Summary
The Fanconi Anemia (FA) pathway is responsible for interstrand crosslink repair. At the heart of this pathway is the FANCI-FAND2 (ID) complex, which, upon ubiquitination by the FA core complex, travels to sites of damage to coordinate repair that includes nucleolytic modification of the DNA surrounding the lesion and translesion synthesis. How the ID complex regulates these events is unknown. Here we describe a shRNA screen that led to the identification of two nucleases necessary for crosslink repair, FAN1 and EXDL2. FAN1 co-localizes at sites of DNA damage with the ID complex in a manner dependent on FAN1’s ubiquitin binding domain (UBZ), the ID complex, and monoubiquitination of FANCD2. FAN1 possesses intrinsic 5′-3′ exonuclease activity and endonuclease activity that cleaves nicked and branched structures. We propose that FAN1 is a repair nuclease that is recruited to sites of crosslink damage in part through binding the ubiquitinated ID complex through its UBZ domain.
Introduction
Cells in all organisms experience massive amounts of spontaneous DNA damage each day. A failure to properly respond to this genotoxic stress can lead to both developmental abnormalities and tumorigenesis. Organisms have evolved a complex signal transduction pathway called the DNA Damage Response (DDR) that senses genotoxic stress and orchestrates a response by activating specific types of repair, arresting the cell cycle and altering transcription. At the core of this signal transduction pathway are two PI-3 kinase-like protein kinases, ATM and ATR (Bakkenist and Kastan, 2004; Bartek et al., 2004; Harper and Elledge, 2007) which support the damage-induced phosphorylation of 100s of substrates to coordinate DNA repair (Matsuoka et al., 2007; Stokes et al., 2007).
A life-threatening lesion is the DNA double-strand crosslink, which covalently connects the Watson and Crick strands of DNA to create a bidirectional polymerase block. A repair pathway known as the Fanconi anemia (FA) pathway has evolved to specifically deal with these types of lesions. FA is a recessive developmental and cancer predisposition syndrome whose patients display multi-organ defects, bone marrow failure in childhood (Fanconi, 1967; Schmid and Fanconi, 1978) and a high incidence of malignancies (Alter et al., 2003). Cells from FA patients are hypersensitive to DNA interstrand crosslinking agents such as mitomycin C (MMC) (Auerbach and Wolman, 1976). To date, 13 proteins have been implicated in FA. At the center of this pathway is the FANCI/FANCD2 (ID) complex, which loads onto sites of cross-links to direct DNA repair. The ID complex is chromatin bound and when it encounters a DNA replication structure stalled due to a DNA crosslink, it becomes phosphorylated by the ATR/ATRIP kinase, which is localized through recognition of RPA at the lesion (Zou and Elledge, 2003). Phosphorylation of both I and D2 are required for ID function (Andreassen et al., 2004; Ho et al., 2006; Ishiai et al., 2008) and leads to the mono-ubiquitination of both subunits by a multisubunit E3 ligase formed by eight Fanconi anemia proteins (FANCA/B/C/E/F/G/L/M) and the E2-conjugating enzyme UBE2T (Cole et al., 2010; Machida et al., 2006; Meetei et al., 2004). Ubiquitinated ID then accumulates at the damage site and directs repair (Smogorzewska et al., 2007).
The repair of a crosslink is thought to involve two incision events on a single DNA strand flanking the lesion, followed by bypass synthesis over the lesion on the remaining intact strand using a translesion polymerase, possibly Rev1 (Niedzwiedz et al., 2004; Simpson and Sale, 2003) in combination with Rev3 and Rev7 (Lehmann et al., 2007; Raschle et al., 2008). A recently identified A-family nuclear DNA polymerase, PolN, might also play a role in this step (Moldovan et al., 2009; Zietlow et al., 2009). After bypass synthesis, two more incision events flanking the lesion occur allowing it to be excised. The initially cleaved strand can then be repaired by gene conversion using homologous recombination. Both the incision step and the bypass polymerase step are dependent upon ubiquitination of the ID complex (Knipscheer et al., 2009; Raschle et al., 2008). The nucleases responsible for the incision and excision events are not precisely known although XPF-ERCC1 and Mus81 complexes have been implicated (Ciccia et al., 2008). Recently, SLX4, a scaffold for various DNA repair nucleases, has been identified to be necessary for resistance to crosslinking agents. SLX4 interacts with both Mus81 and XPF and together with SLX1 forms a Holliday junction resolvase, although it is unclear which SLX4 activity is responsible for conferring resistance to DNA crosslinks (Fekairi et al., 2009; Munoz et al., 2009; Saito et al., 2009; Svendsen et al., 2009). After bypass synthesis the two strands liberated by the first two incision events constitute a double strand break and are repaired by homologous recombination (HR). Since HR requires 3′ overhangs to initiate recombination, it is likely these strands are processed by a 5′-3′ exonuclease to generate the recombination substrate that can gap repair off the strand from which the lesion was removed.
A number of outstanding issues remain unresolved in the FA pathway. First and foremost is how the ID complex orchestrates repair and the role of ubiquitination. Does it play a structural role in simply maintaining the complex at sites of damage, or does it recruit repair factors? In addition, the identities of the enzymatic factors involved in manipulation of the DNA at the site of the lesion are still unknown. Furthermore, it is unknown if there are additional back-up pathways that can also function to repair crosslinks in the absence of FA. The FA pathway appears to be conserved in metazoans, but is missing from prokaryotes and unicellular eukaryotes such as S. cerevisiae and S. pombe. Thus, alternative repair pathways do exist in nature. To address these issues and identify new genes involved in crosslink repair, we performed an RNAi screen for genes required for resistance to the crosslinking agent MMC and discovered two nucleases required for crosslink repair, one of which, FAN1, possesses both endonuclease and exonuclease activities and is recruited to lesions by the mono-ubiquitinated ID complex.
