Abstract
α-Synuclein (αSyn) can self-associate, forming oligomers, fibrils, and Lewy bodies, the pathological hallmark of Parkinson disease. Current dogma suggests that oligomeric αSyn intermediates may represent the most toxic αSyn species. Here, we studied the effect of a potent molecular chaperone, CHIP (carboxyl terminus of Hsp70-interacting protein), on αSyn oligomerization using a novel bimolecular fluorescence complementation assay. CHIP is a multidomain chaperone, utilizing both a tetratricopeptide/Hsp70 binding domain and a U-box/ubiquitin ligase domain to differentially impact the fate of misfolded proteins. In the current study, we found that co-expression of CHIP selectively reduced αSyn oligomerization and toxicity in a tetratricopeptide domain-dependent, U-box-independent manner by specifically degrading toxic αSyn oligomers. We conclude that CHIP preferentially recognizes and mediates degradation of toxic, oligomeric forms of αSyn. Further elucidation of the mechanisms of CHIP-induced degradation of oligomeric αSyn may contribute to the successful development of drug therapies that target oligomeric αSyn by mimicking or enhancing the powerful effects of CHIP.
The clinical symptoms of Parkinson disease (PD)2 are caused by an unknown toxic insult that selectively destroys dopamine-producing cells in the substantia nigra of affected humans (1). α-Synuclein (αSyn)-containing Lewy bodies have long been associated with PD and PD-associated cellular toxicity (2). Recent evidence indicates that αSyn can form prefibrillar, oligomeric assemblies as well as fibrillar cellular aggregates, and it has been suggested that these oligomeric assemblies mediate αSyn neurotoxicity (3–7). Therefore, understanding the molecular mechanisms of αSyn oligomerization will provide insight into targeting therapeutics that can reduce αSyn oligomerization or clear existing αSyn oligomers.
Molecular chaperones such as heat shock proteins (Hsp) 70 and 90 and their associated co-chaperone CHIP (carboxyl terminus of Hsp70-interacting protein) have a well documented ability to reduce toxicity mediated by neurodegenerative disease states, stress, or injury (8–17). CHIP functions as an important link between the molecular chaperone system and the protein degradation system. CHIP has two specialized protein domains, each conferring different yet complementary functions on CHIP activity. The amino terminus domain of CHIP contains a tetratricopeptide (TPR) domain that links it to the molecular chaperones Hsp70 and 90 (18). The carboxyl terminus domain of CHIP contains a U-box domain that confers the E3 ubiquitin ligase function of CHIP.
Our earlier studies suggest that CHIP can enhance degradation of αSyn and protect cells from its toxicity. We have now taken advantage of a bimolecular complementation approach to generate stable αSyn oligomers and demonstrate that CHIP selectively promotes degradation of this potentially more toxic, oligomeric form.
EXPERIMENTAL PROCEDURES
Construct Generation
The two constructs utilized in the present study were GN-αSyn and αSynGC (previously referred to as GN-link-αSyn and αSynGC) (19). αSyn bimolecular fluorescence complementation (BiFC) constructs were generated by PCR, cloned into pcDNA3.1, and verified by DNA sequencing as described previously (19). For CHIP expression, pcDNA3-CHIP, pcDNA3-CHIPΔU (residues 196 –303 deleted), and pcDNA3-CHIPΔTPR (residues 32–144 deleted) constructs were utilized and were kind gifts from Dr. Cam Patterson, University of North Carolina School of Medicine (20).
Cell Culture and Transient Transfections
Human H4 neuroglioma cells (HTB-148; ATCC) were maintained in OPTIMEM medium supplemented with 10% fetal bovine serum (both from Invitrogen) and incubated at 37 °C, 5%CO2. H4 cells were plated 24 h prior to transfection and grown to 80–90% confluency. Cells were transfected with an equimolar ratio of DNA constructs using Superfect (Qiagen) according to the manufacturer’s instructions. Co-transfection of pcDNA3.1+ vector was used as a control.
GN-αSyn/αSynGC Fluorescence Complementation Assay
For optimal fluorophore maturation, transiently transfected cells were incubated overnight at 30 °C (21) after an initial 4-h incubation at 37 °C. 24 h after transfection cells were either observed using a Zeiss LSM510 confocal microscope or harvested for cell lysate preparation.
Live Cell Imaging
Cells were plated on poly-d-lysine-coated coverslipped 35-mm dishes (MatTek Cultureware, Ashland, MA) and transfected the following day. Cells were imaged 24 h post-transfection on a Zeiss LSM510 confocal microscope system. Cells were observed at ×63 for quantification of pixel intensity. For each experimental group the average fluorescence intensity from 50 cells, across three independent experiments, was measured using Adobe Photoshop® software. Adobe Photoshop® translated the average GFP fluorescence to average pixel intensity. Therefore, the average pixel intensity was recorded for each cell. Data were normalized to empty vector (pcDNA3.1+) controls.
