b, or HA-Pat1b. RNA and protein were extracted and analyzed by Northern (top two panels) and Western (bottom) blotting. The sizes of the molecular weight markers (in thousands) are indicated on the left. Ribosomal protein S7 (RPS7) mRNA serves as RNA loading control. (C) HeLa cells were transiently transfected with FLB or FLB-PP7bs reporter together with a Renilla luciferase (RL) reporter and either HA-PP7cp, HA-PP7cp-Pat1a, HA-PP7cp-Pat1b, or HA-PP7cp fused to fragments of Pat1b. Cytoplasmic lysates were prepared after 24 h to measure FL/RL activity. FLB mRNA levels were determined from the same lysates by Northern blot analysis, normalized to RPS7 mRNA, and quantified. Average values ± SD from three repeat experiments were plotted in the graph. (D) HeLa cells were transiently transfected with a T7-tagged β-globin reporter containing 6 copies of PP7bs (7B-PP7bs) together with either the HA tag alone, HA-PP7cp, HA-PP7cp-Pat1b, or HA-Pat1b. Total RNA was extracted at 1-h intervals after blocking transcription with actinomycin D (5 μg/ml). The reporter mRNA was detected by Northern blot analysis; nucleolin mRNA serves as loading control. In the middle panels, deadenylation was visualized by quantifying the signal intensity (in arbitrary units [A.U.]) of 7B-PP7bs mRNA along the length of the signal and plotting it as a function of mRNA size. In the bottom panel, the overall signal intensity of 7B-PP7bs mRNA was quantified and normalized to nucleolin mRNA. Average values ± standard errors (SE) were plotted as a percentage of the initial time point.
We then compared the effect of tethering Pat1a, Pat1b, and fragments of Pat1b to the FLB-PP7bs reporter mRNA. We measured, as a readout, both firefly luciferase activity and reporter mRNA levels (Fig. 3C). The experiment showed that the tethering of Pat1a had no significant effect compared to PP7cp alone. The tethering of full-length Pat1b had the strongest suppressive effect, reducing FL activity by about 3-fold and mRNA levels by about 5-fold. The tethering of the AN fragment, the dH deletion mutant, or the dA deletion mutant resulted in strong inhibition of the FLB-PP7bs reporter, though at the mRNA level the inhibition was less pronounced than with full-length Pat1b. Since the tethering of the HC fragment had a much weaker effect, the AN fragment of Pat1b appears to be most important for the suppressive effect on reporter mRNA expression. In general, there was good agreement between the relative FL activities and mRNA abundance, indicating that tethered Pat1b acts primarily at the level of mRNA expression.
Next, we wanted to determine whether Pat1b would affect the stability of the tethered mRNA. We expressed a β-globin reporter mRNA containing 6 copies of PP7bs (7B-PP7bs) together with the HA tag alone, HA-PP7cp, HA-PP7cp-Pat1b, or HA-Pat1b in HeLa cells (Fig. 3D). Transcription was blocked by the addition of actinomycin D in order to monitor degradation of the reporter mRNA. The analysis showed that 7B-PP7bs mRNA was stable in the presence of coexpressed HA or HA-Pat1b or when HA-PP7cp alone was tethered to the mRNA. In contrast, the tethering of HA-PP7cp-Pat1b caused rapid degradation of the reporter mRNA. Overall signal intensities were quantified to obtain mRNA half-lives (Fig. 3D, bottom). Measurement of the signal intensity along the length of the reporter mRNA shows that the mRNA was deadenylated as a result of Pat1b tethering (Fig. 3D, middle).
Human Pat1b interacts with Rck, Hedls, Xrn1, and Lsm1.
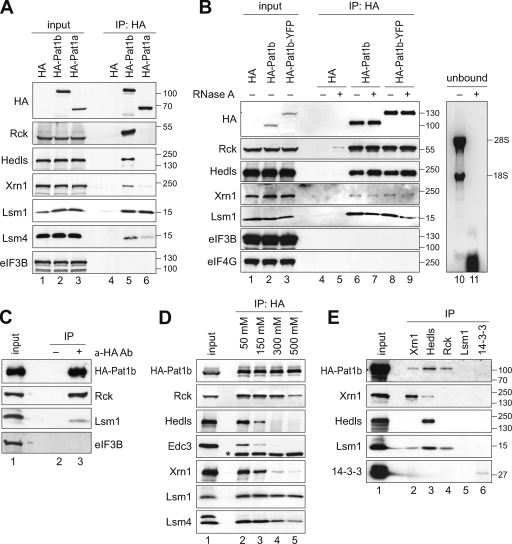
To further explore the connection between human Pat1b and mRNA degradation, we tested whether Pat1b interacts with different mRNA decay factors that localize to P bodies. By expressing HA-tagged Pat1b in HEK293 cells, we found that Pat1b coimmunoprecipitates with the 5′-3′ exoribonuclease Xrn1 and with the activators of decapping Rck, Hedls, Lsm1, and Lsm4 (Fig. 4A, lane 5) as well as Edc3 (Fig. 4D). These interactions were also observed with a YFP-tagged form of Pat1b (HA-Pat1b-YFP) (Fig. 4B). eIF3B and eIF4G, translation initiation factors that do not localize in P bodies, did not interact with Pat1b (Fig. 4A and B). As a negative control, the HA tag alone did not co-IP with any of these proteins. Given that mRNAs are also a component of P bodies, we next tested whether the interaction of Pat1b with its partners is RNA dependent. The addition of RNase A to the IP did not affect the interaction of HA-Pat1b with any of its partners (Fig. 4B). RNA extracted from the unbound fraction showed that RNase A treatment had efficiently degraded rRNA (Fig. 4B, lanes 10 and 11). These results suggest that the association of Pat1b with Rck, Hedls, Xrn1, and Lsm1 is based on protein-protein interactions.
FIG. 4.
