Abstract
The transcription factor C/EBPα plays an important role in granulopoiesis. The tumor suppressor function of C/EBPα is shown by the findings that loss of expression or function of C/EBPα in leukemic blasts contributes to a block in myeloid cell differentiation and to leukemia. C/EBPα mutations are found in around 9% of acute myeloid leukemia (AML) patients. The mechanism by which the mutant form of C/EBPα (C/EBPα-p30) exerts a differentiation block is not well understood. By using a proteomic screen, we have recently reported PIN1 as a target of C/EBPα-p30 in AML. In the present study, we show that C/EBPα-p30 induces PIN1 expression. We observed elevated PIN1 expression in leukemic patient samples. Induction of C/EBPα-p30 results in recruitment of E2F1 in the PIN1 promoter. We demonstrate that the inhibition of PIN1 leads to myeloid differentiation in primary AML blasts with C/EBPα mutations. Overexpression of PIN1 in myeloid cells leads to block of granulocyte differentiation. We also demonstrate that PIN1 increases the stability of the c-Jun protein by inhibiting c-Jun ubiquitination and c-Jun blocks granulocyte differentiation mediated by C/EBPα. Our data suggest the inhibition of PIN1 could be a potential strategy of treating AML patients with C/EBPα mutation.
Keywords: C/EBPα, PIN1, AML
Introduction
The transcription factor CCAAT enhancer binding protein α (C/EBPα) is a critical regulator of the myeloid differentiation programme. Recent studies on animal models and AML patient samples suggest that the loss of function or expression of C/EBPα provides a platform on which AML develops 1. C/EBPα is mutated in about 9% of AML samples 2. The mutations reported in C/EBPα are point mutations in the basic region / leucine zipper domain and frame shift mutations in the N-terminal domain, which result in the expression of the shorter form of C/EBPα, C/EBPα-p303,4. C/EBPα-p30 is produced by alternative transcription initiation. It retains the DNA-binding domain but lacks the N terminal transactivation domain of the longer form of C/EBPα, C/EBPα-p42. C/EBPα-p30 fails to induce differentiation and exhibits a dominant negative function over C/EBPα-p42. The expression of the p30 isoform is high in AML patients with C/EBPα mutations. A recent study has demonstrated the ability of C/EBPα-p30 to induce AML in a mouse model 5.
Using a proteomic approach, we have recently demonstrated that one of the mechanisms behind the dominant negative function of C/EBPα-p30 is through increased sumoylation of C/EBPα-p42 by UBC9 6. In this screen, we also identified the peptidyl-prolyl cis/trans isomerase PIN1 as one of the transcriptional target genes of C/EBPα-p30. PIN1 binds to and isomerizes the peptidyl-prolyl bond in serine or threonine phosphorylated Ser/Thr-Pro motifs7,8. PIN1 appears to be important in tumorigenesis because it has been found to be overexpressed in many cancers including prostate, lung, ovary, cervical, breast, brain and skin cancers 9,10.
Although Pin1 null animals display age-dependent defects, no other phenotypic characteristics related to cancer have been detected 11. Mice lacking Pin1 are resistant to tumorigenesis induced by oncogenic Neu or Ras 12. The inhibition of PIN1 in cancer cells via multiple approaches triggers apoptosis or suppresses the transformed phenotype 13,14.
Indirect evidence for the role of PIN1 in leukemia comes from its positive effect on the transcriptional activity of c-Jun 10,15, a proto-oncogene shown by our lab to be downregulated by C/EBPα-p42 during granulopoiesis 16. We have also shown that c-Jun is overexpressed in AML patients with C/EBPα mutations 17. Furthermore, growing number of studies support the oncogenic potential of PIN1, which is an E2F1 target 18. Interestingly, E2F1 inhibition by C/EBPα is a critical step in myeloid differentiation 19. However the exact role of PIN1 in leukemogenesis remains elusive.
In the present study we investigated the role of PIN1 in AML with C/EBPα mutation. We provide evidence that C/EBPα-p30 upregulates PIN1 protein levels. AML patients show high PIN1 expression. We show that silencing PIN1 leads to granulocytic differentiation of primary AML blasts derived from patients with C/EBPα mutations and also in leukemic cell line. Furthermore, we demonstrate that PIN1 prevents degradation of c-Jun, which in turn blocks C/EBPα induced differentiation.
Materials and methods
Cells, transfections and reagents
Kasumi-6 cells were obtained from ATCC (Manassas, United States). Blast cells from AML patients were obtained from the Laboratory for Leukemia Diagnostics at the University of Munich. All samples were karyotyped and molecular genetics analysis was performed for C/EBPα mutations. Prior to therapy, all patients gave their informed consent for participation in the Acute Myeloid Leukemia Cooperative Group (AMLCG) studies. Details of the study protocol have been published 20.
K562-C/EBPα-p42-ER and K562-C/EBPα-p30-ER cells 21 were maintained in RPMI 1640 without phenol red supplemented with 10% charcoal treated fetal bovine serum, 1% penicillin-streptomycin and 2 μg/ml puromycin; Kasumi-6 cells 22 were cultured in RPMI 1640 supplemented with 20% fetal bovine serum, 1% penicillin-streptomycin and 2 ng/ml GM-CSF; AML blast cells were cultured in IMDM supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin and 20 mM HEPES; U937 cells and NB4 cells were cultured in RPMI supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin. K562-C/EBPα-p30-BRM2-ER cells were established as reported before 21. Briefly, C/EBPα-p30 was amplified by PCR using rat C/EBPα having BRM2 mutation (kind gift from Bruno Calabretta) with BamHI flanking primers (Forward primer 5’ GGG GAT CCG CCA CCA TGT CCG CGG GGG CGC AC 3’ and reverse primer 5’ATG GAT CCG GCG CGC AGT TG 3’) and subsequently cloned into pBabe-ER digested with BamHI with the 30-KDa C/EBPα peptide in frame with the C-terminal ER domain. K562 cells (106 cells) were electroporated with AMAXA transfection method with 3 μg of ScaI linearized plasmid and plated on 6 well plate in phenol red free RPMI/10 % charcoal treated FBS. Selection with 1 μg/ml puromycin began 48 hours after transfection.
293T cells (2 × 104 cells) were transfected with the LipofectAMINE Plus reagent (Invitrogen, Germany) according to manufacturer’s instruction. Transfection of U937 and Kasumi-6 cells was performed with the Nucleofector kit (AMAXA, Germany) as described by the manufacturer. 2 μg DNA plasmid were used for each transfection and the transfection efficiency was analyzed using a plasmid with eGFP marker. U937 and Kasumi-6 cells were transfected with nucleofection programmes V-01 and T-03, respectively. Transfection efficiencies of around 55-70% and 66-75% were observed in these cell lines, respectively.
Peptidylprolyl isomerase-parvulin inhibitor PiB (Calbiochem, United States) was prepared in ethanol and used at 5 μM. C/EBPα siRNA was purchased from Invitrogen.
Proteomics screening
To induce C/EBPα-p30, K562-C/EBPα-p30-ER cells were treated with 5 μM of β-estradiol for 6 hours followed by lysis. The protein identification by mass spectrometry was done essentially as reported before 6.
Promoter assay
293T cells were transiently transfected using LipofectAMINE (Invitrogen, Germany) as described by the manufacturer. Firefly luciferase activities from the PIN1 promoter constructs and Renilla luciferase activity from the internal control plasmid pRL-null were determined 24 h after transfection using the Dual-Luciferase Reporter Assay System (Promega, Germany). Firefly luciferase activities were normalized to the Renilla luciferase values of pRL-null.
Immunoblot analysis
Immunoblot analysis was performed with 50 μg total protein lysates using anti-C/EBPα (sc-61), anti-PIN1 (sc-15340), anti-c-Jun (sc-1694), anti-β-Tubulin (sc-9104) and anti-E2F1 (sc-251) antibodies from Santa Cruz Biotechnology, Germany. The immunodetection was performed with an ECL reagent (Amersham Biosciences, Germany). The band intensities were quantified using ImageJ software.
Chromatin Immunoprecipitation
The crosslinking of proteins to DNA was accomplished by the addition of 1% formaldehyde for 10 min to cultured cells (6×106) at 37°C. After sonication, the chromatin was immunoprecipitated with 5 μg of the antibodies anti-E2F1 (sc-193X) or anti-IgG (SC-2027, Santacruz) at 4°C overnight.