Fluorescent-activated Cell Sorting
Cells were plated and transfected in 10-cm dishes. 24 h post transfection, cells were trypsinized, centrifuged, and the pellet reconstituted in phosphate-buffered saline. The resulting suspension was filtered with cell strainer caps into polypropylene tubes (both from BD Biosciences). Fluorescence was measured on a FACSCanto (BD Biosciences) flow cytometer.
Toxicity Assay
24 h post-transfection levels of toxicity were measured utilizing the ToxiLight™ non-destructive cytotoxicity assay kit (Cambrex, Rockland, ME). This assay quantitatively measures the release of adenylate kinase from damaged cells into the culture medium and was performed according to the manufacturer’s protocol.
Detergent Solubility Fractionation and Gel Electrophoresis
After 24 h, cells were washed with cold phosphate-buffered saline, harvested by scraping in cold lysis buffer with or without detergent for SDS-PAGE and native gels, respectively, (50 mm Tris/HCl, pH 7.4, 175 mm NaCl, 5 mm EDTA, pH 8.0, and protease inhibitor mixture), and sheared once through a 28-gauge needle followed by sonication for 10 s (total cell lysates). Fractionation was performed by adding Triton X-100 (Tx) to total cell lysates (final concentration of 1%) and incubating for 30 min on ice followed by centrifugation (15,000 × g, 60 min, 4 °C). The supernatant was designated as the Tx-soluble fraction, and the pellet was redissolved in 2% SDS-containing lysis buffer and sonicated for 10 s (Tx-insoluble fraction). Protein concentration was determined using a BCA protein assay. 15–40 μg of protein of each cell lysate was loaded onto a 10–20% Tris/glycine gel (Invitrogen) or a Native PAGE Novex 4–16% Bis-Tris gel (Invitrogen) for Western blot analysis. SDS-PAGE was performed with SDS-containing running and sample loading buffer (Invitrogen), whereas native electrophoresis was performed using detergent-free running and sample buffers (Invitrogen). Proteins resolved on the gels were transferred onto polyvinylidene difluoride membrane (Millipore, Billerica, MA) and blocked in 5% milk in Tris-buffered saline (TBS) containing 0.05% Tween 20 (TBS-T) for 2 h prior to the addition of primary antibody, anti-synuclein (610787, 1:1000; BD Biosciences), or anti-GFP (ab6556, 1:3000; Abcam, Cambridge, MA) at room temperature for 2 h or 4 °C overnight. Horseradish peroxidase-labeled anti-mouse or anti-rabbit antibodies (1:2000; Jackson ImmunoResearch, West Grove, PA) were incubated at room temperature for 1 h, processed, and quantified. The results were visualized with ECL (GE Healthcare) or SuperSignal Femto (Pierce) chemiluminescent reagents. Denatured blots were probed with anti-glyceraldehyde-3-phosphate dehydrogenase (ab9485, 1:3000; Abcam) and anti-rabbit horseradish peroxidase (1:2000) as a loading control. A nonspecific protein band at ~150–200 kDa across all samples in the series (Coomassie Blue staining of the gel) served as a loading control for the native blots. ImageJ and ImageQuant were used to quantify optical densities of total, soluble, and insoluble blots and native blots, respectively.
Coimmunoprecipitation
For immunoprecipitation, H4 cells were harvested 24 h after transfection and lysed in lysis buffer: 50 mm Tris/HCl, pH 7.4, 150 mm NaCl, 0.3% CHAPS, 10 mm NaF, 100 μm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (Roche Applied Science) for 1 h on ice. Cell lysates were centrifuged at 10,000 rpm for 10 min at 4 °C, and supernatant was used for immunoprecipitation using protein G-agarose and rat anti-Myc antibody (JAC6; Serotec, Oxford, UK) overnight at 4 °C. Immunoprecipitates were washed with lysate buffer three times and loaded onto 10–20% Tris/glycine gels (Invitrogen). Protein was transferred to polyvinylidene difluoride membrane (Millipore) and blocked in 5% milk in TBS containing 0.05% TBS-T for 2 h prior to the addition of primary antibody, anti-Hsp70 (SPA-812, 1:10000; StressGen, Ann Arbor, MI), or anti-GFP (ab6556, 1:3000; Abcam) at room temperature for 2 h or 4 °C overnight. Horseradish peroxidase-labeled anti-mouse or anti-rabbit antibody (1:2000; Jackson ImmunoResearch) was incubated at room temperature for 1 h, processed, and quantified. The results were visualized with ECL (GE Healthcare).