Results
Genome-wide shRNA screen to identify proteins necessary for resistance to interstrand crosslink (ICL) damage
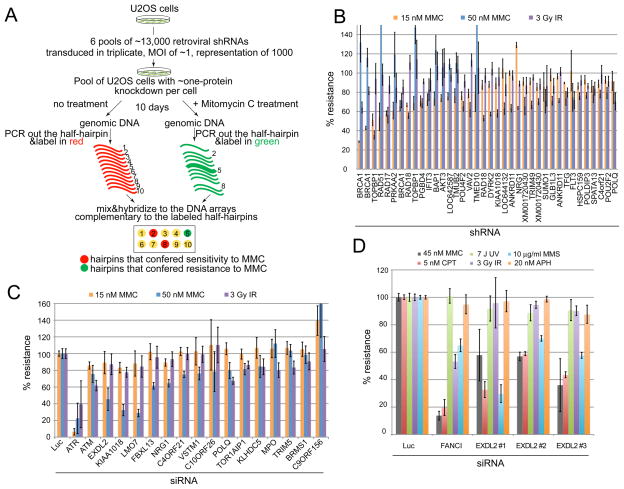
To identify proteins involved in resistance to crosslinking agents, we screened U2OS cells transduced with a library of 74,905 retroviral shRNAs targeting 32,293 unique human transcripts (Figure 1A). Using competitive hybridization of probes that detect the half-hairpins of the shRNAs, we compared the relative abundance of shRNAs between untreated cells and cells treated with low levels of mitomycin C (MMC). Cells bearing shRNAs that conferred sensitivity were depleted from the treated population. About 2,173 hairpins targeting 2,017 genes conferred sensitivity to MMC using the criteria of an average loss of 2-fold from the treated population (log2>1) (Table S1). Among these were previously known DNA damage response proteins including BRCA1, TOPBP1, RAD18, RAD17, RAD51, RAD54, FANCE, and others. We employed the multicolor competition assay (MCA) (Smogorzewska et al., 2007) with 379 shRNAs against a selected group of genes that made the cutoff in the primary screen to retest for MMC sensitivity. Eighty-four shRNAs tested conferred MMC sensitivity (Table S2). Figure 1B shows the top 38 scoring hairpins in which resistance to MMC was below 80% of control shRNA. To further examine damage sensitivity, pools of siRNAs were also tested (Figure 1C). Based on the results of this assay and domain analysis, two genes, EXDL2 and KIAA1018, were chosen for further study.
Figure 1.
Whole genome shRNA screen to identify genes necessary for crosslink resistance. A. Schematic of the primary screen. Changes in hairpin abundance after transduction with the shRNA library and Mitomycin C (MMC) treatment was followed by competitive DNA array hybridization. B. Multicolor competition assay (MCA) in U2OS cells transduced with the indicated shRNAs. Of the tested hairpins only those that showed less than 80% resistance to 15 or 50 nM Mitomycin C treatment are shown. Resistance of cells transduced with a hairpin against Luciferase was set at 100% in all MCA experiments. All hairpins used in MCA are shown in Supplemental Table 2. C. MCA in U2OS cells transfected with the indicated siRNAs (pools of four siGENOME siRNAs from Dharmacon). Cells transfected with siRNAs against ATM and ATR were used as a control. D. MCA in U2OS cells transfected with three separate siRNAs against EXDL2. Cells transfected with siRNAs against FANCI were used as a control. Error bars represent standard deviation across three technical replicates.
EXDL2 - a putative 3′-5′ exonuclease necessary for resistance to MMC
EXDL2 is an uncharacterized 621 amino acid protein with a nuclease domain most similar to the WRN-exo domain in the WRN protein (Figure S1, panels A and B) (Perry et al., 2006). Based on the conservation of the four key negatively-charged residues (DEDD) that serve as ligands for the metal ions, as well as a tyrosine residue which has been shown to be important for the catalysis, EXDL2 is predicted to be a 3′-5′ exonuclease. A mutation in a D. melanogaster ortholog of EXDL2, CG6744, displays a phenotype of hyper-recombination (Cox et al., 2007). In human cells, depletion of EXDL2 using three different siRNAs led to sensitivity to MMC, the Topo1 inhibitor camptothecin (CPT) and the alkylating agent MMS (Figure 1D and S1, panel C). This spectrum of sensitivities is similar to mutants in the Fanconi anemia pathway. Therefore, we tested if FANCD2 ubiquitination was affected in the EXDL2-depleted cells. Based on the normal ubiquitination of FANCD2 before and after damage (Figure S1, panel D) we conclude that EXDL2 is either downstream of FANCD2 in the Fanconi pathway or in a parallel pathway of crosslink repair.
FAN1 (KIAA1018) is required for the resistance to crosslinking agents
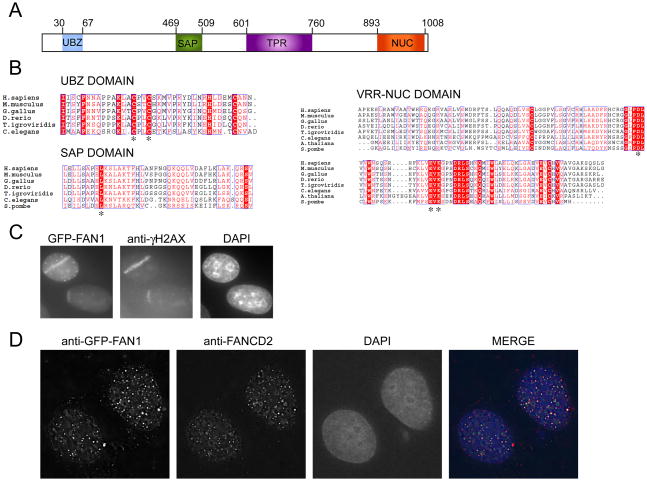
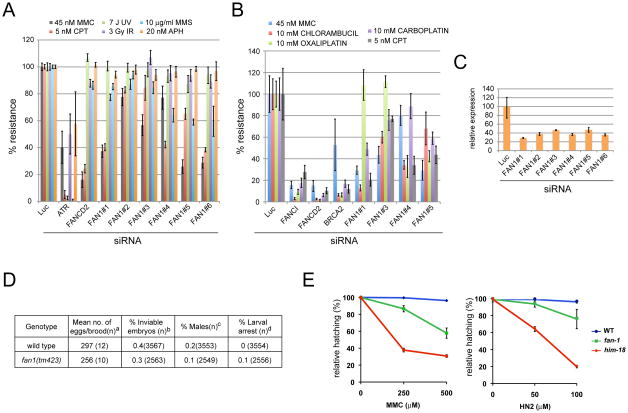
A second protein with an interesting domain structure is KIAA1018, which we renamed FAN1 (Fanconi associated nuclease 1) based on the data presented below. FAN1 has a nuclease-like fold called DUF994 (later renamed VRR-NUC) at its C-terminus, potential DNA-binding (SAP) and protein-protein interaction (TPR) motifs in its mid-section, as well as a Rad18–like ZnF domain at the N-terminus (Kinch et al., 2005) (Figure 4A). Depletion of FAN1 with multiple siRNAs leads to sensitivity to crosslinking agents including MMC, chlorambucil, carboplatin, and oxaliplatin, as well as CPT, and MMS (Figure 2A and B).