Pat1b interacts with P-body proteins. (A) HEK293 cells were transiently transfected with vector alone, HA-tagged Pat1b, or HA-tagged Pat1a. After 1 day, cytoplasmic lysates (input) were prepared for IP with anti-HA antibody. The HA-tagged proteins as well as endogenous Rck, Hedls, Xrn1, Lsm1, Lsm4, and eIF3B were detected by Western blotting. The sizes of the molecular weight markers (in thousands) are indicated on the right. (B) HEK293 cells transiently transfected with vector alone, HA-Pat1b, or HA-Pat1b-YFP were used for IP as described in the legend for panel A. Where indicated, RNase A was added to the lysates during IP. Lanes 10 and 11 show RNA extracted from the unbound fraction and stained with ethidium bromide. (C) HA-Pat1b was immunoprecipitated with HA antibody or without antibody as a control for unspecific precipitation. (D) HA-Pat1b was immunoprecipitated and subjected to increasing NaCl concentrations prior to elution of the protein complexes. *, immunoglobulin heavy chain. (E) Endogenous Xrn1, Hedls, Rck, Lsm1, and 14-3-3 were immunoprecipitated from the cytoplasmic lysate of HEK293 cells transiently transfected with HA-Pat1b. The Western blot for Rck is not shown due to an overlapping signal from the immunoglobulin heavy chain.
Since Pat1b contains an aggregation-prone domain (fragment N) (Fig. 2F), we tested whether Pat1b may aggregate and precipitate unspecifically in our co-IP experiments. From the same HA-Pat1b-containing lysate (input), we carried out the IP procedure with and without the HA antibody (Fig. 4C). In the absence of HA antibody, neither HA-Pat1b nor associated proteins were precipitated (Fig. 4C, lane 2). This shows that precipitation by the antibody was specific.
To further characterize the binding of Pat1b to its partners, we subjected the complexes to increasing salt concentrations (Fig. 4D). Whereas the interaction with Hedls, Edc3, and Xrn1 was severely reduced at a NaCl concentration of 300 mM, the interaction with Rck and Lsm4 was resistant to 300 mM NaCl. Lsm1-Pat1b was the most stable of these interactions and could not be separated by 500 mM NaCl. This indicates that Lsm1/Lsm4 and Rck are more tightly associated with Pat1b than Hedls, Edc3, and Xrn1. Since Xrn1 forms a stable complex with Lsm1-7 in yeast (6), the interaction of human Xrn1 with Pat1b may occur through Lsm1-7. From these co-IP experiments, however, it is not possible to tell whether Pat1b directly binds to Rck and Lsm1.
Interestingly, Lsm1 co-IPs in similar amounts with Pat1a and Pat1b, yet the association of Lsm4 and Xrn1 with Pat1a is much weaker (Fig. 4A, lane 6). Thus, it is conceivable that Pat1a may preferentially bind to “free” Lsm1 that is not associated with the other Lsm proteins or Xrn1.
To obtain additional evidence for the observed interactions, inverse IPs were carried out in HEK293 cells transiently transfected with HA-Pat1b (Fig. 4E). The Xrn1 antibody was able to co-IP HA-Pat1b and Lsm1 (Fig. 4E, lane 2), the Hedls antibody could co-IP HA-Pat1b, Xrn1, and Lsm1 (lane 3), and the Rck antibody could co-IP HA-Pat1b and Lsm1 (lane 4). The Lsm1 antibody was not suitable for IP (Fig. 4E, lane 5), and a 14-3-3 antibody served as a negative control (lane 6). Taken together, these results confirmed the association of Xrn1, Hedls, and Rck with both Pat1b and Lsm1 and provides evidence for multiple interactions within this group of P-body proteins.
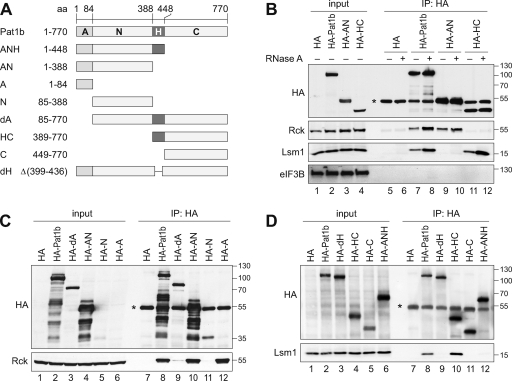
Human Pat1b interacts with enhancers of decapping via separate domains.
Since the above analysis revealed that Rck and Lsm1 are tightly associated with Pat1b, we determined which domains of Pat1b are involved in these interactions. To this end, we expressed different fragments of Pat1b as depicted in Fig. 5A. Figure 5B shows that the AN fragment efficiently co-IPs with Rck (lane 9), whereas the carboxy-terminal (HC) fragment co-IPs Lsm1 (lane 11). Similar to Lsm1, the more loosely associated proteins Hedls, Edc3, and Xrn1 were found to be connected to Pat1b via the HC domain (see Fig. S7 in the supplemental material). As shown in Fig. 4B for full-length Pat1b, the interactions mediated by the AN and HC fragments were resistant to RNase A treatment and are thus likely to be RNA independent (Fig. 5B).
FIG. 5.
Separate interaction domains within Pat1b. (A) Schematic representation of Pat1b fragments. A, acidic domain; N, amino-terminal region; H, homology domain; C, carboxy-terminal region. (B) HEK293 cells were transiently transfected with vector alone, full-length HA-Pat1b, HA-AN, or HA-HC. After 1 day, cytoplasmic lysates (input) were prepared for IP with anti-HA antibody. The HA-tagged proteins as well as endogenous Rck, Lsm1, and eIF3B were detected by Western blotting. *, immunoglobulin heavy chain. The sizes of the molecular weight markers (in thousands) are indicated on the right. (C) HEK293 cells transiently transfected with vector alone, HA-Pat1b, HA-dA, HA-AN, HA-N, or HA-A were used for IP as described in the legend for panel B. The HA-tagged proteins and endogenous Rck were detected by Western blotting. (D) HEK293 cells were transiently transfected with vector alone, HA-Pat1b, HA-dH, HA-HC, HA-C, or HA-ANH and used for IP as described in the legend for panel B. The HA-tagged proteins and endogenous Lsm1 were detected by Western blotting.
