Abstract
Bacillus anthracis is a National Institute of Allergy and Infectious Diseases Category A priority pathogen and the causative agent of the deadly disease anthrax. We applied a transposon mutagenesis system to screen for novel chromosomally encoded B. anthracis virulence factors. This approach identified ClpX, the regulatory ATPase subunit of the ClpXP protease, as essential for both the hemolytic and proteolytic phenotypes surrounding colonies of B. anthracis grown on blood or casein agar media, respectively. Deletion of clpX attenuated lethality of B. anthracis Sterne in murine subcutaneous and inhalation infection models, and markedly reduced in vivo survival of the fully virulent B. anthracis Ames upon intraperitoneal challenge in guinea pigs. The extracellular proteolytic activity dependent upon ClpX function was linked to degradation of cathelicidin antimicrobial peptides, a front-line effector of innate host defense. B. anthracis lacking ClpX were rapidly killed by cathelicidin and α-defensin antimicrobial peptides and lysozyme in vitro. In turn, mice lacking cathelicidin proved hyper-susceptible to lethal infection with wild-type B. anthracis Sterne, confirming cathelicidin to be a critical element of innate defense against the pathogen. We conclude that ClpX is an important factor allowing B. anthracis to subvert host immune clearance mechanisms, and thus represents a novel therapeutic target for prevention or therapy of anthrax, a foremost biodefense concern.
Key Words: Antimicrobial peptides, Bacillus anthracis, Bacterial infection, Cathelicidins, Hemolysis, Innate immunity, Protease, Transposon mutagenesis, Virulence factor
Introduction
Bacillus anthracis is a Gram-positive, spore-forming bacterium and the causative agent of anthrax. Primarily a disease of livestock, anthrax can infect humans through cutaneous, respiratory or gastrointestinal routes of infection. Inhalational anthrax occurs when endospores are introduced to the lung and taken up by resident phagocytes. Escape of B. anthracis from the phagolysosome into regional lymph nodes leads to extracellular proliferation and eventually to high bacterial titers in the blood, shock and death [1]. The high lethality of inhalational anthrax coupled with extended spore viability makes B. anthracis a paramount biodefense concern. This was illustrated in 2001 when a terrorist sent letters containing anthrax spores to government offices and news organizations in the USA, killing 5 people and infecting 17 others.
The best characterized virulence factors of B. anthracis are the anthrax toxins, comprised of lethal factor, edema factor, protective antigen and the antiphagocytic poly-D-glutamate capsule. The structural genes encoding the anthrax toxins and the biosynthetic genes for capsule synthesis are located on the large plasmids, pXO1 and pXO2 respectively, and loss of either plasmid attenuates B. anthracis virulence in certain animal models [2, 3]. While both anthrax toxin and capsule are critical in immune cell invasion and disease progression, evidence indicates that chromosome-encoded genes may also contribute to B. anthracis pathogenicity [4].
To identify novel chromosomal genes involved in virulence, we applied a transposon system using the Himar1 transposase to generate a random chromosomal mutant library of B. anthracis Sterne, then screened individual mutants for loss of hemolytic activity, a potential marker for bacterial cytotoxins. This strategy yielded a mutant in which the transposon insertion mapped to a locus coding for the ClpX ATPase.
Caseinolytic proteases (Clp) are conserved intracellular proteases that eliminate damaged, non-functional proteins and also control the lifespan of transcriptional regulators and rate-limiting enzymes [5]. The ClpXP protease is comprised of a proteolytic core, ClpP, that pairs with the regulatory ATPase ClpX and structurally resembles the eukaryotic 26s proteosome [6]. ClpX is responsible for recognition, unfolding and cleavage of substrate proteins into large peptides to be fed into the proteolytic core. Without ClpX, ClpP is limited to degradation of peptides fewer than 7 amino acids long [7]. ClpX can also function independently of ClpP as a molecular chaperone that assists in protein folding [8]. At least 5 classes of ClpX recognition signals have been identified. Substrates comprise a diverse array of proteins and include enzymes involved in gene regulation, metabolism and stress response [9]. In Bacillus subtilis, ClpX is required for initiation of sporulation [10].
ClpX function proved essential for B. anthracis β-hemolytic and extracellular proteolytic activities in plate-based assays. Further analysis of this mutant uncovered a role for ClpX in B. anthracis resistance to host antimicrobial peptide killing and virulence in several models of systemic anthrax infection.
Materials and Methods
Bacterial Strains and Culture Conditions
B. anthracis Sterne (pXO1+pXO2−) and Ames strains (pXO1+ pXO2+) were propagated in Brain-Heart Infusion (BHI) medium (Sigma) or on BHI agar plates under aerobic conditions at 37°C. When antibiotic selection was indicated, the following concentrations were used: 50 μg/ml of kanamycin (Kan), 5 μg/ml of erythromycin (Em), 3 μg/ml of chloramphenicol (Cm) and 1μg/ml of tetracycline (Tet).
Construction of B. anthracis Sterne Transposon Mutant Library
To develop a Himar1 transposon system for use in B. anthracis, we constructed a delivery plasmid that allowed expression of the Himar1 transposase gene and provided a selectable marker. First, a Himar1-based mini-transposon was constructed by cloning the Ω km-2 cassette from pUTE618 [11] into the unique BglII site in pMMOrf [12]. A hyperactive Himar1 transposase (tnp) gene from pET29C9 [12] was fused to the B. cereus promoter P4325[13]. Finally, the mini-Himar1 cassette and P4325-Himar1-tnp were cloned into pUTE583 [11] to generate pUTE664. The Himar-1 transposon delivery vector pUTE664 was electroporated into B. anthracis using an established protocol [14]. Electroporation mixtures were grown in medium containing Em to select for the transposon delivery plasmid and allow for chromosomal transposition events. Cells were subcultured 6 times without antibiotic selection to facilitate loss of the plasmid. Candidate chromosomal transposon insertion mutants were identified as B. anthracis colonies exhibiting kanamycin resistance and Em sensitivity. Southern blot analysis of selected colonies indicated a highly random distribution of single transposon insertions into the chromosomes of individual mutants. DNA flanking the transposon insertion site was identified by single-primer PCR using outwardly directed primer 5′-GGGAATCATTTGAAGGTT-3′, followed by direct DNA sequencing using the nested primer 5′-TATGCATTTAATACTAGCGACG-3′.
