Abstract
Variation in genes contributing to the host immune response may mediate the relationship between inflammation and prostate carcinogenesis. RNASEL at chromosome 1q25 encodes ribonuclease L, part of the interferon-mediated immune response to viral infection. We therefore investigated the association between variation in RNASEL and prostate cancer risk and progression in a study of 1286 cases and 1264 controls nested within the prospective Physicians’ Health Study. Eleven single-nucleotide polymorphisms (SNPs) were selected using the web-based ‘Tagger’ in the HapMap CEPH panel (Utah residents of Northern and Western European Ancestry). Unconditional logistic regression models assessed the relationship between each SNP and incident advanced stage (T3/T4, T0-T4/M1 and lethal disease) and high Gleason grade (≥7) prostate cancer. Further analyses were stratified by calendar year of diagnosis. Cox proportional hazards models examined the relationship between genotype and prostate cancer-specific survival. We also explored associations between genotype and serum inflammatory biomarkers interleukin-6 (IL-6), C-reactive protein (CRP) and tumor necrosis factor-alpha receptor 2 using linear regression. Individuals homozygous for the variant allele of rs12757998 had an increased risk of prostate cancer [AA versus GG; odds ratio (OR): 1.63, 95% confidence interval (CI): 1.18–2.25), and more specifically, high-grade tumors (OR: 1.90, 95% CI: 1.25–2.89). The same genotype was associated with increased CRP (P = 0.02) and IL-6 (P = 0.05) levels. Missense mutations R462Q and D541E were associated with an increased risk of advanced stage disease only in the pre-prostate-specific antigen era. There were no significant associations with survival. The results of this study support a link between RNASEL and prostate cancer and suggest that the association may be mediated through inflammation. These novel findings warrant replication in future studies.
Introduction
Exposure to infectious agents, hormones or dietary carcinogens may contribute to inflammation in the prostate (1). Intraprostatic inflammation may alter the tissue microenvironment, lead to cellular and genetic damage, and promote proliferation and angiogenesis, which may drive cancer progression (2). Genetic variation in immune and inflammatory response pathways may modulate the relationship between inflammation and prostate cancer pathogenesis and progression. RNASEL, a candidate hereditary prostate cancer gene (HPC1) located at chromosome 1q25, encodes ribonuclease L (RNASEL), an enzyme in the interferon (IFN)-induced antiviral 2-5A system. Upon exposure to IFNs, cells express numerous antiviral genes, including IFN-stimulated genes, which encode 2-5A synthetases (2',5'-oligoadenylate synthetase, OAS) (3). When activated by the presence of dsRNA, OAS proteins produce 2′,5′-linked oligoadenylates (2-5A) from adenosine triphosphate; the short-lived signaling molecules serve to activate latent RNASEL protein (3). RNASEL employs several mechanisms to fight viral infections, including cleavage of viral RNA and induction of apoptosis (4,5). Mutations in the gene may reduce the ability to fight viral infections, as demonstrated by RNASEL knockout mice that have impaired IFN-α activity, suppression of apoptosis and an increased susceptibility to viral infections (6). RNASEL has been linked to hereditary prostate cancer since the mid-1990s when it was included in the first linkage peak identified for prostate cancer in a genome-wide scan of 91 high-risk families (7). However, recent genome-wide association studies of sporadic prostate cancer have not identified RNASEL as a susceptibility locus (8–10). A common missense mutation in RNASEL, R462Q (rs486907, G→A), has been studied extensively in genetic epidemiology studies of prostate cancer risk. The homozygous genotype (AA) has 15% prevalence in Caucasian populations and causes a functionally significant amino acid substitution resulting in a 3-fold decrease in enzymatic activity (4,11,12). Epidemiological studies have been mixed, with some finding an increased risk of prostate cancer with at least one copy of the variant allele (12–14), whereas others report no association (15–19). A recent meta-analysis of seven studies reported no significant association of R462Q with prostate cancer across all ethnic groups or by family history status (20). However, R462Q may still affect prostate cancer progression and outcomes as most prior studies did not report on these potential associations.
A second missense mutation in RNASEL, D541E (rs627928, T→G), has also been examined in studies of prostate cancer risk (21). Results for this single-nucleotide polymorphism (SNP) have been inconsistent across populations and ethnic groups; however, a meta-analysis of six studies reported an increased risk for prostate cancer among Caucasians with at least one copy of the variant (G) allele [odds ratio (OR): 1.27, 95% confidence interval (CI): 1.13–1.44) (20). In addition, a recent study found a complementary 36% decreased risk for men homozygous for the T allele (CI: 0.44–0.92) (14). Both R462Q and D541E variants are located in exonic regions toward the 5′ end of the gene. Several familial studies have identified other less common mutations in RNASEL associated with prostate cancer, including truncating mutation E265X, frameshift mutation 471delAAAG and M1I in the initiation codon (4,22).
In this study, we comprehensively explore RNASEL by capturing genetic variation across a 25.6 kb region, including the gene and 5 kb upstream and downstream. Our analysis builds upon past studies by selecting 11 SNPs and investigating a number of prostate cancer end points, including associations with advanced and high-grade cancers and prostate cancer survival. To our knowledge, no study has yet examined the relationship between RNASEL and prostate cancer survival, although given the inflammatory mechanism, it is a plausible hypothesis. Because RNASEL is involved in innate immunity, and certain mutations may affect the antiviral response, we also explored the possibility that mutations in RNASEL may alter levels of circulating inflammatory biomarkers interleukin-6 (IL-6), C-reactive protein (CRP) and tumor necrosis factor-alpha receptor 2 (TNFR2). We therefore explored the effects of RNASEL genetic variants on prostate cancer risk and progression, as well as inflammatory markers, in the Physicians’ Health Study.