RESULTS
CHIP Reduces αSyn Oligomerization
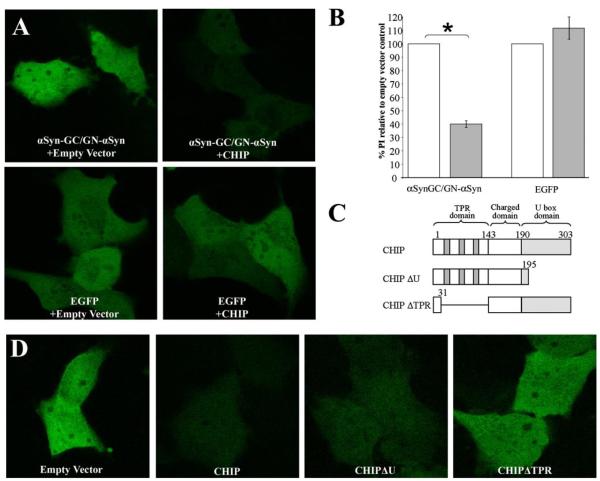
We have previously shown that CHIP modifies and prevents α-Syn inclusion formation by interacting (either directly or indirectly) with αSyn (22). To further investigate the specific αSyn species that is targeted by CHIP, we employed a novel BiFC assay that we have previously demonstrated to be a specific and effective method by which to visualize and monitor αSyn oligomerization in vitro (19). BiFC involves the reconstitution of non-fluorescent fragments of GFP when fused to proteins of interest. Upon protein-protein interaction the two fragments are able to come together and reconstitute the fluorophore. We investigated the ability of CHIP to reduce αSyn oligomerization by transfecting H4 human neuroglioma cells with the αSyn BiFC constructs αSynGC and GN-αSyn in the presence or absence of CHIP (Fig. 1) and found that CHIP dramatically reduced αSyn oligomerization as measured by intensity of GFP fluorescence (Fig. 2, A and B). As a control, we confirmed that CHIP had no effect on fluorescence intensity of enhanced green fluorescent protein (EGFP) (Fig. 2, A and B). To confirm that CHIP’s prevention of αSyn oligomerization is specific to αSyn, and not EGFP, we conducted co-immunoprecipitation experiments and determined that CHIP did not co-immunoprecipitates with EGFP alone (data not shown).
FIGURE 1. A schematic representation of the constructs utilized.

GN-αSyn contains the first 158 amino-terminal residues of green fluorescent protein (GFP) and a poly linker region attached to the amino terminus of full-length αSyn. αSynGC contains the 80 carboxyl-terminal residues of GFP attached to the carboxyl terminus of full-length αSyn.
FIGURE 2. CHIP and CHIPΔU decrease αSynGC/GN-αSyn pixel intensity (PI).
A, H4 cells were transfected with αSynGC/GN-αSyn or enhanced green fluorescent protein (EGFP) and CHIP or an Empty Vector control. CHIP decreased the PI of αSynGC/GN-αSyn, but not EGFP. B, quantification of A. White bars, empty vector; gray bars, CHIP. Data are presented as means + S.E. of three independent experiments. *, p < 0.001 compared with Empty Vector. C, a schematic of the CHIP domain mutant constructs utilized. D, CHIP and the domain mutant CHIPΔU decrease αSynGC/GN-αSyn PI.
To confirm that the effect of CHIP on αSyn oligomerization is specific, we overexpressed domain mutant constructs of CHIP (CHIPΔU and CHIPΔTPR) with αSynGC and GN-αSyn (Fig. 2C) and determined that CHIPΔU, but not CHIPΔTPR, was able to significantly reduce the fluorescence produced by αSyn oligomer formation (Fig. 2D). Because the TPR domain of CHIP is essential for Hsp70 interactions, these data suggest that the CHIP-mediated reduction in αSyn oligomer formation occurs in an Hsp70-associated manner.