Targeted Allelic Replacement Mutagenesis and Complementation Studies
Approximately 1,000 bp of sequence immediately upstream of clpX was amplified with primers UpClpXFwd 5′-CATTAGGATCCATACATGCCAATCATTTCAATACC-3′ and UpClpXRev 5′-TGGCAGGGCGGGGCGTAATGCGAAGGAGAAACA-3′, while 1,000 bp of sequence immediately downstream of clpX was amplified with primers DnClpXFwd 5′-CCAGTGATTTT TTTCTCCATATTTTCACACCCCTTACA-3′ and DnClpXRev 5′-CATGACTCGAGCGGAAAAGGCGAAAACTACTCTC-3′. The UpClpXRev and DnClpXFwd primers were constructed with 25-bp 5′ extensions corresponding to the 5′ and 3′ ends of the cat gene. The upstream and downstream PCR products were then combined with a 650-bp amplicon of the complete cat gene as templates in a second round of PCR using primers UpClpXFwd and DnClpXRev. The resultant amplicon was sub-cloned into pHY304, a temperature-sensitive vector, and transformed into B. anthracis Sterne or Ames. Transformants were grown initially at 30°C and then shifted to 37°C under Em selection to identify single crossover events. Colonies exhibiting single crossovers in both orientations were grown overnight at 30°C in Cm, passed once at 30°C and then plated on BHI Cm plates at an appropriate dilution to generate single colonies and grown overnight at 37°C. Double crossover events were identified as colonies exhibiting Cm resistance but Em sensitivity, and were confirmed by PCR. For the complementation studies in B. anthracis Sterne, clpX plus flanking DNA was PCR amplified from chromosomal DNA using ClpXFwd 5′-ATGCAAGCTTACGACTTAATCCCGTCCTCTTGC-3′ and ClpXRev 5′-GTCAGAATTCCTGTAGACTTCTTAGTAGAAAACGCTGCA-3′ and cloned into the plasmid pUTE29 [14] to yield pClpX. The pClpX plasmid was introduced into B. anthracis Sterne by electroporation.
Hemolytic and Proteolytic Activity
Blood agar plates were prepared by adding 50 ml of washed human red blood cells to 1l of BHI medium containing 1.5% agar held at 50°C. B. anthracis was inoculated onto plates and incubated at 37°C under anaerobic conditions (generated by the AnaeroPouch System, Remel). A liquid phase hemolysis assay was performed as described [15]. To assess proteolytic activity, casein plates were prepared by adding 150 ml of sterile skim milk to 1 l of medium containing 18.5 g of Columbia broth (Acumedia) and 1.5% agar held at 50°C.B. anthracis inoculated onto casein plates were incubated at 37°C under aerobic conditions.
Semi-Quantitative RT-PCR
16-hour bacterial cultures were lysed using Lysing Matrix B beads (MPBiomedical) by pulsing 2 × 40 s in a mini-bead beater (Biospec). RNA was purified using the RNAeasy kit (Invitrogen) followed by DNase treatment using the Turbo DNA-free kit (Ambion). Equal amounts of RNA were reverse transcribed using the Superscript III kit (Invitrogen). PCR was performed using the Platinum Blue Supermix (Invitrogen) and was limited to 26 cycles to maintain semi-quantitative conditions. The following primers were used: ClpX Fwd 5′-gagcatcttcatcaagtggctctagg-3′; ClpX Rev 5′-gagtatgtcatcggacaagataacgcg-3′; Lon Fwd 5′-ccttgccttctttatcgccaagt-3′; Lon Rev 5′-gtacgcttcgtgtccttgtagaagg-3′; FusA Fwd 5′-aagctggtggtgctgaagcac-3′; FusA Rev 5′-ttcccaatcagcttctccttgaag-3′; LEF Fwd 5′-atatccagcataatcatccacagcat-3′; LEF Rev 5′-atggagaatccaattatcaccagatactc-3′; CYA Fwd 5′-taagcgatgatagtgatagtagcgacctt-3′; CYA Rev 5′-ttcatcgctaattctaaattcatatacctgat-3′.
Spore Isolation and Purification
Spores were grown on solid New Sporulation medium (3 g Tryptone, 6 g bacteriological peptone, 3 g yeast extract, 1.5 g Lab Lemco powder, 1 ml 0.1% MnCl2·4H2O, 25 g Bacto agar in 1 l H2O) at 37°C until cultures contained >95% free spores. Spores were scraped off, filtered to remove debris, harvested by centrifugation at 12,000 g for 20 min and washed in PBS to remove contaminating vegetative cells and debris. Microscopic examination established that spore preparations contained nearly 100% phase-bright spores.
Sporulation and Germination Assays
Sporulation was assessed in 2 ways. First, spores were visualized using phase-contrast microscopy after induction of sporulation for up to 7 days. Second, spore formation was quantified by inoculating B. anthracis Sterne into phage assay medium [16] for 24 h before heat-killing vegetative bacteria at 68°C for 40 min and then serially diluting and plating remaining heat-resistant spores. Germination was measured as a loss of heat resistance. Spores were heat activated to synchronize germination and the number of viable spores was determined by serial dilution. Spores were germinated for 20 min in BHI before heat-killing germinated bacteria and determining remaining heat-resistant spores. Percent germination was calculated as [1 – (number of survivors at Tt)/(number of survivors at T0)] × 100 [17].
Antimicrobial Peptide and Lysozyme Assays
Cultures were grown to early log phase, washed in PBS and resuspended in PBS to an optical density of 0.4 at wavelength 600 nm. Bacteria were diluted 1:10 in RPMI 1640 tissue culture medium (Gibco) plus 5% Luria-Bertani (LB) broth (Acumedia) containing either 16 μM of CRAMP or 0.5 μM of LL-37 and incubated at 37°C with nutation. For the EDTA assay, B. anthracis Sterne was incubated with 0.5 mM EDTA in addition to 0.5 μM LL-37. For the 1,10-phenanthroline assay, B. anthracis Sterne was incubated with 0.5 mM 1,10-phenanthroline (Sigma) in addition to 0.25 μM LL-37 or 0.25 μM LL-37 plus the vehicle control (methanol). Samples were incubated at 37°C with nutation and serial dilutions were plated at indicated time points to enumerate surviving cfu. For the HNP-2 and lysozyme assay, 10 μl of bacteria either diluted 1:100 in RPMI plus 5% LB (HNP-2 assay) or undiluted (lysozyme assay) was added to a 96-well plate containing 90 μl of 1.6 or 0.8 μM HNP-2 (Sigma) or 90 μl of 10 mg/ml or 5 mg/ml of lysozyme (Sigma) and allowed to grow for 18 h. Growth was assessed by visual inspection and quantified by reading the OD600. No difference was seen in growth or survival in RPMI + 5% LB without antimicrobial peptides (AMPs) or lysozyme (data not shown). Assays were repeated in triplicate at least twice. Results from 1 representative experiment are shown.
Cathelicidin LL-37 Degradation
B. anthracis Sterne were grown for 18 h in 5 ml of BHI. Culture supernatants were divided into tubes and incubated with LL-37 at a final concentration of 2 μM for 6 h at room temperature. As an activity control, some supernatants were boiled for 10 min prior to assay. At the assay endpoint, all samples were boiled for 5 min to stop the reaction, and subjected to 10% Bis-Tris electrophoresis in MES running buffer (Invitrogen) followed by Western blot analysis using an anti-LL-37 antibody.