Flow cytometry analysis
For flow cytometry analysis, 106 cells were washed twice with PBS and resuspended in 50 μl of PBS with 2 μl of the respective antibody. Incubation was performed for 20 minutes in ice. After the incubation, cells were washed with PBS, resuspended in PBS and analyzed by flow cytometry on a FACScan (Becton Dickinson).
mRNA expression analysis
Total RNA was isolated from leukemic patient samples, processed and analyzed on the Affymetrix HG-U133A and HG-U133B chips as described before 23. The data from Affymetrix analysis were normalized according to the procedure described before 24. Normalized expression data were then analyzed with the R software package and the “boxplot” function (www.r-project.org). Expression signal intensities are expressed on a logarithmic scale.
Real-time RT-PCR
Total RNA was isolated from cells with Trizol reagent (Invitrogen, Germany). 750 ng of RNA was used to synthesize cDNA by Reverse Transcription. Equal amounts of cDNA were taken for a subsequent quantitative real-time PCR (Q-RT-PCR) using the SYBR Green PCR kit (Qiagen, Germany) in a Rotor-Gene RG-3000 cycler (Corbett Research, Australia). The fold change was calculated using delta CT method reported before 25.
Protein stability Assay
293T cells were transfected with 6.6 μg of PIN1 or non silencing siRNA (Qiagen, Germany) using lipofectamine. After 24 hours, cells were treated with cyclohexamide (100 μg/ml) for various periods and lysed. The amount of c-Jun protein was analysed by Western blot and quantified by densitometric scanning.
Ubiquitination Assay
293T cells were transiently transfected with different constructs as described for promoter assay, 24 hours after transfection cells were lysed in RIPA buffer followed by c-Jun immunoprecipitation from 500 μg total protein. The protein samples after immunoprecipitation were analysed in a 10% SDS-PAGE gel and probed with an HA antibody (Roche Applied Science, Germany).
Statistical analysis
We used Student’s t-tests to determine the statistical significance of experimental results. A P value of 0.05 or less was considered significant.
Results
C/EBPα-p30 induces PIN1 expression in leukemia
The K562-ER cell line is an early multipotential cell line derived from the K562 cell line, which was originally obtained from a patient with chronic myeloid leukemia. To derive the K562-ER cell line, K562 cells were stably transfected with a plasmid encoding an estrogen inducible C/EBPα-p30 estrogen receptor fusion protein (C/EBPα-p30-ER)21. K562-ER cells have been shown to be a good model system to study granulopoiesis in the context of C/EBPα proteins (Supplementary figures 1 and 2). We have previously reported a proteomic approach for the identification of C/EBPα-p30 target proteins in C/EBPα-p30-ER cell line 6. In this screen, PIN1 was identified as one of the targets of C/EBPα-p30 (Figure 1a).
Figure 1. C/EBPα-p30 induces PIN1 mRNA and protein expression.
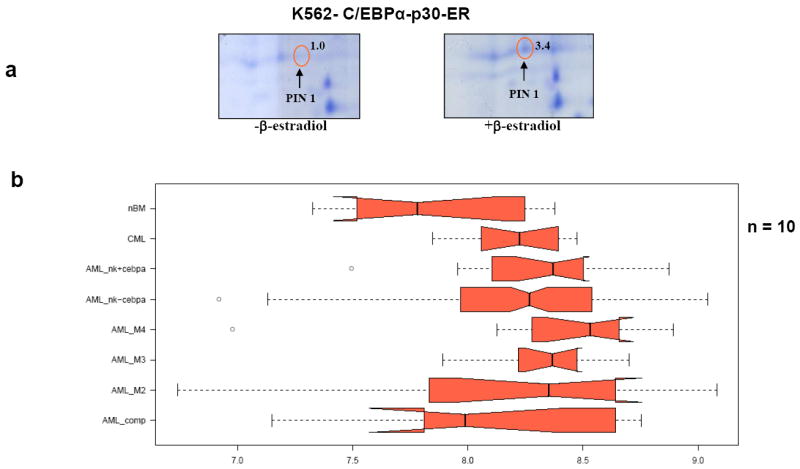
(a) K562- C/EBPα-p30-ER cells were induced with ß-estradiol for 6 hours followed by 2D gel electrophoresis and protein identification by mass spectrometry. The values represented are mean normalized spot volume from the Proteom Weaver software. (b) Affymetrix analysis was done with AML blast cells using total RNA isolated from leukemic patient samples, processed and analyzed on the Affymetrix HG-U133A and HG-U133B chips. Expression signal intensities are expressed in logarithmic scale. 10 samples were used in each subtype of AML. The dark bars represent median and boxes represent 25-75% quantile range. If the notches of the box plots do not overlap there is a strong indication that the medians are different. nBM: normal bone marrow, CML: chronic myeloid leukemia, AML NK+cebpa: Normal karyotype with C/EBPα mutation, AML NK-cebpa: Normal karyotype without C/EBPα mutation, AML Comp: Complex karyotype, AML M4: AML with the CBF/MYH11 fusion gene, AML M3: AML with PML/RARA fusion gene, AML M2: AML with AML1/ETO fusion gene,. (c) Total RNA was isolated from AML blast cells and analysed PIN1 mRNA expression by quantitative real time PCR. (d) K562-C/EBPα-p30-ER cells (left) and K562-C/EBPα-p42-ER cells (right) were induced with ß-estradiol (5 μM) for respective time points and cells were lysed and subjected to Western blot analysis using PIN1 antibody. Values below the gel image indicate the upregulation (fold) of PIN1 protein level normalized to ß-Tubulin.
As a first step to elucidate the importance and role of PIN1 as a target of C/EBPα-p30, we investigated the PIN1 mRNA levels in leukemic patient samples. Affymetrix analysis showed that PIN1 mRNA expression is increased in different subtypes of leukemia compared to normal bone marrow controls (Figure 1b). We also observed elevated PIN1 levels in AML samples with C/EBPα mutation in comparison to AML samples without C/EBPα mutation (Figure 1c). The increased expression of PIN1 in different leukemic subtypes suggests that mechanisms other than C/EBPα mutations can increase PIN1 levels (See Discussion). Next we examined the ability of C/EBPα-p30 to induce PIN1 protein expression in the K562- C/EBPα-p30-ER cell line. Our data revealed that PIN1 protein expression was upregulated by C/EBPα-p30 during ß-estradiol induction (Figure 1d left, supplementary figure 3). These data confirm our proteomic findings and reveal the relevance of PIN1 as a target in leukemia. We also investigated regulation of PIN1 during C/EBPα-p30 overexpression in two AML cell lines - Kasumi-6 (C/EBPα mutation positive) and NB4 (C/EBPα mutation negative),). We observed that C/EBPα-p30 is able to up-regulate PIN1 (Supplementary figure 4). We observed slightly lower expression of PIN1 in Kasumi-6 cells in comparison to NB4 cells (Supplementary figure 4). Similar to the upregulation of PIN1 protein levels, we could observe an upregulation of PIN1 mRNA levels upon C/EBPα-p30 induction (Supplementary figure 5).
C/EBPα-p42 and C/EBPα-p30 have been shown to have different biological properties 3. Based on our finding that C/EBPα-p30 is able to induce PIN1 expression, we asked whether C/EBPα-p42 is able to regulate PIN1 protein levels. Our data show that in contrast to the C/EBPα-p30 form, C/EBPα-p42 repressed PIN1 protein levels (Figure 1d, right). This demonstrates that C/EBPα-p42 and C/EBPα-p30 have opposing effects on PIN1 protein levels. We could not find any regulation of PIN1 during C/EBPα-p42 silencing by siRNA (supplementary Figure 4). So we assume that silencing of C/EBPα-p42 alone is not sufficient for the regulation of PIN1.
We also analysed the role of C/EBPα with mutations in the E2F1 protein binding domain (C/EBPα-BRM2) 21, 26, in regulating PIN1 protein expression. Our data suggests C/EBPα- BRM2 fails to downregulate PIN1 protein levels (data not shown). This denotes E2F1 interaction of C/EBPα-p42 is necessary for its inhibitory action over PIN1.
C/EBPα-p30 induces PIN1 promoter activity in association with E2F1
It has been shown previously that there are three E2F1 binding sites in the PIN1 promoter and E2F1 can transactivate PIN1 promoter activity 27. Since we could observe C/EBPα-p30 induces PIN1 expression, we next investigated how C/EBPα proteins regulate PIN1 transcription. To address this, we performed promoter assays in the human embryonic kidney cell line 293T with different PIN1 promoter constructs 27. Our data revealed that overexpression of E2F1 could transactivate PIN1 promoter as published before (Figure 2a, compare bars 1 and 4). In comparison with E2F1 alone, co-transfection of C/EBPα-p30 enhanced the transactivation of the PIN1 promoter activity to around 2 fold (Figure 2a, compare bars 4 and 6). This indicates that C/EBPα-p30 co-operates with E2F1 to regulate PIN1 mRNA levels. Interestingly, co-transfection of C/EBPα-p42 downregulated PIN1 promoter activity (Figure 2a, compare bars 4 and 5).