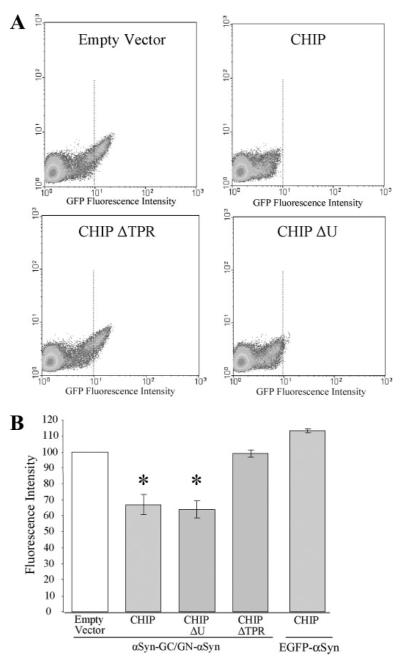
To quantify the CHIP domain mutant effects, we co-transfected H4 human neuroglioma cells with αSynGC, GN-αSyn, and either empty vector, CHIP, CHIPΔU, or CHIPΔTPR and measured GFP fluorescence intensity with fluorescent-activated cell sorting. In agreement with our observations of GFP pixel intensity (Fig. 2), fluorescent-activated cell sorting analysis demonstrated that CHIP and CHIPΔU mediate a reduction in αSyn oligomerization (Fig. 3, A and B). As a control, we verified that CHIP had no effect on αSyn-EGFP fluorescence, confirming that CHIP reduces αSyn oligomer formation in a TPR domain-dependent manner.
FIGURE 3. CHIP and CHIPΔU decrease αSynGC/GN-αSyn fluorescence.
A, density plots illustrating the CHIP- and CHIPΔU-mediated reduction of αSynGC/GN-αSyn-generated green fluorescent protein (GFP) in H4 cells transfected with αSynGC/GN-αSyn or enhanced GFP-αSyn (EGFP-αSyn) and CHIP, CHIPΔU, CHIPΔTPR, or Empty Vector control. The dotted lines indicate the fluorescent intensity of CHIP-treated cells and have been added to assist comparisons between groups. B, quantification of the CHIP- and CHIPΔU-mediated reduction of αSynGC/GN-αSyn and lack of CHIP-mediated EGFP-αSyn reduction. Data from the entire fluorescent signal are presented as means + S.E. of three-five independent experiments. *, p < 0.01 compared with Empty Vector.
CHIP Reduces αSyn Oligomer-induced Toxicity
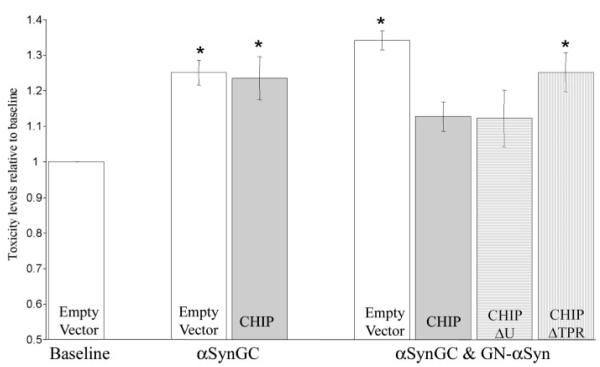
We have shown previously that overexpression of the BiFC pairs αSynGC and GN-αSyn results in increased αSyn oligomerization formation and cytotoxicity (19). To determine whether CHIP can rescue cytotoxicity conferred by oligomeric αSyn, H4 cells were cotransfected with αSynGC, GN-αSyn, and αSynGC and GN-αSyn together, in the presence or absence of CHIP or its domain mutants (CHIPΔU and CHIPΔTPR), and toxicity was monitored. As expected, CHIP and CHIPΔU rescued toxicity conferred by αSynGC and GN-αSyn coexpression (Fig. 4). By contrast, CHIP had no effect on αSyn toxicity induced by overexpression of individual αSyn constructs, i.e. αSynGC expressed alone (Fig. 4). These data suggest that CHIP preferentially reduces cytotoxicity conferred by a stabilized αSyn oligomeric species and not that of the less toxic, transient αSyn constructs. This experiment also confirmed that the TPR domain is critical for the toxicity-rescuing effects of CHIP because CHIPΔTPR was unable to rescue toxicity. These data are in agreement with CHIP preventing the formation of αSyn oligomers (Figs. 2 and 3).
FIGURE 4. Overexpression of CHIP and CHIPΔU reduces toxicity induced by the more toxic, stabilized oligomeric αSyn species (αSynGC & GN-αSyn).
H4 cells were transfected with Empty Vector (Baseline) or equimolar concentrations of either αSynGC or αSynGC/GN-αSyn plus Empty Vector, CHIP, CHIPΔU, or CHIPΔTPR. Data are presented as means + S.E. of three-five independent experiments. *, p < 0.05 compared with baseline.
