SUMMARY
Drosophila embryos are highly sensitive to γ-ray-induced apoptosis at early but not later, more differentiated stages during development. Two proapoptotic genes, reaper and hid, are upregulated rapidly following irradiation. However, in post-stage-12 embryos, in which most cells have begun differentiation, neither proapoptotic gene can be induced by high doses of irradiation. Our study indicates that the sensitive-to-resistant transition is due to epigenetic blocking of the irradiation-responsive enhancer region (IRER), which is located upstream of reaper but is also required for the induction of hid in response to irradiation. This IRER, but not the transcribed regions of reaper/hid, becomes enriched for trimethylated H3K27/H3K9 and forms a heterochromatin-like structure during the sensitive-to-resistant transition. The functions of histone-modifying enzymes Hdac1(rpd3) and Su(var)3-9 and PcG proteins Su(z)12 and Polycomb are required for this process. Thus, direct epigenetic regulation of two proapoptotic genes controls cellular sensitivity to cytotoxic stimuli.
INTRODUCTION
Although caspase activation and apoptosis can proceed without de novo protein synthesis under certain special circumstances, abundant evidence suggests that transcriptional and translational mechanisms play crucial roles in regulating apoptosis induced by cytotoxic stimuli. The genetic requirement of transcription factors such as P53 in irradiation-induced cell death underscores the importance of the transcriptional response. Several proapoptotic genes, including puma (p53 upregulated modulator of apoptosis), are the direct transcriptional targets of P53. In puma knockout mice, irradiation-induced cell death in hematopoietic cells and the developing nervous system is almost completely blocked (Jeffers et al., 2003). Although much has been revealed about the molecular mechanism of P53-mediated proapoptotic gene expression and apoptosis, we understand very little as to why different tissue/cell types can have dramatically different sensitivity to irradiation.
In Drosophila, Inhibitor of Apoptosis Protein (IAP) antagonists play a pivotal role in regulating programmed cell death during development. Upon its initial identification, the IAP antagonist reaper was found to be transcriptionally activated upon irradiation (White et al., 1994). The H99 genomic region, which also includes two other IAP antagonists hid and grim, is required for mediating irradiation-induced cell death in Drosophila. A reporter construct containing the immediate 11 kb sequence upstream of the reaper transcribed region gives a much broader expression pattern in transgenic animals than that of the endogenous reaper mRNA (Nordstrom et al., 1996), suggesting that key inhibitory cis-regulatory function is not present in the reporter construct. This 11 kb reporter construct is responsive to ionizing irradiation and contains at least one putative P53 response element (P53RE) that conforms to the patterns of mammalian P53-binding sites (Brodsky et al., 2000). Correspondingly, genetic analysis indicated that the function of Drosophila P53 (DmP53) is required for mediating ionizing irradiation-induced reaper expression and apoptosis (Brodsky et al., 2004; Lee et al., 2003; Sogame et al., 2003). However, several questions remain to be addressed. First, the sensitivity to irradiation-induced cell death is tissue/cell type specific and restricted to certain developmental stages. The difference in sensitivity has no direct correlation with the availability of DmP53. Rather, the windows of sensitivity seem correlated with developmental marks such as high proliferation. Second, overexpression of DmP53 failed to induce reaper expression or apoptosis in many tissues, indicating that DmP53 alone is not sufficient for inducing reaper expression, or (and) the P53RE is not always accessible.
It has been observed that during development, the sensitivity to irradiation-induced cell death can change rapidly even for the same cell linage. For instance, while proliferating neural precursor cells in the mammalian hippocampus are extremely sensitive to ionizing irradiation, differentiating or differentiated neurons in the same region are resistant (Mizumatsu et al., 2003; Peissner et al., 1999). A similar switch of sensitivity to irradiation was observed during Drosophila embryogenesis. While both reaper and hid are induced to mediate cell death in young embryos with mostly proliferating cells, neither can be induced in embryos developed a few hours further when most cells are differentiating or differentiated. This system offered us a valuable model to explore the molecular mechanisms underlying the sensitive-to-resistant transition accompanying cellular differentiation. In this study, we found that the IRER upstream of the reaper locus, including the putative P53RE, is subject to epigenetic regulation during development. Histone modification and chromatin condensation specific to the IRER, but not the promoter region, are capable of switching off the sensitivity to irradiation-induced proapoptotic gene expression and cell death. To our knowledge, this is the first evidence that direct epigenetic regulation of proapoptotic gene(s) controls cellular sensitivity to cytotoxic stimuli.