Figure 4.
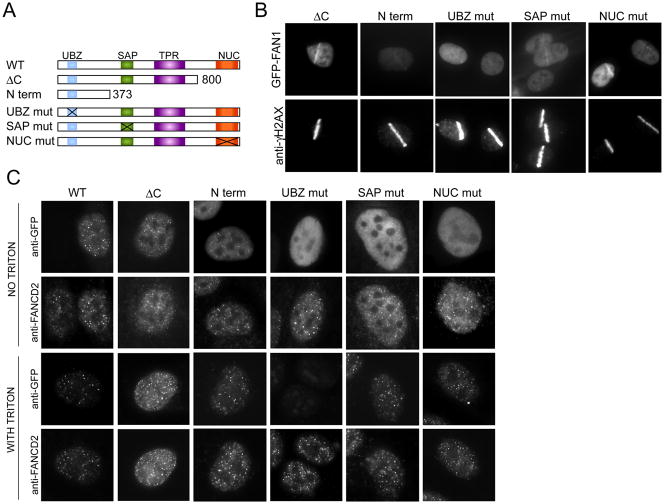
FAN1 is an evolutionarily conserved protein that localizes to sites of DNA damage. A. Schematic of the domain architecture of FAN1. Conserved domains are indicated: UBZ, ubiquitin-binding zinc finger; SAP, SAF-A/B, Acinus and PIAS; TPR, tetratricopeptide repeat; Nuc (VRR-NUC) virus type replication-repair nuclease. B. ClustalW2 alignment of UBZ, SAP and VRR-NUC domains across different species. Note that the S. pombe carries the SAP and NUC domain but no recognizable UBZ domain. Stars indicate residues mutated in subsequent experiments. C. U2OS cells expressing GFP-FAN1 were laser microirradiated and after 30 minutes were processed for imaging of GFP-FAN1 and γ-H2AX. GFP was visualized directly. Nuclei were stained with DAPI. D. U2OS expressing GFP-FAN1 were treated with 1 μM MMC for 24 hours and processed for indirect immunofluorescence with antibodies against GFP and FANCD2. Nuclei are stained with DAPI. Images were captured and deconvolved using the DeltaVision Image Restoration Microscope.
Figure 2.
Sensitivity to crosslinking agents and camptotecin in human cells depleted of FAN1 and in a FAN1 mutant C. elegans strain. A. MCA in U2OS cells transfected with six separate siRNAs against FAN1 and treated with different types of DNA damaging agents. B. MCA in U2OS cells transfected siRNAs against FAN1 and treated with different DNA crosslinking agents. C. RT-qPCR in U2OS cells transfected with the different siRNAs against FAN1. D. Plate phenotypes. Parentheses indicate the total number of: asingled hermaphrodites for which entire brood sizes were scored, bfertilized eggs scored, cadults scored, dL1-L4 worms. ND, not determined due to n=0. Wild type data are from (Saito et al., 2009). E. Relative hatching of wild type, fan-1 and him-18 mutants after treatment with the indicated doses of mitomycin C (MMC) and nitrogen mustard (HN2). Hatching is plotted as a fraction of the hatching observed in untreated animals. Error bars indicate standard error of the mean for at least 20 animals in each of three independent experiments.
To examine the evolutionary conservation of FAN1’s role in crosslink repair, we examined the crosslink sensitivity of a C. elegans FAN1 mutant (tm423) that carries a deletion of the SAP domain (Figure S2). fan-1 mutants lay normal numbers of eggs, and show normal larval development as well as no increase in either embryonic lethality or the % of males among their progeny that would suggest a meiotic phenotype (Figure 2D). However, treatment with either MMC or nitrogen mustard (HN2) results in decreased embryonic viability compared to wild type as judged by decreased hatching (p < 0.0001, respectively, chi-square test) (Figure 2E). Thus, FAN1 plays an evolutionarily conserved role in resistance to crosslink damage. Since the tm423 mutant may be a hypomorph, a meiotic phenotype could emerge in null animals.
FAN1 associates with mismatch repair proteins
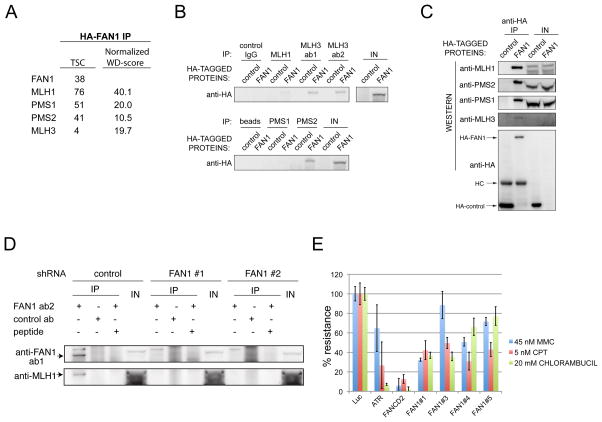
To identify FAN1-associated proteins, a HA-tagged FAN1 was purified from 293TREX cells and interacting proteins were identified by LC-MS/MS. Proteomic data was processed using the Comparative Proteomic Analysis Software Suite (CompPASS) (Sowa et al., 2009). The top-scoring interacting proteins were the mismatch repair proteins MLH1, MLH3, PMS1 and PMS2 (Figure 3A). KIAA1018 was previously detected in a proteomic analysis of MLH1 interacting proteins (Cannavo et al., 2007); however, the interaction was not independently confirmed. To confirm these interaction, we performed immunoprecipitations. HA-FAN1 was identified in immunoprecipitates of MLH1, MLH3, and PMS2 (Figure 3B). Reciprocal immunoprecipitations identified all four proteins in precipitation reactions with anti-HA antibodies (Figure 3C). Endogenous FAN1 immunoprecipitations brought down both FAN1 and MLH1 in a FAN1-dependent manner (Figure 3D). The interaction with the mismatch repair machinery raised the possibility that FAN1’s role in ICL resistance is dependent on mismatch repair. To test this, we depleted FAN1 from the HCT116 cells, which carries inactivating mutations in both alleles of MLH1 (Figure 3E). FAN1 depletion with four different siRNAs still increased sensitivity of these cells to crosslinking agents.
Figure 3.