CHIP Preferentially Degrades αSyn Oligomeric Species
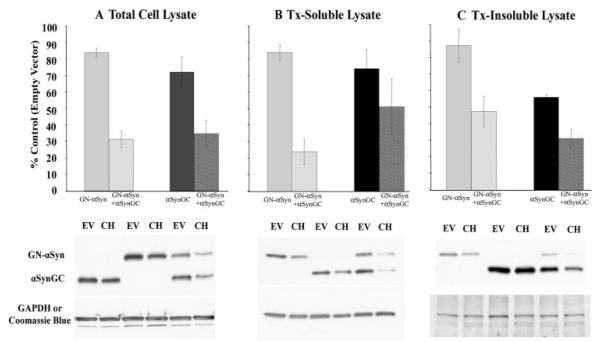
To determine the mechanism of CHIP-mediated effects on αSyn, we examined the expression levels of αSyn in the presence or absence of CHIP under two conditions in which oligomeric species are to determine the mechanism of CHIP-mediated effects on αSyn, we examined the expression levels of αSyn in the presence or absence of CHIP under two conditions in which oligomeric species are, 1) less stabilized: αSynGC or GN-αSyn expressed individually or, 2) more stabilized: αSynGC and GN-αSyn expressed together. Total cell lysate and Tx-soluble and Tx-insoluble lysates were prepared. Interestingly, in the total cell lysate fraction, CHIP had only a modest effect on αSyn levels when αSynGC and GN-αSyn were expressed individually (Fig. 5A). However, when the αSyn BiFC proteins were coexpressed, CHIP significantly reduced the protein levels (Fig. 5A).
FIGURE 5. CHIP preferentially reduces the toxic αSyn oligomers from the total cell lysate and Tx-soluble and Tx-insoluble fractions.
H4 cells were co-transfected with either GN-αSyn or αSynGC alone or with GN-αSyn and αSynGC together, plus CHIP (CH) or Empty Vector (EV). The optical density of αSynGC and GN-αSyn was measured from the total (A), Tx-soluble (B), and Tx-insoluble (C) fractions for EV- or CH-transfected cells. Each bar represents protein levels from cells transfected with CH and are relative to EV. To generate each bar, the ratio between CH and EV was calculated from the optical densities of the Western blot. Representative GFP-probed blots are provided; the loading controls for A/B and C are glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Coomassie blue, respectively.
Previous studies have demonstrated that CHIP can shuttle proteins such as mutant tau, huntingtin, and ataxin to different soluble and insoluble cellular compartments (15–17, 23–25). To determine the effect of CHIP on αSyn solubility we extracted Tx-soluble and -insoluble fractions. We found that CHIP had little or no effect on levels of αSyn in the soluble or insoluble fraction when αSynGC and GN-αSyn were expressed individually (Fig. 5, B and C). However, when coexpressed, CHIP degrades αSyn in both the soluble and insoluble fractions (Figs. 5, B and C).
CHIP Reduces High Molecular Weight αSyn Oligomeric Species
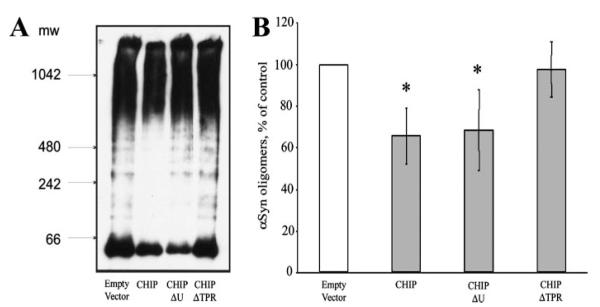
The co-expression of αSynGC and GN-αSyn results in high molecular weight (HMW) oligomeric species when examined by native Western blot. To determine whether CHIP could degrade HMW αSyn oligomers we co-transfected H4 cells with αSynGC and GN-αSyn with an empty vector (pcDNA), CHIP, CHIPΔU, or CHIPΔTPR. Interestingly, CHIP efficiently degrades the HMW αSyn population. Furthermore, consistent with our previous data, the TPR domain, but not the U-box domain, is critical for this oligomeric degradation because CHIPΔTPR is ineffective (Fig. 6, A and B).
FIGURE 6. CHIP and CHIPΔU reduce high molecular weight (HMW) αSyn oligomers.
H4 cells were co-transfected with GN-αSyn and αSynGC plus Empty Vector (pcDNA), CHIP, or the CHIP domain mutants (CHIPΔU or CHIPΔTPR), and the lysates were electrophoresed on Native PAGE Novex 4–16% Bis-Tris gel. A, HMW smears (i.e. oligomers) produced by co-expression of the αSyn constructs together, ±CHIP or domain mutants. B, a quantification of the CHIP-mediated αSyn oligomer reduction. Data are presented as means±S.E. of three-five independent experiments. *, p < 0.01 compared with empty vector.