We further analyzed the association of Rck with the amino-terminal half of Pat1b (Fig. 5C). Removal of the acidic domain (dA) from full-length Pat1b was sufficient to abrogate the interaction with Rck (Fig. 5C, lane 9). The same was observed when the N fragment was expressed alone in comparison to the AN fragment (Fig. 5C, lanes 10 and 11). Indeed, the HA-tagged acidic domain alone, although not visible by Western blot analysis due to its small size, efficiently co-IPs with Rck (Fig. 5C, lane 12).
We then examined in more detail the association of HC with Lsm1. Deletion of the conserved homology domain H from full-length Pat1b (dH) abolished the interaction with Lsm1 (Fig. 5D, lanes 8 and 9). Similarly, comparison of the HC and C fragments indicated that the H domain is required for association with Lsm1 (Fig. 5D, lanes 10 and 11). The ANH fragment, however, did not co-IP with Lsm1 (Fig. 5D, lane 12), indicating that the H domain alone is not sufficient to mediate this interaction. We concluded that Lsm1 binding requires both the H and C fragment of Pat1b.
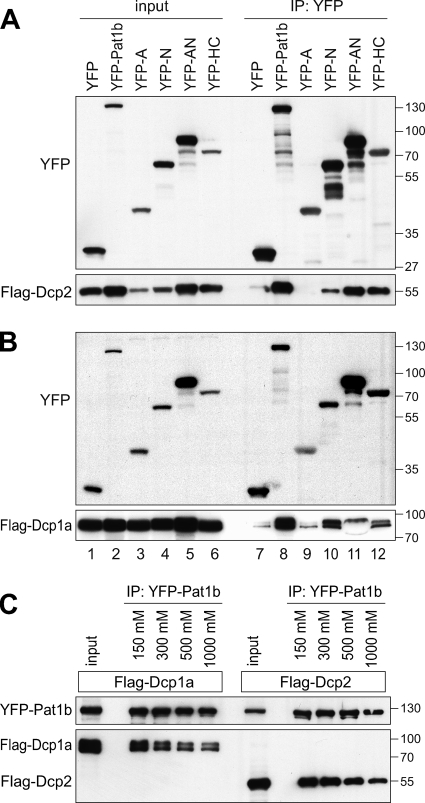
Human Pat1b interacts with the Dcp2-Dcp1a decapping enzyme.
Since we found Pat1b to associate with a number of decapping enhancers (Rck, Hedls, Edc3, and Lsm1), we wanted to know if Pat1b would also interact with the decapping enzyme Dcp2 and its closely associated activator Dcp1a. To this end, we switched to YFP-tagged Pat1b proteins, since the use of GFP-binder (33) overcomes detection problems related to the presence of immunoglobulin in the IP. Full-length Pat1b tagged with YFP was found to co-IP efficiently with Flag-Dcp2 and Flag-Dcp1a (Fig. 6A and B, lanes 8). Interestingly, the two proteins associate with both the N and the HC region of Pat1b (Fig. 6A and B, lanes 10 and 12). The association with Dcp2 and Dcp1a was found to be independent of RNA (see Fig. S8 in the supplemental material). Moreover, the two proteins did not dissociate from Pat1b at 1 M NaCl (Fig. 6C), indicating that Dcp2 and Dcp1a are tightly connected to Pat1b. Taken together, the results show that Pat1b associates with all enhancers of decapping through at least three separate domains: the A fragment associates with Rck, the N fragment with Dcp2 and Dcp1a, and the HC fragment with Dcp2/Dcp1a, Lsm1/Lsm4, Hedls, and Edc3.
FIG. 6.
Pat1b interacts with Dcp2 and Dcp1a. (A) HEK293 cells were transiently transfected with YFP, YFP-Pat1b, YFP-A, YFP-N, YFP-AN, or YFP-HC together with Flag-Dcp2. GFP-binder was used for IP, and Western blot analysis was carried out with anti-GFP and anti-Flag antibody. The sizes of the molecular weight markers (in thousands) are indicated on the right. (B) Same analysis as described in the legend for panel A, except that Flag-Dcp1a was cotransfected. (C) HEK293 cells were transiently transfected with YFP-Pat1b together with either Flag-Dcp1a or Flag-Dcp2. IPs were carried out with GFP-binder and subjected to increasing NaCl concentrations prior to elution.
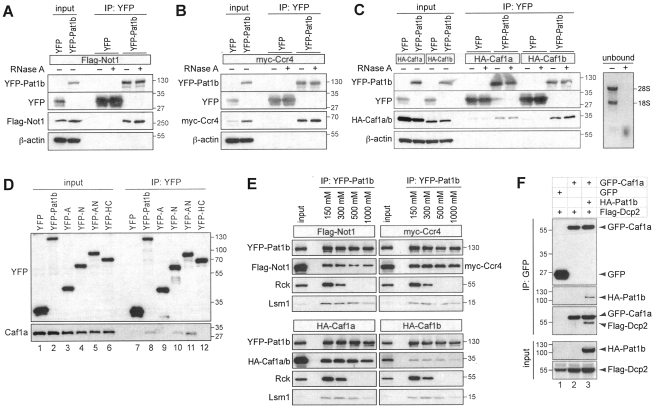
Human Pat1b interacts with the Ccr4-Caf1-Not deadenylation complex.
Since we found that the tethering of Pat1b causes mRNA deadenylation (Fig. 3D), we tested for interactions of Pat1b with the Ccr4-Caf1-Not deadenylation complex. Notably, this complex is also concentrated in P bodies (27, 41, 47). Indeed, we found that YFP-Pat1b co-IPs efficiently with Flag-Not1 (Fig. 7A) and myc-Ccr4 (Fig. 7B), as well as HA-Caf1a and HA-Caf1b (Fig. 7C). In all cases, RNase A treatment did not interfere with binding. We then confirmed the interaction of YFP-Pat1b with endogenous Caf1a (Fig. 7D, lane 8). Analysis of subdomains indicated that Caf1a can interact with the N fragment of Pat1b alone (Fig. 7D, lane 10), yet the acidic domain A enhances Caf1a binding (Fig. 7D, lane 11). Interestingly, the A domain was even more important for association with Flag-Not1 and myc-Ccr4 since only the AN fragment showed a robust interaction with these two proteins (see Fig. S9A and B in the supplemental material). Although Flag-Not1 and myc-Ccr4 also co-IP weakly with the HC domain (Fig. S9A and B), we concluded from these experiments that the Ccr4-Caf1-Not complex associates primarily with the AN domain.