RESULTS
Sensitivity to γ-Ray-Induced Apoptosis Is Developmental Stage Dependent
During the 20 hr of embryogenesis, the sensitivity of fly embryos to irradiation changes dramatically between 7 and 9 hr after egg laying (AEL). When measured by embryonic lethality, embryos before 7 hr AEL (developmental stages 1–11) (Campos-Ortega and Hartenstein, 1985) are extremely sensitive to γ-irradiation (Figure 1A), while embryos after 9 hr AEL (developmental stage 12) become highly resistant. This dramatic change of sensitivity to irradiation was first noticed decades ago (Ashburner, 1989; Wurgler and Ullrich, 1976), but the underlying cellular and molecular mechanisms remain unclear.
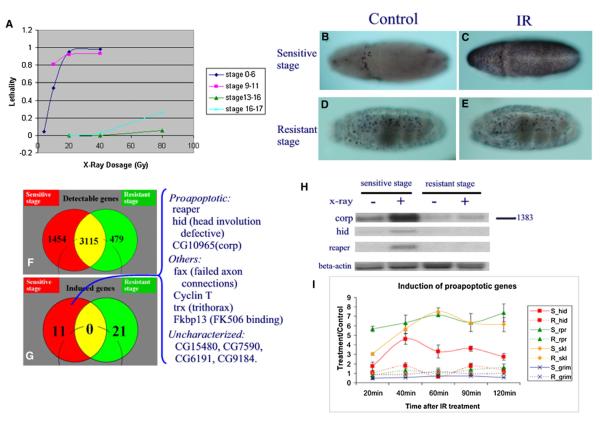
Figure 1. Stage-Specific Sensitivity to γ-ray-Induced Cell Death.
(A) Embryonic lethality induced by γ-rays is dependent on developmental stage. Embryos collected 0–3 hr AEL (developmental stages 0–6), 4–7 hr AEL (stages 9–11), 9–12 hr AEL (stage 13–16), and 14–17 hr AEL (stages 16–17) were irradiated with various dosages of γ-irradiation. Each data point represents the average of two to three treatments. Each time an average of 595 eggs were treated. To count for unfertilized eggs, controls were processed in parallel without γ-ray treatment. Embryos that failed to hatch after a 30 hr incubation at 25°C were counted as lethal.
(B–E) TUNEL labeling of embryos at 75 min after 40 Gy of γ-irradiation (C and E) or control treatment (B and D). (B) and (C) are stage 10–11 embryos, (D) and (E) are stage 16–17 embryos. Note that irradiation induced widespread cell death in stage 10–11 embryos (compare [C] with [B]) but not in stage 16 embryos (compare [E] with [D]).
(F) Venn diagram depicting the overlap of detectable genes in sensitive- and resistant-stage embryos using the pan-genome DNA array.
(G) Venn diagram indicating no overlap between γ-ray-inducible genes detected in sensitive (4–7 hr AEL) and resistant (9–12 hr AEL) embryos. The identity of the 11 genes significantly induced by γ-ray irradiation in sensitive embryos is shown at the right side of the figure.
(H) Northern hybridization analysis confirms the γ-ray responsiveness of the three cell death genes: reaper, hid, and corp (companion of reaper), and β-actin was used as a nonresponsive control.
(I) hid (red square), reaper (green triangle), sickle (yellow diamond), and grim (blue cross) RNA levels (measured by QPCR) in sensitive (continuous lines) and resistant (dashed line) embryos at 20, 40, 60, 90, and 120 min following γ-ray treatment. Data are represented as the fold changes comparing γ-ray-treated with parallel processed control samples (mean ± SD).