Mouse Infection Studies
For the subcutaneous challenge, bacteria were grown to early log phase, washed in PBS and resuspended in PBS to an optical density of 0.4 at wavelength 600 nm. Vegetative bacteria were diluted in PBS to 1 × 106 cfu/ml and 0.1 ml was injected subcutaneously into the shaved hindflank of 8-week-old female C57Bl/6 mice (Charles River) or CRAMP−/− mice [18]. Mice were monitored for survival for 14 days after infection. In a separate experiment, mice were euthanized at day 3, and the site of subcutaneous injection was excised, homogenized and serially diluted to determine the number of surviving cfu. Histology studies were performed at the University of California San Diego Histopathology Core Facility (by N. Varki, Director). For inhalation challenge studies, spores were prepared as above, viable spore count determined by dilution plating, and ΔclpX mutant spores concentrated to normalize viable counts with parent strain spores. Spores were heat-activated at 65°C for 30 min and approximately 5 × 105 spores in 20 μl volume were administered i.n. to 8-week-old A/J mice (Jackson; n = 15, 9 with wild-type bacteria, 6 with mutant bacteria), under ketamine anesthesia. Survival was monitored for 12 days after infection.
Guinea Pig Infection Studies
For i.p. challenge, B. anthracis Ames or its isogenic ΔclpX mutant were grown overnight at 37°C in a media consisting of 50% FBS, 40% BHI and 0.8% sodium bicarbonate, which allowed equivalent growth of each strain but prevented sporulation and long-chain formation. Cells were then washed 3 times in PBS and suspended to approximately 1 × 106 cfu/ml. For single-strain challenges, cells of each strain were diluted to the appropriate concentration in PBS and doses of 102, 103 or 104 cfu were injected into the i.p. cavity of groups of 5 female Hartley guinea pigs (approx. 450 g each). Guinea pigs were monitored for survival up to 14 days after infection. For calculation of a competitive index for the ΔclpX mutant compared to the Ames parental strain, the parental and ΔclpX strains were mixed in a 1:1 ratio after washing and 200 μl (containing 1 × 105 cfu of each strain) was injected into the i.p. cavity of 3 female Hartley guinea pigs (approx. 450 g each). All 3 guinea pigs became moribund between 24 and 28 h after infection. The spleens of all 3 guinea pigs were removed and homogenized and the numbers of B. anthracis Ames and ΔclpX colonies were enumerated by differential plating. Competitive index was calculated as ΔclpX: Ames parent cfu recovered from the spleen divided by ΔclpX: Ames parent cfu in the inoculum. Competitive index of the ΔclpX mutant during growth in vitro was calculated similarly after growing each strain overnight and mixing mutant and parental cells in a 1:1 ratio, inoculating the mixture into BHI broth, incubating overnight at 37°C, and determining ΔclpX and parental cfu by differential plating.
Statistical Analysis
Kaplan-Meier survival plots were evaluated with the log-rank test. CRAMP, LL-37, HNP-2 and lysozyme assays were evaluated with a 1-way ANOVA followed by Tukey's post hoc test using GraphPad Prism software. All other statistics were evaluated by 2-tailed Student's t test using the Microsoft Excel statistical package.
Assurances
All animal experiments were approved by the committee on the use and care of animals, University of California San Diego, and performed using accepted veterinary standards. Experimental protocols were approved by the University of California San Diego biosafety committee. B. anthracis Ames experiments were conducted at the US Army Medical Research Institute of Infectious Diseases, which is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International, in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals.
Results
Nonhemolytic B. anthracis Mutant Identified through Mariner-Based Transposon Mutagenesis
Although not hemolytic on sheep blood, B. anthracis exhibits strong hemolytic activity on human blood agar plates under anaerobic conditions [19]. A candidate agent of this phenotype is the pore-forming cholesterol-dependent cytolysin anthrolysin O (ALO), which is known to lyse human blood cells in solution [15]. We created a precise allelic replacement of the alo gene in B. anthracis Sterne. While the resultant Δalo mutant indeed lost hemolytic activity in 1% human blood solution (fig. 1a), it retained parental levels of hemolytic activity on human blood agar plates grown under anaerobic conditions (fig. 1b). Since ALO was not responsible for the hemolytic phenotype of B. anthracis on blood agar, we sought other candidate genes involved in B. anthracis hemolysis by construction and screening of a random transposon mutant library.
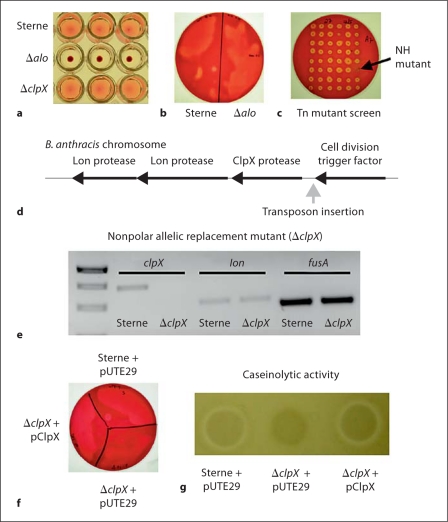
Fig. 1.
B. anthracis β-hemolytic and proteolytic phenotypes are dependent upon ClpX. Hemolytic activity of parent B. anthracis Sterne and the anthrolysin O (ALO)-deficient strain (Δalo) in 1% human blood solution (a) or 5% human blood agar plates incubated under anaerobic conditions (b). c Transposon mutants were screened for loss of hemolytic activity on human blood agar plates under anaerobic conditions. NH = Nonhemolytic. d Schematic representation of the site of transposon insertion within the B. anthracis chromosome. e Semi-quantitative RT-PCR showing expression of the clpX, lon and fusA genes from cDNA prepared from either parental B. anthracis Sterne (Sterne) or an isogenic ΔclpX allelic replacement mutant. f Human blood agar plate showing hemolytic activity of parent B. anthracis Sterne + empty pUTE29, the ΔclpX mutant + empty pUTE29, and the ΔclpX mutant + pUTE29 containing the clpX gene (pClpX). g Proteolytic activity of the same strains on casein agar plates.
We generated a transposon system using the Himar1 transposase gene that is part of the Mariner family of transposable elements. The Himar1 element does not need host factors for transposition and the only sequence-specific insertion requirement is the dinucleotide TA [20]. This makes it ideal for transposon mutagenesis in the AT-rich genome of B. anthracis[21, 22]. Southern blot analysis and DNA sequencing of several individual clones indicated the library to consist of highly random single insertions of the transposon dispersed throughout the B. anthracis chromosome (data not shown).
We screened approximately 5,000 transposon mutants for loss of hemolytic activity on human blood agar plates under anaerobic conditions and identified a nonhemolytic mutant (fig. 1c). We used single-primer PCR amplification to identify the B. anthracis chromosomal DNA sequences immediately flanking the transposon. With NCBI Blast and the National Microbial Pathogen Data Resource, we mapped the transposon insertion to a site immediately upstream of an open reading frame predicted to encode ClpX, the regulatory ATPase subunit of the ClpXP protease (fig. 1d).
B. anthracis ClpX Is Required for Hemolytic and Proteolytic Activities
To determine whether the loss of hemolytic activity in our transposon mutant was specifically attributable to ClpX, we performed precise, in-frame allelic replacement of clpX in B. anthracis Sterne, generating the isogenic mutant ΔclpX. In this procedure, the chloramphenicol acetyltransferase (cat) replacement cassette replaced clpX from the ATG start codon to stop codon without disrupting any flanking regulatory sequences and positioned in the same 5′→3′ orientation to allow read through.