CHIP, but Not CHIPΔTPR, Coimmunoprecipitates with Hsp70
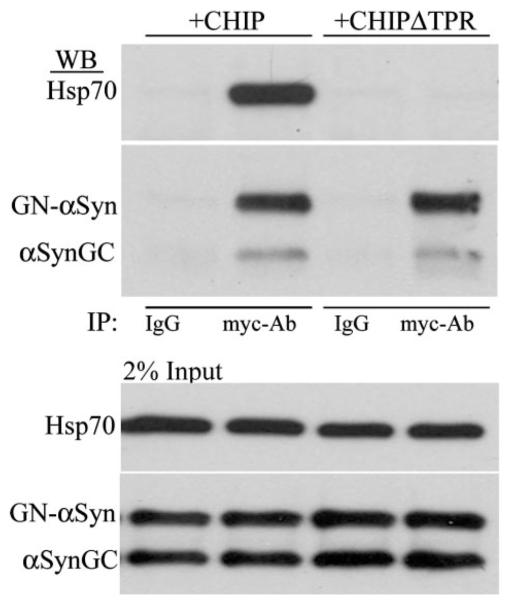
CHIP and Hsp70 form a complex between the amino-terminal TPR domain of CHIP and the carboxyl-terminal EEVD motif of Hsp70. Our data indicate that the TPR domain of CHIP is critical for the CHIP-mediated reduction of αSyn oligomers and associated toxicity. To determine whether the association of Hsp70 with CHIP, αSynGC, and GN-αSyn is critical, we co-transfected H4 cells with Hsp70, αSynGC, GN-αSyn, and CHIP or CHIPΔTPR. Both CHIP and CHIPΔTPR coimmunoprecipitate with GN-αSyn and αSynGC; however, only fulllength CHIP, and not CHIPΔTPR, coimmunoprecipitates with Hsp70 (Fig. 7). These data imply that Hsp70 is required for the CHIP-mediated reduction of αSyn oligomers and their associated toxicity.
FIGURE 7. Hsp70 coimmunoprecipitates with full-length CHIP, GN-αSyn, and αSynGC.
H4 cells were co-transfected with Hsp70, GN-αSyn, αSynGC, and Myc-CHIP or Myc-CHIPΔTPR. 24 h after transfection, cells were harvested and processed for immunoprecipitation (IP) with anti-Myc antibody. Immunoblots were probed with anti-Hsp70 and anti-GFP antibody. IgG, Myc-Ab immunoprecipitation control. Data are representative of two independent experiments.
DISCUSSION
CHIP has been shown to mediate the degradation of misfolded proteins associated with Alzheimer disease, Huntington disease, Spinocerebellar ataxia, and PD (22, 24, 26, 27). This degradation leads to an overall reduction of cellular toxicity and enhancement of cell survival (23, 24, 26, 28). In PD, current thinking identifies αSyn oligomeric species as one of the primary mediators of toxicity (3–7). In the current study we hypothesized that CHIP preferentially targets αSyn oligomers for degradation.
The stabilization of αSyn in an oligomeric conformation using the novel BiFC assay leads to an increase in cytotoxicity, which can be rescued by CHIP in an Hsp70-dependent manner. By contrast, CHIP did not rescue the cytotoxicity associated with the more transient αSyn interactions, which suggests that CHIP targets a specific αSyn conformation for degradation. Moreover, CHIP degrades αSyn species in both Tx-100-soluble and Tx-100-insoluble fractions and significantly reduces the formation of HMW αSyn oligomers. These data are consistent with our previous study where overexpression of Hsp70 prevented αSyn oligomerization (19).
We adapted a BiFC assay (28) to stabilize and detect the formation of αSyn oligomers (19). Here we show that CHIP targets an αSyn oligomeric species for degradation. This activity is associated with cytoprotection, consistent with the idea that oligomeric species are toxic moieties (3, 29). The degradative preference of CHIP for the more toxic protein species is also evident in other models of neurodegeneration. For example, CHIP preferentially degrades the more toxic Aβ42, but not the less toxic soluble APP, holoprotein (28). Furthermore, CHIP also preferentially inhibits toxicity induced by the more toxic four repeat tau compared with the less toxic three repeat tau (23). Thus, the degradative preference of CHIP for a more toxic αSyn species versus a less toxic αSyn species is consistent with other models of neurodegeneration.