This shift of sensitivity to irradiation at the organismic level coincides with changes of sensitivity at the cellular level. Irradiation-induced cell death, as measured by TUNEL, appears about 45–60 min after irradiation and reaches the peak at about 75–90 min. The most dramatic induction of TUNEL-positive cells following γ-ray irradiation was observed in stage 10–11 embryos (Figure 1C versus 1B). In sharp contrast, there is little increase of TUNEL-positive cells in germ-band-retracted embryos after developmental stage 12 (Figure 1E versus 1D). For the clarity of the discussion, we will refer to embryos before or at stage 11 as ‘‘sensitive’’ (stage) embryos, and those after stage 12 as ‘‘resistant’’ (stage) embryos.
To gain a comprehensive picture of genomic responses to γ-ray irradiation, we used Affymetrix DrosGenome1 GeneChips to measure the immediate transcriptional response elicited by γ-rays. For both sensitive- and resistant-stage embryos, total RNA was extracted 15 min after irradiation from treated and parallel processed control samples. Among the 11 genes induced significantly in the sensitive stage, two are known cell death regulatory genes, reaper and hid (Figure 1G). The probability of observing two or more known proapoptotic genes in 11 randomly selected genes from the genome is calculated as 2 × 10−4, indicating that cell death genes are selectively activated following γ-ray treatment in sensitive embryos. In contrast, neither of the two genes, nor any other proapoptotic gene, was significantly induced by γ-rays in resistant embryos (see Table S1 in the Supplemental Data available with this article online).
The specific induction of reaper and hid by γ-rays in sensitive but not resistant embryos was verified by both northern hybridization (Figure 1H) and quantitative PCR (QPCR) (Figure 1I). The QPCR result indicates that, in sensitive embryos, both reaper and hid are induced rapidly (within 20 min) and reach a peak at about 40–60 min after irradiation. In contrast, neither can be significantly induced in resistant-stage embryos at any time points (up to 2 hr). Interestingly, a similar responsive pattern was observed for another IAP antagonist, sickle, which is upstream of reaper (Figure 2A). The other IAP antagonist, grim, showed no radiation induction in either the sensitive or resistant stage.
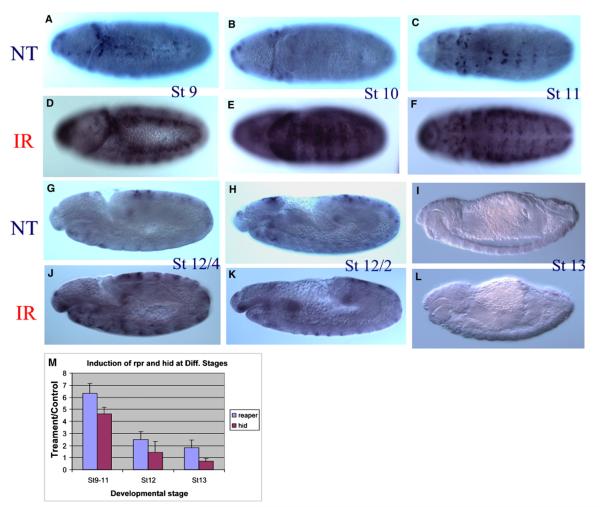
Figure 2. Rapid Transition of reaper Sensitivity to Irradiation between 8 and 9 hr AEL.
Pooled embryos (0–17 hr AEL) were treated with γ-rays or served as nontreatment control. A significant increase of reaper mRNA was observed in stage 7–11 embryos (A–F), with the peak of responsiveness observed in stage 10 embryos (E versus B). This responsiveness is dramatically decreased once the germ band starts to retract, which happens around 7.5 hr AEL ([J] versus [G]). By the time the germ band is half way retracted on the dorsal side, the responsivenessof reaper is almost completely diminished ([K] versus [H]). None of the embryos at the end of stage 12 (8.5–9 hr AEL) or stage 13 has detectable reaper responsiveness ([L] versus [I]). A very similar transition is also observed for hid responsiveness (Figure S1). The sensitive-to-resistant transition was also verified with QPCR (M). The error bars represent standard deviation.