To confirm that deletion of clpX and replacement with cat resulted in loss of clpX expression, but did not affect expression of the other downstream genes in the operon, we performed semi-quantitative RT-PCR for clpX and the lon protease transcripts; fusA, encoding a translation elongation factor, served as a control gene. As expected, in the ΔclpX mutant, clpX transcript cannot be detected; however, genes for both Lon proteases are expressed (fig. 1e, Lon protease gene furthest downstream is shown). To further confirm that deletion of clpX, and not a polar effect on downstream genes, was responsible for these phenotypes, we performed single-gene complementation analysis by inserting the clpX gene in the pUTE29 plasmid [14] to generate the plasmid pClpX. Parental Sterne and the ΔclpX isogenic mutant were also transformed with pUTE29 to control for any effects of the vector itself and are designated as Sterne + pUTE29 and ΔclpX + pUTE29. In contrast to the phenotypes of the Δalo mutant, deletion of clpX resulted in a loss of hemolytic activity on blood agar grown anaerobically (fig. 1f)but not in the liquid culture assay (fig. 1a). Moreover, a clear zone of proteolytic activity surrounding B. anthracis Sterne colonies grown on casein agar was absent in the isogenic ΔclpX mutant (fig. 1g). Complementation of the ΔclpX mutant with pClpX (ΔclpX + pClpX) restored hemolytic and proteolytic activities to those of the parent strain (fig. 1f, g). It is most likely that ClpX contributes indirectly to hemolytic or proteolytic activities through regulation of an extracellular hemolysin or protease, either through its role as the regulatory component of the intracellular ClpXP protease [5] or through its role as a molecular chaperone [8].
Effects of clpX Deletion on B. anthracis Growth, Morphology and Sporulation
No differences were observed for growth kinetics of any of the strains in the enriched media brain heart infusion (BHI) broth (fig. 2a), although we cannot exclude that more extensive testing to induce specific stresses (altered temperature, salt or nutrient deprivation) could reveal altered growth kinetics, as has been seen in other bacterial species [5]. Neither gross changes in vegetative morphology nor alterations in chain length were seen among the parent Sterne, the ΔclpX mutant (fig. 2b) or the complemented mutant (data not shown). Elimination of ClpX reduced sporulation efficiency, as substantially fewer spores were observed in the ΔclpX mutant using phase-bright microscopy even after the bacteria were allowed to sporulate for 7 days (data not shown). The number of heat-resistant spores formed by either the parent Sterne or ΔclpX mutant after 24 h were also quantified and loss of clpX resulted in approximately 100-fold reduction in B. anthracis sporulation efficiency (fig. 2c). Germination efficiency was also affected: after 20 min, nearly 90% of parental Sterne spores had germinated in BHI, as measured by a loss of heat resistance, versus only 60% of ΔclpX spores (fig. 2d). Complementation with pClpX restored both phenotypes.
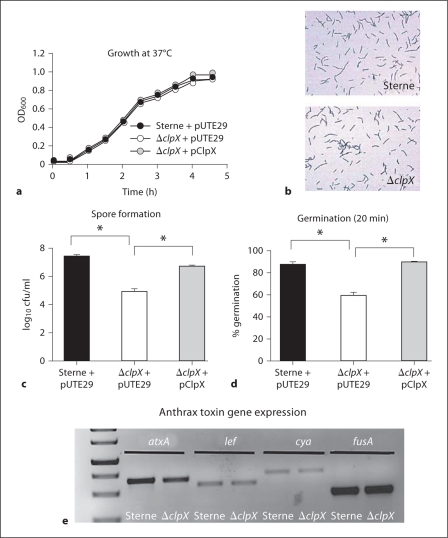
Fig. 2.
Contribution of ClpX to B. anthracis Sterne growth, vegetative morphology and sporulation. a Growth curve at 37°C in BHI for parental B. anthracis Sterne + pUTE29, the ΔclpX + pUTE29, and the ΔclpX complemented in trans by pClpX. b Gram stain of vegetative parent B. anthracis Sterne and ΔclpX in early log growth phase. Magnification × 100. c Sporulation efficiency of parental B. anthracis Sterne + pUTE29, the ΔclpX + pUTE29 and the ΔclpX + pClpX as determined by number of viable spores formed after 24 h growth under nutrient limiting conditions. d Using the same series of strains, germination of spores as determined by loss of heat resistance after 30 min growth in BHI. e Semi-quantitative RT-PCR showing expression of the atxA, lef, cya and fusA genes from cDNA prepared from either parent B. anthracis Sterne (Sterne) or the ΔclpX mutant.
ClpXP is known to degrade transcriptional regulators such as SigmaH [5], which controls the AtxA expression that in turn regulates toxin gene expression in B. anthracis[23]. We tested whether toxin gene expression is affected in the ΔclpX mutant using semi-quantitative RT-PCR. These studies reveal that the atxA, lef (lethal factor) and cya (edema factor) genes are all expressed in the ΔclpX mutant at levels similar to the parent strain Sterne (fig. 2e).
ClpX Promotes Resistance to Cathelicidin Antimicrobial Peptide Killing
In systemic B. anthracis infection, replication in the host demonstrates the organism's capacity to avoid clearance by host innate immune defenses. These defenses include cationic AMPs that disrupt the bacterial membrane. However, many pathogenic bacteria display their own set of defenses against AMPs, including degradation of AMPs with bacterial proteases [24]. Since the ΔclpX mutant lacked proteolytic activity present in the parent strain, we hypothesized that this could make the mutant more susceptible to killing by host AMP-based defenses.
Cathelicidins are a family of mammalian AMPs expressed by epithelial cells and neutrophils that play a critical role in defense against invasive infection by the leading Gram-positive human pathogen, Streptococcus pyogenes[18], and exhibit activity against B. anthracis[25, 26]. Within 15 min of incubation with murine cathelicidin CRAMP, a 100-fold reduction in survival of the ΔclpX mutant compared to the wild-type parent strain was observed, and by 30 min all ΔclpX mutant B. anthracis were killed by the host defense peptide (fig. 3a), although no difference was seen in control media alone (data not shown). Complementation of the ΔclpX mutant with the wild-type clpX gene partially restored cathelicidin resistance (fig. 3a). The ΔclpX mutant also exhibited 50% increased susceptibility to killing with the human cathelicidin LL-37 (p < 0.01); once again complementation partially restored resistance (fig. 3b). Although the unencapsulated Sterne strain was used in these experiments, recent studies have demonstrated that encapsulation does not prevent the antimicrobial activity of cathelicidins [25, 26].
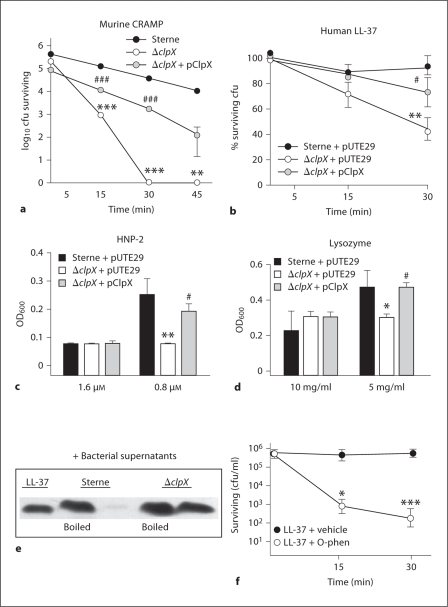
Fig. 3.