Materials and methods
Study population
Our study population is nested within the Physicians’ Health Study I, a completed randomized trial of beta-carotene and aspirin. Study participants included 22 071 USA male physicians aged 40–84 years at enrollment in 1982. All men were free of diagnosed cardiovascular disease and cancer (excluding non-melanoma skin cancers) at enrollment. Before randomization, blood samples were collected from 14 916 study participants (23). Study participants complete annual mailed questionnaires to obtain and update information on medical history and lifestyle factors. They also complete postcards every 6 months to report end points, including prostate cancer. After the end of the aspirin component in 1988 and the beta-carotene component in 1995, the men continued to be followed on an observational basis. Participants provided written consent, and the Human Subjects Committee at Brigham and Women's Hospital approved the study.
We used a nested case–control design from the population that provided a blood sample at baseline. Incident prostate cancer cases (n = 1286) were histologically confirmed and diagnosed from 1982 to 2005. Controls (n = 1264) were frequency matched to cases by age (±1 year, if aged ≤55 years and ±5 years if aged >55 years) and follow-up time. The study cohort is largely Caucasian (94.3% of participants with genetic data). To reduce the influence of population stratification, we restricted our study population to Caucasians.
Outcome ascertainment
Prostate cancer cases are initially identified by self-report and are confirmed through medical record review. Men are followed for mortality (>99% complete), and prostate cancer-specific deaths are confirmed through review of death certificates, medical records and information from families. Men are also followed for development of metastatic disease through annual questionnaires and validated through medical record review through the end of follow-up, 1 March 2008.
Information on disease stage is abstracted from medical records, and advanced cancers are defined as those diagnosed at stage T3–T4 or T0–T4/M1 (distant metastasis) or who developed lethal prostate cancer. Information on Gleason score is available through medical record review and review of tumor specimens from biopsy or prostatectomy. High-grade tumors are defined as those with a total Gleason score of 7 through 10. Cases diagnosed after 1 January 1992 were considered to be in the era of regular testing for prostate-specific antigen (PSA), hereafter referred to as the ‘PSA era’(24).
Genotype and biomarker assessment
Blood samples were collected at baseline. Physicians received blood collection kits in the mail and were instructed to have their blood drawn into vacutainer tubes containing ethylenediaminetetraacetic acid and sent overnight to the laboratory where samples were separated into aliquots (whole blood and plasma) and initially stored at −82°C (later at −140°C) (25). Individuals returning their blood samples >5 days after blood draw were excluded from analyses involving biomarkers.
Eleven SNPs in linkage disequilibrium were selected to sufficiently account for genetic variation in RNASEL using the multimarker aggressive tagging approach in the web-based version of ‘Tagger’ in the HapMap CEPH panel. We captured variation across a 25.6 kb region, including the gene and 5 kb upstream and downstream, with minor allele frequency >5% and coefficient of determination, r2, >0.8. Sequenom GOLD iPlex panels were constructed for genotyping. Genotyping was conducted at the Harvard Medical School-Partners Healthcare Center for Genetics and Genomics, with laboratory personnel blinded to case status.
Levels of circulating blood biomarkers were assayed from prospectively collected stored plasma in the laboratory of Dr Nader Rifai (Boston, MA). IL-6 and TNFR2 levels were determined by a commercially available enzyme-linked immunosorbent assay test (R and D Systems) and CRP was measured by a high-sensitivity assay (Dade Behring). Median coefficients of variation were 0.4% for CRP, 5.3% for IL-6 and 6.8% for TNFR2. Laboratory personnel were blinded to case–control status and case–control pairs were assayed in adjoining wells. Ten percent of samples were included as pooled quality controls randomly distributed across the plates (24). PSA levels for linear regression analyses were determined for controls from baseline blood samples and for cases at diagnosis.
We had available formalin-fixed paraffin-embedded tumor tissue specimens for 161 of the prostate cancer cases on whom we also had genotype data. Tissue samples were obtained from men who underwent radical prostatectomy or transurethral resection of the prostate between 1983 and 2004. The tumor samples were included on tumor tissue microarrays, with three cores of tissue per case on an array. The terminal deoxynucleotide transferase dUTP nick end label assay (TUNEL), using the Apoptag Peroxidase In situ kit (Chemicon International), was used to estimate the percentage of tumor cells undergoing apoptosis in 5 μm sections according to manufacturer instructions. The whole area of each tumor core was evaluated manually to determine the Apoptag score, defined as the number of positive cells among the total number of tumor cells. Two study pathologists independently assessed these samples.
Data analysis
To test each SNP for Hardy–Weinberg equilibrium, we used an exact test with 10 000 permutations among controls and considered a P-value <0.001 to indicate statistically significant disequilibrium. We also compared the observed frequencies of each genotype with those in the HapMap CEPH sample set. Individuals missing data on a particular SNP were left out of analyses involving that SNP but were included in other analyses.
Unconditional logistic regression models adjusted for age, smoking status (current-, former- or never-smoker) and follow-up time were utilized to maximize the number of individuals included in the analysis. The codominant model was employed for genetic analyses assessing the relationship between each SNP with ≥5% of individuals homozygous for the minor allele and total prostate cancer incidence, advanced stage disease and high Gleason grade at diagnosis. SNPs with very low (<5%) frequency of individuals homozygous for the variant allele were analyzed using the dominant model of inheritance, combining rare homozygous and heterozygous individuals into one category. Likelihood ratio tests with 2 d.f. were used to determine statistical significance for codominant models, comparing a model with indicator variables for both heterozygous and rare homozygous genotypes to the null model. P-values <0.05 were considered statistically significant. To assess a possible effect of PSA screening, analyses were also stratified by a binary variable indicating whether diagnosis occurred before or during the PSA era. We also investigated whether any RNASEL genotype may affect baseline PSA levels, using linear regression to model log-transformed PSA levels by genotype, with adjustment for age and body mass index (BMI). Analyses were conducted using SAS v9.1 (SAS Institute, Cary, NC).
We also looked at the relation between RNASEL SNPs and survival following prostate cancer diagnosis among the cases. The men were followed from diagnosis until death from prostate cancer or development of bony metastases or censored at death from other causes or end of follow-up, 1 March 2008. Analysis was conducted by genotype using Cox proportional hazards models adjusted for age at diagnosis.