The ability of CHIP to affect aggregate formation and degrade disease-causing proteins is well documented as is its modulatory effects on these proteins within soluble and insoluble protein fractions (15, 16, 24–26). In models of tau and ataxin-1 overexpression, CHIP increases aggregate formation and accumulation of tau and ataxin-1 within the insoluble fraction (15, 16, 28). However, in instances of huntingtin and αSyn overexpression, CHIP reduces aggregate formation (22, 24). This CHIP-mediated decrease in aggregation leads to a decrease in huntingtin protein from the insoluble fraction. Our data, which also show a CHIP-mediated decrease in αSyn from the insoluble fraction, agree with this finding (24). To our knowledge, these are the first experiments to investigate the effects of CHIP on oligomer formation in neurodegenerative disease. αSyn-induced toxicity is largely attributed to the formation of αSyn oligomers (3–7), and therefore, therapeutically, it will be important to reduce their formation and accumulation. Utilizing the BiFC assay to model αSyn oligomer formation, we determined that CHIP reduces the amount of higher order αSyn oligomers. This result has important implications for CHIP and chaperone proteins as a potential therapeutic for αSyn-associated toxicity and also for other models of neurodegeneration where oligomer formation is a contributing causative agent.
The mechanism of CHIP-induced protein degradation is not completely understood. It is known that CHIP interacts with Hsp70 via the amino-terminal TPR domain and the EEVD motif of Hsp70. However, it has been postulated that CHIP exhibits intrinsic molecular chaperone activities and can associate with misfolded proteins in an Hsp70-independent manner (30). Our data support this hypothesis in that CHIPΔTPR can associate with GN-αSyn and αSynGC in the absence of Hsp70. In further support of this hypothesis, our laboratory has shown that CHIPΔTPR can also associate with misfolded αSyn in the absence of Hsp70 (22). However, while CHIP associates with different αSyn species in an Hsp70-independent manner, we have previously shown that degradation of different αSyn species by CHIP can occur with or without Hsp70 (22). In the present study, CHIP degraded αSyn oligomers and reduced toxicity in an Hsp70-dependent manner. These data suggest that CHIP has the ability to process different species of αSyn via different mechanisms. Further experimentation will be required to elucidate the specific degradation mechanisms involved in the CHIP-mediated degradation of oligomeric αSyn.
In summary, we have shown that CHIP has the ability to preferentially promote degradation of αSyn oligomeric species from both the soluble and insoluble cellular compartments. We postulate that HMW forms of αSyn are toxic and that the CHIP-mediated reduction in αSyn-induced toxicity is produced by the ability of CHIP to preferentially degrade these toxic αSyn species. Understanding the molecular mechanisms of CHIP-induced toxicity will elucidate more effective therapeutic strategies for the treatment of neurodegenerative disease.
Footnotes
- PD
- Parkinson disease
- αSyn
- α-Synuclein
- Hsp
- heat shock protein
- CHIP
- carboxyl terminus of Hsp70-interacting protein
- TPR
- tetratricopeptide
- BiFC
- bimolecular fluorescence complementation
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- GFP
- green fluorescent protein
- EGFP
- enhanced GFP
- GN-αSyn
- GFP amino-terminal plus α-Syn
- αSynGC
- α-Syn plus GFP carboxyl-terminal
- HMW
- high molecular weight
- TX
- Triton X-100
REFERENCES
- 1.Forno LS. J. Neuropathol. Exp. Neurol. 1996;55:259–272. doi: 10.1097/00005072-199603000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Lewy F. In: Handbuch der Neurologie. Lewandowsky M, Abelsdorff G, editors. Springer Verlag; Berlin: 1912. pp. 920–933. [Google Scholar]
- 3.Volles MJ, Lansbury PT., Jr. Biochemistry. 2003;42:7871–7878. doi: 10.1021/bi030086j. [DOI] [PubMed] [Google Scholar]
- 4.Sharon R, Bar-Joseph I, Frosch MP, Walsh DM, Hamilton JA, Selkoe DJ. Neuron. 2003;37:583–595. doi: 10.1016/s0896-6273(03)00024-2. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg MS, Lansbury PT., Jr. Nat. Cell Biol. 2000;2:E115–E119. doi: 10.1038/35017124. [DOI] [PubMed] [Google Scholar]