FAN1 associates with mismatch repair proteins. A. HA-FAN1 was expressed in 293 TREX cells using an inducible retroviral system and cell extracts subjected to immunoprecipitation and LC-MS/MS as described in Experimental Procedures. High confidence candidate interacting proteins are shown as determined using CompPASS to derive normalized WD-score for individual proteins in the immune complex. TSC is total spectral count. This experiment was done in duplicate and average values are reported. In both experiments all listed proteins were identified B. Cell extracts of 293 TREX cells expressing HA-FAN1 were subjected to immunoprecipitation using the indicated antibodies. HA-FAN1 was identified by immunoblotting with an anti-HA antibody. IN represents 5% input. C. Cell extracts of 293 TREX cells expressing HA-FAN1 were subjected to immunoprecipitation using an anti-HA antibody and immunoprecipitates were probed with the indicated antibodies. HC-heavy chain of the antibody used in the immunoprecipitation. D. Endogenous FAN1 was immunoprecipitated (+/−antigenic peptide) from HeLa cells with or without FAN1 depletion using two separate shRNAs and immunoblotted for FAN1 and MLH1. shRNAs #1-739 and shRNA#2-600. Note that the FAN1 antibody does not recognize endogenous protein in straight western, only in IPs. E. Mitomycin C and camptotecin sensitivity caused by depletion of FAN1 is not dependent on MLH1 or MSH3. MCA in HCT116 cells transfected with the indicated siRNAs.
FAN1 co-localizes to sites of damage with FANCD2 via its UBZ domain
The domain structure of FAN1 with its well conserved UBZ, SAP, and nuclease domains (Figure 4, panels A and B) led us to test FAN1 localization to sites of DNA damage. We subjected GFP-tagged FAN1 cells to laser microirradiation, which results in localized DNA damage tracks (Bekker-Jensen et al., 2006). Within 15 minutes of microirradiation, GFP-FAN1 localized to sites of DNA damage along with γ-H2AX, a marker of DNA damage (Figure 4C) (Rogakou et al., 1999). The GFP tracks were also seen in the absence of γ-H2AX staining (data not shown). We next asked if FAN1 localized to sites of crosslinked DNA damage caused by MMC. Most MMC-induced foci contained both FAN1 and FANCD2 (Figure 4D). We next tested various FAN1 domain mutants for their ability to localize to sites of laser microirradiation (Figure 5A). Mutants in the nuclease domain behaved like wild type. The N-terminal 373 amino acids lacking SAP or a protein with a SAP domain mutation (L477P) still localized to sites of damage but the strength of the GFP-FAN1 signal was substantially diminished. Only the mutant with two conserved cysteine residues of the UBZ domain substituted by alanines (C44A, C47A) completely failed to localize to microirradiation tracks (Figure 5B). The same UBZ mutant did not form foci in MMC-treated cells (Figure 5C) but was expressed at similar levels as the other alleles (Figure 5C, top panel “no triton”). Several other mutant alleles localized to sites of damage with reduced efficiency (Figure S3). Interestingly, the N-terminal 90 amino-acids containing the UBZ domain was able to co-localize with FANCD2, although with diminished efficiency. Based on these experiments we conclude that the UBZ domain is critical for localizing FAN1 to sites of damage. However, other parts of the protein also assist in localization. Careful kinetic analysis of the behavior of the different mutants will be necessary to resolve this issue.
Figure 5.
Localization of FAN1 to damage sites depends on the UBZ domain. A. Schematic of mutant proteins used in FAN1 localization experiments. B. U2OS cells expressing indicated GFP-FAN1 mutants were laser microirradiated and after 30 minutes were processed for imaging of GFP-FAN1 and γ-H2AX. C. U2OS expressing indicated GFP-FAN1 mutants were treated with 1 μM MMC for 24 hours and processed for indirect immunofluorescence with antibodies against GFP and FANCD2. Cells were either directly fixed with formaldehyde or pre-extracted with TritonX-100 before fixation. Exposures were adjusted to show the presence of foci even if foci were dim. For the comparison of the intensity of foci see Figure S3.
FANCI and FANCD2 are required for FAN1 localization to sites of DNA damage
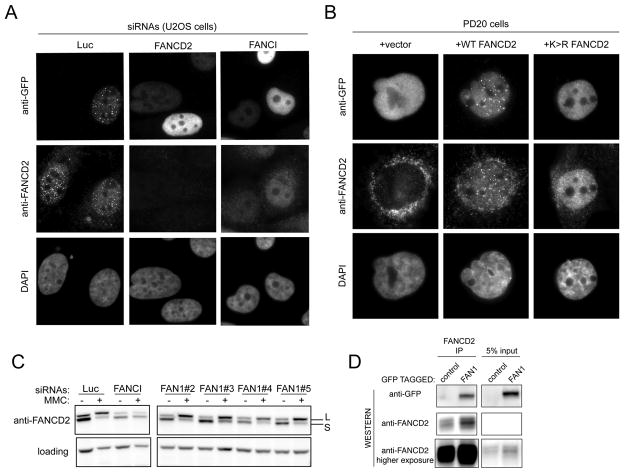
To examine possible dependency on the ID complex, we assessed formation of GFP-FAN1 foci after MMC treatment in cells depleted of FANCD2 and FANCI. In FANCI or FANCD2 depleted cells, FAN1 was no longer able to form foci (Figure 6A and S4B). The percentage of cells with foci in cells depleted of either FANCI or FANCD2 was 28% and 24%, respectively while 93% of control depleted cells showed FAN1 foci. FAN1 also fails to form foci in PD20 cells, which lack FANCD2 protein (Figure 6B), but reappear upon complementation with WT FANCD2. PD20 cells expressing the monoubiquitination-defective FANCD2 K561R mutant also fail to form FAN1 foci (Figure 6B). We next asked if FAN1 is required for FANCD2 monoubiquitination. Cells transfected with 4 different siRNAs against FAN1 were treated with MMC. FANCD2 in these cells was ubiquitinated to the same extent as in cells treated with siRNAs against luciferase (Figure 6C). We conclude that FA proteins and specifically FANCD2 monoubiquitination is necessary for the recruitment of FAN1 to sites of damage and that the crosslink sensitivity of FA defective cells is in part due to a failure to recruit FAN1 to sites of DNA damage.
Figure 6.
Localization of FAN1 depends on the Fanconi anemia proteins FANCI and FANCD2. A. U2OS cells expressing GFP-FAN1 and transfected with the indicated siRNAs were treated with 1 μM MMC for 24 hours, fixed and processed for indirect immunofluorescence with antibodies against GFP and FANCD2. The corresponding TritonX-100 experiment is shown in Figure S4, panel B. B. PD20 cells (FANCD2 negative) complemented with vector, wild type FANCD2 and FANCD2K561R mutant (which cannot be ubiquitinated) and expressing GFP-FAN1 were treated with 1 μM MMC for 24 hours, fixed and processed for indirect immunofluorescence with antibodies against GFP and FANCD2. The corresponding TritonX-100 experiment is shown in Figure S4, panel C. C. U2OS cells transfected with the indicated siRNAs were treated with 1 μM MMC for 24 hours and collected for western blotting with anti-FANCD2 antibody. D. Cell extracts of 293 cells expressing GFP-FAN1 were subjected to immunoprecipitation using FANCD2 antibody. GFP-FAN1 and FANCD2 were identified by immunoblotting.
