Abstract
Norovirus GII/4 is a leading cause of acute viral gastroenteritis in humans. We examined here how the GII/4 virus evolves to generate and sustain new epidemics in humans, using 199 near-full-length GII/4 genome sequences and 11 genome segment clones from human stool specimens collected at 19 sites in Japan between May 2006 and February 2009. Phylogenetic studies demonstrated outbreaks of 7 monophyletic GII/4 subtypes, among which a single subtype, termed 2006b, had continually predominated. Phylogenetic-tree, bootscanning-plot, and informative-site analyses revealed that 4 of the 7 GII/4 subtypes were mosaics of recently prevalent GII/4 subtypes and 1 was made up of the GII/4 and GII/12 genotypes. Notably, single putative recombination breakpoints with the highest statistical significance were constantly located around the border of open reading frame 1 (ORF1) and ORF2 (P ≤ 0.000001), suggesting outgrowth of specific recombinant viruses in the outbreaks. The GII/4 subtypes had many unique amino acids at the time of their outbreaks, especially in the N-term, 3A-like, and capsid proteins. Unique amino acids in the capsids were preferentially positioned on the outer surface loops of the protruding P2 domain and more abundant in the dominant subtypes. These findings suggest that intersubtype genome recombination at the ORF1/2 boundary region is a common mechanism that realizes independent and concurrent changes on the virion surface and in viral replication proteins for the persistence of norovirus GII/4 in human populations.
Norovirus (NoV) is a nonenveloped RNA virus that belongs to the family Caliciviridae and can cause acute gastroenteritis in humans. The NoV genome is a single-stranded, positive-sense, polyadenylated RNA that encodes three open reading frames, ORF1, ORF2, and ORF3 (68). ORF1 encodes a long polypeptide (∼200 kDa) that is cleaved in the cells by the viral proteinase (3Cpro) into six proteins (4). These proteins function in NoV replication in host cells (19). ORF2 encodes a viral capsid protein, VP1. The capsid gene evolved at a rate of 4.3 × 10−3 nucleotide substitutions/site/year (7), which is comparable to the substitution rates of the envelope and capsid genes of human immunodeficiency virus (30). The capsid protein of NoV consists of a shell (S) and two protruding (P) domains: P1 and P2 (47). The S domain is relatively conserved within the same genetic lineages of NoVs (38) and is responsible for the assembly of VP1 (6). The P1 subdomain is also relatively conserved (38) and has a role in enhancing the stability of virus particles (6). The P2 domain is positioned at the most exposed surface of the virus particle (47) and forms binding clefts for putative infection receptors, such as human histo-blood group antigens (HBGA) (8, 13, 14, 60). The P2 domain also contains epitopes for neutralizing antibodies (27, 33) and is consistently highly variable even within the same genetic lineage of NoVs (38). ORF3 encodes a VP2 protein that is suggested to be a minor structural component of virus particles (18) and to be responsible for the expression and stabilization of VP1 (5).
Thus far, the NoVs found in nature are classified into five genogroups (GI to GV) and multiple genotypes on the basis of the phylogeny of capsid sequences (71). Among them, genogroup II genotype 4 (GII/4), which was present in humans in the mid-1970s (7), is now the leading cause of NoV-associated acute gastroenteritis in humans (54). The GII/4 is further subclassifiable into phylogenetically distinct subtypes (32, 38, 53). Notably, the emergence and spread of a new GII/4 subtype with multiple amino acid substitutions on the capsid surface are often associated with greater magnitudes of NoV epidemics (53, 54). In 2006 and 2007, a GII/4 subtype, termed 2006b, prevailed globally over preexisting GII/4 subtypes in association with increased numbers of nonbacterial acute gastroenteritis cases in many countries, including Japan (32, 38, 53). The 2006b subtype has multiple unique amino acid substitutions that occur most preferentially in the protruding subdomain of the capsid, the P2 subdomain (32, 38, 53). Together with information on human population immunity against NoV GII/4 subtypes (12, 32), it has been postulated that the accumulation of P2 mutations gives rise to antigenic drift and plays a key role in new epidemics of NoV GII/4 in humans (32, 38, 53).
Genetic recombination is common in RNA viruses (67). In NoV, recombination was first suggested by the phylogenetic analysis of an NoV genome segment clone: a discordant branching order was noted with the trees of the 3Dpol and capsid coding regions (21). Subsequently, many studies have reported the phylogenetic discordance using sequences from various epidemic sites in different study periods (1, 10, 11, 16, 17, 22, 25, 40, 41, 44-46, 49, 51, 57, 63, 64, 66). These results suggest that genome recombination frequently occurs among distinct lineages of NoV variants in vivo. However, the studies were done primarily with direct sequencing data of the short genome portion, and information on the cloned genome segment or full-length genome sequences is very limited (21, 25). Therefore, we lack an overview of the structural and temporal dynamics of viral genomes during NoV epidemics, and it remains unclear whether NoV mosaicism plays a role in these events.
To clarify these issues, we collected 199 near-full-length genome sequences of GII/4 from NoV outbreaks over three recent years in Japan, divided them into monophyletic subtypes, analyzed the temporal and geographical distribution of the subtypes, collected phylogenetic evidence for the viral genome mosaicism of the subtypes, identified putative recombination breakpoints in the genomes, and isolated mosaic genome segments from the stool specimens. We also performed computer-assisted sequence and structural analyses with the identified subtypes to address the relationship between the numbers of P2 domain mutations at the times of the outbreaks and the magnitudes of the epidemics. The obtained data suggest that intersubtype genome recombination at the ORF1/2 boundary region is common in the new GII/4 outbreaks and promotes the effective acquisition of mutation sets of heterogeneous capsid surface and viral replication proteins.
MATERIALS AND METHODS
Stool specimens.
The Norovirus Surveillance Group of Japan collected stool specimens from NoV-GII- or GII/4-positive individuals with acute gastroenteritis (n = 247). Most of the specimens were from NoV outbreaks around the collection sites. The group collected the specimens in spring, summer, autumn, and winter for 3 years: the 2006/2007 season (May 2006 to January 2007), 2007/2008 season (March 2007 to February 2008), and 2008/2009 season (May 2008 to February 2009). The collection sites were located at 20 different regional public health institutes in Japan (five samples from each institute per year). The genogroup of NoVs was evaluated by real-time reverse transcription-PCR (RT-PCR) (23). In some cases, the genotype of NoVs was evaluated by sequencing of the reverse transcription-PCR products of the ORF1 and ORF2 bordering region (29). Near-full-length genome sequences were obtained with 199 of the 247 specimens. Epidemiological information on 37 of the 199 samples from the 2006/2007 season was described previously (38). Information on the rest (n = 162) is described in Tables S1 and S2 in the supplemental material. Briefly, the 162 specimens were from outbreaks (n = 90), sporadic infection cases (n = 15), and undescribed cases (n = 57) during December 2006 to February 2009 in Japan. The major sites of the incidences were a nursing care center (n = 19), restaurant (n = 17), kindergarten (n = 15), hotel (n = 8), hospital (n = 7), sports event (n = 1), self-defense force (n = 1), family home (n = 1), elementary school (n = 1), and bank (n = 1), and one was undescribed (n = 91). The viral RNA copy numbers in the specimens ranged from 5.0 × 104 to 1.9 × 1011 copies/g stool (average, 6.1 × 109 copies/g stool) as judged by the real-time quantitative reverse transcription-PCR assay (23). All stool specimens were stored at −80°C until use.
Viral genome sequencing.
NoV GII/4 genome sequencing was done as described previously (38). Briefly, two overlapping fragments (approximately 5.2 and 2.5 kb) were amplified by RT-PCR from stool specimens. The PCR products were purified and used as a template for sequencing in a 96-well scale using an ABI 3730 xl DNA analyzer (Applied Biosystems, Foster City, CA). The sequences of 5.2-kb and 2.5-kb segments from the same individual were used to reconstruct near-full-length genome sequences (about 7.5 kb) by alignment at an overlapping region using the Staden Package (http://staden.sourceforge.net). The 5.2-kb fragment covers the complete ORF1 and the 5′ end of ORF2. The 2.5-kb fragment covers the 3′ end of ORF1, complete ORF2 and ORF3, and 3′-end noncoding region of the genome. The primers used for reverse transcription and nested PCR for the 5.2-kb fragment were GII4-1F/GII4r5412 (outer primer pair) and GII4-2F/GII4r5295 (inner primer pair) (38). Those for the 2.5-kb fragment were COG-2F/Tx30SXN (outer primer pair) and G2SKF/Tx30SXN (inner primer pair) (38). The initial 22 nucleotides at the 5′ ends of the reconstructed genomes were from PCR primers. The final 45 nucleotides at the 3′ ends of the genome were excluded from analysis because of the low levels of sequence accuracy. We obtained 199 near-full-length genome sequences from 247 GII-positive specimens. The 199 sequences included 37 GII/4 sequences previously reported between May 2006 and January 2007 (38) and 162 sequences newly obtained between December 2006 and February 2009.