ClpX function protects B. anthracis Sterne against antimicrobial peptide killing. a Killing kinetics of parent Sterne, the ΔclpX mutant, and ΔclpX + pClpX incubated with 16 μM of murine cathelicidin CRAMP. b Survival of parent Sterne + pUTE29, the isogenic ΔclpX mutant + pUTE29, or ΔclpX complemented by pClpX incubated with 0.5 μM of human cathelicidin LL-37. c Growth of parent Sterne + pUTE29, the isogenic ΔclpX mutant + pUTE29 or ΔclpX complemented by pClpX in 0.8 or 1.6 μM of human α-defensin-2 (HNP-2). d Growth of parent Sterne + pUTE29, the isogenic ΔclpX mutant + pUTE29 or the ΔclpX complemented by pClpX in 5 mg/ml or 10 mg/ml μM of lysozyme. e Western blot analysis of LL-37 (2 μM final concentration) exposed to culture supernatants from B. anthracis Sterne or its isogenic ΔclpX mutant. f B. anthracis Sterne incubated with either 0.25 μM LL-37 or 0.25 μM LL-37 plus 0.5 m M o-phenanthroline (o-phen). Statistically significant changes between parent Sterne and the ΔclpX mutant: ∗p < 0.05; ∗∗ p < 0.01; ∗∗∗ p < 0.001. Statistically significant differences between complemented strain and ΔclpX mutant: # p < 0.05 or ### p < 0.001.
To determine whether the resistance mediated by ClpX was restricted to cathelicidins, sensitivity to another class of mammalian antimicrobial peptides, the defensins, was assessed. Like cathelicidins, defensins are small cationic peptides produced by leukocytes and epithelial cells [27]. Growth of the ΔclpX mutant was inhibited by human α-defensin-2 (HNP-2) compared to the parental Sterne strain (p < 0.01), and the wild-type growth level was restored by complementation with clpX on a plasmid vector (fig. 3c); growth in media alone was comparable for all strains (data not shown). Doses for all 3 AMPs tested were within the physiological levels generated by induction through infection or inflammation [28, 29]. The ΔclpX mutant did not differ from the parent strain in susceptibility to a cationic AMP of bacterial origin, polymyxin B (data not shown). Lastly, we tested another host antimicrobial factor, lysozyme, a muramidase that degrades bacterial cell walls. The ΔclpX mutant was again inhibited in comparison to the parent Sterne or the complemented strain (fig. 3d) demonstrating that several areas of bacterial resistance against innate immune factors are affected by the loss of clpX.
It is unlikely that ClpX directly degrades AMPs. Rather, ClpX probably indirectly affects AMP resistance by mediating expression of an extracellular protease that is directly acting on AMPs. To ascertain if bacterial extracellular proteolytic activity per se was responsible for inactivation of the cathelicidin peptide, we performed Western blot analysis of human LL-37 exposed to fresh culture supernatants prepared from the parent or isogenic ΔclpX mutant strains. Degradation of the LL-37 peptide was observed following exposure to the parent B. anthracis supernatant, but this activity was absent in culture supernatant of the ΔclpX mutant (fig. 3e). Extracellular metalloproteases are known to contribute to the caseinolytic and collagenolytic activities of B. anthracis[30] and there is evidence that metalloproteases may protect B. anthracis from killing by LL-37 [25]. Therefore, we hypothesized that metalloprotease inhibitors would increase sensitivity of the bacterium to AMP killing. When bacteria were incubated with 5 mM of the metalloprotease inhibitor 1,10-phenanthroline (also known as o-phenanthroline), they became much more sensitive to LL-37 killing (fig. 3g); no differences were seen between the vehicle control and o-phenanthroline or LL-37 alone at the time points and concentrations used (data not shown). A similar effect to increase B. anthracis sensitivity to LL-37 killing was seen upon addition of EDTA to chelate metal ions required for metalloprotease activity (data not shown)
ClpX is Required for B. anthracis Virulence
As ClpX was found to be important for B. anthracis hemolytic activity and antimicrobial peptide resistance in vitro, we hypothesized ClpX could promote anthrax disease progression in vivo through membrane injury to host cells and/or innate immune resistance. Beginning with a subcutaneous infection model, we injected 1 × 105 cfu from an early log-phase culture of the parent strain, the ΔclpX mutant, or the complemented mutant into the flank of 8-week-old female C57Bl6 mice. Between days 3 and 5 after challenge, all mice injected with the parent strain became moribund, whereas all mice injected with the ΔclpX mutant survived (fig. 4a). Complementation of the mutant with the clpX gene in trans partially restored virulence with 40% of mice succumbing to infection (fig. 4a). The lack of full complementation in this as well as the antimicrobial peptide assays may have several causes. It is possible that inappropriately high levels of ClpX, as may be seen using a multi-copy complementation plasmid, are detrimental to overall bacterial survival function, although less so than complete loss of ClpX. Another possibility is that loss of the plasmid during the course of infection may have contributed to the delayed time to death in the mice injected with the complemented strain. Increased mouse survival was associated with decreased survival of the B. anthracis ΔclpX mutant in vivo, since at day 3 after challenge 100-fold fewer bacterial cfu were recovered from the injection site of mice inoculated with the mutant than those inoculated with the parent strain (fig. 4b). Similarly, many fewer bacteria were observed on Gram stain of the inoculation site lesions of animals challenged with the ΔclpX mutant compared to the parent strain (fig. 4c, left). As seen in hematoxylin and eosin (HE) stain of the skin lesions (fig. 4c, right), an inflammatory response was elicited in both parent- and ΔclpX-infected mice, but the amount and magnitude of tissue necrosis found at the injection site was greatly diminished in those mice infected with the mutant strain (fig. 4c).
Fig. 4.
ClpX is required for B. anthracis Sterne virulence. a Kaplan-Meyer survival curve of C57Bl/6 mice after s.c. injection with parent Sterne (11 mice), ΔclpX (11 mice) or ΔclpX + pClpX (10 mice). b Enumeration of surviving cfu of parent or ΔclpX mutant at the site of inoculation 3 days after s.c. challenge. c Histopathologic analysis of lesional biopsies by Gram stain (left) and hematoxylin-eosin (HE) stain (right) from mice challenged s.c. with parent B. anthracis Sterne or ΔclpX mutant 3 days after challenge. d Kaplan-Meyer survival curve of A/J mice following inhalation of parent B. anthracis Sterne (9 mice) or ΔclpX mutant (6 mice); B. anthracis spores administered intranasally.
The spore is the infectious agent of B. anthracis and spore inhalation triggers the most lethal form of disease produced by the pathogen. To determine the contribution of ClpX to B. anthracis virulence upon inhalational challenge, we delivered parent or ΔclpX spores to A/J mice by intranasal inoculation. Once again, while 90% of B. anthracis Sterne-infected mice succumbed to the spore inhalation challenge, 100% of mice infected with the isogenic ΔclpX mutant survived (fig. 4d), illustrating a second infection model in which no mortality resulted following loss of clpX expression.