FAN1 interacts with FANCD2
The dependence of FAN1 foci formation on the presence of monoubiquitinated FANCD2 raised a possibility that FANCD2 and FAN1 interact. Cells expressing GFP-FAN1 were subjected to immunoprecipitation with anti-FANCD2 antibodies. GFP-FAN1 strongly co-immunoprecipitated with FANCD2 (Figure 6D). Therefore, FAN1 and FANCD2 interact in vivo.
FAN1 has an endonuclease and 5′exonuclease activity
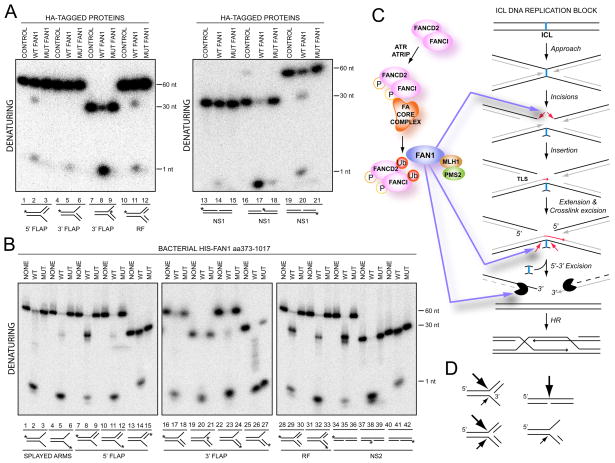
FAN1 contains a conserved nuclease domain. To assess its activity, we examined whether HA-FAN1 complexes immunoprecipitated from 293TREX cells could act on radiolabled substrates including 3′ and 5′ flaps, replication forks, and nicked substrates. Two nuclease activities were observed. One was a 5′ to 3′ exonuclease especially active on 3′FLAP substrate (Figure 7A, lane 8) as well as a nicked substrate (Figure 7A, lane 17). The second activity was an endonuclease activity on a 5′FLAP substrate (Figure 7A, lane 2), replication fork substrate (Figure 7A, lane 11) and nicked substrate (Figure 7A, lane 20). The endonuclease activity on the 5′FLAP substrate was seen on the top strand at the junction of the two DNA duplexes, not on the single strand flap itself. Neither the exonuclease nor the endonuclease activities were seen in the immunoprecipitates from cells expressing control HA protein or a mutant FAN1 with the key residues predicted to be necessary for the catalysis substituted to alanines (Q864A, D960A, E975A, K977A) (Figure 7A, lanes 3, 9, 12, 18, and 21), although the WT and mutant proteins were immunoprecipitated to the same extent (Figure S5A). To further show that this activity is intrinsic to FAN1 as opposed to a factor like MLH1/PMS2 associated with FAN1 in mammalian cells, we expressed the last 644 amino acids of human FAN1 (aa373-1017) as a His6 fusion protein in bacteria (Figure 7B, Figure S5B and C). Indeed, the purified protein had the activities seen with the mammalian FAN1. More robust activity of FAN1 purified from bacteria gave us an opportunity to define the substrate specificity. Using a 3′-labeled substrate, we have confirmed that the exonuclease activity was a 5′- to 3′ activity (Figure 7B, lane 26). The major endonuclease activity of FAN1 was observed on the top strand of the 5′FLAP substrate at the junction of the DNA duplexes (Figure 7B, lane 8) and across from a nick (Figure 7B, lane 35). A weak 5′FLAP endonuclease activity was seen on the bottom strand (Figure 7B, lane 11). On a replication fork, the major activity was on a top strand (Figure 7B, lane 29) with some activity seen on the bottom strand (Figure 7B, lane 32). FAN1 activity was stimulated by ATP (Figure S5E). We noted a well-conserved motif in FAN1 (GFDQGIHGEGST, amino acids 826-837 of human protein) that may be able to bind to ATP. The significance of this motif remains to be determined. A mutant FAN1 with the key residues predicted to be necessary for the catalysis substituted to alanines (Q864A, D960A, E975A, K977A) (Figure 7B) or a mutant with just two (E975A, K977A) (Figure S5F) lacked the 5′-3′ exonuclease and the endonuclease activities. The bacterially-purified proteins did have some contaminating 3′ exonuclease activity (Figure 7B, lane 27). Based on these in vitro experiments, we conclude that FAN1 possesses an intrinsic nuclease activity that participates in DNA repair.
Figure 7.
FAN1 possesses an intrinsic endonuclease and exonuclease activity. A. Control HA (CONTROL), HA-FAN1 or HA-FAN1 nuclease mutant (MUT FAN1) (Q864A_D960A_E975A_K977A) complexes were precipitated from 293 TREX cells and incubated with the indicated 32P-end labeled substrates prior to electrophoresis on native or denaturing gels. Asterisk (*) indicates the position of label on the labeled strand. B. Bacterial His6-tagged FAN1aa373-1017 (WT or mutant Q864A_D960A_E975A_K977A) were incubated with 32P-end labeled substrates prior to electrophoresis on native or denaturing gels. C. A model of FAN1 at the interstrand crosslink. Right-hand side is based on (Knipscheer et al., 2009; Raschle et al., 2008). See text for details
D. Summary of FAN1’s endonucleaolytic activity. The size of the arrows corresponds tothe strength of the endonuclease.
Discussion
An shRNA screen identifies many putative players in crosslink resistance
Interstrand crosslinks are among the most lethal lesions to cells and their repair pathway(s) remain poorly understood. Given the difficulty that ICLs create during replication and transcription, it is critical to identify all the participants in the repair process. Therefore we performed an shRNA screen to identify new components of crosslink repair. Despite the limitations of RNAi, we were able to confirm a number of proteins involved in resistance to crosslinking agents using shRNAs and siRNAs. Among these were C4orf21, which has both a ZF-GRF, a presumed DNA binding domain, and a domain with similarities to helicases and FLJ25006, which has a kinase domain with predicted serine/threonine activity. Among the known DNA repair proteins was POLQ, an error prone translesion DNA polymerase (Arana et al., 2008; Seki et al., 2004). POLQ in chicken cells has been shown to be involved in repair of oxidative damage but not in crosslink repair (Yoshimura et al., 2006). However, in C. elegans, POLQ has been implicated in ICL repair, in a pathway distinct from the Fanconi anemia pathway but in the same pathway as the C. elegans BRCA1 ortholog (Muzzini et al., 2008).