Figure 2. C/EBPα-p30 recruits E2F1 to the PIN1 promoter.
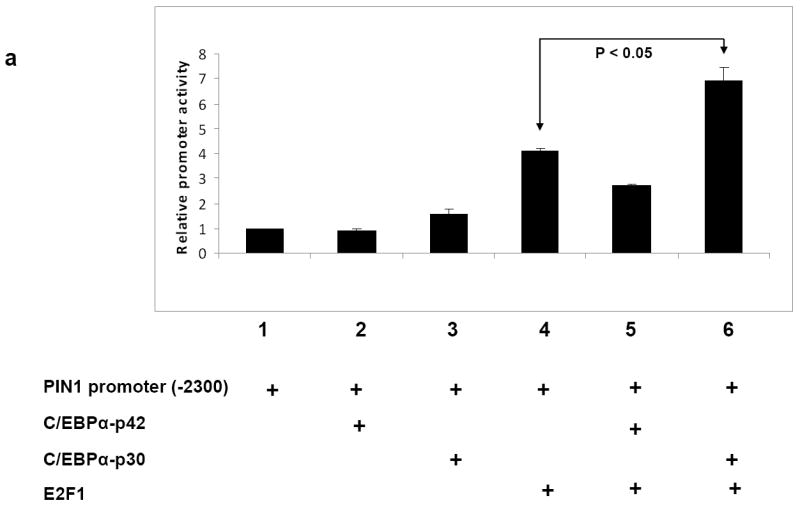
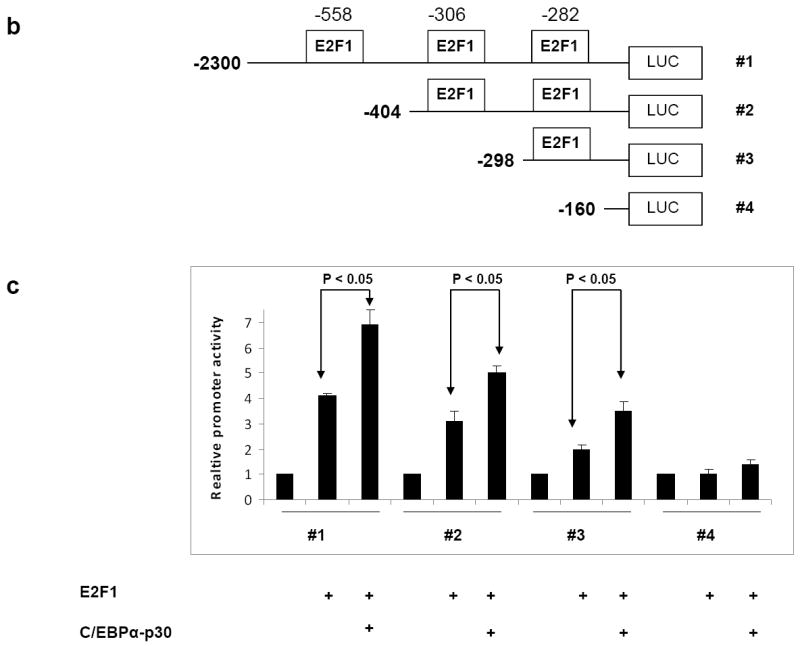
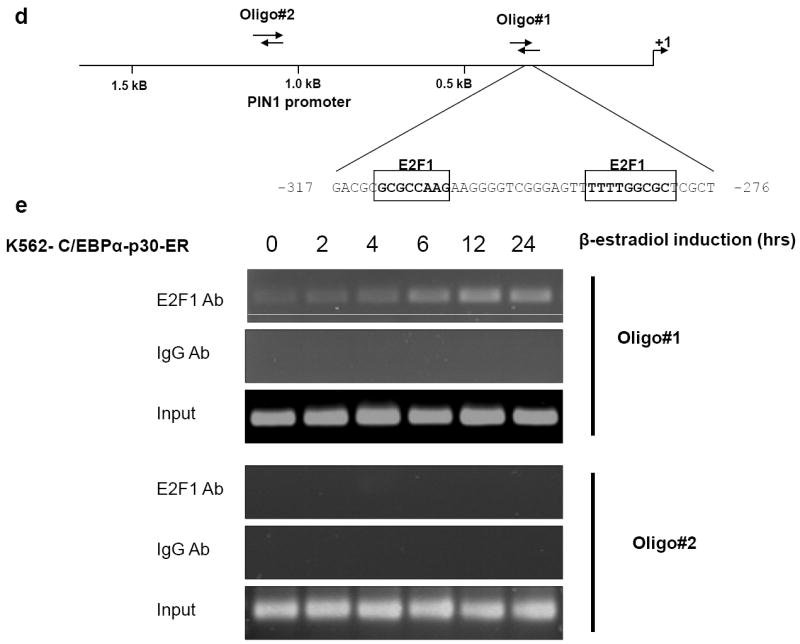
(a) Transient transfection in 293T cells with a reporter construct of 2.3 kb of the human PIN1 promoter cloned in pGL3 basic vector and expression plasmids for C/EBPα-p42, C/EBPα-p30 and E2F1. Luciferase activities were measured 24 hr after transfection and values were normalized by using Renilla luciferase PRL0. The results are the means from three independent experiments, and error bars represent the standard deviation. (b) Schematic presentation of various PIN1 promoter 5’ deletion constructs used for promoter assays. (c) 293T cells were transfected with different PIN1 promoter constructs in combination with C/EBPα̃-p30 and E2F1. (d) Human PIN1 promoter sequence with putative E2F1 binding sites. (e) K562-C/EBPα-p30-ER cells were induced with ß-estradiol for respective time points and chromatin was immunoprecipitated with anti-E2F1 or IgG antibodies, and the recovered DNA was PCR amplified with primers specific for E2F1 binding sites in the PIN1 promoter as shown in figure 2d.
To determine the importance of the three E2F1 binding sites for the C/EBPα-p30 co-operation with E2F1 in regulating PIN1 promoter activity, we performed promoter assays with different deletion mutants of PIN1 promoter (Figure 2b). Compared to the wild type PIN1 promoter vector (-2300 Luc), deletion of the E2F1 binding sites (sites located at -557 to -550, -312 to -305 and at -288 to -281) exhibited decrease in the PIN1 promoter activity in a site dependent manner (Figure 2c). These data suggest that E2F1 binding sites are important for the association of C/EBPα-p30 with E2F1.
To gain further insights into the mechanism of association of C/EBPα-p30 with E2F1 in regulating PIN1 expression, we performed chromatin immunoprecipitation assays in K562-C/EBPα-p30-ER cells. Cells were treated with β-estradiol to induce C/EBPα-p30 and chromatin fragments were immunoprecipitated with an anti-E2F1 antibody. DNA from the immunoprecipitates was PCR amplified using primers flanking the two E2F1 binding sites PIN1 promoter region shown in figure 2d. We observed increased binding of E2F1 during C/EBPα-p30 induction in a time dependent manner (Figure 2e). We analysed whether increased binding of E2F1 in the PIN1 promoter is associated with induction of E2F1. Our data shows that C/EBPα-p30 does not induce E2F1 protein (Supplementary figure 6). It has been previously shown that C/EBPα-p30 is able to interact with E2F protein 21. To evaluate interaction of C/EBPα-p30 with E2F1 plays a role in the induction of PIN1 by C/EBPα-p30, we analysed induction of PIN1 in a cell line which has mutation for E2F1 interacting domain of C/EBPα-p30. Our data shows that that E2F1 interaction is not needed for PIN1 induction (Supplementary figure 7). Taken together, our data demonstrate that induction of C/EBPα-p30 leads to a recruitment of E2F1 to the PIN1 promoter and this is independent of interaction of C/EBPα-p30 with E2F1. These data suggest that PIN1 might represent an important target of E2F1 activation in the C/EBPα-p30 induced differentiation block. We also analysed binding of C/EBPα-p30 to the PIN1 promoter in a similar experimental setting. We didn’t observe any binding of C/EBPα-p30 to the PIN1 promoter (data not shown).
To find the possibility of C/EBPα-p30 co-operation with E2F1 in regulating other E2F1 target genes, we performed chromatin immunoprecipitation in K562-C/EBPα-p30-ER cells for c-Myc promoter (Supplementary Figure 8). Even though we could observe a slight increase in E2F1 binding in c-Myc promoter during C/EBPα-p30 induction, the effects were minor. This suggests co-operation of C/EBPα-p30 with E2F1 is more effective in the regulation of PIN1 expression.