Because RNASEL is a candidate gene in an inflammation pathway, we looked at the mean levels of plasma inflammatory biomarkers IL-6, CRP and TNFR2 by genotype. Values of IL-6, CRP and TNFR2 were log10 transformed to ensure a normal distribution. Linear regression models examined log-transformed levels of the biomarkers by genotype, adjusting for age at randomization, case status and baseline BMI, due to the strong correlation between adipose tissue and these inflammatory markers. Individuals with <5 years between blood draw and diagnosis were removed from biomarker analyses in an attempt to ensure that preclinical disease did not affect levels of inflammatory markers. For statistically significant associations, we also explored potential interactions between that genotype and median biomarker levels in relation to prostate cancer incidence and mortality. We used unconditional logistic regression models that included the matching factors, indicator variables for the heterozygous and rare homozygous genotypes, a dichotomous variable to indicate biomarker levels above the median and the product terms of genotype with biomarker level. A likelihood ratio test with 2 d.f. assessed statistical significance, comparing the above model with one without the product terms.
As mutations in RNASEL may affect the cell’s ability to induce apoptosis in infected cells (26), we used linear regression to determine if any genotype predicted the percent of tumor tissue undergoing apoptosis. TUNEL results were log transformed to improve normality, and models were adjusted for matching factors and case status.
Results
There were 1286 cases and 1264 controls with data on RNASEL included in the analyses. Characteristics of study participants can be found in Table I. Eleven tagging SNPs were chosen to account for genetic variation in RNASEL (rs12034888, rs12757998, rs635261, rs12729828, rs10911099, rs627839, rs11807829, rs533259, rs627928, rs486907 and rs682585). One SNP, rs12729828, violated Hardy–Weinberg equilibrium, with P = 0.0002. Although we included the SNP in our analyses, results should be interpreted with caution. All other SNPs were in Hardy–Weinberg equilibrium and were in line with observed frequencies reported in the HapMap CEPH sample set.
Table I.
Selected characteristics for prostate cancer cases and controls, Physicians' Health Study (1982–2008)
| Cases (n = 1286) | Controls (n = 1264) | |
| Age at randomization | ||
| Mean (±SD) | 57.9 (±8.4) | 57.5 (±8.4) |
| Follow-up time | ||
| Mean (± SD) | 12.2 (±5.1) | 12.1 (±5.0) |
| BMI (kg/m2) | ||
| <25 | 746 (58%) | 759 (60%) |
| 25 to <30 | 499 (39) | 465 (37) |
| ≥30 | 41 (3) | 40 (3) |
| Among cases only | ||
| Age at diagnosis | ||
| Mean (±SD) | 70 (±7.4) | |
| Stage, N (%) | ||
| T1/T2 | 1079 (84) | |
| T3 | 63 (5) | |
| T4/N1/M1 | 79 (6) | |
| Not available | 65 (5) | |
| Gleason score, N (%) | ||
| 2–6 | 720 (56) | |
| 7–10 | 448 (35) | |
| Not available | 118 (9) | |
| Outcomes, N | ||
| Cases with bone metastases | 23 | |
| Prostate cancer deaths | 159 | |
| Deaths from other causes | 298 | |
Among the cases, 70% were diagnosed during the PSA era. We observed a significant increased risk of prostate cancer with variant rs12757998, located downstream of the gene (AA versus GG; OR: 1.63, 95% CI: 1.18–2.25, Table II). Unlike previous reports in the literature, we found no increased risk of prostate cancer with R462Q (AA versus GG; OR: 0.96, 95% CI: 0.75–1.24) or D541E (GG versus TT; OR: 1.03, 95% CI: 0.82–1.28). Dominant models were employed for four SNPs with <5% rare homozygous genotypes but produced the same results as the codominant models (Table II).
Table II.
Association between RNASEL SNPs and incident prostate cancer
| SNP | Genotype | Positiona | HapMap frequency | Prostate cancer risk |
||||
| Cases, n (%) | Controls, n (%) | ORb | 95% CI | Pc | ||||
| rs682585 | GG (referent) | 180826133 | 0.45 | 489 (39.3) | 468 (38.3) | 1.00 | Referent | 0.74 |
| GA | 0.43 | 554 (44.5) | 542 (44.4) | 0.99 | (0.83–1.17) | |||
| AA | 0.12 | 202 (16.2) | 211 (17.3) | 0.91 | (0.73–1.15) | |||
| rs486907 (R462Q) | GG (referent) | 180821180 | 0.32 | 529 (42.8) | 505 (41.7) | 1.00 | Referent | 0.91 |
| GA | 0.55 | 547 (44.3) | 546 (45.1) | 0.97 | (0.81–1.14) | |||
| AA | 0.13 | 159 (12.9) | 159 (13.1) | 0.96 | (0.75–1.24) | |||
| rs627928 (D541E) | TT (referent) | 180817960 | 0.12 | 277 (22.8) | 282 (23.6) | 1.00 | Referent | 0.76 |
| TG | 0.52 | 560 (46.1) | 536 (44.9) | 1.08 | (0.88–1.32) | |||
| GG | 0.37 | 378 (31.1) | 376 (31.5) | 1.03 | (0.82–1.28) | |||
| rs533259 | CC (referent) | 180815642 | 0.87 | 1088 (87.6) | 1075 (87.8) | 1.00 | Referent | 0.85 |
| CT/TT | 0.13 | 154 (12.4) | 150 (12.2) | 1.02 | (0.80–1.30) | |||
| rs11807829 | AA (referent) | 180811438 | 0.50 | 571 (47) | 529 (44.3) | 1.00 | Referent | 0.43 |
| AG | 0.40 | 508 (41.8) | 523 (43.8) | 0.90 | (0.76–1.07) | |||