FIG. 7.
Pat1b interacts with the Ccr4-Caf1-Not complex. (A) HEK293 cells were transiently transfected with YFP or YFP-Pat1b together with Flag-Not1. GFP-binder was used for IP, and Western blot analysis was carried out with anti-GFP and anti-Flag antibody. The sizes of the molecular weight markers (in thousands) are indicated on the right. Where indicated, RNase A was added during IP. (B) IP was carried out as described in the legend for panel A using YFP or YFP-Pat1b together with myc-Ccr4; anti-myc was used for Western blot analysis. (C) IP was carried out as described in the legend for panel A using YFP or YFP-Pat1b together with HA-Caf1a or HA-Caf1b; anti-HA was used for Western blot analysis. On the right side, RNA was extracted from unbound fractions and stained with ethidium bromide. (D) HEK293 cells were transiently transfected with YFP, YFP-Pat1b, YFP-A, YFP-N, YFP-AN, or YFP-HC and processed for IP with GFP-binder. The YFP-tagged proteins and endogenous Caf1a were detected by Western blotting. (E) HEK293 cells were transiently cotransfected with YFP-Pat1b together with either Flag-Not1, myc-Ccr4, HA-Caf1a, or HA-Caf1b. IPs were carried out with GFP-binder and subjected to increasing NaCl concentrations prior to elution. (F) HEK293 cells were transiently transfected with Flag-Dcp2 together with either GFP alone, GFP-Caf1a alone, or GFP-Caf1a together with HA-Pat1b. Cytoplasmic lysates were processed for IP with GFP-binder and then subjected to Western blot analysis. In the IP samples, the Flag antibody cross-reacted with GFP-Caf1a.
By challenging these interactions with increasing salt concentrations, we found that Flag-Not1, myc-Ccr4, and HA-Caf1a/b are very stably associated with YFP-Pat1b (Fig. 7E). At a concentration of 1 M NaCl, these proteins remained in a complex with YFP-Pat1b, whereas binding of Lsm1 was reduced and binding of Rck was lost. Thus, it appears that Pat1b associates tightly with the Ccr4-Caf1-Not deadenylase complex and may act as a physical bridge between deadenylases and decapping enzymes.
To provide further evidence for such a bridging function, we tested whether Pat1b would promote the association between Caf1a and Dcp2. By IP of GFP-Caf1a, we found that the co-IP of Flag-Dcp2 is strongly enhanced by coexpression of HA-Pat1b (Fig. 7F, lane 3). This result shows that Pat1b can simultaneously interact with Caf1a and Dcp2 and thereby promote the association between the deadenylation-and-decapping complex.
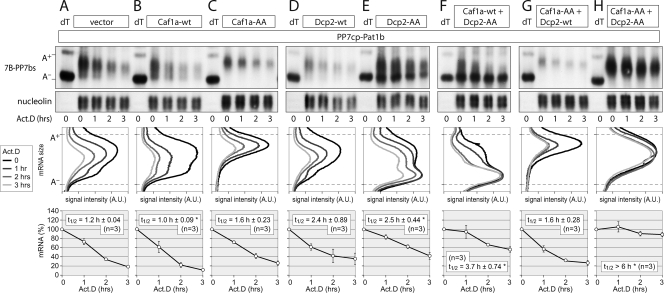
Tethering of Pat1b causes both deadenylation and mRNA decapping.
In order to test whether both deadenylation and decapping activities were associated with Pat1b, we again turned to the tethering assay. A dominant negative Caf1a-AA (D40A/E42A) deadenylase similar to a mutant described by Zheng et al. (47) was generated. In comparison to the vector control, coexpression of Caf1a-wt slightly accelerated the degradation rate of the reporter mRNA tethered to Pat1b (Fig. 8A and B). Quantification of the mRNA decay rates is shown at the bottom of Fig. 8 and summarized in Table S3 in the supplemental material. In contrast to that of Caf1a-wt, coexpression of dominant negative Caf1a-AA prevented deadenylation of the mRNA (Fig. 8C and G). The lack of the A− mRNA population is clearly visible in the middle panels of Fig. 8C and G, in which the signal intensity was quantified along the length of the mRNA. This result suggests that the tethering of Pat1b causes Caf1-dependent mRNA deadenylation. Interestingly, the A+ mRNA still underwent degradation without shortening of its size, suggesting that tethered Pat1b could promote decapping of the A+ mRNA independently of deadenylation.
FIG. 8.
Tethering of Pat1b causes mRNA deadenylation and decapping. HeLa cells were transiently transfected with the T7-tagged β-globin reporter containing 6 copies of the PP7bs (7B-PP7bs) together with HA-PP7cp-Pat1b and vector alone (A), Caf1a-wt (B), dominant negative Caf1a-AA (D40A/E42A) (C), Dcp2-wt (D), dominant negative Dcp2-AA (E147A/E148A) (E), Caf1a-wt plus Dcp2-AA (F), Caf1a-AA plus Dcp2-wt (G), and Caf1a-AA plus Dcp2-AA (H). Total RNA was extracted at 1-h intervals after blocking transcription with actinomycin D (5 μg/ml). The reporter mRNA was detected by Northern blot analysis; nucleolin mRNA served as a loading control. RNA samples marked dT were treated with oligo(dT) and RNase H and served as a size marker for deadenylated (A−) reporter mRNA. In the middle panels, deadenylation was visualized by quantifying the signal intensity of 7B-PP7bs mRNA along the length of the signal and plotting it as a function of mRNA size. In the bottom panels, the overall signal intensity of 7B-PP7bs mRNA was quantified and normalized to nucleolin mRNA. Average values ± SE were plotted as a percentage of the initial time point. An asterisk indicates a significant difference in the mRNA half-life (t1/2) (P < 0.05 by two-tailed Student's t test) compared to the vector control depicted in panel A. Statistical analysis of the mRNA t1/2 is summarized in Table S3 in the supplemental material.