Molecular cloning and sequencing of genome segments.
The 5.2-kb, 1.0-kb, and 2.8-kb genome segments were amplified by RT-PCR products as described above and cloned into pPCR-XL-TOPO vectors (Invitrogen, Carlsbad, CA). Each of the segments covers a junction of putative recombination breakpoints around the 5′ end of ORF2: the 5.2-kb segment contains the near-full-length ORF1 and 5′-end portion of ORF2, the 2.8-kb segment contains the 3′-end portion of ORF1, complete ORF2, and complete ORF3, and the 1.0 kb segment contains the 3′-end portion of ORF1 and 5′-end portion of ORF2. The primers used for the nested PCR of the 5.2-kb segment were the same ones described above: GII4-1F/GIIr5412 (outer primer pair) and GII4-2F/GIIr5295 (inner primer pair) (38). The primers used for the nested PCR of the 2.8-kb fragment were GII4f4117 (5′-CTGACAAAATTTATGGTAAGATCAAGAAGAGG-3′)/Tx30 SXN (outer primer pair) and GII4f4762 (5′-GACCCAGCTGGTTGGTTTGGAAAA-3′)/GII4r7516 (5′-ATAGTTTAGCGGCCGCATTCTTATCACATTACACCCGTGACTCCCCTCG-3′) (inner primer pair). The primers used for the nested PCR of the 1.0-kb fragment were GII4f4117/GII4r5412 (outer primer pair) and GII4f4223 (5′-GGTATGAATATGAATGAGGATG-3′)/GII4r5295 (inner primer pair). The single clones of the 5.2-kb, 2.8-kb, and 1.0-kb genome segments of the GII/4 subtypes were randomly chosen and sequenced in 96-well plates using an ABI 3730 xl DNA analyzer as described above.
Phylogenetic analysis.
Phylogenetic trees were constructed using the neighbor-joining method and maximum-likelihood method. Briefly, the near-full-length genome sequences from this study were aligned with the available GII/4 genome sequences from past NoV epidemics occurring over the past 3 decades, using CLUSTAL W software included in the MEGA software package, version 4.0 (58) (http://evolgen.biol.metro-u.ac.jp/MEGA/) and the MAFFT multiple sequence alignment software program, version 6.0 (26) (http://align.bmr.kyushu-u.ac.jp/mafft/software/). The neighbor-joining trees were constructed with the nucleotide substitution values estimated with the maximum composite likelihood model (59) using MEGA. The maximum-likelihood trees were inferred on the basis of the general time reversible models (31) using the PHYML software program included in the RDP3 software package (35) (http://darwin.uvigo.es/rdp/rdp.html). The reliability of interior branches in the phylogenetic tree was assessed by the bootstrap method with 1,000 resamplings. The GII/4 genome reference sequences were from samples taken before 1990 (<1990) (6 sequences, CHDC591-1974, CHDC2490-1974, CHDC4871-1977, CHDC4108-1987, Lordsdale, MD145-12/US/1987, and CHDC3967-1988), before 2000 (<2000) (2 sequences, and Dresden174/US/1997), in 2002/2003 (6 sequences, Famington Hill, B2S16/2002/UK, B5S22/2002/UK, Langen1061/2002/DE, YURI32073/2002/JPN, and MD-2004/2004/US), and in 2004/2005 (4 sequences, Guangzhou/NVgz01/CHN/2006, Chiba/04-1050/2005/JP, Sakai/04-179/2005/JP, and Ehime/05-30/2005/JP). Accession numbers for the reference genome sequences are given elsewhere (7, 38).
We initially constructed the phylogenetic trees with 199 genome sequences from the Japanese variants from 2006 to 2009 and 6 representative sequences of GII genotypes whose complete genome sequences were available in GenBank in October 2009 (GII/1, GII/3, GII/4, GII/6, GII/10, and GII/12; accession no. U07611, AB067542, X86557, AB039776, AY237415, and AB039775, respectively). The trees showed that the 199 genome sequences reproducibly grouped with the GII/4 reference sequences outside other GII references. The GII/4 cluster was positioned most closely to the GII/12 reference (Saitama U1/JP). Therefore, we used GII/12 as an outgroup in the present study for a better grasp of the relationship of the phylogeny among the Japanese GII/4 variant subgroups and between GII/4 and GII/12 variants.
Bootscanning-plot analysis.
Bootscanning-plot analysis was performed as described previously (69). Briefly, each query sequence was aligned with three NoV reference sequences using CLUSTAL W software, version 1.4 (62). The bootstrap values were plotted for a window of 300 bp, moving in increments of 10 bp along the alignment using the software program Simplot (48) (version 3.5.1; http://sray.med.som.jhmi.edu/SCRoftware/simplot/). Thus far, 19 genotypes of the NoV GII variants have been reported on the basis of complete capsid sequences (65, 71). Among them, only 7 genotypes have been fully sequenced at the genome level (GII/1, GII/3, GII/4, GII/6, GII/8, GII/10, and GII/12; accession numbers U07611, AB067542, X86557, AB039776, AB067543, AY237415, and AB039775, respectively). To search for sequences that are phylogenetically relevant to the query sequences, we constructed phylogenetic trees of the complete ORF1, ORF2, and ORF3 sequences using all available representatives of the 19 genotypes in the GenBank database. We also used the automated exploratory analysis tool included in the RDP3 software package (35). The genome sequence set used for the analysis consisted of 7 query sequences (2004/05, 2006a, 2006b, 2007a, 2007b, 2008a, and 2008b), all available GII genotype representatives, and all available GII/4 variant subgroups which caused epidemics over the past 34 years (7, 31). Two putative parent sequences with the best confidence values and a single distantly related sequence were used for the bootscanning plots with MEGA. The confidence values of the recombination events were also assessed with tools included in the RDP3 software package, such as RDP, GENECONV, Maxchi, Chimera, 3seq, and Siscan. The query sequences used in this study were Sakai2/2006/JP for the 2004/05 subtype (accession no. AB447448) and representative genomes of the 2007a (Osaka1/2007/JP), 2007b (Iwate5/2007/JP), 2008a (Hokkaido5/2008/JP), and 2008b (Hokkaido4/2008/JP) subtypes obtained in this study. The reference sequences were Saitama_U1/JP (GII/12 genotype [25], accession no. AB039775), B2S16/2002/UK (2002/03 subtype [38], accession no. AY587989), Saitama_U3/JP (GII/6 genotype [25], accession no. AB039776), Sakai2/2006/JP (2004/05 subtype [38], accession no. AB447448), Aomori1/2006/JP (2006a subtype [38], accession no. AB447432), Aichi3/2006/JP (2006b subtype [38], accession no. AB447446), and Hokkaido5/2008/JP and Hokkaido4/2008/JP (2008a and 2008b subtypes, respectively, obtained in this study).
Informative-site analysis.
The informative-site analysis was performed as described previously (50). Briefly, each query sequence was aligned with two putative parental sequences and an outgroup sequence. The alignments were used to identify informative sites that support alternative tree topologies between downstream and upstream regions using the Simplot software program (48), version 3.5.1. This information allowed identification of genome regions that were assigned as chimeras of heterologous sequences of distinct evolutionary origins. The statistical significance of the resultant division by the informative sites was evaluated by the maximal χ2 test using in-house programs. The programs were designed to execute the calculation algorithms described by Robertson et al. (50, 55).
Molecular modeling.
Three-dimensional (3-D) structural models of the capsid P-domain dimers were constructed by homology modeling as described previously (38). Briefly, the P-domain monomer models were first constructed using the crystal structure of the NoV capsid P domain of the GII/4 VA387 strain at a resolution of 2.00 Å (PDB code 2OBS [13]) as the template. The P domains of the GII/4 subtypes described in this study have sequence similarities of greater than 90% to that of VA387, high enough to construct models with a root mean square distance (RMSD) of ∼1 Å for the main chain between the predicted and actual structures (3). The P-domain monomer models were used to construct the P-domain dimer models by superimposing the chains A and B using the crystal structure of the NoV capsid dimer (PDB code 1IHM [47]).
Nucleotide sequence accession numbers.
The DDBJ database accession numbers for the nucleotide sequences of NoV genomes for the 2006/2007 season (n = 37) have been reported elsewhere (38). The DDBJ database accession numbers for the nucleotide sequences of NoV genomes for the 2007/2008 and 2008/2009 seasons (n = 162) are AB541201 to AB541362. The DDBJ database accession numbers for the nucleotide sequences of NoV genome segment clones (n = 11) are AB541190 to AB541200.
RESULTS
Phylogenetic classification of NoV GII/4 subtypes in Japan during 2006 and 2009.