Among the validated genes with strong phenotypes were two putative nucleases. We validated involvement of one, EXDL2, in crosslink sensitivity using multiple siRNAs. EXDL2 has a WRN-like exonuclease domain and promises to shed light on the mechanism of crosslink resistance. The other protein FAN1, with an intriguing domain structure, has now been placed as a bona fide nuclease necessary for crosslink repair in the Fanconi anemia pathway.
Evolutionary conservation of FAN1 function
FAN1 is easily identifiable in Dictyostelium discoideum, which has orthologs of several Fanconi anemia proteins (Zhang et al., 2009) but no obvious orthologs are apparent in Drosophila or Xenopus. Interestingly, an S. pombe ortholog of FAN1 has a SAP domain, TRP domain and a nuclease domain, thus it is predicted to also function in DNA repair transactions. Fission yeast does not possess the classical FA proteins except for an ortholog of FANCM, Fml1, which promotes Rad51-dependent gene conversion at stalled replication forks and limits crossing over during mitotic double-strand break repair (Sun et al., 2008). It will be interesting to test if the S. pombe FAN1 mutant is sensitive to crosslinking agents and if so, how it functions without the other proteins present in human cells. This could shed light on alternative pathways for crosslink repair in human cells.
Interaction of FAN1 with mismatch repair (MMR) proteins
We identified all human MutL proteins in nearly stoichiometric complexes with FAN1, suggesting a highly conserved function. Despite the strong interaction with MutLs, we have yet to identify a mismatch repair defect in extracts derived from cells depleted of FAN1 using shRNAs. This could be due to the limitations of RNAi to create the equivalent of a null mutation. Alternatively, the MutL complexes may play roles outside of mismatch repair.
During mismatch repair in bacteria, MutL complexes are recruited to sites of mismatches by MutS complexes that sense the mismatch lesions. MutL complexes then recruit the UvrD helicase and the MutH endonuclease, which it also activates, to initiate repair. MutL complexes have also been shown to have an endogenous endonuclease activity that could participate in mismatch repair (Kadyrov et al., 2006). It is possible that MutL complexes could play a similar role in ICL repair. In place of MutS complexes, the monoubiquitinated ID complex would play the analogous role as an ICL lesion sensor to recruit the FAN1-MutL complex to introduce one or more of the four incisions needed for ICL repair. In addition, this complex could recruit additional factors to aid in repair. The MutLα (MLH1-PMS2 heterodimer) complex has been identified as an interactor of FANCJ (BRIP1), a helicase involved in crosslink repair (Peng et al., 2007). A direct MLH1-FANCJ interaction was required for FANCJ to complement the crosslink sensitivity of a patient cell line with a mutation in the FANCJ gene. Thus, it is possible that FAN1 and MutLα might act as a bridge between the ID complex and FANCJ allowing the nuclease activity of FAN1 to pair with FANCJ’s helicase activity during the crosslink repair process. It will be important to determine if the interaction of MLH1 with FAN1 and FANCJ is mutually exclusive and whether FAN1 or MLH1 are required to localize FANCJ to the sites of crosslinks.
FAN1 localization to sites of crosslink damage and interaction with FANCD2
FAN1 localizes to laser microirradiation sites as well as to crosslink-induced damage foci where it co-localized with FANCD2. Localization of FAN1 to foci was dependent on the UBZ domain of FAN1, the presence of FANCI and FANCD2, and the monoubiquitination of FANCD2. Based on these findings, we hypothesize that FAN1 is recruited to sites of damage by the ID complex using monoubiquitinated FANCD2 and FANCI as an interaction platform for FAN1’s UBZ domain.
Since several mutant alleles of FAN1 examined, including the truncation allele lacking the SAP domain and the TPR domain, showed decreased GFP-FAN1 staining at the sites of DNA damage, it is very likely that in addition to the interaction between the ubiquitinated ID complex and the UBZ domain, FAN1 is stabilized in damage foci by interactions with the DNA or other proteins.
FAN1 was also identified by our group as a phospho-protein in a proteomic analysis of DNA damage response (Zhou and Elledge, in preparation). The identified phosphorylated SQ site, S210, in the human protein is evolutionarily conserved in mouse, chicken and fish and is the only SQ/TQ site in FAN1 with such high extent of conservation. Phosphorylation plays a pivotal role in activating FA pathway and it is likely that the activity of enzymes participating in repair will be tightly regulated. Therefore, it will be important to establish the functional consequences of abolishing FAN1 phosphorylation.
FAN1 is a nuclease in the Fanconi anemia pathway
Biochemical analysis of FAN1 isolated from human cells revealed that FAN1 acts as both an endonuclease and as an exonuclease. The major endonucleolytic activity of FAN1 appears to act opposite a nick and at branched structures with a 3′ end at the branch point (Figure 7C). FAN1’s minor activities are towards a 3′ FLAP and towards a replication forks across a 5′ end at the branch point. The 5′ to 3′ exonuclease activity is active on most 5′ ends although its strength varies depending on the specific substrate, with those substrates processed efficiently by the endonuclease activity being poorer substrates for the exonuclease than those inefficiently processed by the endonuclease. The ultimate test of FAN1’s activity will come from examining its function on crosslinked substrates in the setting of DNA replication, since this is where ATR is activated and where the interaction with the monoubiquitinated ID complex positions FAN1 at the sites of damage. However, the in vitro activities of FAN1 seen so far fit well with the repair activities hypothesized to be present at the crosslink and other lesions that necessitate replication restart (Figure 7D). FAN1 may be involved in the unhooking of the interstrand crosslink in the setting of replication. The ID complex is required for this process and since it is also necessary for FAN1 localization at the site of DNA damage, FAN1 is a good candidate for this activity. The exonuclease activity of FAN1 might also be important for the processing of the DNA strands to generate 3′ overhangs for the homologous recombinational repair that is necessary to restart the replication fork after the crosslink is removed or to gap repair unreplicated regions when both forks reach the ICL. Lastly, the final steps of removal of the fully unhooked crosslink might also rely on FAN1 activity. This is among the first instances for which we have an understanding of how a nuclease important for crosslink resistance localizes to sites of damage. Although other nucleases including XPF-ERCC1, MUS81-EME1 and SLX1 have been implicated in crosslink repair (Ciccia et al., 2008; Munoz et al., 2009; Svendsen et al., 2009) we still do not understand how they are recruited to the sites of DNA damage and the nature of their relationship to Fanconi anemia proteins.