Irradiation-induced cell death is largely blocked in the H99 deficiency mutant that lacks reaper, hid, and grim (White et al., 1994). The selective and rapid induction of reaper and hid after irradiation indicates that the two genes are responsible for mediating irradiation-induced cell death in sensitive-stage embryos. Their coordinated induction is likely essential for the rapid induction of apoptosis, as has been demonstrated before (Zhou et al., 1997). The IAP antagonists sickle is not deleted in the H99 mutant; however, it is also induced upon ionizing irradiation (Brodsky et al., 2004; Christich et al., 2002). In this study, we focus on the immediate induction of reaper and hid in sensitive embryos and the sensitive-to-resistant transition of the responsiveness of these two genes.
Rapid Sensitive-to-Resistant Transition of Proapoptotic Gene Responsiveness during Developmental Stage 12
To pinpoint the timing of the sensitive-to-resistant transition during development, pooled embryos (0–16 hr AEL) were treated with 20 Gy γ-ray irradiation and then monitored for expression of proapoptotic genes at 20–30 min following irradiation. The pooled embryos were collected overnight, irradiated on the same plate, fixed together, and processed for in situ hybridization in the same tube. Our data indicated that both reaper and hid can be induced by γ-ray irradiation in embryos developed beyond developmental stage 6, when gastrulation begins. The responsiveness of the two proapoptotic genes peaks at stage 10 and remains responsive at developmental stage 11 (Figures 2D versus 2A; 2E versus 2B; 2F versus 2C). However, once the germ band starts to retract at early stage 12, the responsiveness begins to diminish rapidly (Figures 2G–2L). By the time most of the germ band has retracted to the ventral side (late stage 12), the responsiveness of both genes is totally lost (Figures 2K versus 2H; 2L versus 2I; Figure S1). The contrast of reaper or hid in situ hybridization (ISH) signals in sensitive versus resistant embryos following irradiation is most apparent when viewed under low magnification (Figure S1). Increasing the γ-ray dosage up to 120 Gy failed to induce any detectable increase of reaper and hid expression at 30 min after irradiation (data not shown). The rapid sensitive-to-resistant transition was also verified independently with QPCR (Figure 2M). It is clear that compared to embryos at stages 9–11 (4–7 hr AEL), there is little induction of reaper or hid at stage 12 or 13.
The change of radiation responsiveness of proapoptotic genes is unlikely due to a reduced amount of DNA damage or suppressed cellular signaling response in resistant-stage embryos. Two DNA repair genes, ku70 and ku80, were also significantly induced by irradiation in sensitive-stage embryos through a DmP53-dependent mechanism (Brodsky et al., 2004). However, in sharp contrast to the proapoptotic genes, not only did the two genes remain responsive to irradiation at the resistant stage, but their induction levels were significantly higher in the resistant stage than in the sensitive stage (Figure S2A). This suggests that the loss of responsiveness is specific to the proapoptotic genes.
Mapping the Genomic Region Responsible for Mediating γ-ray Responsiveness
To map the genomic region responsible for mediating the γ-ray responsiveness of reaper and hid, we took advantage of the insertional mutants generated by Exelixis (Thibault et al., 2004). These insertion lines were generated in an isogenic background with transposon vectors containing the Su(Hw) insulator sequences. If the insertion is located between the promoter and the enhancer region mediating γ-ray responsiveness, it could interrupt the responsiveness of the proapoptotic genes. In addition, these transposons have FRT sequences that can be used for making well-defined genomic deletions (Parks et al., 2004).
Both reaper and hid reside in the 75C1-2 region, together with the other two IAP antagonists grim and sickle. The organization of the genomic region harboring the four genes is depicted in Figure 3A. Interestingly, all four proapoptotic genes are transcribed in the same direction. Remarkably long gene-less regions surround the reaper locus: from grim to reaper is approximately 93 kb and from reaper to sickle is about 40 kb. In contrast, left of hid and right of sickle are gene-dense regions (more than five genes in 50–60 kb). When compared to homologous regions in D. Pseudoobscura (D. pseu) and D. virilis (D. viri), the intergenic genomic region is better conserved than the transcribed and coding region of reaper at the nucleotide level (Figure 3B). The two species diverged from D. mela between 40 and 60 million years ago, respectively. The exceptional conservation of nontranscribed sequences around reaper suggests that vital regulatory functions may reside in these regions.