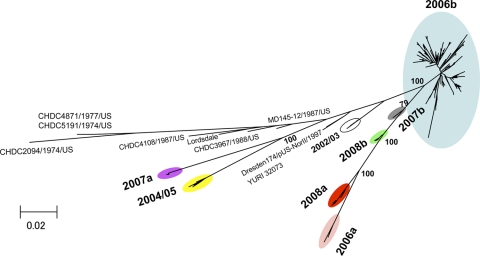
First, we investigated the phylogeny of the NoV near-full-length genome sequences (about 7.5 kb). For this study, we used sequences obtained in this study from 19 sites in Japan between May 2006 and February 2009 (n = 199), various reported GII/4 reference sequences of past global or Japanese epidemics, and various reported outgroup sequences of other NoV genotypes. Figure 1 shows a maximum-likelihood tree constructed with the 199 Japanese genome sequences and the 19 GII/4 reference sequences from past NoV epidemics throughout the world during the 1970s and 1980s (7), <2000, in 2002/2003, and in 2004/2005 (38). The tree shows that the 199 Japanese sequences are divisible into 7 distinct lineage groups within a GII/4 cluster with a high bootstrap value (the 7 colored ovals in Fig. 1). The monophyly of the 7 GII/4 groups was reproducible independently of the algorithms to infer the phylogeny and reference sequences used. We tentatively named the 7 monophyletic subtypes of GII/4 variants 2004/05, 2006a, 2006b, 2007a, 2007b, 2008a, and 2008b.
FIG. 1.
Phylogenetic classification of the NoV GII/4 subtypes in Japan during 2006 and 2009. The maximum-likelihood tree was constructed with the near-full-length genome sequences (about 7.5 kb) obtained from stool specimens collected at 19 sites in Japan between May 2006 and February 2009 in this study (n = 199) and GII/4 reference genome sequences from past epidemics in Japan and other countries in the <2000, 2002/2003, and 2004/2005 winter seasons (7, 38) (n = 18). The sequence clusters enclosed by colored ovals indicate the 7 monophyletic GII/4 subtypes identified in Japan in previous (38) and present studies.
The 2004/05 genome sequences were first obtained in Japan in the winter of 2004-2005 (accession no. AB220921 to AB220923 [42]). The geographic distribution of the 2004/05 sequences seemed to be restricted to East Asia (54). The 2006a and 2006b genome sequences were first obtained in Japan during the winter of 2006-2007 (accession no. AB447427 to AB447463 [38]). The 2006a and 2006b sequences were detected in many countries in Europe, North America, and East Asia during 2006-2007, wherein the 2006b subtype was generally more dominant than the 2006a subtype (54). The 2007a, 2007b, 2008a, and 2008b genome sequences were newly obtained in this study. Phylogenetic tree analyses showed that the nucleotide sequences of ORF2 of the 2008a subtype were genetically closely related to the ORF2 sequence obtained in the Netherlands in 2008 (accession no. AB445395), and together these sequences formed a single monophyletic group with a high bootstrap value (data not shown). These results suggest that at least 4 of the 7 GII/4 subtypes identified in Japan during 2006 and 2009, i.e., the 2004/05, 2006a, 2006b, and 2008a subtypes, caused NoV infections outside Japan.
We estimated the genetic divergence within and between the 7 monophyletic groups on the basis of the maximum composite likelihood model using MEGA software. The intragroup divergence was comparably high in the 2006b subtype among the 7 groups (see Table S3, diagonal lines, in the supplemental material), suggesting that the diversity of the 2006b genome is higher than that of the other subtypes. This is consistent with the epidemiological data that 2006b had predominated for 3 years in Japan whereas the others emerged only temporally. The intergroup divergence was comparably high between 2004/05 and the other groups and between 2007a and the other groups, and about 12 to 15% sequence divergence existed in the genomes (see Table S3, bottom left portion, in the supplemental material).
Temporal and geographical distribution of Nov GII/4 subtypes in Japan.
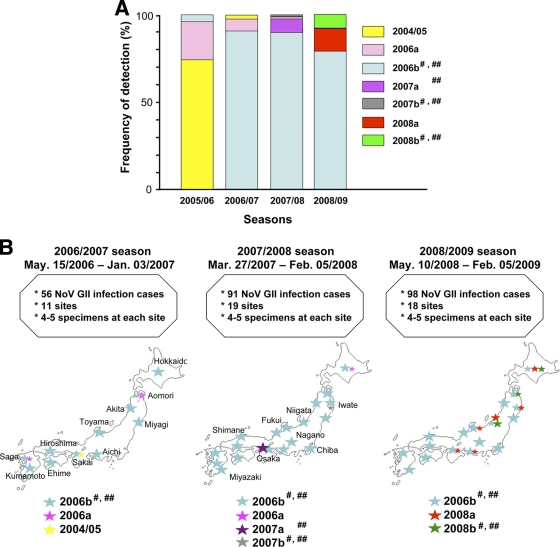
We then analyzed the temporal and geographical distribution of the 7 GII/4 subtypes in Japan. The 199 near-full-length genome sequences were divided into 3 groups according to the collection periods: the 2006/2007 (May 2006 to January 2007) (n = 39), 2007/2008 (March 2007 to February 2008) (n = 78), and 2008/2009 (May 2008 to February 2009) (n = 82) seasons. The frequencies of detection of particular NoV subtypes were obtained for each of the three seasons. We also used published subtyping data for the analysis of the previous winter season in Japan (November 2005 to March 2006) (n = 38) (38, 43).
The 2004/05 and 2006a sequences were detected at multiple collection sites and were prevalent in the 2005/2006 season (38, 43) (Fig. 2 A and B, 2004/05 and 2006a). However, they became minor in the 2006/2007 season and were hardly detected thereafter. The 2006b sequences were minor in the 2005/2006 season (38) (Fig. 2A, 2006b). However, they rapidly became dominant in the 2006/2007 season and continually predominated in most of the collection sites in Japan, representing 176 of the 199 genome sequences (88.4%) during the study period. This result is consistent with the data of partial capsid sequences obtained during December 2007 to January 2008 in Japan (28). The 2007a and 2007b sequences were detected only at single collection sites in the 2007/2008 season (Fig. 2A and B, 2007a and 2007b). The 2008a and 2008b sequences were detected most recently at multiple collection sites in the 2008/2009 season (Fig. 2A and B, 2008a and 2008b). These data indicate that the 2006b subtype displaced the 2004/05 subtype in the 2006/2007 season and continued to predominate for the next 2 years in Japan. During the period of the 2006b predominance, however, several GII/4 subtypes caused NoV outbreaks in Japan, and the frequencies and sites of non-2006b outbreaks increased slightly in the 2008/2009 season.
FIG. 2.
Temporal and geographical distribution of the NoV GII/4 subtypes in Japan. The 199 near-full-length genome sequences were divided into 3 subgroups according to the collection periods: the 2006/2007 (May 2006 to January 2007) (n = 39), 2007/2008 (March 2007 to February 2008) (n = 78), and 2008/2009 (May 2008 to February 2009) (n = 82) seasons. For the analysis of the 2005/2006 season, published subtyping data (38, 43) were used (n = 38). (A) Frequencies of detection of particular NoV GII/4 subtypes in each season in Japan. (B) Geographic locations of the GII/4 subtype outbreaks. Colored stars indicate the locations of sample collection sites. Larger stars indicate the collection sites with greater frequencies of detection. #, ORF2s were classified as the same phylogenetic group (see Fig. 3A, ORF2). ##, ORF3s were classified as the same phylogenetic group (see Fig. 3A, ORF3).
Phylogenetic evidence for NoV genome mosaicism.