FAN1 is a candidate tumor suppressor
Based on FAN1’s function in ICL resistance, co-localization, dependence on FANCI and FANCD2 function, as well as interaction with FANCD2, FAN1 is a candidate Fanconi anemia gene. Since three of the FA genes, FANCD1 (BRCA2), FANCN (PALB2), and FANCJ (BRIP1) are also mutated in familial breast cancer predisposition syndrome (Rahman et al., 2007; Wooster et al., 1995) FAN1 should be sequenced in the appropriate cohort of patients who display familiar predisposition to breast cancer but lack identified predisposing mutations.
The identification of FAN1 brings us closer to an understanding of the biochemical pathway involved in DNA crosslink repair and sets the stage for more precise examination of the repair process in reconstituted crosslink repair systems.
Experimental procedures
Cell Culture, plasmids, antibodies, RNAi, RT-qPCR
U2OS, DR-U2OS, HeLa, 293T, 293TREX, and HCT116 cells were grown in Dulbecco Modified Eagle medium (DMEM) supplemented with 10% (v/v) FBS (Invitrogen), 100 units of penicillin per ml, and 0.1 mg streptomycin per ml. PD20 cells were grown as above but with 15% FBS. Plasmids were constructed using recombinational cloning via the Gateway system (Invitrogen). KIAA1018 clone was obtained from Origene and the WT or truncation mutants were amplified and recombined into pDONR223 (Lamesch et al., 2007). pDONR223 derivatives were recombined into appropriate recipient vectors using LR clonase (Invitrogen). Mutagenesis was performed using multisite mutagenesis kit (Agilent) (primers are listed in Table S4). Antibodies against FAN1 were raised in rabbits against peptide CGQSDSAKREVKQKIS (YenZym) and affinity purified using the antigenic peptide. The other antibodies were FANCD2 (Novus NB100-182), GFP (Roche 11814460001), HA (Covance MMS-101R), MLH1 (Santa Cruz sc-582), MLH3 (Bethyl A301-849A & A301-850A), PMS1 (Santa Cruz sc-615), and PMS2 (BD Pharmigen 556415). siRNA transfections were performed using Lipofectamine RNAiMAX as suggested by the manufacturer with the final siRNA concentration of 50 nM. siRNA sequences are listed in Table S5. For RT-qPCR, Supperscript III reverse transcriptase followed by Platinum cybergreen super mix (Invitrogen) were used according to the instructions. GAPDH or Actin were used as controls.
Whole genome shRNA screen
The pool-based shRNA screen using half-hairpin (HH) barcode deconvolution was performed as described before (Schlabach et al., 2008).
Protein purification and mass spectrometry
293 TREX cells expressing FAN1 were lysed and immunoprecipitated using anti-HA antibodies (Sowa et al., 2009; Svendsen et al., 2009). Complexes were either used for DNA cleavage assays, subjected to immunoblotting, or eluted with HA peptide and trypsinized prior to mass spectrometry (Sowa et al., 2009; Svendsen et al., 2009). Processing of samples for mass spectrometry as well as analysis of proteomic data using CompPASS was as described (Sowa et al., 2009). For immunoprecipitations with FANCD2, chromatin fraction was prepared as described (Moldovan et al., 2009). For DNA cleavage assays, the immune complexes were washed 3 times in buffer containing 20 mM Tris HCl pH 8.0, 5 mM MgCl2, 1 mM DTT. Bacterial proteins (FAN1 373-1017) were expressed using pDEST17 (Invitrogen), induced with L-arabinose and purified on a Ni-NTA column. The proteins were dialyzed against a buffer containing 50 mM Tris HCl pH7.5, 100 mM NaCl, 0.01 %NP40, 10% glycerol, 1 mM DTT, 0.5 mM EDTA and were stored at −80°C.
Multicolor competition assay
Experiments were done as described (Smogorzewska et al., 2007).
Laser-induced damage and immunofluorescence
Microirradiation was performed as described previously (Bekker-Jensen et al., 2006). Immunofluorescence experiments were done as described (Smogorzewska et al., 2007).
In vitro cleavage assays
In vitro cleavage of DNA substrates was performed using the FAN1 immune complexes or bacterially-purified FAN1 in conjunction with previously described DNA substrates (Ciccia et al., 2003; Ip et al., 2008; Rass and West, 2006; Svendsen et al., 2009). DNA cleavage assays were performed using 5′-32P or 3′ -32P-end labeled DNA substrates. Substrates were generated by annealing oligonucleotides and were purified by polyacrylamide gel electrophoresis as described previously (Ciccia et al., 2003; Ip et al., 2008; Rass and West, 2006; Svendsen et al., 2009). The sequences of substrates are provided in Table S4. Radiolabeled substrates were incubated with the indicated immune complexes or bacterially-purified FAN1. DNA cleavage assays were performed in 20 mM Tris HCl pH 8.0, 5 mM MgCl2, 1 mM ATP, and 1 mM DTT. For bacterially-purified FAN1, 20 ng of WT protein or 40 ng of mutant protein was used with each substrate. After 30 minutes (for the bacterial protein) or 2 hours (for the immunoprecipitated proteins) at 37°C, reaction mixtures were treated with 1% proteinase K in SDS prior to electrophoresis on either 12% polyacrylamide gels (native) or 16% polyacrylamide-urea gels (denaturing). Reaction products were visualized by autoradiography.
C. elegans Genetics
C. elegans strains were cultured at 20°C under standard conditions (Brenner, 1974). The N2 Bristol strain was used as the wild-type background. The following mutations and chromosome rearrangements were used in this study: LGIII: him-18(tm2181) (Saito et al., 2009), qC1[dpy-19(e1259) glp-1(q339) qIs26] (III); LGIV: fan-1(tm423), nT1[unc-?(n754) let-? qIs50] (IV; V), nT1[qIs51] (IV; V).
The fan-1(tm423) mutant, obtained from the Japanese National Bioresource Project, carries a 411 bp in-frame deletion encompassing parts of exons 5 to 8. This deletion results in the loss of the predicted SAP motif.