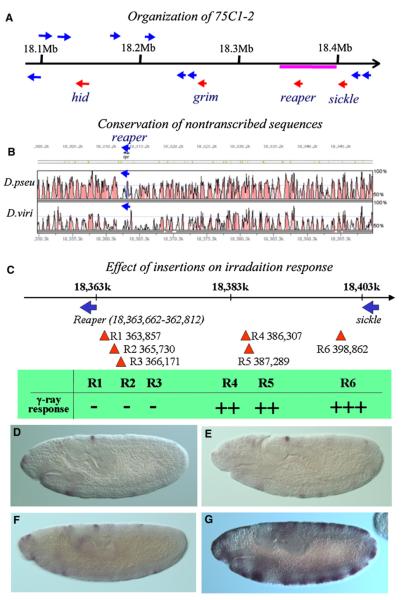
Figure 3. Mapping of the Irradiation-Responsive Region.
(A) Organization of the 75C1-2 region that harbors four IAP antagonist genes (red arrows). Other annotated genes in this region were marked with blue arrows. The region underlined by the red line is represented in (B).
(B) Conservation of the intergenic region around the reaper locus. The figure was drawn with Vista (Mayor et al., 2000), the curve indicating the percent of identity (window size 100 bp). The region is colored if the identity is higher than 75%. Color code: pink, untranscribed or intronic region; light blue, untranslated transcribed region; dark blue, coding region.
(C) The Exelixis insertions localized between reaper and sickle, R1(P{XP}d11052), R2(P{XP}d00909), R3(PBac{WH}f02826), R4(PBac{WH}f03056), R5(PBac{WH}f07603), and R6(PBac{WH}f03389). Induction of reaper by γ-ray irradiation was totally blocked by R1, R2, or R3 but is only slightly attenuated by R4 and R5 and is not at all affected by R6. +++, WT responsiveness; –, no response. For insertion site information, refer to Table S3.
(D and E) reaper ISH of control and irradiated homozygous R3 embryos, respectively.
(F and G) reaper ISH of control and irradiated homozygous R6 embryos, respectively.
From the Exelixis collections, we obtained a total of 45 strains that were recorded as having a single insertion in the 75C1-2 region. Their insertion sites were verified by inverse PCR (Table S3). Analysis of these strains indicated that a 20 kb region between 3L:18,366,171 and 18,386,107 is required for the γ-ray responsiveness of reaper. Three insertions (R1, R2, and R3) between this region and the reaper promoter all blocked the γ-ray responsiveness of reaper (Figures 3C–3E). In contrast, insertions (R4, R5, and R6) after this region did not block γ-ray responsiveness (Figures 3F and 3G). Significant γ-ray responsiveness of reaper transcription was clearly visible in homozygous R4 and R5 embryos, indicating that the essential γ-ray-responsive region for reaper transcriptional regulation is located between R3(18,366,171) and R4(18,386,107). However, there may be additional enhancer element(s) in the DNA region between R5(18,387,288) and R6(18,398,861), as the γ-ray responsiveness of reaper is conceivably stronger in homozygous R6 embryos than that in R5 and R4. In terms of reaper transcriptional response to irradiation, there is no detectable difference between R6 and wild-type embryos, indicating that all essential elements are on the left side of R6.
We then generated deletions that removed the interval 3L:18,365,736–18,398,898 between R2 and R6 [referred to as Df(3L:18,366–398)], the interval 3L:18,365,736–18,386,300 between R2 and R4 [Df(3L:18,366–386)], and the interval 3L:18,386,300–18,398,898 between R4 and R6 [Df(3L:18,386–398)]. For each deficiency, 5–10 independent deletion strains were obtained. The span of the deletion was verified by PCR using primers flanking the deletion, and the breaking point was verified by sequencing the PCR product. None of these deficiencies removed the transcribed region of reaper or sickle. The left breaking points for the deficiencies are more than 2 kb away from the reaper transcription starting site.