PIN1 inhibition by PiB can overcome the differentiation block observed in human myeloid cells
To further understand how silencing of the PIN1 activity is biologically significant in the context of C/EBPα mutation, we carried out myeloid differentiation experiments in AML blast cells with C/EBPα mutation and in Kasumi-6 cells. Kasumi-6 is a myeloid leukemia cell line established from the bone marrow cells of an individual with acute myeloid leukemia, subtype M2 having C/EBPα mutation and expressing both the p42 and the p30 isoforms of the C/EBPα protein endogenously 22. For silencing PIN1, we used the PIN1 inhibitor, PiB 28. PiB has been shown to inhibit the PIN1 activity by binding to the PPIase domain of PIN1. The myeloid cell differentiation was assessed by CD11b and CD15 expression by flow cytometry analysis as well as by the level of G-CSFR expression by Real Time RT-PCR. Treatment of AML blast cells and the Kasumi-6 cells with PiB induced myeloid differentiation (Figure 3). We could not observe any significant change in granulocyte morphology during PIN1 inhibition by PiB (data not shown). So we assume PIN1 inhibition by PiB is not sufficient to induce terminal stages of differentiation. We investigated the role of PIN1 silencing in NB4 cells, which do not have any CEBPA mutation. We could observe that PIN1 inhibition in NB4 cells leads to granulocytic differentiation (Supplementary figure 9). How ever, compared to Kasumi-6, the differentiation effects were modest during PIN1 silencing in NB4 cells. We also analysed the effect of PIN1 silencing in normal bone marrow cells. We could not observe any change in granulocytic differentiation (data not shown). These data suggest that CEBPA mutation status (presence of C/EBPa-p30) is significant in granulocytic differentiation during PIN1 silencing. The granulocytic differentiation during PIN1 inhibition by PiB was found independent of apoptosis (Supplementary figure 10) and cell proliferation (data not shown).
Figure 3. PIN1 inhibition using PiB can overcome the differentiation block observed in human myeloid cells.
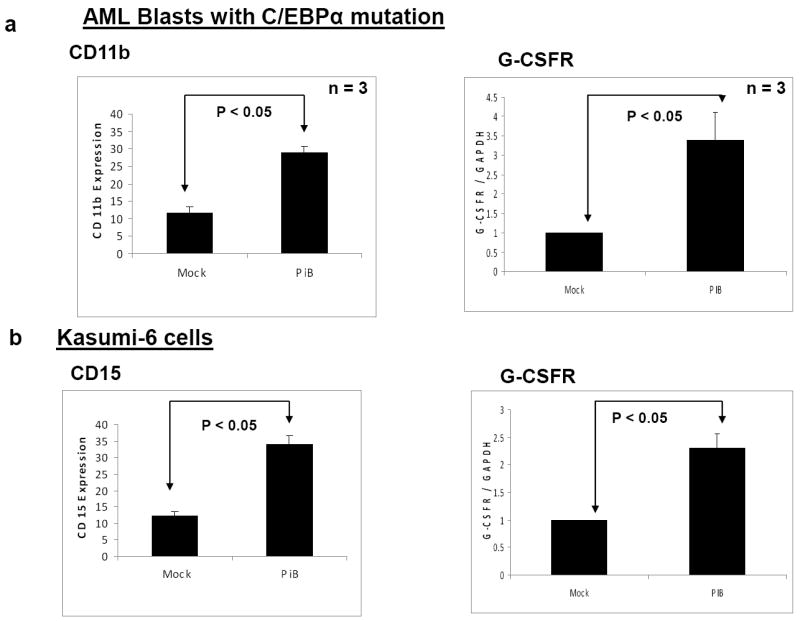
(a,b) AML blast cells with C/EBPα mutation (a) and Kasumi-6 cells (b) were treated with PiB (5 μM) for 6 days and myeloid cell differentiation was assessed by FACS analysis using CD11b and CD15 expression as well G-CSFR expression by Real-time RT-PCR analysis. The results are the means from three independent experiments, and error bars represent the standard deviation.
PIN1 blocks granulocytic differentiation
C/EBPα-p30 has been shown to block granulocytic differentiation mediated by C/EBPα 3 as well as to be able to induce AML 5. Based on our data that C/EBPα-p30 induces PIN1 in AML (Figure 1) and silencing PIN1 leads to granulocytic differentiation (Figure 3), we assessed the ability of PIN1 to block granulocytic differentiation. We overexpressed PIN1 in myeloid U937 cells and treated the cells with retinoic acid to induce granulocytic differentiation. Overexpression of PIN1 blocked granulocytic differentiation as shown by around 50% reduction in CD15 expression as well as reduction in G-CSFR expression (Figure 4). Taken together, these data demonstrates that PIN1 could have significant role in granulocytic differentiation block observed in AML.
Figure 4. PIN1 inhibits granulocytic differentiation.
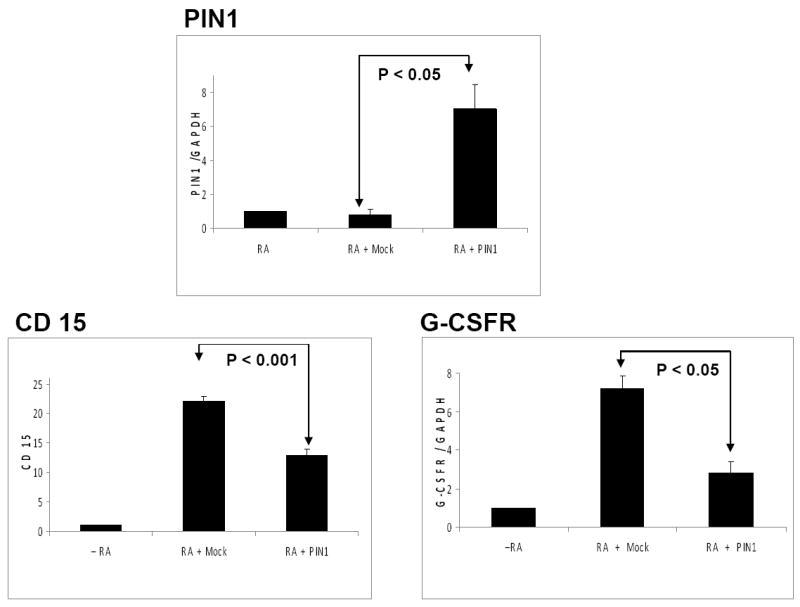
U937 cells were transfected with mock vector or PIN1 vector followed by treatment with retinoic acid (1μM). Three days after transfection, CD15 expression was analysed with FACS analysis and PIN1 and G-CSFR expressions were analysed with Real-time RT-PCR. The results are the means from three independent experiments, and error bars represent the standard deviation.
PIN1 controls the stability of the c-Jun protein
We have previously reported that c-Jun expression is high in AML patient samples with C/EBPα mutation 17. Upon JNK activation, PIN1 binds to c-Jun which is phosphorylated on Ser63/73-Pro motifs 10. Due to the fact that C/EBPα-p30 could induce PIN1 and PIN1 regulates the protein stability of many targets with which it interacts 8, we hypothesized that PIN1 could have a significant role in regulating c-Jun protein stability. To test this hypothesis, PIN1 expression was silenced in 293T cells by RNA interference and c-Jun protein stability was then assessed by blocking protein synthesis with cyclohexamide (CHX) and measuring the c-Jun protein remaining at various time points. Our data shows that the stability of c-Jun was reduced when PIN1 expression was knocked down by a specific siRNA (Figure 5a and supplementary figure 11). These results indicate that PIN1 is an important factor in regulating the stability of c-Jun protein.
Figure 5. PIN1 stabilises c-Jun protein by inhibiting c-Jun ubiquitination and c-Jun blocks the granulocytic differentiation.