| GG | 0.10 | 136 (11.2) | 142 (11.9) | 0.90 | (0.69–1.17) | |||
| rs627839 | CC (referent) | 180808704 | 0.25 | 364 (29.2) | 363 (29.8) | 1.00 | Referent | 0.76 |
| CA | 0.59 | 599 (48) | 571 (46.8) | 1.06 | (0.88–1.28) | |||
| AA | 0.15 | 285 (22.8) | 285 (23.4) | 0.99 | (0.80–1.24) | |||
| rs10911099 | AA (referent) | 180806772 | 0.83 | 949 (76.4) | 947 (77.9) | 1.00 | Referent | 0.29 |
| AG/GG | 0.17 | 294 (23.7) | 268 (22.1) | 1.12 | (0.92–1.36) | |||
| rs12729828 | GG (referent) | 180806741 | 0.70 | 922 (76.9) | 941 (78.5) | 1.00 | Referent | 0.37 |
| GA/AA | 0.30 | 277 (23.1) | 258 (21.5) | 1.09 | (0.90–1.33) | |||
| rs635261 | GG (referent) | 180805664 | 0.42 | 492 (40.5) | 449 (37.6) | 1.00 | Referent | 0.38 |
| CG | 0.44 | 525 (43.2) | 542 (45.4) | 0.89 | (0.74–1.06) | |||
| CC | 0.14 | 199 (16.4) | 203 (17.0) | 0.90 | (0.71–1.13) | |||
| rs12757998 | GG (referent) | 180805101 | 0.64 | 632 (52.8) | 627 (53.9) | 1.00 | Referent | 0.004 |
| GA | 0.27 | 452 (37.8) | 469 (40.3) | 0.95 | (0.80–1.13) | |||
| AA | 0.09 | 113 (9.4) | 68 (5.8) | 1.63 | (1.18–2.25) | |||
| rs12034888 | CC (referent) | 180804419 | 0.86 | 1008 (83.8) | 986 (84.3) | 1.00 | Referent | 0.64 |
| CT/TT | 0.14 | 195 (16.2) | 183 (15.7) | 1.06 | (0.85–1.32) | |||
Genomic positions from National Center for Biotechnology Information database, build 36.3.
Odds ratios adjusted for age, smoking status and follow-up time.
P-values from likelihood ratio test.
We further assessed subgroups of cases defined by Gleason grade or stage at diagnosis (Table III). Variant rs12757998 (AA) was associated with a significantly increased risk of having a high-grade tumor (OR: 1.90, 95% CI: 1.25–2.89), although not with advanced stage disease (OR: 1.44, 95% CI: 0.79–2.63). R462Q was not significantly associated with advanced stage disease overall or in the PSA era; however, there was a significantly increased risk in the pre-PSA era with the rare homozygous genotype (OR: 1.93, 95% CI: 1.05–3.56). Similar results were observed with D541E, which was null overall and in the PSA era, yet significantly increased risk of advanced stage disease in individuals with one (OR: 2.18, 95% CI: 1.20–4.00) or two (OR: 2.04, 95% CI: 1.08–3.85) copies of the variant allele among cases diagnosed before 1992 (supplementary Table I is available at Carcinogenesis Online). The remaining SNPs do not appear to be significantly associated with high-grade or advanced stage disease.
Table III.
Association of RNASEL SNPs with high grade and advanced stage disease
| SNPa | Genotype | High-grade disease (Gleason ≥7) |
Advanced stage (T3/T4/N1/M1 or PCa mortality) |
||||||
| Cases (n) | ORb | 95% CI | Pc | Cases (n) | ORb | 95% CI | Pc | ||
| rs682585 | GG (referent) | 173 | 1.00 | Referent | 0.24 | 93 | 1.00 | Referent | 0.12 |
| GA | 203 | 1.03 | (0.81–1.31) | 106 | 0.97 | (0.71–1.33) | |||
| AA | 61 | 0.78 | (0.56–1.10) | 28 | 0.63 | (0.40–1.01) | |||
| rs486907 (R462Q) | GG (referent) | 190 | 1.00 | Referent | 0.72 | 86 | 1.00 | 0.17 | |
| GA | 185 | 0.91 | (0.71–1.15) | 100 | 1.05 | (0.76–1.45) | |||
| AA | 56 | 0.95 | (0.67–1.36) | 39 | 1.51 | (0.98–2.34) | |||
| rs627928 (D541E) | TT (referent) | 90 | 1.00 | Referent | 0.56 | 39 | 1.00 | Referent | 0.18 |
| TG | 200 | 1.17 | (0.88–1.57) | 108 | 1.43 | (0.95–2.15) | |||
| GG | 132 | 1.10 | (0.80–1.50) | 73 | 1.42 | ( 0.92–2.19) | |||
| rs533259 | CC (referent) | 377 | 1.00 | Referent | 0.62 | 202 | 1.00 | Referent | 0.91 |
| CT/TT | 57 | 1.09 | (0.78–1.51) | 26 | 1.03 | (0.65–1.63) | |||
| rs11807829 | AA (referent) | 204 | 1.00 | Referent | 0.34 | 103 | 1.00 | Referent | 0.23 |
| AG | 175 | 0.87 | (0.68–1.10) | 82 | 0.76 | (0.55–1.05) | |||
| GG | 43 | 0.80 | (0.55–1.17) | 25 | 0.98 | (0.60–1.60) | |||
| rs627839 | CC (referent) | 125 | 1.00 | Referent | 0.61 | 63 | 1.00 | Referent | 0.78 |
| CA | 214 | 1.12 | (0.86–1.45) | 113 | 1.13 | (0.80–1.60) | |||
| AA | 98 | 1.00 | (0.73–1.36) | 50 | 1.09 | (0.72–1.66) | |||
| rs10911099 | AA (referent) | 330 | 1.00 | Referent | 0.26 | 165 | 1.00 | Referent | 0.20 |
| AG/GG | 109 | 1.16 | (0.90–1.50) | 60 | 1.25 | (0.89–1.77) | |||
| rs12729828 | GG (referent) | 317 | 1.00 | Referent | 0.43 | 172 | 1.00 | Referent | 0.99 |
| GA/AA | 99 | 1.12 | (0.85–1.46) | 46 | 1.00 | (0.70–1.45) | |||
| rs635261 | GG (referent) | 177 | 1.00 | Referent | 0.45 | 90 | 1.00 | Referent | 0.39 |
| CG | 187 | 0.89 | (0.70–1.13) | 92 | 0.80 | (0.57–1.10) | |||
| CC | 65 | 0.83 | (0.59–1.15) | 38 | 0.89 | (0.58–1.36) | |||
| rs12757998 | GG (referent) | 208 | 1.00 | Referent | 0.003 | 109 | 1.00 | Referent | 0.22 |
| GA | 161 | 1.02 | (0.80–1.30) | 72 | 0.84 | (0.60–1.17) | |||
| AA | 43 | 1.90 | (1.25–2.89) | 16 | 1.44 | (0.79–2.63) | |||
| rs12034888 | CC (referent) | 353 | 1.00 | Referent | 0.42 | 182 | 1.00 | Referent | 0.25 |
| CT/TT | 74 | 1.13 | (0.84–1.53) | 42 | 1.26 | (0.86–1.85) | |||
SNPs listed by genomic position.