In embryos homozygous for either Df(3L:18,366–386) or Df(3L:18,366–398) (identified with a GFP balancer), the responsiveness of reaper to γ-ray irradiation was totally abolished (Figures 4A–4H), indicating that essential enhancer elements are located in the Df(3L:18,366–386) interval. Homozygous Df(3L:18,386–398) showed a significantly decreased level of reaper responsiveness (Figures 4C and 4G), which reconfirms that the region between R4 and R6 has nonessential enhancer(s). These results are in perfect agreement with our insertion mapping data described above. The insulators in the original insertions were removed during the deletion generation process, so there is no Su(Hw) insulator in Df(3L:18,366–386) or Df(3L:18,366–398). There is one remaining insulator left in Df(3L:18,386–398), but that should not affect the conclusion of the results.
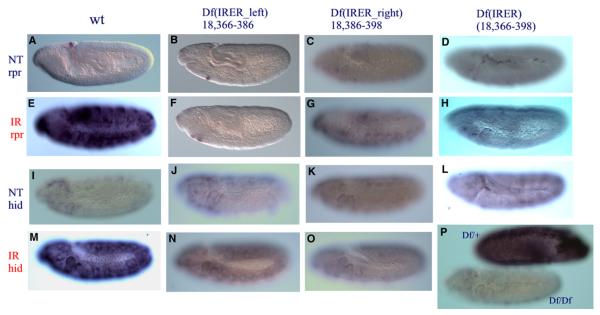
Figure 4. The IRER Is Required for the Responsiveness of reaper and hid.
Df(IRER_left) abolished the responsiveness of reaper to irradiation (B and F). hid responsiveness to γ-ray irradiation was also significantly reduced (J and N). Df(IRER_right) reduced reaper responsiveness ([C] and [G] versus [A] and [E]) but blocked hid responsiveness ([K] and [O] versus [I] and [M]). Df(IRER) blocked the responsiveness of both reaper and hid (D, H, L, and P). In (P), the dark embryo is a heterozygous (Df(IRER)/TM3ubi-GFP) embryo that is also stained for GFP.
Thus both approaches unequivocally indicated that the enhancer region responsible for mediating reaper irradiation responsiveness resides in the interval between R2 and R6, i.e., 3L:18,365,736–18,398,861. We named this region the irradiation responsive enhancer region (IRER). The previously identified putative P53RE (18,368,516) is within this region and close to the left boundary. Since overexpression of P53 alone was not sufficient to induce reaper expression in the embryo, it is very likely that another enhancer element(s) in this region is (are) also required for mediating irradiation-induced reaper expression. In addition, our data indicate that there is (are) nonessential enhancer element(s) in the region between R5 and R6. To facilitate the discussion, we will refer to the region between R2 and R6 as IRER, and the deletion of this region as Df(IRER). Correspondingly, we will refer to the genomic region between R2 and R4 as ‘‘IRER_left,’’ and the region between R5 and R6 as ‘‘IRER_right.’’ For the responsiveness of reaper, IRER_left is essential, and IRER_right is supplemental.
An unexpected result from the deletion analysis is that the responsiveness of hid to γ-ray irradiation was also significantly reduced in the Df(IRER_left) mutant and abolished in the Df(IRER) mutant (Figures 4I–4P). This is surprising since the insulator-containing insertions (R1, R2, R3) did not have any effect on γ-ray-induced hid expression (data not shown). It indicates that there may exist a high-order arrangement which enables the IRER to interact with the hid promoter. The essential region mediating this interaction most likely resides in the interval between R4 and R6 (IRER_right). In homozygous Df(IRER_right), hid responsiveness is lost even though reaper is still responsive (albeit reduced). To rule out the possibility of unintended damage to the hid locus in the process of the FLP/FRT-mediated deletion, we performed complementation tests between Df(IRER) and hid mutant alleles, including [05014], [A206], and [8d]. All of the hid mutant alleles are homozygous lethal, and homozygous Df(IRER) has greatly reduced viability. Invariably, the lethality of the hid alleles was complemented by the Df(IRER) chromosome, indicating that the developmental function of hid is intact in the Df(IRER) mutant. In addition, we tested hid responsiveness in the X38 deletion mutant, which removes the reaper transcription unit and all of the IRER region (Peterson et al., 2002). In both X38/X38 and Df(IRER)/X38, the responsiveness of hid is abolished, indicating that indeed hid responsiveness to irradiation is mediated by the IRER.