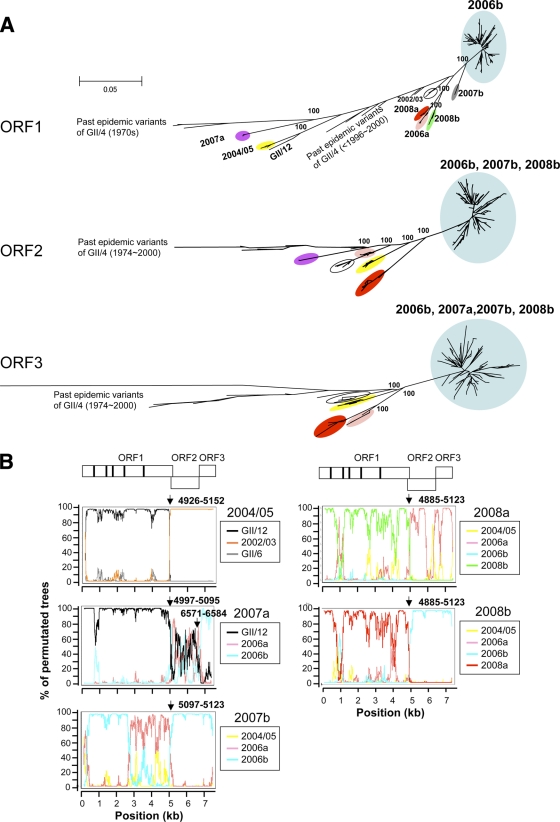
Next, we investigated the possibility of genome mosaicism of the 7 GII/4 subtypes. For this purpose, we first compared the branching orders of the subtype clusters in the maximum-likelihood and neighbor-joining trees of the ORF1, ORF2, and ORF3 sequences using representative sequences of the 19 GII genotypes (GII/1 to GII/19) reported to date in the GenBank database. Figure 3A shows the maximum-likelihood trees, in which most of the non-GII/4 sequences were positioned far from the GII/4 cluster and were therefore excluded for a better grasp of the relationship of the phylogeny among the GII/4 variant subgroups. The exception was the ORF1 tree, in which the GII/12 sequence branched inside the GII/4 cluster. The comparisons of the three trees revealed that there was marked inconsistency in the branching orders of the GII/4 subgroups. The inconsistency was reproducible independently of the algorithms to infer the phylogeny and reference sequences used. First, the ORF1 sequences of the 2006b, 2007a, 2007b, and 2008b subtypes formed independent monophyletic clusters, whereas the ORF2 sequences of the 2006b, 2007a, and 2008b subtypes formed a single cluster and the ORF3 sequences of these four subtypes formed the same cluster (Fig. 3A, light blue circles). Second, the ORF1 sequences of the 2004/05 subtype were clustered near the ORF1 sequence of a GII/12 strain (Saitama_U1/JP [25]) and relatively distant from the reported GII/4 reference sequences, whereas the ORF2 and ORF3 sequences of the 2004/05 subtype were very distantly related to the Saitama_U1/JP sequence and closely related to the GII/4 reference sequences (Fig. 3A, yellow circles). Third, the branching orders of the 2008a sequences were also different in the ORF1, ORF2, and ORF3 trees (Fig. 3A, red circles). These results suggested that most subtypes identified in this study had mosaic genomes.
FIG. 3.
Phylogenetic evidence for NoV genome mosaicism. (A) Maximum-likelihood trees of the nucleotide sequences of the complete ORF1 (about 5.1 kb), ORF2 (about 1.6 kb), and ORF3 (about 0.8 kb). The trees were constructed with the sequences obtained in previous (38) and present studies (n = 199) and the reference sequences described in Fig. 1. The GII/12 sequence (Saitama_U1/JP [25]) was used as an outgroup sequence in each tree but is shown only in the ORF1 tree. In the ORF2 and ORF3 trees, the GII/12 sequence was located far apart from the GII/4 cluster and is not shown for simplicity. (B) Bootscanning plots of nucleotide sequences of near-full-length NoV genomes. A query genome sequence (2004/05, 2007a, 2007b, 2008a, or 2008b) was aligned with three reference sequences, two sequences that were positioned relatively closely to the query sequence in the neighbor-joining trees and a sequence that was distantly related to the query sequence, using CLUSTAL W software, version 1.4 (62). The bootstrap values are plotted for a window of 300 bp moving in increments of 10 bp along the alignment using the program Simplot (48). Informative-site analyses (50) were performed using the same query and reference sequence set. Arrows indicate putative recombination breakpoints with the highest statistical significance (P ≤ 0.000001) in the informative-site analysis.
To further assess this possibility, we performed bootscanning-plot analyses as described previously (69). For each bootscanning plot, we used a query genome sequence of a given subtype, two to three reference sequences that were positioned relatively closely to the query sequence in the neighbor-joining trees, and a distantly related outgroup sequence. The analyses showed that the genomes of the 2004/05, 2007a, 2007b, 2008a, and 2008b subtypes were indeed composed of multiple segments from recently prevalent or as-yet-undefined genogroups, genotypes, and subtypes of NoVs in this and previous reports (2, 7, 25, 38, 53, 65, 71) (Fig. 3B; see also Fig. 3A). The 2004/05 genome (Sakai2/2006/JP) was comprised of the ORF1 related to GII/12 (Saitama_U1/JP) and the ORF2/3 related to GII/4 2002/03 (B2S16/2002/UK). The 2007a genome (Osaka1/2007/JP) was made up of the ORF1 related to GII/12, the ORF2 of as-yet-undefined classes of GII/4, and the ORF3 related to GII/4 2006b (Aichi3/2006/JP). The 2007b genome (Iwate5/2007/JP) was made up of the ORF1 related to GII/4 2006b and 2006a (Aomori1/2006/JP) and the ORF2 and ORF3 related to GII/4 2006b. The 2008a genome (Hokkaido4/2008/JP) was comprised of the ORF1 related to GII/4 2008b (Hokkaido5/2008/JP) and as-yet-undefined classes of GII/4 and the ORF2 and ORF3 of GII/4 2006a and as-yet-undefined classes of GII/4. The 2008b genome was made up of the ORF1 of 2008a and the ORF2 and ORF3 related to GII/4 2006b. We also investigated possible genome mosaicism for the 2006a and 2006b subtypes, but we could not identify putative ancestral sequences of ORF1, ORF2, and ORF3 that were genetically closely related to 2006a and 2006b when we used the available NoV sequences in the public database as references.
To define statistically the possible recombination breakpoints of the 2004/05, 2007a, 2007b, 2008a, and 2008b genomes, we performed informative-site analysis (50) using the same reference sequences used in the bootscanning-plot analysis. With this approach, we identified several patches of genome regions that were assigned with statistical significance as putative recombination breakpoints. Notably, a putative breakpoint located around the junction of ORF1 and ORF2 constantly gave the highest statistical significance, i.e., maximum χ2 values, in the 2004/05, 2007a, 2007b, 2008a, and 2008b genomes (P ≤ 0.000001) (Fig. 3B, arrows). The results were in good agreement with the phylogenetic-tree and bootscanning-plot analyses. These data consistently suggest that the new GII/4 subtypes identified in Japan were mostly hybrid viruses composed of viral protein elements from distinct genetic lineages of NoVs.
We further assessed possible genome recombination events using other tools included in the RDP3 software package (30). The analysis again identified single recombination breakpoints with the best or second-best confidence values around the junction of ORF1 and ORF2 in the 2004/05, 2007a, 2007b, 2008a, and 2008b genomes (P < 0.001). The analysis also identified additional putative breakpoints around the junction of ORF2 and ORF3 of 2007a. However, we could not obtain evidence for genome mosaicism with 2006a and 2006b using a selected sequence data set of the NoV GII genotypes reported to date (GII/1 to GII/19) (25, 65, 71) and GII/4 subtypes (7, 38, 53). Because information on the entire genome sequences of NoV is very limited, it remains to be determined whether 2006a and 2006b also have mosaic genomes.
Isolation of NoV mosaic genome segments.
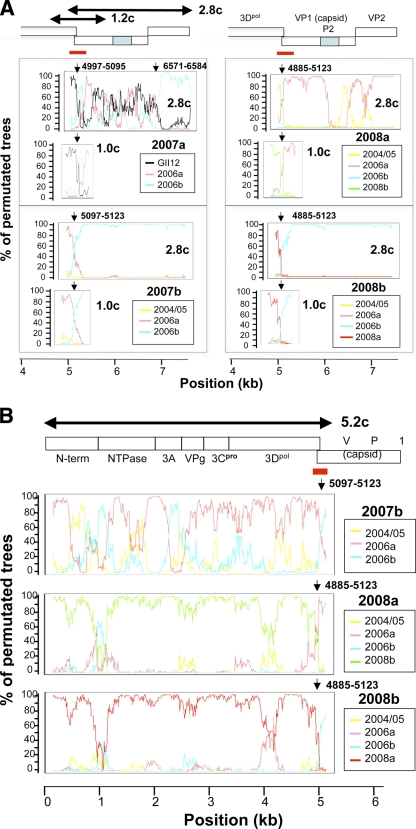
To clarify the presence of the mosaic viral genomes in nature, we cloned and sequenced the genomes of the 2007a, 2007b, 2008a, and 2008b subtypes. For this study, we cloned the genome segments, i.e., the 5.2-kb, 2.8-kb, and 1.0-kb genome segments, that presumably contain a junction of putative recombination breakpoints around the ORF1/ORF2 junction (Fig. 4). The subtype 2004/5 was not included in the cloning analysis because we did not have sufficient amounts of clinical specimens for the cloning. Likewise, the subtypes 2006a and 2006b were not included because the major parental sequences are not clear in the present study. We successfully obtained the molecular clones except for the 5.2-kb fragment of 2007a. We could amplify but failed to clone the 5.2-kb fragment of 2007a. Although the precise reason for the failure is unclear at present, it might be due to the decreased cloning efficiency of the larger insert by the TA-cloning method. Because the appropriate restriction enzyme sites for the cloning were absent in the 2007a 5.2-kb fragment, we did not include this fragment in further analyses. Nucleotide sequences of the segments were used for the bootscanning-plot analysis using the same sets of reference sequences described in Fig. 3B, and the statistically significant putative recombination breakpoints were assessed by informative-site analysis.
FIG. 4.