Odds ratios adjusted for age, smoking status and follow-up time.
P-values from likelihood ratio test.
We confirmed 176 lethal prostate cancers (prostate cancer-specific deaths or bony metastases) among the cases in our analysis by the end of follow-up. On average, those who developed bony metastases or died from prostate cancer had 6.5 years of follow-up since diagnosis compared with 10.6 years for non-fatal cases of prostate cancer. Survival analyses did not yield significant associations between any SNP and survival following prostate cancer diagnosis in this population, including R462Q (hazard ratio: 1.05, 95% CI: 0.66–1.67) and D541E (hazard ratio: 1.02, 95% CI: 0.66–1.57). The results of the survival analysis can be found in supplementary Table II (available at Carcinogenesis Online).
For cases in the analyses of inflammatory biomarkers, the median time between blood collection and prostate cancer diagnosis was 9.9 years (range 0.3–17.9 years). We excluded 24 men who returned blood samples >5 days after blood draw, as well as 69 cases and 50 matched controls with <5 years between blood draw and diagnosis from biomarker analyses, leaving 538 cases and 420 controls with genetic, plasma biomarker and BMI data.
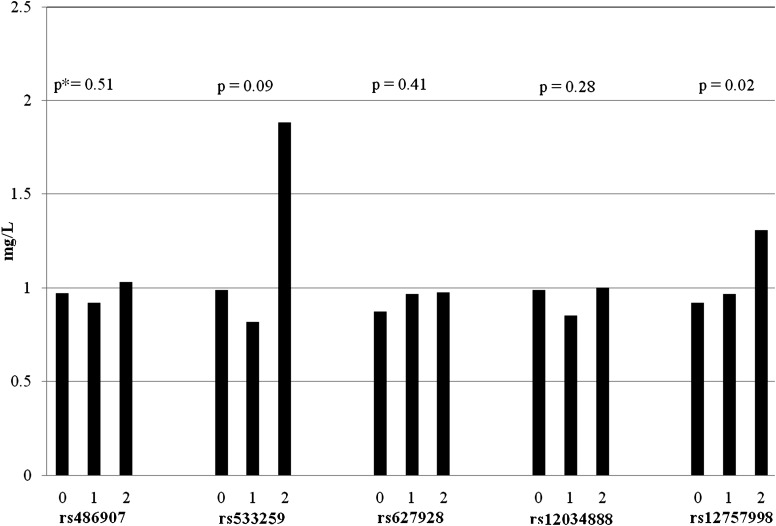
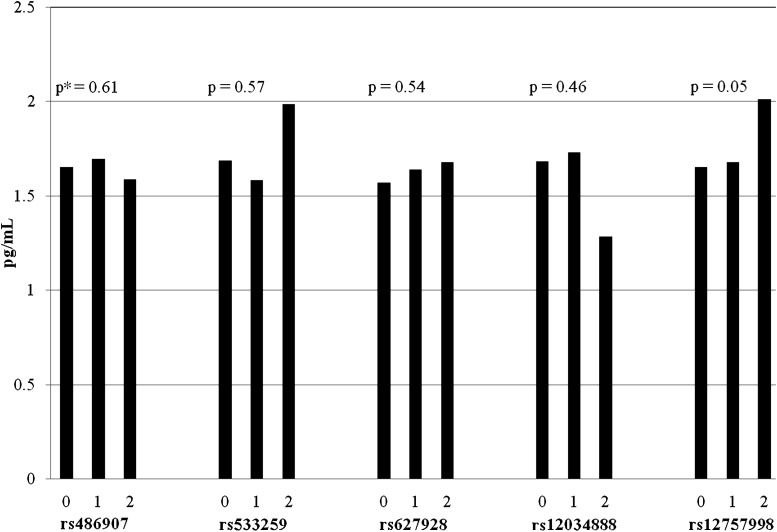
RNASEL variants appear to affect circulating levels of plasma inflammatory biomarkers CRP (Figure 1) and IL-6 (Figure 2). Variant rs12757998, which increased risk of both overall prostate cancer and high-grade tumors in this analysis, was also associated with elevated serum levels of both CRP (P = 0.02) and IL-6 (P = 0.05) in linear regression models. Participants with two copies of the variant allele had on average 14% higher plasma CRP levels (1.12 versus 0.96 mg/l) and 18% higher plasma IL-6 levels (1.78 versus 1.46 pg/ml) than individuals with the wild-type form (GG). We also observed a significant association between variant rs12034888, located downstream of the gene, and reduced TNFR2 levels (P = 0.03), although there are very few individuals who are homozygous for the variant allele (T) in this study (n = 8). We did not detect any significant interactions between median levels of biomarkers and genotype in relation to prostate cancer risk or progression.
Fig. 1.
Mean levels of serum CRP by selected genotypes. *P-values from linear regression models adjusted for age at randomization, case status and baseline BMI. 0 = homozygous dominant; 1 = heterozygous and 2 = homozygous recessive.
Fig. 2.
Mean levels of serum IL-6 by selected genotypes. *P-values from linear regression models adjusted for age at randomization, case status and baseline BMI. 0 = homozygous dominant; 1 = heterozygous and 2 = homozygous recessive.