To test this possibility, we generated Dcp2-AA (E147A/E148A) in analogy to a published mutation that inactivates the decapping enzyme Dcp2 (44). Whereas coexpression of Dcp2-wt did not have a significant effect on mRNA degradation (Fig. 8D), Dcp2-AA caused accumulation of the A− mRNA (Fig. 8E). Strong accumulation of A− mRNA was also visible when we cotransfected Caf1a-wt and Dcp2-AA (Fig. 8F). From this we concluded that the tethering of Pat1b also leads to Dcp2-dependent mRNA decapping. Only by coexpressing both Caf1a-AA and Dcp2-AA did we achieve full stabilization of the Pat1b-tethered mRNA (Fig. 8H). Taken together, these experiments demonstrate that Pat1b is tightly associated with both deadenylation and decapping activities in the cell.
N is the primary effector domain of Pat1b.
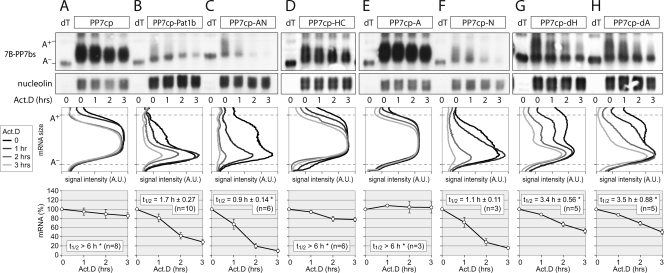
To determine which domain of Pat1b mediates deadenylation and decapping, we tested the effect of tethering Pat1b fragments to the reporter mRNA. The tethering of AN (Fig. 9C) or the N domain alone (Fig. 9F) very efficiently caused mRNA deadenylation and decapping, comparable to the effect of tethering full-length Pat1b (Fig. 9B). The mRNA decay rates are quantified in Fig. 9, bottom, and summarized in Table S4 in the supplemental material. In contrast to that of AN or N, the tethering of the HC fragment (Fig. 9D) or the A domain (Fig. 9E) did not induce mRNA decay and was similar to the tethering of PP7cp alone (Fig. 9A). These data suggest that N is the most potent fragment of Pat1b, and its tethering causes both deadenylation and decapping. This is in good agreement with our observation that the N fragment interacts with both Caf1a (Fig. 7D) and Dcp2 (Fig. 6A). We finally tested the effect of removing the A and the H domain (Fig. 9G and H). Compared to full-length Pat1b, the tethering of either the dA or dH mutant led to a significantly reduced mRNA decay rate. Both mutants caused mRNA deadenylation but resulted in the accumulation of A− mRNA. This suggests that the A domain, possibly by interacting with Rck (Fig. 5C), and the H domain, possibly by enhancing the binding of Lsm1 (Fig. 5D), activate decapping of the tethered mRNA. These enhancing activities require the presence of the N domain since the A and HC fragments on their own were inactive in the tethering assay.
FIG. 9.
Tethering of Pat1b fragments. HeLa cells were transiently transfected with the 7B-PP7bs reporter and HA-tagged PP7cp (A), PP7cp-Pat1b (B), PP7cp-AN (C), PP7cp-HC (D), PP7cp-A (E), PP7cp-N (F), PP7cp-dH (G), or PP7cp-dA (H). Degradation of the reporter mRNA was analyzed and quantified as described in the legend to Fig. 8. Average values ± SE were plotted as a percentage of the initial time point. An asterisk indicates a significant difference in the mRNA t1/2 (P < 0.05 by two-tailed Student's t test) compared to tethering of PP7cp-Pat1b depicted in panel B. Statistical analysis of mRNA t1/2 is summarized in Table S4 in the supplemental material.
DISCUSSION
In this study we provide evidence that human Pat1b connects two major steps in mRNA degradation: deadenylation and decapping. In yeast, the major pathway by which mRNA is degraded is deadenylation-dependent decapping followed by 5′-3′ exonucleolytic decay (15, 29). While this pathway has been established through numerous genetic experiments, the entity that physically connects the deadenylation with the decapping step has not been identified. Pat1 and the Lsm1-7 complex were likely suspects since both are activators of decapping and were found to be preferentially associated with oligoadenylated mRNAs (5, 8, 18, 42). Our analysis reveals that human Pat1b is tightly associated with both deadenylation and decapping enzymes. By co-IP experiments, we found that Pat1b interacts with the Ccr4-Caf1-Not deadenylase complex (Fig. 7), with the decapping enzyme Dcp2 and its enhancers Dcp1a, Rck, Lsm1, Lsm4, Hedls, and Edc3, and with the exoribonuclease Xrn1 (Fig. 4 and 6). Thus, the entire deadenylation-decapping-5′-3′ decay machinery appears to be connected to Pat1b. These interactions are functional within a cell since tethering of Pat1b to a reporter mRNA caused both rapid deadenylation and decapping (Fig. 8). By challenging these interactions with increasing salt concentrations, we observed that Pat1b binds most tightly to Ccr4-Caf1-Not1 and to Dcp2/Dcp1a followed by Lsm1/Lsm4 and Rck. Although co-IP experiments cannot provide conclusive evidence for direct binding, our data suggest that Pat1b is in close proximity to the Ccr4-Caf1-Not complex, Dcp2/Dcp1a, the Lsm1-7 heptamer and to Rck. Association with Xrn1, Hedls, and Edc3 was much weaker and is likely to be indirect.
Within this complex, Pat1b appears to have a scaffold function since it interacts with its partners through at least three different domains (summarized in Fig. 10A). The first interaction domain is the very amino-terminal A region of Pat1b that mediates binding to Rck (Fig. 5C). Given that this domain has an astonishing number of 32 acidic amino acids within its 84 residues and a net charge of −27.7 at pH 7.0, it will be interesting to determine whether the Pat1b A domain has an influence on the nucleotide binding or helicase activity of Rck.