Isolation of NoV mosaic genome segments. Three genome segments (5.2, 1.0, and 2.8 kb) were amplified from the 2007a, 2007b, 2008a, and 2008b stool specimens, cloned into plasmid vectors, and sequenced. Nucleotide sequences of the cloned segments were subjected to the bootscanning-plot analysis using the same sets of reference sequences described in Fig. 3B, and the putative recombination breakpoints were assessed by informative-site analysis. (A) Results for the 2.8-kb and 1.0-kb genome segment clones (2.8c and 1.0c). (B) Results for the 5.2 kb-genome segment clones (5.2c). Red bars indicate the ORF1/ORF2 bordering region. Arrows indicate the putative recombination breakpoints with the highest statistical significance (P ≤ 0.000001) in the informative-site analysis.
Figure 4 shows representative results of the bootscanning-plot and informative-site analyses with the 5.2-, 1.0-, and 2.8-kb segment clones. Importantly, all 11 clones from the 2007a, 2007b, 2008a, and 2008b stool specimens had the same putative recombination breakpoints, with the highest statistical significance around the ORF1/ORF2 junction region identified with direct sequencing analyses (Fig. 4A and B, arrows). In addition, the patterns of the bootscanning plots were almost identical over the viral genomes examined between the sequences of the uncloned and cloned genome segment except for the 5′-half region of the 2007b ORF1 (Fig. 3B and 4). Although the precise reason for the discrepancy is unclear at present, it might be due to the cloning of the minor population of the 2007b quasispecies in the stool specimens. The overall good agreement of the results by the two sequencing strategies strongly suggests that the genome mosaicisms we found by analysis of the direct sequencing data were intrinsic rather than an artifact of the analysis. Taken together, these data indicate that the NoV mosaic genomes were present in the human stool specimens and that the ORF1/ORF2 junction region is the common hot spot for generating the mosaic genomes in GII/4 subtypes in nature.
Amino acid signatures of the NoV GII/4 subtypes.
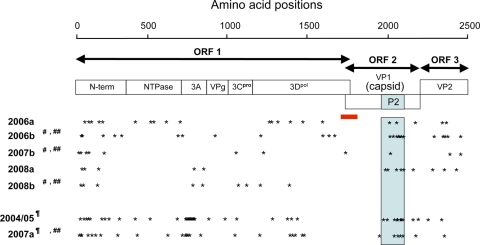
We then investigated sequential characteristics of the proteins of the 7 GII/4 subtypes by searching for unique amino acid signatures in viral proteins. The deduced amino acid sequences of ORF1, ORF2, and ORF3 of a given subtype were aligned with reference sequences of the past GII/4 subtypes (38) that were identified before detection of the query subtype. Amino acids specific to the query subtype were extracted and referred to as amino acid signatures of the new epidemic subtype. In the case of the 2006b subtype, we also analyzed the changes in the signatures in the capsid protein VP1 between 2006 and 2009, because information on the structure and function is more abundant for the capsid than for other viral proteins.
The amino acid signatures of the 7 GII/4 subtypes were distributed throughout the three ORFs (Fig. 5, asterisks). 2004/5 and 2007a had more substitutions in ORF1 than the others because their ORF1s seemed to have originated with the GII/12 relatives (Fig. 3A and 4). When they were compared with the two available complete ORF1 sequences of GII/12, they still had many amino acid substitutions (27 and 63 for 2004/05 and 2007a, respectively). 2007b and 2008b had fewer substitutions in ORF2s and ORF3s than the others because these regions seemed to have originated from the 2006b relatives.
FIG. 5.
Amino acid signatures of the NoV GII/4 subtypes. The deduced amino acid sequences of ORF1, ORF2, and ORF3 of a given GII/4 subtype were aligned with the GII/4 sequences identified before the outbreak season of the subtype. Amino acids specific to each subtype at the time of its first outbreak season were extracted and referred to as amino acid signatures of the new epidemic subtype. Asterisks illustrate approximate locations of the amino acid signatures in ORF1, ORF2, and ORF3. A light-blue box denotes approximate locations of the capsid P2 domain in ORF2. A red bar indicates the ORF1/2 boundary region where the single putative recombination breakpoint was assigned for each subtype genome by informative-site analyses (50). ¶, ORF1s were similar to those for GII/12 (see Fig. 4, ORF2). 2004/05 and 2007a had 27 and 63 amino acid substitutions, respectively, in ORF1s compared to the two available complete ORF1 sequences of GII/12 (accession numbers AB045603 and AB039775). #, ORF2s were classified as the same phylogenetic group (see Fig. 3A, ORF2). ##, ORF3s were classified as the same phylogenetic group (see Fig. 3A, ORF3).
As seen in the 2006b variants in the 2006/2007 season (38), the capsid protein signatures were preferentially distributed on the P2 domain in other GII/4 subtypes (Fig. 5, blue box). All 7 capsid signatures identified in the 2006b variants in the 2006/2007 season were highly conserved during the 2006/2007 season, although two of them (P357 and N412) were gradually lost in the 2006b variant population during 2007 and 2009. Instead, other amino acid substitutions were sporadically accumulated in the P2 domain of the later 2006b variants (data not shown). The 7 signatures in the P2 domain were also well retained in the 2007b and 2008b subtypes, whose genomes had capsid gene segments from the 2006b relatives (Fig. 3B). These data indicate that (i) all of the 7 GII/4 subtypes had unique amino acid substitutions in viral capsid and replication proteins at the time of their outbreaks in Japan, (ii) the dominant 2006b subtype retained the capsid signatures during its persistence between 2006 and 2009, and (iii) some GII/4 subtypes acquired unique mutation sets of the 2006b capsid P2 domain by putative genome recombination events.
3-D locations of the subtype-specific amino acids in the capsid P domain dimer.
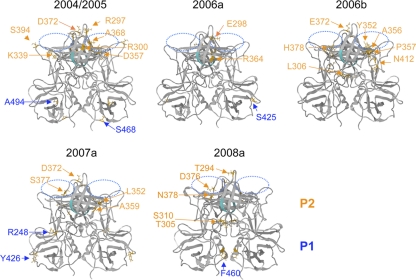
To clarify 3-D locations of the capsid signatures, we constructed structure models of the VP1 P-domain dimer of the GII/4 subtypes by the homology modeling method as described previously (38). The 2007b and 2008b models were not included for the study because their capsid proteins had no signature or a single signature in the P2 domain due to putative genome recombination with 2006b (Fig. 3, 4, and 5). The thermodynamically and sterically optimized structural models of the P-domain dimer of the 2004/05, 2006a, 2006b, 2007a, and 2008a subtypes showed no major differences in the folding of the main chains (Fig. 6). This result suggests that the capsid amino acid substitutions primarily influenced physicochemical properties around the substitution sites by changing the size and chemical properties of the side chains. These models were then used to map the 3-D locations of the P2 domain mutations.
FIG. 6.
3-D locations of the subtype-specific amino acids in the capsid P domain dimer. Structural models of the capsid P domain dimers of recent NoV subtypes were constructed by homology modeling as described previously (38). The 2007b and 2008b capsid models were not included because their ORF2s were classified as belonging to the same phylogenetic group as ORF2 of 2006b due to putative genome recombination (Fig. 3 and 4), and their capsid proteins had no signature or only a single signature in the P2 domain with 2006b (Fig. 5). Orange arrows and letters indicate locations and types of the unique amino acids in each GII/4 subtype at the time of its first outbreak season. Putative functional sites for virus entry into the cells are highlighted. Blue-dotted ovals, the fucose ring binding sites formed by the P domain dimer (8, 13); cyan chain, an RGD motif (60) on the β2 sheet of the P domain.
Importantly, the capsid P2 domain signatures were mostly mapped on the outer surface loops in all of the GII/4 subtypes examined (Fig. 6). These loops form an accessible protein surface with which host proteins, such as a cellular receptor(s) and antibodies, can directly interact. The P2 mutations are often positioned near the putative functional sites for virus entry into the cells: the fucose ring binding sites formed by the P-domain dimer (8, 13) (blue-dotted ovals) and an RGD motif (60) on the β2 sheet of the P domain (cyan chain). Notably, the P2 mutations were more abundant in the widely prevalent subtypes, 2004/05 and 2006b, than in the others (Fig. 2 and 6). The 2008a subtype, which was detected most recently and caused NoV epidemics at multiple sites in the 2008/2009 season, also had 5 unique substitutions in the P2 domain (Fig. 2 and 6). These data indicate that the GII/4 subtypes that were dominant between 2004 and 2009 in Japan had a greater number of unique amino acids preferentially positioned on their capsid surfaces at the time of their first outbreaks.