DNA interstrand crosslink sensitivity assay in C. elegans
Young adult worms were treated with 0, 250 or 500 μM of mitomycin C (MMC; Sigma) in M9 buffer containing E. coli OP50 with slow shaking in the dark for 19 hours. Treatment with nitrogen mustard (mechlorethamine hydrochloride; Sigma) was similar, but with doses of 0, 50 or 100 μM. Following treatment with MMC or HN2, animals were plated to allow recovery for 3 hours. 20 animals were plated 5 per plate and hatching was assessed for the time period 22–26 hours from the start of treatment. Each damage condition was replicated at least three times in independent experiments. him-18/slx-4 mutants, shown previously to be extremely sensitive to ICL inducing agents (Saito et al. 2009), were used as a control. Since untreated him-18(tm2181) mutants have reduced hatching, embryonic viability after DNA damage treatment was plotted as a percentage of the hatching after DNA damage normalized by that in untreated animals (relative hatching) (Saito et al., 2009).
Highlights.
FAN1 (KIAA1018) and EXDL2 were identified in a genetic screen in mammalian cells
Loss of FAN1 or EXDL2 results in DNA crosslink sensitivity
FAN1 localization to sites of DNA damage depends on the Fanconi anemia pathway and a UBZ domain of FAN1
FAN1 possesses an intrinsic endonuclease and exonuclease activity that might be important for crosslink repair and replication restart.
Supplementary Material
Acknowledgments
We thank Jen Svendsen and members of the Elledge lab for protocols and discussion. Special thanks to Alberto Ciccia for insightful suggestions and comments. This work is supported by a grant from the National Institutes of Health and CMCR grant #1U19A1067751-01 to S.J.E, a National Institutes of Health grant R01GM072551, and a Giovanni Armenise-Harvard Foundation award to M.P.C., a National Institutes of Health grant to J.W.H. and by Project Z01 ES065089 to T.A.K., in the Division of Intramural Research of the National Institute of Environmental Health Sciences, National Institutes of Health. A.S. was supported by T32CA09216 to the Pathology Department at the Massachusetts General Hospital and by Burroughs Wellcome Fund Career Award for Medical Scientists and is an Irma T. Hirschl scholar. S.J.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alter BP, Greene MH, Velazquez I, Rosenberg PS. Cancer in Fanconi anemia. Blood. 2003;101:2072. doi: 10.1182/blood-2002-11-3597. [DOI] [PubMed] [Google Scholar]
- Andreassen PR, D’Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18:1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach AD, Wolman SR. Susceptibility of Fanconi’s anaemia fibroblasts to chromosome damage by carcinogens. Nature. 1976;261:494–496. doi: 10.1038/261494a0. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Gerrits B, Marra G, Schlapbach R, Jiricny J. Characterization of the interactome of the human MutL homologues MLH1, PMS1, and PMS2. J Biol Chem. 2007;282:2976–2986. doi: 10.1074/jbc.M609989200. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Constantinou A, West SC. Identification and characterization of the human mus81-eme1 endonuclease. J Biol Chem. 2003;278:25172–25178. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- Cole AR, Lewis LP, Walden H. The structure of the catalytic subunit FANCL of the Fanconi anemia core complex. Nat Struct Mol Biol. 2010;17:294–298. doi: 10.1038/nsmb.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LS, Clancy DJ, Boubriak I, Saunders RD. Modeling Werner Syndrome in Drosophila melanogaster: hyper-recombination in flies lacking WRN-like exonuclease. Ann N Y Acad Sci. 2007;1119:274–288. doi: 10.1196/annals.1404.009. [DOI] [PubMed] [Google Scholar]
- Fanconi G. Familial constitutional panmyelocytopathy, Fanconi’s anemia (F.A.). I. Clinical aspects. Semin Hematol. 1967;4:233–240. [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Ho GP, Margossian S, Taniguchi T, D’Andrea AD. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol Cell Biol. 2006;26:7005–7015. doi: 10.1128/MCB.02018-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, Saberi A, Kinoshita E, Kinoshita-Kikuta E, Koike T, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15:1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Kinch LN, Ginalski K, Rychlewski L, Grishin NV. Identification of novel restriction endonuclease-like fold families among hypothetical proteins. Nucleic Acids Res. 2005;33:3598–3605. doi: 10.1093/nar/gki676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D’Andrea AD, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Yan Z, Wang W. FANCL replaces BRCA1 as the likely ubiquitin ligase responsible for FANCD2 monoubiquitination. Cell Cycle. 2004;3:179–181. [PubMed] [Google Scholar]
- Moldovan GL, Madhavan MV, Mirchandani KD, McCaffrey RM, Vinciguerra P, D’Andrea AD. DNA polymerase POLN participates in crosslink repair and homologous recombination. Mol Cell Biol. 2009 doi: 10.1128/MCB.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Muzzini DM, Plevani P, Boulton SJ, Cassata G, Marini F. Caenorhabditis elegans POLQ-1 and HEL-308 function in two distinct DNA interstrand cross-link repair pathways. DNA Repair (Amst) 2008;7:941–950. doi: 10.1016/j.dnarep.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Peng M, Litman R, Xie J, Sharma S, Brosh RM, Jr, Cantor SB. The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 2007;26:3238–3249. doi: 10.1038/sj.emboj.7601754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JJ, Yannone SM, Holden LG, Hitomi C, Asaithamby A, Han S, Cooper PK, Chen DJ, Tainer JA. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat Struct Mol Biol. 2006;13:414–422. doi: 10.1038/nsmb1088. [DOI] [PubMed] [Google Scholar]
- Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass U, West SC. Synthetic junctions as tools to identify and characterize Holliday junction resolvases. Methods Enzymol. 2006;408:485–501. doi: 10.1016/S0076-6879(06)08030-X. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TT, Youds JL, Boulton SJ, Colaiacovo MP. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 2009;5:e1000735. doi: 10.1371/journal.pgen.1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid W, Fanconi G. Fragility and spiralization anomalies of the chromosomes in three cases, including fraternal twins, with Fanconi’s anemia, type Estren-Dameshek. Cytogenet Cell Genet. 1978;20:141–149. doi: 10.1159/000130845. [DOI] [PubMed] [Google Scholar]
- Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson LJ, Sale JE. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell. 2008;32:118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, et al. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Langenick J, Traynor D, Babu MM, Kay RR, Patel KJ. Xpf and not the Fanconi anaemia proteins or Rev3 accounts for the extreme resistance to cisplatin in Dictyostelium discoideum. PLoS Genet. 2009;5:e1000645. doi: 10.1371/journal.pgen.1000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietlow L, Smith LA, Bessho M, Bessho T. Evidence for the involvement of human DNA polymerase N in the repair of DNA interstrand cross-links. Biochemistry. 2009;48:11817–11824. doi: 10.1021/bi9015346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.