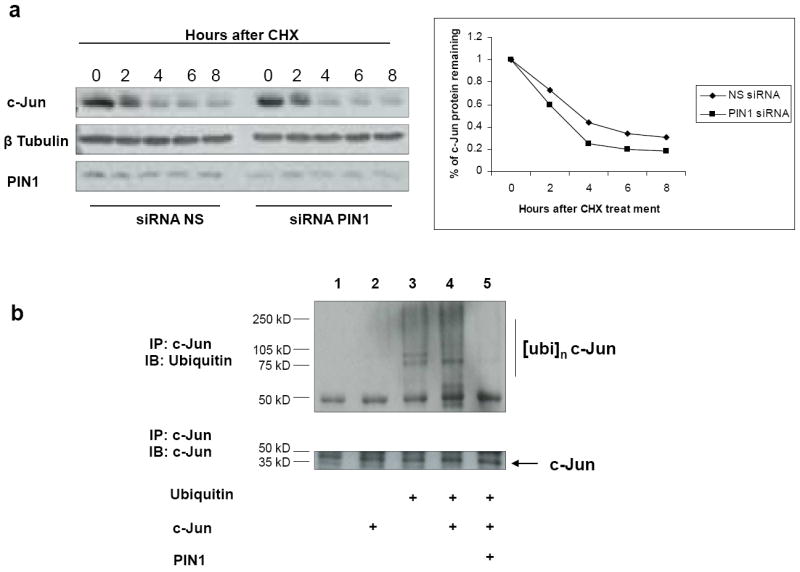
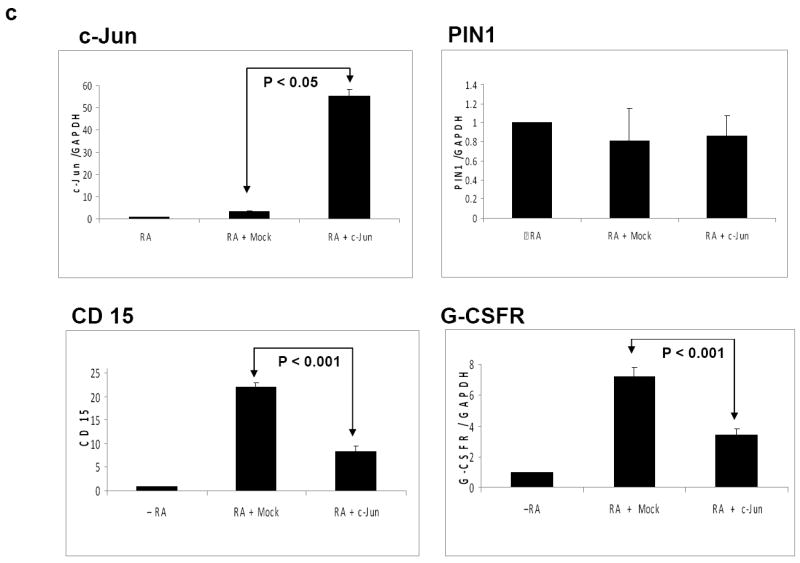
(a) 293T cells were transfected with PIN1 and control siRNA. After 24 hours protein synthesis was blocked with cyclohexamide (CHX) and cells were harvested at different time points. The amount of c-Jun protein remaining was analyzed by Western blot followed by densitometric scanning. (b) Ubiquitination assay was performed by transfecting 293T cells with the expression plasmids for HA-ubiquitin, c-Jun and PIN1 as indicated, 24 hrs post transfection cells were lysed and c-Jun was immunoprecipitated and immunoblot against HA antibody (upper panel) was done. Membrane was stripped and and reprobed for c-Jun (lower panel) as control for c-Jun immunoprecipitation (c) U937 cells were transfected with mock vector or c-Jun vector followed by treatment with retinoic acid (1μM). Three days after transfection, CD15 expression was analysed with FACS analysis and c-Jun, PIN1 and GCSFR expressions were analysed with Real-time RT-PCR. The results are the means from three independent experiments, and error bars represent the standard deviation.
PIN1 protects c-Jun from ubiquitination mediated protein degradation
c-Jun has been shown to be subjected to ubiquitin-mediated degradation in a JNK dependent manner 29, 30. Based on our finding that PIN1 regulated c-Jun protein stability (Figure 5A), we tested the role of PIN1 in the degradation of c-Jun. We performed ubiquitination assays in 293T cells by overexpressing c-Jun, PIN1 and HA-tagged ubiquitin. When c-Jun was co-transfected with HA-tagged ubiquitin, higher molecular weight bands were detected in the immunoprecipitation with the HA antibody which reflects c-Jun ubiquitination (Figure 5b, lane 4). Interestingly, when PIN1 was co-transfected, the ubiquitination of c-Jun was prevented suggesting a role for PIN1 in the regulation of c-Jun degradation (Figure 5b, lane 5).
Our data suggest that PIN1 prevents the degradation of c-Jun protein (Figure 5a, b). The Jun proto-oncogene is positively autoregulated by its product, Jun/AP-1 31. Based on these findings, we hypothesized that PIN1 could have a major role in regulating c-Jun mRNA levels. We overexpressed PIN1 in Kasumi-6 cells and analysed c-Jun mRNA levels by quantitative Real-Time RT-PCR. We observed that overexpression of PIN1 leads to an increase in c-Jun mRNA levels (Supplementary figure 12a). We next investigated the effect of silencing PIN1 on the c-Jun mRNA levels. We found that silencing PIN1 with the inhibitor PiB downregulated the c-Jun mRNA level (Supplementary figure 12b). These data demonstrate that PIN1 stabilizes c-Jun protein, which in turn induces c-Jun mRNA by auto regulation.
c-Jun blocks granulocytic differentiation
We have previously shown that downregulation of the proto-oncogene c-Jun by C/EBPα is important for granulocytic lineage commitment 16. We have also reported that c-Jun expression is high in different subtypes of AML including AML samples with C/EBPα mutation and that c-Jun is able to block the DNA binding of C/EBPα 17. Based on these findings, we hypothesized that c-Jun is able to block the granulocytic differentiation induced by C/EBPα. To assess this, we overexpressed c-Jun in human myeloid U937 cells and treated the cells with retinoic acid. The granulocyte differentiation was assessed by FACS analysis for CD15 expression and quantitative Real-Time RT-PCR for G-CSFR. The expression of CD15 during granulocytic differentiation was downregulated to around 3 fold in the presence of c-Jun (Figure 5c). The myeloid markers such as G-CSFR was also found downregulated during overexpression of c-Jun (Figure 5c). These data suggests that c-Jun is able to block granulocytic differentiation induced by C/EBPα.
Discussion
Initially, it was suggested that the dominant negative effect of C/EBPα-p30 is the result of heterodimerization of the mutant form with the C/EBPα-p42. A recent report shows that a C/EBPα-p30 with modification in the leucine zipper, which can not form heterodimers with C/EBPα-p42, still exhibits dominant negative properties over C/EBPα-p42 32. This suggests that rather than heterodimerization, regulatory networks activated by C/EBPα-p30 could play a critical role in its dominant negative function. One recent study showed that C/EBPα-p30 has quite distinct functional properties compared to the C/EBPα-p42 33. This study showed that C/EBPα-p30 is able to bind to a unique set of target genes with higher affinity than C/EBPα-p42. Mice with targeted disruption of C/EBPα-p42, which express C/EBPα-p30, develop leukemia 5. This finding is of particular interest since C/EBPα knock-out mice did not develop leukemia even though they exhibited a block of granulocytic differentiation 34. Taken together, these findings demonstrate that the disruption of C/EBPα-p42 alone is not sufficient to initiate leukemogenesis, but that pathways modulated by C/EBPα-p30 might be critical in the development of AML.
By using a proteomic screen, we have recently shown that C/EBPα-p30 is able to upregulate the expression of a number of proteins including PIN1 6. Our finding that PIN1 is upregulated in leukemic patient samples including those with C/EBPα mutations (Figure 1b,c) is intriguing since PIN1 overexpression was shown to amplify multiple signaling pathways important in oncogenesis 35. We have shown that C/EBPα-p42 is able to downregulate PIN1 protein levels (Figure 1d, right). One possible explanation for the upregulation of PIN1 in different leukemic samples could be that C/EBPα has been shown to be downregulated by diverse mechanisms in different subtypes of leukemia 36. This down-regulation of C/EBPα function might then lead to PIN1 overexpression in leukemia (Figure 1b). However, we didn’t find any significant upregulation of PIN1 during C/EBPα silencing in cell lines (Supplementary Figure 4). It has been observed that BRCA1 tumor suppressor (breast cancer associated gene 1) represses PIN137. Women with germline heterozygous mutations in BRCA1 are at increased risk of developing breast cancers38 and PIN1 is overexpressed in breast cancer10. These findings suggest the possibility that other tumor suppressors could be also regulating PIN1 expression in granulopoiesis. Further studies are needed to show how C/EBPα as well as other tumor suppressors regulates PIN1 and how PIN1 is overexpressed in AML without C/EBPα mutation.
A recent study proposes that C/EBPα could be a PIN1 target 39. This study points out that several S/T-P motifs in C/EBPα could be regulated by PIN1 mediated isomerization. Most of the S/T-P motifs in the C/EBPα-p42 are also present in C/EBPα-p30. Even though C/EBPα has been shown to be phosphorylated, there is no report about PIN1 mediated post phosphorylation mechanisms regulating the C/EBPα function. Further studies are needed to explain how PIN1 regulates C/EBPα mediated transactivation and whether CEBPA might be a target of PIN1-mediated proline isomerization.