DISCUSSION
In this study, we have examined the possible involvement of genome recombination in the generation of new outbreaks of the NoV GII/4 variants. We first analyzed the evolutionary lineage of the GII/4 variants that were present in Japan during 2006 to 2009 and clarified their temporal and geographical distribution. We showed the following: (i) that at least 7 monophyletic GII/4 subtypes were present in humans during the 3-year period, (ii) that 3 to 4 subtypes were cocirculated in each NoV season, and (iii) that the 2006b subtype had spread and persisted more effectively in human communities than the other GII/4 subtypes during the study period (Fig. 1 and 2). These and other findings on the recent GII/4 subtypes (32, 38, 53, 54) consistently support the notion that the 2006b subtype had some selective advantages over the other GII/4 subtypes, which allowed it to quickly spread throughout human communities at the time of its initial appearance in the early winter of 2006. Our study additionally suggests that the possible advantages of the 2006b subtype remained effective over the subsequent 2 years in Japan. However, we could not obtain evidence for genome mosaicism with the 2006b subtype using the available sequence data set of the NoV genogroups, genotypes, and subtypes reported to date (2, 7, 25, 38, 53, 65, 71). Therefore, it is not clear whether genome recombination played a significant role in the generation of new large-scale outbreaks. Because information on the entire genome sequences of NoV is very limited at present, further genome study of NoVs is necessary to clarify this issue.
We then analyzed the GII/4 subtypes to determine whether they showed genome mosaicisms. We demonstrated clearly and for the first time that intersubtype genome recombination is common in the new NoV GII/4 outbreaks: 4 of the 7 new GII/4 subtypes (2007a, 2007b, 2008a, and 2008b) were mosaics comprised primarily of sequences of the recently prevalent distinct GII/4 subtypes, and 1 (2004/05) was made up of GII/4 and GII/12 genotypes (Fig. 3 and 4). Because of the genome mosaicism, the number of monophyletic clusters of the new variants in the phylogenetic trees differed depending on the region of the genome studied; the numbers of clusters were 7, 7, 5, and 4 for the near-full-length genome, ORF1, ORF2, and ORF3, respectively. Sequences of 11 randomly selected genome segment clones all exhibited bootscanning-plot profiles identical to those obtained with the direct sequencing data except in one case, suggesting dominance of the specific mosaic genomes in the corresponding stool specimens. Phylogenetic-tree, bootscanning-plot, and informative-site analyses consistently provided the same conclusions in regard to genome mosaicism: these virus genomes encoded capsid proteins whose evolutionary lineages were distinct from those of nonstructural proteins. The good agreement of the results by the two sequencing strategies and by three evolutionary methods strongly suggests that the mosaic genomes made from multiple GII/4 subtypes were indeed constantly arising in vivo and became the dominant species in infected individuals in some of the NoV GII/4 outbreaks.
We failed to find evidence for the genome mosaicism of the 2006a and 2006b subtypes by using available NoV sequences. These subtypes are unlikely to be the intergenotype or intergenogroup recombinants, because their ORF1, ORF2, and ORF3 sequences constantly showed the strong monophyly within the GII/4 cluster out of the other genotypes and genogroups at their first (38) and successive outbreaks (Fig. 3A). However, the possibility of intersubtype recombination among as-yet-defined classes of GII/4 subtypes remains undetermined for the 2006a and 2006b subtypes.
Interestingly, the mosaic genomes that caused the new NoV GII/4 outbreaks all had the putative recombination breakpoints with the highest statistical significance in the ORF1/2 boundary region (P ≤ 0.000001) (Fig. 3B and 4). This breakpoint location is consistent with previous reports on intergenogroup and intergenotype recombination (1, 10, 11, 17, 21, 22, 25, 40, 41, 44-46, 49, 57, 63, 64, 66), suggesting the presence of a common hot spot for generation and survival of recombinant NoVs in nature. To a lesser extent, a putative recombination event around the ORF2/3 boundary was identified in 1 of 7 new variant subgroups (Fig. 4A, 2007a). A recombination event around the ORF2/3 junction has also been reported for GII/4 variants circulating in Cairo, Egypt, between 2006 and 2007 (24). The ORF1/2 boundary region is highly conserved in NoV GII/4, as shown previously by very low scores of Shannon entropy within the reported GII/4 sequences (38). This and our present findings on the presence of the putative parent GII/4 sequences of the mosaic genomes suggest that the ORF1/2 mosaic genomes we identified were generated by homologous recombination, as seen in other single-stranded, positive-sense RNA viruses, including poliovirus (20), foot-and-mouth disease virus (36), brome mosaic virus (9, 39), turnip crinkle virus (70), and tomato ringspot virus (52). If this were the case, the intersubtype recombination at the ORF1/2 boundary region would occur and generate variable recombinant viruses in vivo more frequently than the intergenotype and intergenogroup recombination would, because the boundary region and neighboring sequences are more similar within the NoV subtype than within the genotype and genogroup. Our results are consistent with this possibility.
The presence of putative recombination at the ORF1/2 boundary region has a direct impact on the modes of NoV subtype evolution in vivo. First, the presence of the breakpoint at this region drives independent evolution of ORF1 and ORF2/3 nucleotide sequences and thus of nonstructural and capsid proteins (Fig. 3), leading to divergent evolution of the NoV GII/4 genome (Fig. 1). Second, the presence of the breakpoint allows concurrent acquisition of new mutation sets that arise independently in ORF1 and ORF2/3 among distinct GII/4 subtypes. However, further study is necessary to clarify whether the genome recombination indeed confers any fitness advantage to the virus within a mixed NoV variant population in nature.
The high levels of sequence homology of the ORF1/2 boundary region (38) also suggest that the region is functionally and/or structurally very important for NoV replication and receives strong selective constraints against diversity for NoV survival in nature. Consistently, this region is indicated to contain an important functional motif that regulates capsid expression from a full-length genome in bovine NoV (37). Thus, the ORF1/2 boundary region may be a multifunctional region critical for both replication and evolution of NoVs.
The relatively high detection frequency of the ORF1/2 mosaic genomes in the new GII/4 subtypes (5 of 7) was rather unexpected, because multiple factors, such as retention of virion stability, viral infectivity, and viral replication capabilities in human cells, should restrict the generation of viable hybrid viruses. The present findings therefore raise the possibility of large-scale coinfections by distinct lineage groups of NoVs and of natural selection for the particular ORF1/2 hybrid viruses. The former possibility remains to be clarified but is feasible (57) if one considers the high stability of the NoV virion outside the host, as well as NoV transmission modes, i.e., ingestion of contaminated food and water, direct person-to-person contact, and exposure to contaminated airborne vomitus droplets in a semiclosed community (15).
The latter possibility of natural selection also remains to be clarified. However, it is possible that some of the unique mutations identified in each ORF1/2 hybrid genome at the time of their outbreaks (Fig. 5) may be involved in the survival of the hybrid viruses. In this regard, it is noteworthy that the hybrid viruses had multiple mutations in the N-term, NTPase, 3A-like, Vpg, 3Cpro, and 3Dpol proteins. These proteins are likely to function primarily in NoV replication in host cells (19). Therefore, acquisition of an appropriate mutation set in ORF1 might confer some advantages in replication of the hybrid viruses in particular hosts. It should also be noted that the 2007b and 2008b subtypes encoded the VP1 and VP2 proteins from 2006b (Fig. 3 and 4). VP1 plays critical roles in binding to the putative infection receptors (8, 60, 61) and antibody neutralization (33, 34). The VP2 protein is also essential for the production of infectious virions in caliciviruses (56). Therefore, acquisition of an appropriate mutation set in ORF2 and ORF of 2006b might confer some advantages in infection and/or immune escape of the hybrid viruses at some outbreaks.
Computer-assisted modeling studies provide a structural basis for addressing the potential selective advantages of the capsids of the new GII/4 subtypes. We showed that unique capsid amino acids of the 7 GII/4 subtypes identified in this study were preferentially positioned on the outer surface loops of the protruding P2 domain and were more abundant in the dominant subtypes (Fig. 6). This is also a common characteristic of the past epidemic GII/4 subtypes (32, 38, 53). These findings suggest that physicochemical changes in the capsid surface are a prerequisite for effective virus spread of NoV GII/4 in humans. The specific mutations around the outer surface loops of the protruding P2 domain can modulate the local electrostatic environment and shape of the exposed capsid surface by changing the chemical properties and the size of side chains, respectively. Therefore, acquisition of an adequate set of capsid P2 mutations might be able to decrease antibody affinity without decreasing affinity to the infection receptors of GII/4. This would confer an advantage to the variants that would allow them to spread in human communities in the presence of immunity against precirculated variants. To effectively gain such a set of P2 domain mutations, as well as those of nonstructural proteins, genetic recombination around the ORF1/2 boundary region may be an ideal mechanism. Establishment of a tissue culture system to support effective replication of human NoVs, as well as a reverse genetics system to study the roles of mutations in NoV infection and replication, will be critical to clarify each of these possibilities.