Formation of DNase-I-Resistant Structure in the IRER, but Not the Promoter and Transcribed Region of reaper, in Post-Stage-12 Embryos
Like its mammalian ortholog, DmP53 is required for mediating irradiation- and DNA-damage-induced cellular responses including apoptosis and/or DNA repair. However, the sensitive-to-resistant transition we observed for the induction of proapoptotic genes is unlikely due to the unavailability of DmP53, since it is ubiquitously expressed in the embryo at both sensitive and resistant stages (Jin et al., 2000; Figure S2B). DmP53-mediated induction of DNA repair genes ku70 and ku80 is not diminished, and actually increased, after the transition observed for the proapoptotic genes (Figure S2A). We tried overexpressing DmP53 using UAS-DmP53 but it failed to convey any detectable radiation sensitivity in resistant-stage embryos (data not shown). Previous studies using reporter constructs containing part of IRER_left have found that the reporter remained responsive to X-rays till the end of embryogenesis (Qi et al., 2004). All of this evidence suggests that the transition is not due to unavailability or lack of activation of DmP53; rather, they point to epigenetic regulation of the IRER that controls its accessibility.
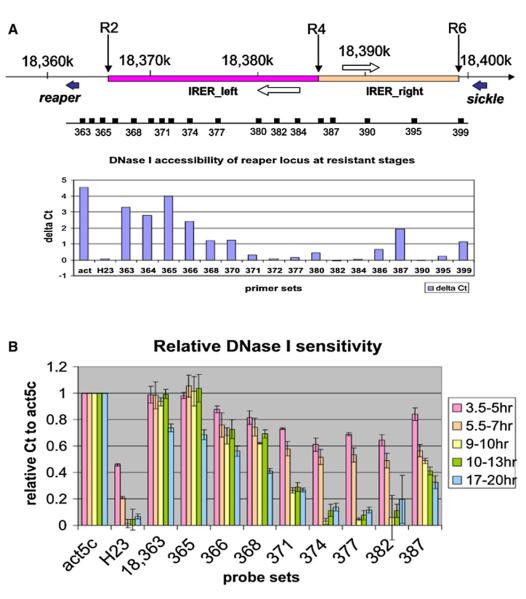
A DNaseI sensitivity assay was performed to scan the DNA accessibility around the IRER. A primer set was designed and verified for a selected 1,000 bp interval, e.g., 18,363,000–18,363,999 (referred to as ‘‘363’’ in the paper) (Table S2). Unless otherwise stated, the ΔCt values mentioned thereafter refer to Ct(50U) – Ct(0U). For constitutively active genes such as the act5c transcribed region, the ΔCt value is between 4 and 5 in resistant-stage embryos. In contrast, heterochromatin areas such as the H23 (22,000–24,000 of chr2 heterochromatin) locus are refractory to the DNase I treatment, with the ΔCt value close to zero (Figure 5A and Figure S3). In resistant-stage embryos, the reaper transcribed region and proximal promoter and enhancer regions (363–365) remain as sensitive to DNase I as the constitutively active act5c locus. In sharp contrast, most of the IRER is almost as inaccessible as the heterochromatin locus (H23). The only region in the IRER that remains relatively open at the resistant stage is 18,386–387, which is probably the shared enhancer/promoter region of two putative noncoding RNAs that are transcribed in opposite directions (represented by EST sequences RE73107 [3L:18,383–379] and RE07245 [3L:18,388–392], respectively). It is also where R4 and R5 insertions are located. This is unlikely just a coincidence; rather, we believe the relative open-ness of this region allowed R4 and R5 to be recovered from the mutagenesis.