We did not detect significant associations between any of the genotypes and PSA levels among cases diagnosed >5 years after blood draw and controls together, or among controls alone, with P-values from linear regression models ranging from 0.32 (rs533259) to 0.92 (rs12034888). Despite reports of diminished capacity to induce apoptosis with mutated RNASEL protein, there were no apparent associations between RNASEL genotype and percent of tumor cells undergoing apoptosis (mean 2.3%; range 0–30%). The results of linear regression analysis of the TUNEL data resulted in non-significant P-values ranging from 0.09 (rs12034888) to 0.84 (rs682585). The R462Q missense mutation was not associated with apoptosis (P = 0.84).
Discussion
In our study expanding coverage of genetic variation in RNASEL, we identified new associations with the SNP rs12757998, which appeared marginally associated with overall prostate cancer risk and high-grade disease. Located downstream of the gene, rs12757998 is not strongly correlated with either missense mutation R462Q (r2 = 0.08) or D541E (r2 = 0.02), suggesting an independent association with prostate cancer. In addition to increasing the risk of overall and high-grade disease by >50% when compared with the wild-type genotype, variant rs12757998 was also linked to higher levels of circulating inflammatory biomarkers CRP and IL-6. Taken together, these results suggest a potential inflammatory mechanism linking the mutation to prostate cancer, although an exact biological mechanism is currently unknown. Significant SNPs downstream of the gene could be involved in gene regulation or could be in linkage disequilibrium with a functional SNP within the gene that we did not directly genotype. The SNP rs12757998 tags nearby SNP rs12723593 with r2 = 1.0, suggesting the putative SNP could be either of these with equal probability, as well as any other known or unknown SNP in strong linkage disequilibrium.
The associations between variant 12757998 and circulating inflammatory biomarkers are novel findings. IL-6, a cytokine with both pro- and anti-inflammatory properties, plays a key role in mediating inflammation, including the acute-phase response, which promotes CRP production. One recent study found a significant positive association between IL-6 levels and prostate cancer incidence among men with BMI <25 kg/m2 (24). Elevated prediagnostic CRP, a non-specific marker of systemic inflammation, has been associated with cancer risk and survival in some (27,28) but not all (29,30) studies. CRP levels have also been associated with poor survival among men with androgen-independent prostate cancer (31). Levels of both inflammatory biomarkers have been observed to be higher in men with prostate cancer when compared with healthy men or men with benign conditions (32,33). Relationships between variation in RNASEL and these biomarkers have not been reported in the literature.
We did not replicate previously reported associations for missense mutations R462Q and D541E with overall prostate cancer risk in this study population. However, the potential modification by calendar year of diagnosis is a novel finding. Cases diagnosed after 1992 during the ‘PSA era’ are more probably to be diagnosed by regular PSA screening and thus at an earlier stage. In contrast, men diagnosed before PSA screening was commonplace are more probably to have presented with symptoms of their disease and thus at a more advanced stage. Furthermore, pre-PSA era cases may better reflect the natural history of prostate cancer and the subsequent shift toward PSA-detected early stage cancers could have diluted any association between the well-studied missense mutations and advanced stage disease. However, because we are looking at a subset of the data, these findings could be due to chance.
The SNP rs12757998 also appeared recently in an analysis of the San Antonio Center for Biomarkers of Risk of Prostate Cancer (SABOR) cohort, although with somewhat conflicting results (34). In the SABOR cohort, having at least one copy of the variant (A) allele of rs12757998 was associated with reduced prostate cancer risk only among Hispanic men with a family history of the disease (OR: 0.41, 95% CI: 0.19–0.88); separate results for men carrying two copies of the variant allele were not shown. They did not report significant findings for rs12757998 among Caucasian men. The study also investigated 10 other SNPs in RNASEL, including rs635261, rs10911099 and rs533259, which also appear in our analysis. Similar to our results, they did not find any significant associations between prostate cancer and these three SNPs (34). A recent study of genetic variants among familial prostate cancer cases also reported null findings for rs533259, as well as for rs11807829, which is consistent with the results of our study (14). Although several genome-wide association studies failed to identify SNPs in RNASEL as significantly associated with sporadic prostate cancer, genetic variation in RNASEL may still impact disease risk. Genome-wide association studies have lower power to detect rare alleles and associations of smaller magnitude, and thus, our study design may be more appropriate in these cases.
There are several strengths to consider when assessing the findings of this study. The prospective nature of our analysis allowed us to ascertain exposure information before disease onset. In particular, this is important for the biomarker analyses, where disease and treatment could affect circulating levels of inflammatory markers. We have long and complete follow-up, with reasonably accurate reporting by physician participants. Residual confounding from variations in socioeconomic status and access to healthcare is unlikely in this population and is not probably to influence results of a genetic study. We have also restricted our study sample to self-reported Caucasians to reduce the influence of population stratification.
Our large study population gives us sufficient power (>80%) to detect an association of OR ≥1.4 for each of the 11 SNPs. The large sample size also allows us to conduct subgroup analyses and investigate relationships with disease stage and grade. However, although we have considerable power in this study, we remain underpowered to detect very small effects of RNASEL on prostate cancer or interactions with plasma biomarkers. The relatively small number of prostate cancer deaths may have restricted the power of the survival analysis to detect associations in the more rare genotypes; no clear relation between RNASEL and prostate cancer-specific survival emerged from our data.
As with other genetic studies, multiple testing is of concern. A conservative approach to correct for multiple testing is the Bonferroni correction, where the alpha value (0.05) is divided by the number of tests. With the 11 SNPs in this study, the corrected alpha value would be 0.005. After applying this conservative measure, the result for variant rs12757998 and total prostate cancer would maintain statistical significance, with a P-value of 0.004 from the likelihood ratio test, as would the association between variant rs12757998 and high Gleason grade tumors, with a likelihood ratio P-value of 0.003. However, the associations between rs12757998 and inflammatory biomarkers CRP and IL-6 would not remain statistically significant after correcting for multiple testing, with original P-values of 0.02 and 0.05, respectively. Because the associations between variant rs12757998 and prostate cancer remain significant after conservative correction for multiple testing, the SNP deserves replication in future studies.