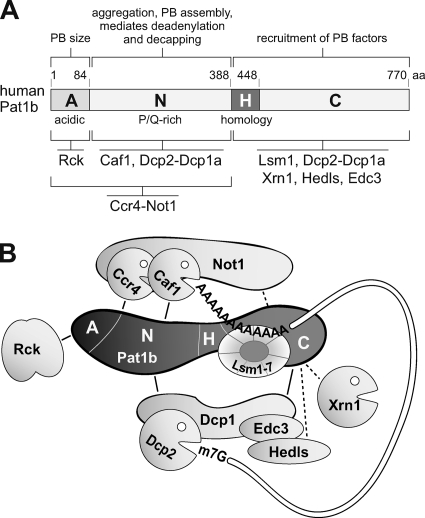
FIG. 10.
Role of Pat1b in P-body assembly and mRNA degradation. (A) Schematic representation human Pat1b subdomains, together with interacting proteins and associated functions. All the interactions depicted are RNA independent yet may be direct or indirect. (B) Hypothetical model of the Pat1b-associated complex that connects deadenylation with mRNA decapping. The acidic region (A) associates with Rck, the AN fragment with the Ccr4-Not complex, and the N fragment with Caf1 and Dcp2-Dcp1a. The homology region (H) is required for Lsm1 binding, and the HC fragment further associates with Dcp2-Dcp1a. The HC fragment also shows weak association with Edc3, Hedls, and Xrn1.
The N region that follows the acidic A domain mediates interaction with Dcp2 and Dcp1a (Fig. 6) as well as Caf1a (Fig. 7). Interestingly, Ccr4 and Not1 do not interact with Pat1b through the N region alone but also require the A domain (see Fig. S9 in the supplemental material). Although binding of all the deadenylation complex components appeared to be very tight, as estimated by their resistance to salt, this difference may suggest that Caf1a is positioned particularly close to the N domain of Pat1b. Importantly, the N domain is also the fragment that can potently induce deadenylation and decapping in the tethering assay (Fig. 9F).
On the carboxy-terminal half of Pat1b, HC co-IPs efficiently with Lsm1, Dcp2, and Dcp1a (Fig. 5 and 6), whereas the interaction of HC with Not1, Ccr4, Xrn1, Hedls, and Edc3 is weaker and possibly indirect (see Fig. S7 and S9 in the supplemental material). The H domain that shows the highest homology to the yeast Pat1 sequence appears to contribute in particular to Lsm1 binding (Fig. 5D). This is in good agreement with the interactions in S. cerevisiae, in which Pat1 occurs in a stable complex with Lsm1-7 and Xrn1 (5, 6) and was found to interact with Lsm1 via a middle domain that contains the H region (30). Edc3 and Dcp1 were shown by two-hybrid analysis to bind yeast Pat1 via its carboxy-terminal domain (30), which we find to be true for human Pat1b as well. In addition, and different from the situation in yeast, human Pat1b also interacts very efficiently with Dcp2 and Dcp1a through the N domain (Fig. 6). Moreover, Dcp2 in yeast was found to interact with Pat1 and Lsm1 in an RNA-dependent manner by co-IP experiments (42). This is not the case for human Pat1b, in which the interaction with Dcp2 is RNA independent (see Fig. S8 in the supplemental material). Thus, human Pat1b differs substantially from its yeast counterpart: it makes very tight and RNA-independent contacts with both the Ccr4-Caf1-Not deadenylase and the Dcp2-Dcp1 decapping complexes and thereby enhances the interaction between Caf1a and Dcp2 (Fig. 6 and 7). Based on our findings, a speculative model of the human Pat1b complex is presented in Fig. 10B. While the manuscript was in revision, Haas et al. published that the homologous protein in Drosophila, HPat, also forms a tight link between the Ccr4-Not complex and the Dcp2 decapping enzyme (17).
It is interesting to note that the couplings of subsequent steps in mRNA decay appear to be different in human and yeast cells. While deletion of Xrn1 in yeast causes the accumulation of mRNAs lacking both a cap and a poly(A) tail (20, 26), knockdown of Xrn1 in human cells causes the accumulation of fully adenylated mRNAs (38). This indicates that coupling of deadenylation, decapping, and 5′-3′ decay is more tight in human cells than in yeast. It is tempting to speculate that the close association of human Pat1b with the deadenylase and decapping complexes may contribute to this tight coupling.
Analysis of Pat1b localization indicated that this protein plays an important role in P-body assembly. First, Pat1b localizes to P bodies (34) (Fig. 1) similarly to Pat1 in yeast (40), HPat in Drosophila (12), and PATR-1 in C. elegans (4). Second, knocking down Pat1b strongly reduces the number of P bodies in HeLa (34) and U2OS cells (Fig. 1; see also Fig. S5 in the supplemental material). Third, overexpression of Pat1b dramatically increases the number of P bodies in COS7 (Fig. 1) and U2OS cells (see Fig. S3 in the supplemental material). Many cells expressing YFP-Pat1b or HA-Pat1b had 50 or more P bodies with a “salt”-like appearance. We noticed that some of the small Pat1b foci did not contain other P-body proteins, whereas larger Pat1b foci consistently colocalized with all P-body proteins tested (Hedls, Lsm1, Lsm4, Xrn1, and Rck). This observation suggests that Pat1b could serve as a seed for P-body assembly. Pat1b may initially self-assemble into small foci and subsequently serve as a scaffold for additional P-body proteins as the aggregates grow in size. Other proteins, such as Dcp1a, can increase the size of P bodies when overexpressed (data not shown), yet the ability to serve as a “seed” and augment P-body numbers has not been reported for any other protein and thus appears to be a unique feature of Pat1b. Our finding that Pat1b co-IPs with itself (Fig. 2H) supports the idea of self-assembly. In addition, the ability of Pat1b-induced P bodies to resist disassembly in the presence of CHX (see Fig. S6 in the supplemental material) emphasizes that Pat1b plays a role in stabilizing P bodies. The fact that Pat1b interacts with other P-body proteins via at least three separate domains (Fig. 5 and 6) reflects its scaffolding function.