Cathelicidin Contributes to Innate Immune Defense against B. anthracis
Our in vitro data (fig. 3a, b, e, f) suggested that enhanced resistance to cathelicidin antimicrobial peptide killing could be 1 of several underlying mechanisms by which ClpX promotes B. anthracis virulence in vivo, along with the observed proteolytic/hemolytic injury to host cells and defensin resistance. This hypothesis requires that cathelicidin indeed serves a critical innate immune defense function limiting B. anthracis pathogenicity. To examine this concept directly, we s.c. injected a lower dose (8 × 103 cfu) of B. anthracis Sterne from an early log-phase culture into the flank of male CRAMP−/− knockout mice or age-matched C57Bl6 wild-type controls. In this challenge experiment, 75% of knock-out mice (3 of 4) died versus only 10% of wild-type controls (1 of 10; p = 0.01; fig. 5a). Erythematous and swollen skin lesions developed at the injection site in all 4 CRAMP−/− mice, even the 1 that survived (fig. 5b, c). In contrast, 9 of 10 survivors among the wild-type mice developed no lesions, swelling or erythema at the injection site; this finding was only present to a reduced degree in the single wild-type mouse that died following low-dose challenge (fig. 5b, c). This study confirms a role for the murine cathelicidin in defense against B. anthracis.
Fig. 5.
Cathelicidin contributes to innate immune defense against B. anthracis. a Kaplan-Meyer survival curve of wild type (WT; 10 mice) and CRAMP−/− (4 mice) challenged subcutaneously s.c. with B. anthracis Sterne. b Magnitude of inflammatory changes on gross examination of site of s.c. inoculation in wild-tpye and CRAMP−/− mice 5 days after s.c. challenge with wild-type B. anthracis. 0 = Absent; + = trace; ++ = mild; +++ = moderate; ++++ = severe. c Representative images of individual mice.
ClpX Contributes to in vivo Survival of Fully Virulent B. anthracis Ames
To this point, our studies of B. anthracis ClpX function were conducted under BSL-2 conditions using the B. anthracis Sterne strain, which is attenuated due to the absence of pXO2 encoding poly-D-glutamate capsule synthesis. To ascertain the requirement of ClpX for the pathogenicity of encapsulated B. anthracis, we reproduced the precise, targeted allelic replacement of the clpX gene in B. anthracis Ames (pXO1+, pXO2+). Again as with clpX deletion in B. anthracis Sterne, no differences in chain length, morphology or growth in BHI media were observed in the Ames ΔclpX mutant; however, sporulation of this mutant was completely abrogated compared to the partial sporulation defect observed with the Sterne ΔclpX mutant (data not shown).
Virulence of the ΔclpX B. anthracis Ames derivative was assessed in 2 ways. First, the ability of the mutant to compete with the parental strain during the infection, as a measure of its relative fitness in the host, was assessed with a mixed infection of the 2 strains. Secondly, the ability of the ΔclpX mutant to cause lethality in the absence of the parental strain was assayed and subsequent determination of the mean time to death after challenge. Since mice are extremely sensitive to the encapsulated strain, guinea pigs are the standard animal model for work done with the fully virulent strains and were used in both in vivo experiments.
To analyze the relative fitness of the ΔclpX mutant by competitive index, vegetative cells of wild-type and isogenic ΔclpX B. anthracis Ames were mixed in a 1:1 ratio (105 cfu of each) and introduced by i.p. injection into guinea pigs. Spleens were harvested from moribund animals and the relative ratio of ΔclpX mutant to wild-type Ames bacilli calculated by differential plating. The competitive index of the ΔclpX mutant strain was 0.007 (p < 0.0002), indicating a greater than 100-fold decrease in fitness compared to the parental B. anthracis Ames strain (fig. 6a). This decrease in fitness was specific to the host environment since the competitive index of the ΔclpX mutant grown in BHI was 0.97 (fig. 6a). The capacity of the ΔclpX mutant to initiate a productive infection was then observed directly by single strain challenges using a dose of 102, 103 or 104 cfu of either the mutant or parental strain. 100% of the guinea pigs challenged with 103 or 104 cfu and 60% of the guinea pigs challenged with 102 cfu of the parental B. anthracis Ames strain died within 6 days of inoculation. In contrast, 100% of the guinea pigs challenged with 102 or 103 cfu and 80% of the guinea pigs challenged with 104 cfu of the ΔclpX derivative survived throughout the course of the experiment (fig. 6b; p < 0.003 for 104 dose, p < 0.002 for 103 dose, p = 0.05 for 102 dose). These results confirm a significant role for ClpX in pathogenesis of a fully virulent, encapsulated B. anthracis strain.
Fig. 6.
ClpX contributes to in vivo survival of fully virulent B. anthracis Ames. a Competitive index of the ΔclpX mutant compared to the parental strain, B. anthracis Ames, grown overnight in BHI or after infection in guinea pigs. Results are the mean ± standard deviation of 3 overnight growth experiments or 3 spleens. b Kaplan-Meier survival curves of guinea pigs after i.p. injection with B. anthracis Ames or the isogenic ΔclpX mutant. For statistical analysis, p values are positioned below the curves corresponding to animals given B. anthracis Ames and indicate the significance by log-rank test between the outcomes of that group and the group given a similar dose of the ΔclpX mutant.
Discussion
We have identified a role for the ClpX, in B. anthracis β-hemolytic and proteolytic phenotypes, germination and sporulation, and resistance to cathelicidin and defensin antimicrobial peptides. As a consequence, ClpX promotes B. anthracis virulence in s.c., i.p. and inhalational challenge models. After the manganese transporter MntA [31], ClpX is only the second chromosome-encoded gene product shown to have a role in virulence in fully virulent (pXO1+, pXO2+) B. anthracis.
ClpX expression in B. anthracis is present throughout the life cycle, peaking 1–3 h after sporulation, along with other proteins associated with rapid growth and response to toxic environments [32]. ClpX likely regulates a variety of proteins within B. anthracis. Loss of clpX does not affect growth in BHI, gross morphological appearance or toxin gene expression. It does, however, affect a number of other phenotypes including hemolytic and proteolytic activity, sporulation and germination, sensitivity to antimicrobial peptides and lysozyme, and overall virulence of the pathogen. ClpX has also been demonstrated to be important for sporulation in B. subtilis[10].
The precise mechanism for the loss of hemolytic and proteolytic activity in ClpX-deficient B. anthracis is not known. Distinct from ALO, our observed hemolytic phenotype is absent in liquid culture, distinct on solid blood agar, and accentuated under anaerobic growth. The zone of hemolysis associated with colonies of parent, ΔclpX mutant and complemented B. anthracis strains mirrors the zone of proteolysis produced on casein agar plates. It is possible that these are 2 independent activities under the control of ClpX. However, another possibility is that the hemolytic activity is a ‘pseudo-hemolysin’ phenotype attributable to proteolytic action.