- 6.Volles MJ, Lansbury PT., Jr. Biochemistry. 2002;41:4595–4602. doi: 10.1021/bi0121353. [DOI] [PubMed] [Google Scholar]
- 7.Li W, Lee MK. J. Neurochem. 2005;93:1542–1550. doi: 10.1111/j.1471-4159.2005.03146.x. [DOI] [PubMed] [Google Scholar]
- 8.Tetzlaff J, Tanzer L, Jones KJ. J. Neuroendocrinol. 2007;19:383–389. doi: 10.1111/j.1365-2826.2007.01538.x. [DOI] [PubMed] [Google Scholar]
- 9.McLean PJ, Klucken J, Shin Y, Hyman BT. Biochem. Biophys. Res. Commun. 2004;321:665–669. doi: 10.1016/j.bbrc.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 10.Klucken J, Shin Y, Masliah E, Hyman BT, McLean PJ. J. Biol. Chem. 2004;279:25497–25502. doi: 10.1074/jbc.M400255200. [DOI] [PubMed] [Google Scholar]
- 11.Klucken J, Shin Y, Hyman BT, McLean PJ. Biochem. Biophys. Res. Commun. 2004;325:367–373. doi: 10.1016/j.bbrc.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 12.Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 13.Evans CG, Wisen S, Gestwicki JE. J. Biol. Chem. 2006;281:33182–33191. doi: 10.1074/jbc.M606192200. [DOI] [PubMed] [Google Scholar]
- 14.Novoselova TV, Margulis BA, Novoselov SS, Sapozhnikov AM, van der Spuy J, Cheetham ME, Guzhova IV. J. Neurochem. 2005;94:597–606. doi: 10.1111/j.1471-4159.2005.03119.x. [DOI] [PubMed] [Google Scholar]
- 15.Petrucelli L, Dickson D, Kehoe K, Taylor J, Snyder H, Grover A, De Lucia M, McGowan E, Lewis J, Prihar G, Kim J, Dillmann WH, Browne SE, Hall A, Voellmy R, Tsuboi Y, Dawson TM, Wolozin B, Hardy J, Hutton M. Hum. Mol. Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 16.Shimura H, Schwartz D, Gygi SP, Kosik KS. J. Biol. Chem. 2004;279:4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 17.Sahara N, Murayama M, Mizoroki T, Urushitani M, Imai Y, Takahashi R, Murata S, Tanaka K, Takashima A. J. Neurochem. 2005;94:1254–1263. doi: 10.1111/j.1471-4159.2005.03272.x. [DOI] [PubMed] [Google Scholar]
- 18.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. Mol. Cell. Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Outeiro TF, Putcha P, Tetzlaff JE, Spoelgen R, Koker M, Carvalho F, Hyman BT, McLean PJ. PLoS ONE. 2008;3:e1867. doi: 10.1371/journal.pone.0001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. Nat. Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 21.Hu CD, Kerppola TK. Nat. Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin Y, Klucken J, Patterson C, Hyman BT, McLean PJ. J. Biol. Chem. 2005;280:23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- 23.Hatakeyama S, Matsumoto M, Kamura T, Murayama M, Chui DH, Planel E, Takahashi R, Nakayama KI, Takashima A. J. Neurochem. 2004;91:299–307. doi: 10.1111/j.1471-4159.2004.02713.x. [DOI] [PubMed] [Google Scholar]
- 24.Miller VM, Nelson RF, Gouvion CM, Williams A, Rodriguez-Lebron E, Harper SQ, Davidson BL, Rebagliati MR, Paulson HL. J. Neurosci. 2005;25:9152–9161. doi: 10.1523/JNEUROSCI.3001-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi JY, Ryu JH, Kim HS, Park SG, Bae KH, Kang S, Myung PK, Cho S, Park BC, Lee do H. Mol. Cell. Neurosci. 2007;34:69–79. doi: 10.1016/j.mcn.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Al-Ramahi I, Lam YC, Chen HK, de Gouyon B, Zhang M, Perez AM, Branco J, de Haro M, Patterson C, Zoghbi HY, Botas J. J. Biol. Chem. 2006;281:26714–26724. doi: 10.1074/jbc.M601603200. [DOI] [PubMed] [Google Scholar]
- 27.Adachi H, Waza M, Tokui K, Katsuno M, Minamiyama M, Tanaka F, Doyu M, Sobue G. J. Neurosci. 2007;27:5115–5126. doi: 10.1523/JNEUROSCI.1242-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar P, Ambasta RK, Veereshwarayya V, Rosen KM, Kosik KS, Band H, Mestril R, Patterson C, Querfurth HW. Hum. Mol. Genet. 2007;16:848–864. doi: 10.1093/hmg/ddm030. [DOI] [PubMed] [Google Scholar]
- 29.Ding TT, Lee SJ, Rochet JC, Lansbury PT., Jr. Biochemistry. 2002;41:10209–10217. doi: 10.1021/bi020139h. [DOI] [PubMed] [Google Scholar]
- 30.Rosser MF, Washburn E, Muchowski PJ, Patterson C, Cyr DM. J. Biol. Chem. 2007;282:22267–22277. doi: 10.1074/jbc.M700513200. [DOI] [PubMed] [Google Scholar]