Our analysis of Pat1b subfragments suggests that the N region is critical for P-body assembly. By overexpression of YFP-AN or YFP-N, we observed a strong tendency of these fragments to form aggregates, many of which did not contain other P-body proteins (Fig. 2E and F and data not shown). Moreover, we observed that the AN and N fragments strongly reduced the number of P bodies in transfected cells. This may indicate that by forming autonomous aggregates, the N region when expressed alone can compete with the aggregation process within P bodies. This capacity is reminiscent of the C-terminal aggregation-prone domain of TIA-1, which when expressed on its own antagonizes stress granule formation (22). Similar to the C-terminal domain of TIA-1 that is strongly enriched in glutamine residues (21.6%), the N region of Pat1b contains a sequence of 120 aa (aa 261 to 380) with a high proportion of glutamine and asparagine (17%) as well as proline (17%) residues. This sequence is preceded by a stretch of 80 aa (aa 180 to 260) that is highly enriched in proline (28%). Glutamine/asparagine- and proline-rich sequences are characteristic of many aggregation-prone proteins, including yeast prion proteins as well as RNA-binding proteins that promote stress granule and P-body assembly (11, 16, 32, 36).
In addition to the N region, we found that the acidic domain (A) at the very amino terminus of Pat1b (aa 1 to 84) influences the size of P bodies. When mutant Pat1b lacking the A domain (dA) was overexpressed, Pat1b foci became smaller and more numerous, giving the protein a “pepper”-like distribution (Fig. 2D). These small foci still colocalized with P-body markers, such as Hedls. Thus, the A domain seems to control the size of P bodies, maybe by promoting the fusion of small aggregates into larger P bodies. Since we found that the A domain interacts with Rck (Fig. 5C), it is possible that Rck may be an important driver of the aggregation process within P bodies. Indeed, Rck was shown to be essential for P-body formation in human cells (1).
In contrast to A and N, the H and C regions of Pat1b do not appear to play an active role in P-body assembly. The H, C, and HC fragments showed a mostly diffuse localization in the cell, with a rather weak accumulation in P bodies (Fig. 2G; see also Table S2 in the supplemental material). Localization of these fragments in P bodies presumably occurs through interactions with Lsm proteins, Xrn1, or decapping factors (Fig. 5 and 6). Since the overexpression of the H, C, or HC fragments did not affect the size or number of P bodies, it is likely that the HC region is recruited passively to P bodies. This may be different in yeast Pat1, in which the carboxy-terminal region (aa 422 to 797) was found to induce P bodies (30).
While Pat1b is clearly a central component of the RNA decay machinery, its paralog Pat1a does not share any of its functional characteristics: it does not localize to P bodies, does not augment P-body number or size, and has no influence on the expression level of a tethered reporter mRNA. In the cell lines we have tested so far, Pat1a mRNA was expressed at very low levels (data not shown). Rather, Pat1a seems to play an important role in the germ line since the Xenopus ortholog of Pat1a was recently shown to suppress translation during oocyte maturation (28).
In S. cerevisiae, Pat1 is a regulator of both mRNA decay and translation. Besides its role in decapping, yeast Pat1, together with the helicase Dhh1, is required for suppression of translation upon glucose starvation (9). This second function is probably linked to the finding that yeast Pat1 interacts with Pab1, eIF4E, and eIF4G in an RNA-dependent manner (30, 42). In our tethering experiments, human Pat1b did not have a strong effect on translation since luciferase activity and mRNA levels were reduced to similar degrees (Fig. 3C). We also measured global translation rates by [35S]methionine-cysteine incorporation and polysome profile analysis but could not see changes after knocking down or overexpressing human Pat1b (data not shown). Finally, we did not observe an interaction of human Pat1b with either eIF4G (Fig. 4B) or eIF4E (data not shown). Thus, the role of human Pat1b appears to be restricted to mRNA degradation. One might speculate that the two functions of yeast Pat1 have segregated into two gene products in the course of vertebrate evolution: Pat1a as a regulator of translation and Pat1b as a scaffold of the mRNA decay machinery. During this process, human Pat1b seems to have acquired a much closer association with both the Ccr4-Caf1-Not deadenylase and the Dcp2-Dcp1 decapping complexes, thereby providing a tight physical link between deadenylation and decapping.
Supplementary Material
Acknowledgments
We thank Jochen Kreth, Julia Luther, Oksana Seibert, and Gizem Olmezer for technical assistance, Sahil Sharma for cloning Caf1a expression vectors, Stefan Wiemann (German Cancer Research Center, Heidelberg, Germany) for providing the human Pat1b cDNA clone, Ann-Bin Shyu (University of Texas, Houston, TX) for the Caf1a antibody, Mark Timmers (University of Utrecht) for Flag-Not1, Jens Lykke-Andersen (University of California at San Diego) for myc-Ccr4, Flag-Dcp1, and Flag-Dcp2, and Ulrike Engel (Nikon Imaging Center, University of Heidelberg) for generous support and advice on microscopy, as well as Witold Filipowicz (Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland) and Nancy Kedersha (Brigham and Woman's Hospital, Boston, MA) for helpful discussions on the manuscript.