The generalizability of our study results across racial or ethnic groups may be limited as our population was restricted to Caucasians. There have been discrepant findings across ethnic groups for carriage of the variant allele of R462Q. Several studies report an increased risk (13,35) or no association (16) in African-Americans, inconsistent observations in Caucasian men (12,13,35), an increased risk among Hispanic men (34,35) and an inverse association in a Japanese population (36). The recent analysis of the SABOR cohort also found an increased risk of prostate cancer among Hispanic men with two copies of the variant (G) allele of D541E. The same study also saw a 42% reduced risk of prostate cancer limited to Hispanic men with two copies of the variant (A) allele of rs682585, a SNP which was also selected in our analysis, although results were non-significant (34). The results from the SABOR cohort were not replicated among Caucasians, suggesting that the relationship between prostate cancer and RNASEL may vary by ethnic group.
Furthermore, our biomarker data are based upon a single baseline blood sample and may not be representative of long-term levels of inflammatory biomarkers. Multiple samples over the course of time could be more informative, possibly reducing within-person variation. In addition, CRP and IL-6 are markers of systemic inflammation and not exclusive to the prostate. The findings of increased CRP and IL-6 levels with variant rs12757998 suggest that an association between rs12757998 and prostate cancer may be mediated through inflammation; however, these results could also indicate that men with two copies of the variant allele have higher levels of systemic inflammation not necessarily intraprostatic inflammation. From our analysis of PSA levels, we found that the SNPs in RNASEL do not raise PSA, suggesting that the SNPs are probably associated with prostate cancer for a reason other than increasing PSA. We did not find any significant interactions between biomarkers and genotype in relation to prostate cancer risk or progression. It is possible that biomarker levels drawn closer to date of diagnosis may be more informative in this regard.
Recently, the novel xenotropic murine leukemia virus-related virus (XMRV) was discovered in prostate cancer tissue differentially by R462Q genotype (37). Men with the QQ genotype were more probably to have evidence of an infection than men who were heterozygous or wild-type, indicating the mutation might impede viral clearance (37). Although this result was not replicated in a subsequent study of 105 non-familial prostate cancer cases (38), future studies on RNASEL may expand the analysis to explore the relation of xenotropic murine leukemia virus-related virus infection with other SNPs.
The results from this study further support the proposed link between RNASEL and prostate cancer. The additional association between variant rs12757998 and levels of inflammatory biomarkers CRP and IL-6 lends credence to the findings and suggests inflammation as a possible mechanism linking rs12757998 to prostate cancer risk and progression. These results should be validated in other populations.
Supplementary material
Supplementary Tables I and II can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute (CA-058684 to M.J.S.); the National Research Service Award Training Program in Cancer Epidemiology from the National Institutes of Health (T32 CA-09001 to J.R.S. and K.L.P.); the Harvard University Franklin Reynolds Fund (to M.S.M.); L.A.M. is a Milken Scholar, Prostate Cancer Foundation. The Physicians’ Health Study is supported by the National Cancer Institute (CA-34944, CA-40360, CA-097193); the National Heart, Lung, and Blood Institute (HL-26490, HL-34595), Bethesda, MD.
Supplementary Material
Acknowledgments
Conflict of Interest Statement: Dr Kurth has received within the last 2 years investigator-initiated research funding from the French National Research Agency, the National Institutes of Health, Merck and the Migraine Research Foundation. Furthermore, he is a consultant to i3 Drug Safety and World Health Information Science Consultants, LLC; he has received honoraria from Genzyme, Merck and Pfizer for educational lectures; he has received travel funds from the Restless Legs Syndrome Foundation. The other authors report no conflicts of interest.
Glossary
Abbreviations
- BMI
body mass index
- CI
confidence interval
- CRP
C-reactive protein
- IFN
interferon
- IL-6
interleukin-6
- OR
odds ratio
- PSA
prostate-specific antigen
- SABOR
San Antonio Center for Biomarkers of Risk of Prostate Cancer
- SNP
single-nucleotide polymorphism
- TNFR2
tumor necrosis factor-alpha receptor 2
References
- 1.De Marzo AM, et al. Inflammation in prostate carcinogenesis. Nat. Rev. Cancer. 2007;7:256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palapattu GS, et al. Prostate carcinogenesis and inflammation: emerging insights. Carcinogenesis. 2005;26:1170–1181. doi: 10.1093/carcin/bgh317. [DOI] [PubMed] [Google Scholar]
- 3.Silverman RH. A scientific journey through the 2-5A/RNase L system. Cytokine Growth Factor Rev. 2007;18:381–388. doi: 10.1016/j.cytogfr.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carpten J, et al. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat. Genet. 2002;30:181–184. doi: 10.1038/ng823. [DOI] [PubMed] [Google Scholar]
- 5.Summers K, et al. Molecular evolution of the prostate cancer susceptibility locus RNASEL: evidence for positive selection. Infect. Genet. Evol. 2008;8:297–301. doi: 10.1016/j.meegid.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Zhou A, et al. Interferon action and apoptosis are defective in mice devoid of 2',5'-oligoadenylate-dependent RNase L. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith JR, et al. Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science. 1996;274:1371–1374. doi: 10.1126/science.274.5291.1371. [DOI] [PubMed] [Google Scholar]