It should be noted that despite the prolonged dominance of the 2006b subtype, the magnitudes of NoV epidemics in Japan have gradually declined since 2007: the total numbers of reported NoV infection cases during October and March of 2007-2008 and 2008-2009 showed more than 2- and 5-fold decreases, respectively, compared with the same period in 2006-2007 under the same surveillance system (Infectious Disease Surveillance Center [http://idsc.nih.go.jp/iasr/index.html]). These observations may imply that biological niches within human communities that support replication of the 2006b subtype have gradually been shrinking in Japan since 2007. A possible explanation for this phenomenon is that immunity against the 2006b subtype has been gradually strengthened in human populations due to the persistence of the 2006b infections in Japan. Nevertheless, none of the new GII/4 variant subtypes were able to replace the 2006b epidemic in the 2007/2008 and 2008/2009 seasons. In addition, two of the four new putative recombinants (2007b and 2008b), which appeared in the 2007/2008 and 2008/2009 seasons, gained ORF2/3 of 2006b. These observations may imply that 2006b still had some selective advantages over other GII/4 variant subgroups in the 2008/2009 seasons. Further follow-up study is necessary to address these possibilities.
Our findings on genome mosaicism may have an impact on epidemiological and virological studies of NoVs. For example, mosaicism could influence the validity of NoV classification, which is based on the sequences of parts of the NoV genome. Because hybrid viruses that cause epidemics seem to share a recombination breakpoint around the ORF1/2 boundary region, this junction segment may be useful for monitoring the prevalence of hybrid NoVs in nature. The genome mosaicism could also impact measurement of the mutation rates of NoVs in nature: careful selection of the genome segments that contain no recombination breakpoints would be critical to measure the nucleotide substitution rates. Continual accumulation of information on the complete genome sequences of NoVs in natural and living environments will provide genetic bases for dealing with these issues and illustrate mechanisms by which NoV evolves to generate and sustain new epidemics in human populations.
Supplementary Material
Acknowledgments
We thank T. Shiino for helpful comments and suggestions.
This work was supported by grants for Research on Food Safety from the Ministry of Health, Labor, and Welfare, Japan, and for Research on Publicly Essential Drugs and Medical Devices from the Japan Health Sciences Foundation.
Footnotes
Published ahead of print on 9 June 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ambert-Balay, K., F. Bon, F. Le Guyader, P. Pothier, and E. Kohli. 2005. Characterization of new recombinant noroviruses. J. Clin. Microbiol. 43:5179-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181(Suppl. 2):S336-S348. [DOI] [PubMed] [Google Scholar]
- 3.Baker, D., and A. Sali. 2001. Protein structure prediction and structural genomics. Science 294:93-96. [DOI] [PubMed] [Google Scholar]
- 4.Belliot, G., S. V. Sosnovtsev, T. Mitra, C. Hammer, M. Garfield, and K. Y. Green. 2003. In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J. Virol. 77:10957-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertolotti-Ciarlet, A., S. E. Crawford, A. M. Hutson, and M. K. Estes. 2003. The 3′ end of Norwalk virus mRNA contains determinants that regulate the expression and stability of the viral capsid protein VP1: a novel function for the VP2 protein. J. Virol. 77:11603-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertolotti-Ciarlet, A., L. J. White, R. Chen, B. V. Prasad, and M. K. Estes. 2002. Structural requirements for the assembly of Norwalk virus-like particles. J. Virol. 76:4044-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bok, K., E. J. Abente, M. Realpe-Quintero, T. Mitra, S. V. Sosnovtsev, A. Z. Kapikian, and K. Y. Green. 2009. Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J. Virol. 83:11890-11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bu, W., A. Mamedova, M. Tan, M. Xia, X. Jiang, and R. S. Hegde. 2008. Structural basis for the receptor binding specificity of Norwalk virus. J. Virol. 82:5340-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bujarski, J. J., and P. Kaesberg. 1986. Genetic recombination between RNA components of a multipartite plant virus. Nature 321:528-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bull, R. A., G. S. Hansman, L. E. Clancy, M. M. Tanaka, W. D. Rawlinson, and P. A. White. 2005. Norovirus recombination in ORF1/ORF2 overlap. Emerg. Infect. Dis. 11:1079-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bull, R. A., M. M. Tanaka, and P. A. White. 2007. Norovirus recombination. J. Gen. Virol. 88:3347-3359. [DOI] [PubMed] [Google Scholar]
- 12.Cannon, J. L., L. C. Lindesmith, E. F. Donaldson, L. Saxe, R. S. Baric, and J. Vinje. 2009. Herd immunity to GII.4 noroviruses is supported by outbreak patient sera. J. Virol. 83:5363-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao, S., Z. Lou, M. Tan, Y. Chen, Y. Liu, Z. Zhang, X. C. Zhang, X. Jiang, X. Li, and Z. Rao. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi, J. M., A. M. Hutson, M. K. Estes, and B. V. Prasad. 2008. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. U. S. A. 105:9175-9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estes, M. K., B. V. Prasad, and R. L. Atmar. 2006. Noroviruses everywhere: has something changed? Curr. Opin. Infect. Dis. 19:467-474. [DOI] [PubMed] [Google Scholar]
- 16.Etherington, G. J., J. Dicks, and I. N. Roberts. 2006. High throughput sequence analysis reveals hitherto unreported recombination in the genus Norovirus. Virology 345:88-95. [DOI] [PubMed] [Google Scholar]
- 17.Fukuda, S., Y. Sasaki, S. Takao, and M. Seno. 2008. Recombinant norovirus implicated in gastroenteritis outbreaks in Hiroshima Prefecture, Japan. J. Med. Virol. 80:921-928. [DOI] [PubMed] [Google Scholar]
- 18.Glass, P. J., L. J. White, J. M. Ball, I. Leparc-Goffart, M. E. Hardy, and M. K. Estes. 2000. Norwalk virus open reading frame 3 encodes a minor structural protein. J. Virol. 74:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyde, J. L., S. V. Sosnovtsev, K. Y. Green, C. Wobus, H. W. Virgin, and J. M. Mackenzie. 2009. Mouse norovirus replication is associated with virus-induced vesicle clusters originating from membranes derived from the secretory pathway. J. Virol. 83:9709-9719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarvis, T. C., and K. Kirkegaard. 1992. Poliovirus RNA recombination: mechanistic studies in the absence of selection. EMBO J. 11:3135-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, X., C. Espul, W. M. Zhong, H. Cuello, and D. O. Matson. 1999. Characterization of a novel human calicivirus that may be a naturally occurring recombinant. Arch. Virol. 144:2377-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin, M., H. P. Xie, Z. J. Duan, N. Liu, Q. Zhang, B. S. Wu, H. Y. Li, W. X. Cheng, S. H. Yang, J. M. Yu, Z. Q. Xu, S. X. Cui, L. Zhu, M. Tan, X. Jiang, and Z. Y. Fang. 2008. Emergence of the GII4/2006b variant and recombinant noroviruses in China. J. Med. Virol. 80:1997-2004. [DOI] [PubMed] [Google Scholar]
- 23.Kageyama, T., S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, N. Takeda, and K. Katayama. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamel, A. H., M. A. Ali, H. G. El-Nady, A. de Rougemont, P. Pothier, and G. Belliot. 2009. Predominance and circulation of enteric viruses in the region of Greater Cairo, Egypt. J. Clin. Microbiol. 47:1037-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katayama, K., H. Shirato-Horikoshi, S. Kojima, T. Kageyama, T. Oka, F. Hoshino, S. Fukushi, M. Shinohara, K. Uchida, Y. Suzuki, T. Gojobori, and N. Takeda. 2002. Phylogenetic analysis of the complete genome of 18 Norwalk-like viruses. Virology 299:225-239. [DOI] [PubMed] [Google Scholar]
- 26.Katoh, K., G. Asimenos, and H. Toh. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 537:39-64. [DOI] [PubMed] [Google Scholar]
- 27.Katpally, U., C. E. Wobus, K. Dryden, H. W. Virgin IV, and T. J. Smith. 2008. Structure of antibody-neutralized murine norovirus and unexpected differences from viruslike particles. J. Virol. 82:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khamrin, P., S. Takanashi, W. Chan-It, M. Kobayashi, S. Nishimura, N. Katsumata, S. Okitsu, N. Maneekarn, O. Nishio, and H. Ushijima. 2009. Immunochromatography test for rapid detection of norovirus in fecal specimens. J. Virol. Methods 157:219-222. [DOI] [PubMed] [Google Scholar]
- 29.Kojima, S., T. Kageyama, S. Fukushi, F. B. Hoshino, M. Shinohara, K. Uchida, K. Natori, N. Takeda, and K. Katayama. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 100:107-114. [DOI] [PubMed] [Google Scholar]