This work was supported by young investigator grant HZ-NG-210 from the Helmholtz Gemeinschaft and research grant STO 859/2-1 from the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 28 June 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andrei, M. A., D. Ingelfinger, R. Heintzmann, T. Achsel, R. Rivera-Pomar, and R. Luhrmann. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11:717-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bashkirov, V. I., H. Scherthan, J. A. Solinger, J. M. Buerstedde, and W. D. Heyer. 1997. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 136:761-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharyya, S. N., R. Habermacher, U. Martine, E. I. Closs, and W. Filipowicz. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125:1111-1124. [DOI] [PubMed] [Google Scholar]
- 4.Boag, P. R., A. Atalay, S. Robida, V. Reinke, and T. K. Blackwell. 2008. Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J. Cell Biol. 182:543-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnerot, C., R. Boeck, and B. Lapeyre. 2000. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol. Cell. Biol. 20:5939-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouveret, E., G. Rigaut, A. Shevchenko, M. Wilm, and B. Seraphin. 2000. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 19:1661-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chekulaeva, M., and W. Filipowicz. 2009. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 21:452-460. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury, A., J. Mukhopadhyay, and S. Tharun. 2007. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 13:998-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coller, J., and R. Parker. 2005. General translational repression by activators of mRNA decapping. Cell 122:875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cougot, N., S. Babajko, and B. Seraphin. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165:31-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decker, C. J., D. Teixeira, and R. Parker. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179:437-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eulalio, A., I. Behm-Ansmant, D. Schweizer, and E. Izaurralde. 2007. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27:3970-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenger-Gron, M., C. Fillman, B. Norrild, and J. Lykke-Andersen. 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell 20:905-915. [DOI] [PubMed] [Google Scholar]
- 14.Franks, T. M., and J. Lykke-Andersen. 2007. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 21:719-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garneau, N. L., J. Wilusz, and C. J. Wilusz. 2007. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8:113-126. [DOI] [PubMed] [Google Scholar]
- 16.Gilks, N., N. Kedersha, M. Ayodele, L. Shen, G. Stoecklin, L. M. Dember, and P. Anderson. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15:5383-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas, G., J. E. Braun, C. Igreja, F. Tritschler, T. Nishihara, and E. Izaurralde. 2010. HPat provides a link between deadenylation and decapping in metazoa. J. Cell Biol. 189:289-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatfield, L., C. A. Beelman, A. Stevens, and R. Parker. 1996. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:5830-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, W., and R. Parker. 2001. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics 158:1445-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu, C. L., and A. Stevens. 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13:4826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingelfinger, D., D. J. Arndt-Jovin, R. Luhrmann, and T. Achsel. 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8:1489-1501. [PMC free article] [PubMed] [Google Scholar]
- 22.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147:1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim, F., T. P. Downey, and D. S. Peabody. 2001. Translational repression and specific RNA binding by the coat protein of the Pseudomonas phage PP7. J. Biol. Chem. 276:22507-22513. [DOI] [PubMed] [Google Scholar]
- 24.Liu, J., M. A. Valencia-Sanchez, G. J. Hannon, and R. Parker. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7:719-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lykke-Andersen, J., and E. Wagner. 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 19:351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muhlrad, D., C. J. Decker, and R. Parker. 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 8:855-866. [DOI] [PubMed] [Google Scholar]
- 27.Muhlrad, D., and R. Parker. 2005. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 24:1033-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura, Y., K. J. Tanaka, M. Miyauchi, L. Huang, M. Tsujimoto, and K. Matsumoto. 21 May 2010. Translational repression by the oocyte-specific protein P100 in Xenopus. Dev. Biol. [Epub ahead of print.] doi: 10.1016/j.ydbio.2010.05.006. [DOI] [PubMed]
- 29.Parker, R., and H. Song. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11:121-127. [DOI] [PubMed] [Google Scholar]
- 30.Pilkington, G. R., and R. Parker. 2008. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol. Cell. Biol. 28:1298-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pillai, R. S., S. N. Bhattacharyya, C. G. Artus, T. Zoller, N. Cougot, E. Basyuk, E. Bertrand, and W. Filipowicz. 2005. Inhibition of translational initiation by Let-7 microRNA in human cells. Science 309:1573-1576. [DOI] [PubMed] [Google Scholar]
- 32.Reijns, M. A., R. D. Alexander, M. P. Spiller, and J. D. Beggs. 2008. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 121:2463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothbauer, U., K. Zolghadr, S. Muyldermans, A. Schepers, M. C. Cardoso, and H. Leonhardt. 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics 7:282-289. [DOI] [PubMed] [Google Scholar]
- 34.Scheller, N., P. Resa-Infante, S. de la Luna, R. P. Galao, M. Albrecht, L. Kaestner, P. Lipp, T. Lengauer, A. Meyerhans, and J. Diez. 2007. Identification of PatL1, a human homolog to yeast P body component Pat1. Biochim. Biophys. Acta 1773:1786-1792. [DOI] [PubMed] [Google Scholar]
- 35.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shorter, J., and S. Lindquist. 2005. Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 6:435-450. [DOI] [PubMed] [Google Scholar]
- 37.Stoecklin, G., and P. Anderson. 2006. Posttranscriptional mechanisms regulating the inflammatory response. Adv. Immunol. 89:1-37. [DOI] [PubMed] [Google Scholar]
- 38.Stoecklin, G., T. Mayo, and P. Anderson. 2006. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 7:72-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoecklin, G., P. Stoeckle, M. Lu, O. Muehlemann, and C. Moroni. 2001. Cellular mutants define a common mRNA degradation pathway targeting cytokine AU-rich elements. RNA 7:1578-1588. [PMC free article] [PubMed] [Google Scholar]
- 40.Teixeira, D., and R. Parker. 2007. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 18:2274-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Temme, C., S. Zaessinger, S. Meyer, M. Simonelig, and E. Wahle. 2004. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 23:2862-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tharun, S., and R. Parker. 2001. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell 8:1075-1083. [DOI] [PubMed] [Google Scholar]
- 43.van Dijk, E., N. Cougot, S. Meyer, S. Babajko, E. Wahle, and B. Seraphin. 2002. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21:6915-6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, Z., X. Jiao, A. Carr-Schmid, and M. Kiledjian. 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. U. S. A. 99:12663-12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkler, G. S., K. W. Mulder, V. J. Bardwell, E. Kalkhoven, and H. T. Timmers. 2006. Human Ccr4-Not complex is a ligand-dependent repressor of nuclear receptor-mediated transcription. EMBO J. 25:3089-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamasaki, S., G. Stoecklin, N. Kedersha, M. Simarro, and P. Anderson. 2007. T-cell intracellular antigen-1 (TIA-1)-induced translational silencing promotes the decay of selected mRNAs. J. Biol. Chem. 282:30070-30077. [DOI] [PubMed] [Google Scholar]
- 47.Zheng, D., N. Ezzeddine, C. Y. Chen, W. Zhu, X. He, and A. B. Shyu. 2008. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 182:89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.