In granulopoiesis, the inhibition of the E2F1 activity has been proven to be the unique mechanism for the anti-mitotic activity of C/EBPα 19, 40. E2F1 activates the transcription of the MYC oncogene, which had been shown to block granulopoiesis. C/EBPα inhibition of E2F1 has been shown to result in the downregulation of c-Myc, leading to granulopoiesis41. This raises the possibility that other E2F1 targets could be also regulated by C/EBPα in a similar mechanism as observed for c-Myc. In this context it should be noted that PIN1 has been shown to be an E2F1 target gene 27 and lack of E2F inhibition by C/EBPα has been shown to be a key event in AML 5, 42. Our finding that C/EBPα interferes with the E2F1 transactivation of PIN1 promoter (Figure 2a) suggests that the downregulation of PIN1 by C/EBPα could be an important event in granulopoiesis. Our observation points out intrinsic differences between C/EBPα-p42 and C/EBPα-p30 in regulating PIN1 expression. C/EBPα-p30 co-operates with E2F1 in regulating PIN1 promoter activity (Figure 2 a, c) and C/EBPα-p30 is capable of recruiting E2F1 to the PIN1 promoter (Figure 2e). This suggests regulatory networks co-ordinated by C/EBPα-p30 and E2F1 could play an important role in AML with C/EBPα mutations.
A recent study demonstrates C/EBPα-p30 not only blocks granulopoiesis, but also has the potential to induce AML 5. This study also indicates that loss of C/EBPα function alone is not sufficient for the development of AML. Rather, a restricted modulation of C/EBPα-p42 function by C/EBPα-p30 is a pre-requirement for AML to develop. Our findings that C/EBPα-p30 induces PIN1 (Figure 1), inhibition of PIN1 leads to granulocytic differentiation (Figure 3) and PIN1 blocks granulopoiesis (Figure 4) point out that PIN1 might have a major role in contributing to the differentiation block observed during C/EBPα mutation.
PIN1 is overexpressed in breast cancer and increases the transcriptional activity of c-Jun 10. PIN1 regulates the protein degradation of many targets with which it interacts including p53, p73, NF-κB and cyclin D1 8. In addition, PIN1 has been demonstrated to bind to c-Jun 10 and has been proposed to increase the protein stability of c-Jun8. Our data show that PIN1 increases the stability of c-Jun protein via inhibition of its ubiquitination (Figure 5a,b). As observed for other PIN1 targets, we assume stabilization of c-Jun by PIN1 is a general phenomenon in cancer. c-Jun is an effector of many signal cascades and has crucial functions in diverse mechanisms of oncogenesis 43. An oncogenic role for c-Jun in AML was suggested by the finding that c-Jun is overexpressed in different subtypes of AML 17, 44. One explanation for the upregulation of c-Jun expression in AML might be the downregulation of C/EBPα by different mechanisms36. Furthermore, leukemic fusion proteins such as BCR-ABL 45 and AML1-ETO44 have been shown to induce c-Jun expression through the JNK signaling pathway. We can not rule out the possibility that loss of activity of other factors that negatively regulate PIN1 could also upregulate c-Jun in AML. Our lab had demonstrated that c-Jun inactivates C/EBPα via direct protein-protein interaction through their leucine zipper domains 17. One recent study suggested that C/EBPα - c-Jun interaction has the potential to induce monocytic differentiation 46, pointing out how protein-protein interactions of C/EBPα are critical in lineage committement decisions. Another report showed that c-Jun has the ability to promote monocytic differentiation at the expense of granulocytic differentiation 47. Our data also indicate that overexpression of c-Jun induces monocytic differentiation (Supplementary Figure 13). Our findings that PIN1 can stabilize c-Jun (Figure 5a) and prevent c-Jun ubiquitination (Figure 5b) suggest that PIN1 might have an important role in regulating the c-Jun protein turn over. The stabilized c-Jun might bind to its own promoter and increase its expression in a positive feed back loop 31. The ability of c-Jun to block DNA binding of C/EBPα 17 as well as our finding that c-Jun is able to inhibit granulocytic differentiation (Figure 5d) suggests a model in which c-Jun promotes proliferation and leads to differentiation block by inactivating C/EBPα. In other words, the relative protein concentrations of C/EBPα and c-Jun regulate granulopoiesis - higher C/EBPα levels favor differentiation while higher c-Jun levels favor proliferation.
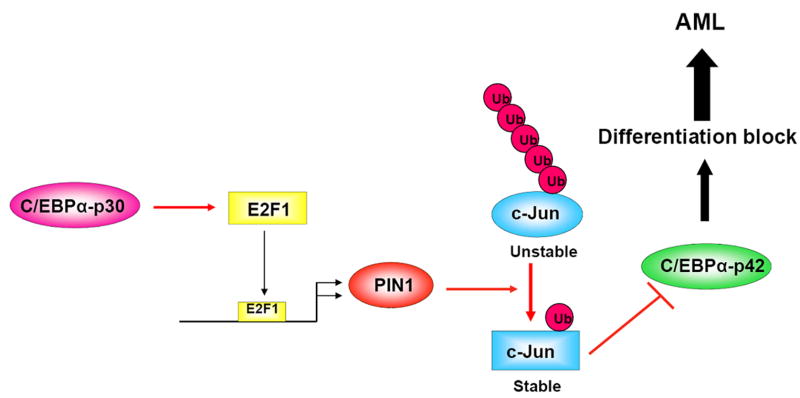
In summary, our study identifies PIN1 as an important player that might contribute to myeloid leukemia development through the inhibition of the C/EBPα function. Here we demonstrate that PIN1 is upregulated by C/EBPα-p30 and that silencing of PIN1 is able to overcome the differentiation block mediated by C/EBPα-p30. Our observations suggest a model in which C/EBPα-p30 induces PIN1 expression and increases the stability of c-Jun, which in turn inhibits the function of C/EBPα-p42 (Figure 6). Inhibiting PIN1 function could provide a novel strategy in the treatment of myeloid leukemic patients.
Figure 6.
Model depicting the PIN1 mediated dominant negative role of C/EBPα-p30 in AML with C/EBPα mutation. C/EBPα–p30 cooperates with E2F1 and increases the PIN1 mRNA and protein levels. PIN1 stabilises the c-Jun protein, which in turn blocks C/EBPα functions and leads to differentiation block and AML.
Supplementary Material
K562-C/EBPα-p30-ER and K562-C/EBPα-p42-ER cells were induced with ß-estradiol for 6 hours, cytoplasmic fraction (CF) and nuclear extract (NE) were prepared and analyzed for C/EBPα proteins by Western blot.
Kasumi-6 and NB4 cells were treated with ethanol or PiB (5 μM) for 6 days. Apoptotic cells were analysed by dual staing with FITC labeled Annexin- V and PI. Numbers in the lower right quadrant represents percentage of cells in early apoptosis (Annexin-V+ and PI−). Numbers in the top right quadrant represents percentage of cells in late apoptosis (Annexin-V+ and PI+).
293T cells were transfected with PIN1 and control siRNA. After 24 hours protein synthesis was blocked with cyclohexamide (CHX) and cells were harvested at different time points. The amount of c-Jun protein remaining was analyzed by Western blot followed by densitometric scanning.
(a) Kasumi-6 cells were transfected with PIN1 expression vector. Total RNA was isolated after 24 hrs with TRIZOL reagent and proceeded for reverse transcription followed by Real-time RT-PCR for c-Jun. Values were normalized with GAPDH mRNA. (b) Kasumi-6 cells were treated with PIN1 inhibitor, PiB for various time points and proceeded for Real-time RT-PCR for c-Jun as described. The results are the means from three independent experiments, and error bars represent the standard deviation.
Cord blood derived human CD34+ cells transfected with c-Jun or mock vectors. After 24 hours total RNA was isolated and analysed CD14 and M-CSFR mRNA by real time RT-PCR. The results are the means from three independent experiments, and error bars represent the standard deviation.
K562-C/EBPα-p30-ER and K562-C/EBPα-p42-ER cells were induced with ß-estradiol for 2 days and granulocytic differentiation was analysed by FACS analysis using CD11b antibody.
K562-C/EBPα-p30-ER cells were induced with ß-estradiol for respective time points and cells were lysed and subjected to Western blot analysis using PIN1 antibody. Values below the gel image indicate the upregulation (fold) of PIN1 protein level normalized to ß-Tubulin.
Kasumi-6 and NB4 cells were transfected with described DNA / siRNA and 24 hours later cells were lysed and subjected to Western blot analysis using PIN1 antibody. Values below the gel image indicate the upregulation (fold) of PIN1 protein level normalized to ß-Tubulin.
K562- C/EBPα-p30-ER cells were induced with ß-estradiol (5 μM) for respective time points to induce C/EBPα-p30 and total RNA was isolated at respective time points, proceeded for reverse transcription followed by quantitative real time PCR for PIN1.