Limited prior data are available on mechanisms of B. anthracis sensitivity or resistance to mammalian AMPs. A B. anthracis Sterne dlt operon mutant lacking D-alanyl modification of cell wall teichoic acids is rapidly killed by certain AMPs including the human defensins HNP-1, HNP-2 and HBD-2 [33], and it is possible that this sensitivity profile would extend to cathelicidin peptides. Several strains of B. anthracis (including Sterne and Ames) were observed to be more resistant to human cathelicidin LL-37 than other Bacillus species [25]. In this study, a chromosomally-encoded AMP neutralizing activity in B. anthracis culture supernatants was postulated, since it was present in the UM23-C12 strain lacking both pXO1 and pXO2 [25]. This likely corresponds to the extracellular protease activity lost in our ΔclpX mutant.
We hypothesize that ClpX increases expression of extracellular protease(s) and perhaps hemolysin(s) through regulation of a transcriptional or post-transcriptional regulator. ClpX could accomplish this either through its role in protein folding and activation as a molecular chaperone or via directed proteolysis in conjunction with ClpP. Precedent for both these models exists in Staphylococcus aureus, where ClpX has shown to effect transcriptional regulators by itself and as part of the ClpXP protease. This in turn regulates expression levels of extracellular proteases, α-hemolysins and the important surface protein, Protein A [34, 35]. Our observation also sheds light on an interesting finding that animals infected with B. anthracis and treated with a combination of antibiotics and metalloprotease inhibitors showed improved survival compared to those treated with antibiotics alone [30].
Exogenously administered cathelicidins can be used therapeutically in a B. anthracis spore infection model [26]. Here we directly demonstrate the critical function of these defense peptides in innate immunity to anthrax using cathelicidin-deficient mice. Previous work in the knockout mouse model has identified roles for cathelicidin in restricting bacterial infection of the skin [18], conjunctivae [36], gastrointestinal tract [37], urinary tract [38] and bloodstream [39]. Ours is the first study to demonstrate that host expression of cathelicidin protects against mortality in a lethal infectious challenge model.
Our data strongly suggest the marked attenuation of ΔclpX mutant in vivo is in significant part due to increased sensitivity to host AMPs. However, given the pleiotropic effects of ClpX, it is likely that additional systems contributing to survival within the host are also affected. The ΔclpX mutant is more susceptible to the lysozyme and this could be due to subtle differences in cell wall constitution and integrity rather the lack of extracellular proteolytic activity. Furthermore, the extracellular protease activity controlled by ClpX could produce direct cellular and tissue injury, as suggested by its toxicity to erythrocyte membranes. Finally, in the case of the murine inhalational challenge model, delays in the kinetics of spore germination may synergize with AMP sensitivity to attenuate virulence of the ΔclpX mutant.
The ClpXP proteases are highly conserved among bacteria and have a diverse array of functions. Although initially identified for their role in degrading cytosolic proteins in E. coli, our studies join other work in highlighting their contribution to the virulence of important pathogens [5]. A new class of antibiotics, acyldepsipeptides, specifically targets and dysregulates this proteolytic complex [40], leading to unregulated proteolysis that is lethal to cells. Our data suggest that ClpX may represent a novel target for therapy of B. anthracis infection, but through a different mechanism. Effective neutralization of B. anthracis ClpX activity would serve not to directly kill the pathogen, but rather to render it exquisitely susceptible to clearance by the normal innate immune system.
Acknowledgements
This research was supported by NIH grants AR052728 (R.L.G., V.N.), AI52453 (R.L.G., V.N.), AI065993 (R.V.A., V.N.) and AI33537 (T.M.K.). S.M.M. was supported by an IRACDA postdoctoral fellowship from NIGMS (GM068524) and a Biomedical Research Fellowship from The Hartwell Foundation. The opinions, interpretations, conclusions and recommendations in this paper are those of the authors and are not necessarily endorsed by the US Army.
References
- 1.Dixon TC, Meselson M, Guillemin J, Hanna PC. Anthrax. N Engl J Med. 1999;341:815–826. doi: 10.1056/NEJM199909093411107. [DOI] [PubMed] [Google Scholar]
- 2.Banks DJ, Ward SC, Bradley KA. New insights into the functions of anthrax toxin. Expert Rev Mol Med. 2006;8:1–18. doi: 10.1017/S1462399406010714. [DOI] [PubMed] [Google Scholar]
- 3.Ezzell JW, Welkos SL. The capsule of Bacillus anthracis, a review. J Appl Microbiol. 1999;87:250. doi: 10.1046/j.1365-2672.1999.00881.x. [DOI] [PubMed] [Google Scholar]
- 4.Read TD, Peterson SN, Tourasse N, Baillie LW, Paulsen IT, Nelson KE, Tettelin H, Fouts DE, Eisen JA, Gill SR, Holtzapple EK, Okstad OA, Helgason E, Rilstone J, Wu M, Kolonay JF, Beanan MJ, Dodson RJ, Brinkac LM, Gwinn M, DeBoy RT, Madpu R, Daugherty SC, Durkin AS, Haft DH, Nelson WC, Peterson JD, Pop M, Khouri HM, Radune D, Benton JL, Mahamoud Y, Jiang L, Hance IR, Weidman JF, Berry KJ, Plaut RD, Wolf AM, Watkins KL, Nierman WC, Hazen A, Cline R, Redmond C, Thwaite JE, White O, Salzberg SL, Thomason B, Friedlander AM, Koehler TM, Hanna PC, Kolsto AB, Fraser CM. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature. 2003;423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- 5.Frees D, Savijoki K, Varmanen P, Ingmer H. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol Microbiol. 2007;63:1285–1295. doi: 10.1111/j.1365-2958.2007.05598.x. [DOI] [PubMed] [Google Scholar]
- 6.Kessel M, Maurizi MR, Kim B, Kocsis E, Trus BL, Singh SK, Steven AC. Homology in structural organization between E. coli ClpAP protease and the eukaryotic 26 S proteasome. J Mol Biol. 1995;250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- 7.Woo KM, Chung WJ, Ha DB, Goldberg AL, Chung CH. Protease Ti from Escherichia coli requires ATP hydrolysis for protein breakdown but not for hydrolysis of small peptides. J Biol Chem. 1989;264:2088–2091. [PubMed] [Google Scholar]
- 8.Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J. 1995;14:1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Cosby WM, Zuber P. Role of Lon and ClpX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation in Bacillus subtilis. Mol Microbiol. 1999;33:415–428. doi: 10.1046/j.1365-2958.1999.01489.x. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Tenover FC, Koehler TM. Beta-lactamase gene expression in a penicillin-resistant Bacillus anthracis strain. Antimicrob Agents Chemother. 2004;48:4873–4877. doi: 10.1128/AAC.48.12.4873-4877.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ, Robertson HM. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc Natl Acad Sci USA. 1999;96:11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn AK, Handelsman J. A vector for promoter trapping in Bacillus cereus. Gene. 1999;226:297–305. doi: 10.1016/s0378-1119(98)00544-7. [DOI] [PubMed] [Google Scholar]