- 30.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- 31.Lanave, C., G. Preparata, C. Saccone, and G. Serio. 1984. A new method for calculating evolutionary substitution rates. J. Mol. Evol. 20:86-93. [DOI] [PubMed] [Google Scholar]
- 32.Lindesmith, L. C., E. F. Donaldson, A. D. Lobue, J. L. Cannon, D. P. Zheng, J. Vinje, and R. S. Baric. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lochridge, V. P., and M. E. Hardy. 2007. A single-amino-acid substitution in the P2 domain of VP1 of murine norovirus is sufficient for escape from antibody neutralization. J. Virol. 81:12316-12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lochridge, V. P., K. L. Jutila, J. W. Graff, and M. E. Hardy. 2005. Epitopes in the P2 domain of norovirus VP1 recognized by monoclonal antibodies that block cell interactions. J. Gen. Virol. 86:2799-2806. [DOI] [PubMed] [Google Scholar]
- 35.Martin, D. P., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260-262. [DOI] [PubMed] [Google Scholar]
- 36.McCahon, D., and W. R. Slade. 1981. A sensitive method for the detection and isolation of recombinants of foot-and-mouth disease virus. J. Gen. Virol. 53:333-342. [DOI] [PubMed] [Google Scholar]
- 37.McCormick, C. J., O. Salim, P. R. Lambden, and I. N. Clarke. 2008. Translation termination reinitiation between open reading frame 1 (ORF1) and ORF2 enables capsid expression in a bovine norovirus without the need for production of viral subgenomic RNA. J. Virol. 82:8917-8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Motomura, K., T. Oka, M. Yokoyama, H. Nakamura, H. Mori, H. Ode, G. S. Hansman, K. Katayama, T. Kanda, T. Tanaka, N. Takeda, and H. Sato. 2008. Identification of monomorphic and divergent haplotypes in the 2006-2007 norovirus GII/4 epidemic population by genomewide tracing of evolutionary history. J. Virol. 82:11247-11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagy, P. D., and J. J. Bujarski. 1996. Homologous RNA recombination in brome mosaic virus: AU-rich sequences decrease the accuracy of crossovers. J. Virol. 70:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nayak, M. K., G. Balasubramanian, G. C. Sahoo, R. Bhattacharya, J. Vinje, N. Kobayashi, M. C. Sarkar, M. K. Bhattacharya, and T. Krishnan. 2008. Detection of a novel intergenogroup recombinant Norovirus from Kolkata, India. Virology 377:117-123. [DOI] [PubMed] [Google Scholar]
- 41.Nayak, M. K., D. Chatterjee, S. M. Nataraju, M. Pativada, U. Mitra, M. K. Chatterjee, T. K. Saha, U. Sarkar, and T. Krishnan. 2009. A new variant of norovirus GII.4/2007 and intergenotype recombinant strains of NVGII causing acute watery diarrhoea among children in Kolkata, India. J. Clin. Virol. 45:223-229. [DOI] [PubMed] [Google Scholar]
- 42.Okada, M., T. Tanaka, M. Oseto, N. Takeda, and K. Shinozaki. 2006. Genetic analysis of noroviruses associated with fatalities in healthcare facilities. Arch. Virol. 151:1635-1641. [DOI] [PubMed] [Google Scholar]
- 43.Ozawa, K., T. Oka, N. Takeda, and G. S. Hansman. 2007. Norovirus infections in symptomatic and asymptomatic food handlers in Japan. J. Clin. Microbiol. 45:3996-4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phan, T. G., K. Kaneshi, Y. Ueda, S. Nakaya, S. Nishimura, A. Yamamoto, K. Sugita, S. Takanashi, S. Okitsu, and H. Ushijima. 2007. Genetic heterogeneity, evolution, and recombination in noroviruses. J. Med. Virol. 79:1388-1400. [DOI] [PubMed] [Google Scholar]
- 45.Phan, T. G., T. Kuroiwa, K. Kaneshi, Y. Ueda, S. Nakaya, S. Nishimura, A. Yamamoto, K. Sugita, T. Nishimura, F. Yagyu, S. Okitsu, W. E. Muller, N. Maneekarn, and H. Ushijima. 2006. Changing distribution of norovirus genotypes and genetic analysis of recombinant GIIb among infants and children with diarrhea in Japan. J. Med. Virol. 78:971-978. [DOI] [PubMed] [Google Scholar]
- 46.Phan, T. G., S. Nishimura, K. Sugita, T. Nishimura, S. Okitsu, and H. Ushijima. 2007. Multiple recombinant noroviruses in Japan. Clin. Lab. 53:567-570. [PubMed] [Google Scholar]
- 47.Prasad, B. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 48.Ray, S. C. 1999. Simplot for Windows, version 2.5. Johns Hopkins Medical Institutions, Baltimore, MD.
- 49.Reuter, G., K. Krisztalovics, H. Vennema, M. Koopmans, and G. Szucs. 2005. Evidence of the etiological predominance of norovirus in gastroenteritis outbreaks—emerging new-variant and recombinant noroviruses in Hungary. J. Med. Virol. 76:598-607. [DOI] [PubMed] [Google Scholar]
- 50.Robertson, D. L., B. H. Hahn, and P. M. Sharp. 1995. Recombination in AIDS viruses. J. Mol. Evol. 40:249-259. [DOI] [PubMed] [Google Scholar]
- 51.Rohayem, J., J. Munch, and A. Rethwilm. 2005. Evidence of recombination in the norovirus capsid gene. J. Virol. 79:4977-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rott, M. E., J. H. Tremaine, and D. M. Rochon. 1991. Comparison of the 5′ and 3′ termini of tomato ringspot virus RNA1 and RNA2: evidence for RNA recombination. Virology 185:468-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siebenga, J. J., H. Vennema, B. Renckens, E. de Bruin, B. van der Veer, R. J. Siezen, and M. Koopmans. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 81:9932-9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siebenga, J. J., H. Vennema, D. P. Zheng, J. Vinje, B. E. Lee, X. L. Pang, E. C. Ho, W. Lim, A. Choudekar, S. Broor, T. Halperin, N. B. Rasool, J. Hewitt, G. E. Greening, M. Jin, Z. J. Duan, Y. Lucero, M. O'Ryan, M. Hoehne, E. Schreier, R. M. Ratcliff, P. A. White, N. Iritani, G. Reuter, and M. Koopmans. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001-2007. J. Infect. Dis. 200:802-812. [DOI] [PubMed] [Google Scholar]
- 55.Smith, J. M. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126-129. [DOI] [PubMed] [Google Scholar]
- 56.Sosnovtsev, S. V., G. Belliot, K. O. Chang, O. Onwudiwe, and K. Y. Green. 2005. Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 79:4012-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Symes, S. J., I. C. Gunesekere, J. A. Marshall, and P. J. Wright. 2007. Norovirus mixed infection in an oyster-associated outbreak: an opportunity for recombination. Arch. Virol. 152:1075-1086. [DOI] [PubMed] [Google Scholar]
- 58.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 59.Tamura, K., M. Nei, and S. Kumar. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030-11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan, M., P. Huang, J. Meller, W. Zhong, T. Farkas, and X. Jiang. 2003. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: evidence for a binding pocket. J. Virol. 77:12562-12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan, M., M. Xia, Y. Chen, W. Bu, R. S. Hegde, J. Meller, X. Li, and X. Jiang. 2009. Conservation of carbohydrate binding interfaces: evidence of human HBGA selection in norovirus evolution. PLoS One 4:e5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsugawa, T., K. Numata-Kinoshita, S. Honma, S. Nakata, M. Tatsumi, Y. Sakai, K. Natori, N. Takeda, S. Kobayashi, and H. Tsutsumi. 2006. Virological, serological, and clinical features of an outbreak of acute gastroenteritis due to recombinant genogroup II norovirus in an infant home. J. Clin. Microbiol. 44:177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vidal, R., P. Roessler, V. Solari, J. Vollaire, X. Jiang, D. O. Matson, N. Mamani, V. Prado, and M. L. O'Ryan. 2006. Novel recombinant norovirus causing outbreaks of gastroenteritis in Santiago, Chile. J. Clin. Microbiol. 44:2271-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, Q. H., M. G. Han, S. Cheetham, M. Souza, J. A. Funk, and L. J. Saif. 2005. Porcine noroviruses related to human noroviruses. Emerg. Infect. Dis. 11:1874-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waters, A., S. Coughlan, and W. W. Hall. 2007. Characterisation of a novel recombination event in the norovirus polymerase gene. Virology 363:11-14. [DOI] [PubMed] [Google Scholar]
- 67.Worobey, M., and E. C. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 80(Pt. 10):2535-2543. [DOI] [PubMed] [Google Scholar]
- 68.Xi, J. N., D. Y. Graham, K. N. Wang, and M. K. Estes. 1990. Norwalk virus genome cloning and characterization. Science 250:1580-1583. [DOI] [PubMed] [Google Scholar]
- 69.Yang, R., S. Kusagawa, C. Zhang, X. Xia, K. Ben, and Y. Takebe. 2003. Identification and characterization of a new class of human immunodeficiency virus type 1 recombinants comprised of two circulating recombinant forms, CRF07_BC and CRF08_BC, in China. J. Virol. 77:685-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, C. X., P. J. Cascone, and A. E. Simon. 1991. Recombination between satellite and genomic RNAs of turnip crinkle virus. Virology 184:791-794. [DOI] [PubMed] [Google Scholar]
- 71.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312-323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.