(top) K562-C/EBPα-p30-ER cells were induced with ß-estradiol for respective time points and cells were lysed and subjected to Western blot analysis using E2F1 antibody. Values below the gel image indicate the upregulation (fold) of E2F1 protein level normalized to ß-Tubulin. (bottom) 293T cells were transfected with C/EBPα-p30. 12 and 14 hours after transfection, cells were lysed and subjected to Western blot analysis using E2F1 antibody. Values below the gel image indicate the upregulation (fold) of E2F1 protein level normalized to ß-Tubulin.
K562-C/EBPα-p30-BRM2-ER cells were induced with ß-estradiol for respective time points and cells were lysed and subjected to Western blot analysis using PIN1 antibody. Values below the gel image indicate the upregulation (fold) of PIN1 protein level normalized to ß-Tubulin.
C/EBPα-p30-ER cells were induced with ß-estradiol for respective time points and chromatin was immunoprecipitated with anti-E2F1 or IgG antibodies, and the recovered DNA was PCR amplified with primers specific for E2F1 binding sites in the c-Myc promoter.
NB4 cells were treated with PiB (5 μM) for 6 days and myeloid cell differentiation was assessed by FACS analysis using CD11b expression as well as G-CSFR expression by Real-time RT-PCR analysis. The results are the means from three independent experiments, and error bars represent the standard deviation.
Acknowledgments
We thank Dr. Alan D. Friedman for BRM2 cell line, Dr. Giannino Del Sal, Dr. Lu KP and Dr. Bruno Calabretta for DNA constructs and Dr. George Bornkamm for valuable discussions. This work was supported by grants from Deutsche Josẻ Leukämie-Stiftung (F06/03) to J.A.P; National Institute of Health (R01 HL56745) to D.G.T and Deutsche Josẻ Leukämie-Stiftung, DFG and Krebshilfe to G.B.
Footnotes
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
References
- 1.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 2.Nerlov C. C/EBPalpha mutations in acute myeloid leukaemias. Nat Rev Cancer. 2004;4:394–400. doi: 10.1038/nrc1363. [DOI] [PubMed] [Google Scholar]
- 3.Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 4.Gombart AF, Hofmann WK, Kawano S, Takeuchi S, Krug U, Kwok SH, et al. Mutations in the gene encoding the transcription factor CCAAT/enhancer binding protein alpha in myelodysplastic syndromes and acute myeloid leukemias. Blood. 2002;99:1332–1340. doi: 10.1182/blood.v99.4.1332. [DOI] [PubMed] [Google Scholar]
- 5.Kirstetter P, Schuster MB, Bereshchenko O, Moore S, Dvinge H, Kurz E, et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13:299–310. doi: 10.1016/j.ccr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Geletu M, Balkhi MY, Peer Zada AA, Christopeit M, Pulikkan JA, Trivedi AK, et al. Target proteins of C/EBPalphap30 in AML: C/EBPalphap30 enhances sumoylation of C/EBPalphap42 via up-regulation of Ubc9. Blood. 2007;110:3301–3309. doi: 10.1182/blood-2007-01-071035. [DOI] [PubMed] [Google Scholar]
- 7.Lu KP. Prolyl isomerase Pin1 as a molecular target for cancer diagnostics and therapeutics. Cancer Cell. 2003;4:175–180. doi: 10.1016/s1535-6108(03)00218-6. [DOI] [PubMed] [Google Scholar]
- 8.Wulf G, Finn G, Suizu F, Lu KP. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nat Cell Biol. 2005;7:435–441. doi: 10.1038/ncb0505-435. [DOI] [PubMed] [Google Scholar]
- 9.Bao L, Kimzey A, Sauter G, Sowadski JM, Lu KP, Wang DG. Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am J Pathol. 2004;164:1727–1737. doi: 10.1016/S0002-9440(10)63731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, et al. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. Embo J. 2001;20:3459–3472. doi: 10.1093/emboj/20.13.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liou YC, Ryo A, Huang HK, Lu PJ, Bronson R, Fujimori F, et al. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc Natl Acad Sci U S A. 2002;99:1335–1340. doi: 10.1073/pnas.032404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wulf G, Garg P, Liou YC, Iglehart D, Lu KP. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. Embo J. 2004;23:3397–3407. doi: 10.1038/sj.emboj.7600323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 14.Rippmann JF, Hobbie S, Daiber C, Guilliard B, Bauer M, Birk J, et al. Phosphorylation-dependent proline isomerization catalyzed by Pin1 is essential for tumor cell survival and entry into mitosis. Cell Growth Differ. 2000;11:409–416. [PubMed] [Google Scholar]
- 15.Rinehart-Kim J, Johnston M, Birrer M, Bos T. Alterations in the gene expression profile of MCF-7 breast tumor cells in response to c-Jun. Int J Cancer. 2000;88:180–190. doi: 10.1002/1097-0215(20001015)88:2<180::aid-ijc6>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 16.Rangatia J, Vangala RK, Treiber N, Zhang P, Radomska H, Tenen DG, et al. Downregulation of c-Jun expression by transcription factor C/EBPalpha is critical for granulocytic lineage commitment. Mol Cell Biol. 2002;22:8681–8694. doi: 10.1128/MCB.22.24.8681-8694.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rangatia J, Vangala RK, Singh SM, Peer Zada AA, Elsasser A, et al. Elevated c-Jun expression in acute myeloid leukemias inhibits C/EBPalpha DNA binding via leucine zipper domain interaction. Oncogene. 2003;22:4760–4764. doi: 10.1038/sj.onc.1206664. [DOI] [PubMed] [Google Scholar]
- 18.Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 19.Porse BT, Pedersen TA, Xu X, Lindberg B, Wewer UM, Friis-Hansen L, et al. E2F repression by C/EBPalpha is required for adipogenesis and granulopoiesis in vivo. Cell. 2001;107:247–258. doi: 10.1016/s0092-8674(01)00516-5. [DOI] [PubMed] [Google Scholar]
- 20.Buchner T, Berdel WE, Schoch C, Haferlach T, Serve HL, Kienast J, et al. Double induction containing either two courses or one course of high-dose cytarabine plus mitoxantrone and postremission therapy by either autologous stem-cell transplantation or by prolonged maintenance for acute myeloid leukemia. J Clin Oncol. 2006;24:2480–2489. doi: 10.1200/JCO.2005.04.5013. [DOI] [PubMed] [Google Scholar]
- 21.D’Alo F, Johansen LM, Nelson EA, Radomska HS, Evans EK, Zhang P, et al. The amino terminal and E2F interaction domains are critical for C/EBP alpha-mediated induction of granulopoietic development of hematopoietic cells. Blood. 2003;102:3163–3171. doi: 10.1182/blood-2003-02-0479. [DOI] [PubMed] [Google Scholar]
- 22.Asou H, Gombart AF, Takeuchi S, Tanaka H, Tanioka M, Matsui H, et al. Establishment of the acute myeloid leukemia cell line Kasumi-6 from a patient with a dominant-negative mutation in the DNA-binding region of the C/EBPalpha gene. Genes Chromosomes Cancer. 2003;36:167–174. doi: 10.1002/gcc.10161. [DOI] [PubMed] [Google Scholar]
- 23.Schoch C, Kohlmann A, Schnittger S, Brors B, Dugas M, Mergenthaler S, et al. Acute myeloid leukemias with reciprocal rearrangements can be distinguished by specific gene expression profiles. Proc Natl Acad Sci U S A. 2002;99:10008–10013. doi: 10.1073/pnas.142103599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;4:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Wang QF, Cleaves R, Kummalue T, Nerlov C, Friedman AD. Cell cycle inhibition mediated by the outer surface of the C/EBPalpha basic region is required but not sufficient for granulopoiesis. Oncogene. 2003;22:2548–2557. doi: 10.1038/sj.onc.1206360. [DOI] [PubMed] [Google Scholar]
- 27.Ryo A, Liou YC, Wulf G, Nakamura M, Lee SW, Lu KP. PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells. Mol Cell Biol. 2002;22:5281–5295. doi: 10.1128/MCB.22.15.5281-5295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchida T, Takamiya M, Takahashi M, Miyashita H, Ikeda H, Terada T, et al. Pin1 and Par14 peptidyl prolyl isomerase inhibitors block cell proliferation. Chem Biol. 2003;10:15–24. doi: 10.1016/s1074-5521(02)00310-1. [DOI] [PubMed] [Google Scholar]