- 8.Gudmundsson J, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas G, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 10.Yeager M, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Packer BR, et al. SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 2006;34:D617–D621. doi: 10.1093/nar/gkj151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casey G, et al. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat. Genet. 2002;32:581–583. doi: 10.1038/ng1021. [DOI] [PubMed] [Google Scholar]
- 13.Rennert H, et al. Association of susceptibility alleles in ELAC2/HPC2, RNASEL/HPC1, and MSR1 with prostate cancer severity in European American and African American men. Cancer Epidemiol. Biomarkers Prev. 2005;14:949–957. doi: 10.1158/1055-9965.EPI-04-0637. [DOI] [PubMed] [Google Scholar]
- 14.Breyer JP, et al. Genetic variants and prostate cancer risk: candidate replication and exploration of viral restriction genes. Cancer Epidemiol. Biomarkers Prev. 2009;18:2137–2144. doi: 10.1158/1055-9965.EPI-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shea PR, et al. RNASEL and RNASEL-inhibitor variation and prostate cancer risk in Afro-Caribbeans. Prostate. 2008;68:354–359. doi: 10.1002/pros.20687. [DOI] [PubMed] [Google Scholar]
- 16.Robbins CM, et al. Association of HPC2/ELAC2 and RNASEL non-synonymous variants with prostate cancer risk in African American familial and sporadic cases. Prostate. 2008;68:1790–1797. doi: 10.1002/pros.20841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang MH, et al. Association of IL10 and Other immune response- and obesity-related genes with prostate cancer in CLUE II. Prostate. 2009;69:874–885. doi: 10.1002/pros.20933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daugherty SE, et al. RNASEL Arg462Gln polymorphism and prostate cancer in PLCO. Prostate. 2007;67:849–854. doi: 10.1002/pros.20537. [DOI] [PubMed] [Google Scholar]
- 19.Cybulski C, et al. DNA variation in MSR1, RNASEL and E-cadherin genes and prostate cancer in Poland. Urol. Int. 2007;79:44–49. doi: 10.1159/000102913. [DOI] [PubMed] [Google Scholar]
- 20.Li H, et al. RNASEL gene polymorphisms and the risk of prostate cancer: a meta-analysis. Clin. Cancer Res. 2006;12:5713–5719. doi: 10.1158/1078-0432.CCR-05-2799. [DOI] [PubMed] [Google Scholar]
- 21.Noonan-Wheeler FC, et al. Association of hereditary prostate cancer gene polymorphic variants with sporadic aggressive prostate carcinoma. Prostate. 2006;66:49–56. doi: 10.1002/pros.20320. [DOI] [PubMed] [Google Scholar]
- 22.Rennert H, et al. A novel founder mutation in the RNASEL gene, 471delAAAG, is associated with prostate cancer in Ashkenazi Jews. Am. J. Hum. Genet. 2002;71:981–984. doi: 10.1086/342775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gann PH, et al. Prospective study of sex hormone levels and risk of prostate cancer. J. Natl Cancer Inst. 1996;88:1118–1126. doi: 10.1093/jnci/88.16.1118. [DOI] [PubMed] [Google Scholar]
- 24.Stark JR, et al. Circulating prediagnostic interleukin-6 and C-reactive protein and prostate cancer incidence and mortality. Int. J. Cancer. 2009;124:2683–2689. doi: 10.1002/ijc.24241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fall K, et al. No association between a polymorphic variant of the IRS-1 gene and prostate cancer risk. Prostate. 2008;68:1416–1420. doi: 10.1002/pros.20797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malathi K, et al. HPC1/RNASEL mediates apoptosis of prostate cancer cells treated with 2',5'-oligoadenylates, topoisomerase I inhibitors, and tumor necrosis factor-related apoptosis-inducing ligand. Cancer Res. 2004;64:9144–9151. doi: 10.1158/0008-5472.CAN-04-2226. [DOI] [PubMed] [Google Scholar]
- 27.Erlinger TP, et al. C-reactive protein and the risk of incident colorectal cancer. JAMA. 2004;291:585–590. doi: 10.1001/jama.291.5.585. [DOI] [PubMed] [Google Scholar]
- 28.Pierce BL, et al. Correlates of circulating C-reactive protein and serum amyloid A concentrations in breast cancer survivors. Breast Cancer Res. Treat. 2009;114:155–167. doi: 10.1007/s10549-008-9985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang SM, et al. C-reactive protein levels are not associated with increased risk for colorectal cancer in women. Ann. Intern. Med. 2005;142:425–432. doi: 10.7326/0003-4819-142-6-200503150-00008. [DOI] [PubMed] [Google Scholar]
- 30.Pierce BL, et al. C-reactive protein, interleukin-6, and prostate cancer risk in men aged 65 years and older. Cancer Causes Control. 2009;20:1193–1203. doi: 10.1007/s10552-009-9320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beer TM, et al. C-reactive protein as a prognostic marker for men with androgen-independent prostate cancer: results from the ASCENT trial. Cancer. 2008;112:2377–2383. doi: 10.1002/cncr.23461. [DOI] [PubMed] [Google Scholar]
- 32.Adler HL, et al. Elevated levels of circulating interleukin-6 and transforming growth factor-beta1 in patients with metastatic prostatic carcinoma. J. Urol. 1999;161:182–187. [PubMed] [Google Scholar]
- 33.Lehrer S, et al. C-reactive protein is significantly associated with prostate-specific antigen and metastatic disease in prostate cancer. BJU Int. 2005;95:961–962. doi: 10.1111/j.1464-410X.2005.05447.x. [DOI] [PubMed] [Google Scholar]
- 34.Beuten J, et al. Single and multivariate associations of MSR1, ELAC2, and RNASEL with prostate cancer in an ethnic diverse cohort of men. Cancer Epidemiol. Biomarkers Prev. 2010 doi: 10.1158/1055-9965.EPI-09-0864. 19, 588–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shook SJ, et al. Association of RNASEL variants with prostate cancer risk in Hispanic Caucasians and African Americans. Clin. Cancer Res. 2007;13:5959–5964. doi: 10.1158/1078-0432.CCR-07-0702. [DOI] [PubMed] [Google Scholar]
- 36.Nakazato H, et al. Role of genetic polymorphisms of the RNASEL gene on familial prostate cancer risk in a Japanese population. Br. J. Cancer. 2003;89:691–696. doi: 10.1038/sj.bjc.6601075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urisman A, et al. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Fischer N, et al. Prevalence of human gammaretrovirus XMRV in sporadic prostate cancer. J. Clin. Virol. 2008;43:277–283. doi: 10.1016/j.jcv.2008.04.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.