- 14.Koehler TM, Dai Z, Kaufman-Yarbray M. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J Bacteriol. 1994;176:586–595. doi: 10.1128/jb.176.3.586-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shannon JG, Ross CL, Koehler TM, Rest RF. Characterization of anthrolysin O, the Bacillus anthracis cholesterol-dependent cytolysin. Infect Immun. 2003;71:3183–3189. doi: 10.1128/IAI.71.6.3183-3189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green BD, Battisti L, Koehler TM, Thorne CB, Ivins BE. Demonstration of a capsule plasmid in Bacillus anthracis. Infect Immun. 1985;49:291–297. doi: 10.1128/iai.49.2.291-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholson WL, Setow P. Sporulation, germination and outgrowth. In: Harwood CR, Cutting SM, editors. Molecular biological methods for Bacillus. Chichester: John Wiley & Sons; 1990. pp. 391–450. [Google Scholar]
- 18.Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- 19.Klichko VI, Miller J, Wu A, Popov SG, Alibek K. Anaerobic induction of Bacillus anthracis hemolytic activity. Biochem Biophys Res Commun. 2003;303:855–862. doi: 10.1016/s0006-291x(03)00440-6. [DOI] [PubMed] [Google Scholar]
- 20.Lampe DJ, Grant TE, Robertson HM. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics. 1998;149:179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tam C, Glass EM, Anderson DM, Missiakas D. Transposon mutagenesis of Bacillus anthracis strain Sterne using Bursa aurealis. Plasmid. 2006;56:74–77. doi: 10.1016/j.plasmid.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Wilson AC, Perego M, Hoch JA. New transposon delivery plasmids for insertional mutagenesis in Bacillus anthracis. J Microbiol Methods. 2007;71:332–335. doi: 10.1016/j.mimet.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadjifrangiskou M, Chen Y, Koehler TM. The alternative sigma factor SigmaH is required for toxin gene expression by Bacillus anthracis. J Bacteriol. 2007;189:1874–1883. doi: 10.1128/JB.01333-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peschel A. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 2002;10:179–186. doi: 10.1016/s0966-842x(02)02333-8. [DOI] [PubMed] [Google Scholar]
- 25.Thwaite JE, Hibbs S, Titball RW, Atkins TP. Proteolytic degradation of human antimicrobial peptide LL-37 by Bacillus anthracis may contribute to virulence. Antimicrob Agents Chemother. 2006;50:2316–2322. doi: 10.1128/AAC.01488-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lisanby MW, Swiecki MK, Dizon BL, Pflughoeft KJ, Koehler TM, Kearney JF. Cathelicidin administration protects mice from Bacillus anthracis spore challenge. J Immunol. 2008;181:4989–5000. doi: 10.4049/jimmunol.181.7.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klotman ME, Chang TL. Defensins in innate antiviral immunity. Nat Rev Immunol. 2006;6:447–456. doi: 10.1038/nri1860. [DOI] [PubMed] [Google Scholar]
- 28.Kurosaka K, Chen Q, Yarovinsky F, Oppenheim JJ, Yang D. Mouse cathelin-related antimicrobial peptide chemoattracts leukocytes using formyl peptide receptor-like 1/mouse formyl peptide receptor-like 2 as the receptor and acts as an immune adjuvant. J Immunol. 2005;174:6257–6265. doi: 10.4049/jimmunol.174.10.6257. [DOI] [PubMed] [Google Scholar]
- 29.Panyutich AV, Panyutich EA, Krapivin VA, Baturevich EA, Ganz T. Plasma defensin concentrations are elevated in patients with septicemia or bacterial meningitis. J Lab Clin Med. 1993;122:202–207. [PubMed] [Google Scholar]
- 30.Popov SG, Popova TG, Hopkins S, Weinstein RS, MacAfee R, Fryxell KJ, Chandhoke V, Bailey C, Alibek K. Effective antiprotease-antibiotic treatment of experimental anthrax. BMC Infect Dis. 2005;5:25. doi: 10.1186/1471-2334-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gat O, Mendelson I, Chitlaru T, Ariel N, Altboum Z, Levy H, Weiss S, Grosfeld H, Cohen S, Shafferman A. The solute-binding component of a putative Mn(II) abc transporter (MntA) is a novel Bacillus anthracis virulence determinant. Mol Microbiol. 2005;58:533–551. doi: 10.1111/j.1365-2958.2005.04848.x. [DOI] [PubMed] [Google Scholar]
- 32.Bergman NH, Anderson EC, Swenson EE, Niemeyer MM, Miyoshi AD, Hanna PC. Transcriptional profiling of the Bacillus anthracis life cycle in vitro and an implied model for regulation of spore formation. J Bacteriol. 2006;188:6092–6100. doi: 10.1128/JB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fisher N, Shetron-Rama L, Herring-Palmer A, Heffernan B, Bergman N, Hanna P. The dltABCD operon of Bacillus anthracis Sterne is required for virulence and resistance to peptide, enzymatic, and cellular mediators of innate immunity. J Bacteriol. 2006;188:1301–1309. doi: 10.1128/JB.188.4.1301-1309.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frees D, Qazi SN, Hill PJ, Ingmer H. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol Microbiol. 2003;48:1565–1578. doi: 10.1046/j.1365-2958.2003.03524.x. [DOI] [PubMed] [Google Scholar]
- 35.Frees D, Sorensen K, Ingmer H. Global virulence regulation in Staphylococcus aureus: Pinpointing the roles of ClpP and ClpX in the sar/agr regulatory network. Infect Immun. 2005;73:8100–8108. doi: 10.1128/IAI.73.12.8100-8108.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang LC, Reins RY, Gallo RL, McDermott AM. Cathelicidin-deficient (CnlP–/–) mice show increased susceptibility to Pseudomonas aeruginosa keratitis. Invest Ophthalmol Vis Sci. 2007;48:4498–4508. doi: 10.1167/iovs.07-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iimura M, Gallo RL, Hase K, Miyamoto Y, Eckmann L, Kagnoff MF. Cathelicidin mediates innate intestinal defense against colonization with epithelial adherent bacterial pathogens. J Immunol. 2005;174:4901–4907. doi: 10.4049/jimmunol.174.8.4901. [DOI] [PubMed] [Google Scholar]
- 38.Chromek M, Slamova Z, Bergman P, Kovacs L, Podracka L, Ehren I, Hokfelt T, Gudmundsson GH, Gallo RL, Agerberth B, Brauner A. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat Med. 2006;12:636–641. doi: 10.1038/nm1407. [DOI] [PubMed] [Google Scholar]
- 39.Bergman P, Johansson L, Wan H, Jones A, Gallo RL, Gudmundsson GH, Hokfelt T, Jonsson AB, Agerberth B. Induction of the antimicrobial peptide cramp in the blood-brain barrier and meninges after meningococcal infection. Infect Immun. 2006;74:6982–6991. doi: 10.1128/IAI.01043-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brotz-Oesterhelt H, Beyer D, Kroll HP, Endermann R, Ladel C, Schroeder W, Hinzen B, Raddatz S, Paulsen H, Henninger K, Bandow JE, Sahl HG, Labischinski H. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat Med. 2005;11:1082–1087. doi: 10.1038/nm1306. [DOI] [PubMed] [Google Scholar]