- 29.Gao M, Labuda T, Xia Y, Gallagher E, Fang D, Liu YC, et al. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- 30.Fang D, Kerppola TK. Ubiquitin-mediated fluorescence complementation reveals that Jun ubiquitinated by Itch/AIP4 is localized to lysosomes. Proc Natl Acad Sci U S A. 2004;101:14782–14787. doi: 10.1073/pnas.0404445101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 32.Niebuhr B, Iwanski GB, Schwieger M, Roscher S, Stocking C, Cammenga J. Investigation of C/EBPalpha function in human (versus murine) myelopoiesis provides novel insight into the impact of CEBPA mutations in acute myelogenous leukemia (AML) Leukemia. 2008;23:978–983. doi: 10.1038/leu.2008.332. [DOI] [PubMed] [Google Scholar]
- 33.Wang C, Chen X, Wang Y, Gong J, Hu G. C/EBPalphap30 plays transcriptional regulatory roles distinct from C/EBPalphap42. Cell Res. 2007;17:374–383. doi: 10.1038/sj.cr.7310121. [DOI] [PubMed] [Google Scholar]
- 34.Zhang P, Iwasaki-Arai J, Iwasaki H, Fenyus ML, Dayaram T, Owens BM, et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21:853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Ryo A, Liou YC, Lu KP, Wulf G. Prolyl isomerase Pin1: a catalyst for oncogenesis and a potential therapeutic target in cancer. J Cell Sci. 2003;116:773–783. doi: 10.1242/jcs.00276. [DOI] [PubMed] [Google Scholar]
- 36.Schuster MB, Porse BT. C/EBPalpha: a tumour suppressor in multiple tissues? Biochim Biophys Acta. 2006;1766:88–103. doi: 10.1016/j.bbcan.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 37.MacLachlan TK, Somasundaram K, Sgagias M, Shifman Y, Muschel RJ, Cowan KH, et al. BRCA1 effects on the cell cycle and the DNA damage response are linked to altered gene expression. J Biol Chem. 2000;275:2777–2785. doi: 10.1074/jbc.275.4.2777. [DOI] [PubMed] [Google Scholar]
- 38.Wooster R, Weber BL. Breast and ovarian cancer. N Engl J Med. 2000;348:2339–2347. doi: 10.1056/NEJMra012284. [DOI] [PubMed] [Google Scholar]
- 39.Miller M. Phospho-Dependent Protein Recognition Motifs Contained in C/EBP Family of Transcription Factors: in Silico Studies. Cell Cycle. 2006;5:2501–2508. doi: 10.4161/cc.5.21.3421. [DOI] [PubMed] [Google Scholar]
- 40.Porse BT, Bryder D, Theilgaard-Monch K, Hasemann MS, Anderson K, Damgaard I, et al. Loss of C/EBP alpha cell cycle control increases myeloid progenitor proliferation and transforms the neutrophil granulocyte lineage. J Exp Med. 2005;202:85–96. doi: 10.1084/jem.20050067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johansen LM, Iwama A, Lodie TA, Sasaki K, Felsher DW, Golub TR. c-Myc is a critical target for c/EBPalpha in granulopoiesis. Mol Cell Biol. 2001;21:3789–3806. doi: 10.1128/MCB.21.11.3789-3806.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castilla LH. C/EBPalpha in leukemogenesis: a matter of being in the right place with the right signals. Cancer Cell. 2008;13:289–291. doi: 10.1016/j.ccr.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 43.Vogt PK. Jun, the oncoprotein. Oncogene. 2001;20:2365–2377. doi: 10.1038/sj.onc.1204443. [DOI] [PubMed] [Google Scholar]
- 44.Elsasser A, Franzen M, Kohlmann A, Weisser M, Schnittger S, Schoch C, et al. The fusion protein AML1-ETO in acute myeloid leukemia with translocation t(8;21) induces c-jun protein expression via the proximal AP-1 site of the c-jun promoter in an indirect, JNK-dependent manner. Oncogene. 2003;22:5646–5657. doi: 10.1038/sj.onc.1206673. [DOI] [PubMed] [Google Scholar]
- 45.Burgess GS, Williamson EA, Cripe LD, Litz-Jackson S, Bhatt JA, Stanley K, et al. Regulation of the c-jun gene in p210 BCR-ABL transformed cells corresponds with activity of JNK, the c-jun N-terminal kinase. Blood. 1998;92:2450–2460. [PubMed] [Google Scholar]
- 46.Cai DH, Wang D, Keefer J, Yeamans C, Hensley K, Friedman AD. C/EBP alpha:AP-1 leucine zipper heterodimers bind novel DNA elements, activate the PU.1 promoter and direct monocyte lineage commitment more potently than C/EBP alpha homodimers or AP-1. Oncogene. 2008;27:2772–2779. doi: 10.1038/sj.onc.1210940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuler A, Schwieger M, Engelmann A, Weber K, Horn S, Muller U, et al. The MADS transcription factor Mef2c is a pivotal modulator of myeloid cell fate. Blood. 2008;111:4532–4541. doi: 10.1182/blood-2007-10-116343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
K562-C/EBPα-p30-ER and K562-C/EBPα-p42-ER cells were induced with ß-estradiol for 6 hours, cytoplasmic fraction (CF) and nuclear extract (NE) were prepared and analyzed for C/EBPα proteins by Western blot.
Kasumi-6 and NB4 cells were treated with ethanol or PiB (5 μM) for 6 days. Apoptotic cells were analysed by dual staing with FITC labeled Annexin- V and PI. Numbers in the lower right quadrant represents percentage of cells in early apoptosis (Annexin-V+ and PI−). Numbers in the top right quadrant represents percentage of cells in late apoptosis (Annexin-V+ and PI+).
293T cells were transfected with PIN1 and control siRNA. After 24 hours protein synthesis was blocked with cyclohexamide (CHX) and cells were harvested at different time points. The amount of c-Jun protein remaining was analyzed by Western blot followed by densitometric scanning.
(a) Kasumi-6 cells were transfected with PIN1 expression vector. Total RNA was isolated after 24 hrs with TRIZOL reagent and proceeded for reverse transcription followed by Real-time RT-PCR for c-Jun. Values were normalized with GAPDH mRNA. (b) Kasumi-6 cells were treated with PIN1 inhibitor, PiB for various time points and proceeded for Real-time RT-PCR for c-Jun as described. The results are the means from three independent experiments, and error bars represent the standard deviation.
Cord blood derived human CD34+ cells transfected with c-Jun or mock vectors. After 24 hours total RNA was isolated and analysed CD14 and M-CSFR mRNA by real time RT-PCR. The results are the means from three independent experiments, and error bars represent the standard deviation.
K562-C/EBPα-p30-ER and K562-C/EBPα-p42-ER cells were induced with ß-estradiol for 2 days and granulocytic differentiation was analysed by FACS analysis using CD11b antibody.
K562-C/EBPα-p30-ER cells were induced with ß-estradiol for respective time points and cells were lysed and subjected to Western blot analysis using PIN1 antibody. Values below the gel image indicate the upregulation (fold) of PIN1 protein level normalized to ß-Tubulin.
Kasumi-6 and NB4 cells were transfected with described DNA / siRNA and 24 hours later cells were lysed and subjected to Western blot analysis using PIN1 antibody. Values below the gel image indicate the upregulation (fold) of PIN1 protein level normalized to ß-Tubulin.
K562- C/EBPα-p30-ER cells were induced with ß-estradiol (5 μM) for respective time points to induce C/EBPα-p30 and total RNA was isolated at respective time points, proceeded for reverse transcription followed by quantitative real time PCR for PIN1.
(top) K562-C/EBPα-p30-ER cells were induced with ß-estradiol for respective time points and cells were lysed and subjected to Western blot analysis using E2F1 antibody. Values below the gel image indicate the upregulation (fold) of E2F1 protein level normalized to ß-Tubulin. (bottom) 293T cells were transfected with C/EBPα-p30. 12 and 14 hours after transfection, cells were lysed and subjected to Western blot analysis using E2F1 antibody. Values below the gel image indicate the upregulation (fold) of E2F1 protein level normalized to ß-Tubulin.
K562-C/EBPα-p30-BRM2-ER cells were induced with ß-estradiol for respective time points and cells were lysed and subjected to Western blot analysis using PIN1 antibody. Values below the gel image indicate the upregulation (fold) of PIN1 protein level normalized to ß-Tubulin.
C/EBPα-p30-ER cells were induced with ß-estradiol for respective time points and chromatin was immunoprecipitated with anti-E2F1 or IgG antibodies, and the recovered DNA was PCR amplified with primers specific for E2F1 binding sites in the c-Myc promoter.
NB4 cells were treated with PiB (5 μM) for 6 days and myeloid cell differentiation was assessed by FACS analysis using CD11b expression as well as G-CSFR expression by Real-time RT-PCR analysis. The results are the means from three independent experiments, and error bars represent the standard deviation.