Figure 5. Formation of Closed Heterochromatin Structure in the IRER.
(A) DNase I sensitivity assay of the IRER in resistant-stage embryos. In resistant embryo, most of the IRER is as resistant to DNase I as the pericentromeric heterochromatin locus H23. The only exception is a relatively open island around 18,387, flanked by two putative noncoding RNAs (open arrows).
(B) Change of DNase I sensitivity in the IRER in staged embryos. There is a dramatic transition of DNase I sensitivity around 18,368–382 between 7 and 9 hr AEL. Data were represented as relative ΔCt, which is ΔCt (target region)/ΔCt (act5c). The ΔCt (act5c) values for different stages are: 6.420 ± 0.424 (3.5–5 hr), 7.278 ± 0.797 (5.5–7 hr), 5.043 ± 0.34 (9–10 hr), 4.460 ± 0.339 (10–13 hr), 4.988 ± 0.256 (17–20 hr). Data are represented as mean ± SD; n = 3 or 4 for all age groups.
To monitor the dynamics of the accessibility of the IRER, we performed the DNase I sensitivity assay in staged embryos that were 3.5–5 hr, 5.5–7.0 hr, 9–10 hr, 10–13 hr, and 14–17 hr AEL (Figure 5B). A significant decrease of DNase I sensitivity was found in the IRER between 7 and 9 hr AEL, consistent with the sensitive-to-resistant transition observed for irradiation-induced reaper/hid expression and cell death. When the ΔCt values of different loci were normalized against the ΔCt value of the act5c locus, it was apparent that the reaper transcribed region and immediate promoter and enhancer region (primer sets 363 and 365, respectively) remained as open as the act5c locus throughout embryogenesis. However, the IRER (detected with primer sets 368, 370, 372, 377, and 382) underwent a dramatic shift of accessibility between 7 and 9 hr AEL (Figure 5B). Within the IRER, it seems that the center of IRER_left, represented by probe sets 371–382, becomes inaccessible first, while the left boundary of the IRER, represented by probe sets 368–370, becomes inaccessible to DNase I at relatively later stages. The control H23 heterochromatic region also changes dramatically in DNase I sensitivity between sensitive and resistant stages, which is consistent with the timing of heterochromatin formation in Drosophila embryogenesis (Lu et al., 1998).
Histone Modifications in the IRER Region
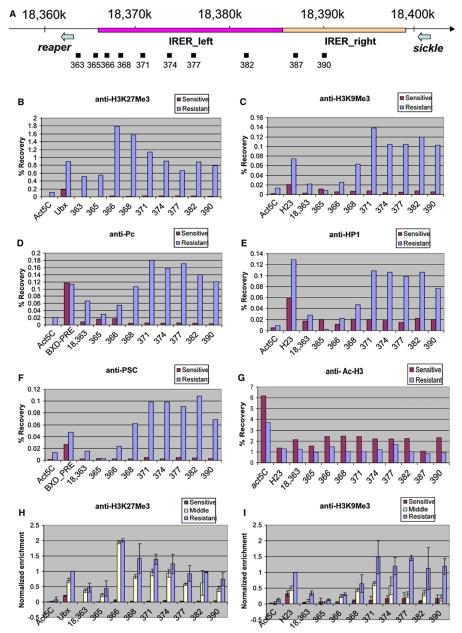
The formation of heterochromatin-like structure is associated with posttranslational modification of histones (Jenuwein and Allis, 2001). To monitor histone modification in and around the IRER, chromatin immunoprecipitation (ChIP) experiments were performed in parallel with sensitive- and resistant-stage embryos (Figures 6B and 6C) using antibodies against trimethylated H3K9 and H3K27 (a gift from T. Jenuwein). As shown in Figure 6B, we observed a dramatic increase of H3K27 trimethylation in the IRER at the resistant stage. For the region 18,366–368, the recovery rates of resistant embryos are over 100-fold higher than those of sensitive embryos. As expected, the level of H3K27Me3 in the positive control Ultrabithorax (Ubx) promoter region also increased at the resistant stage. However, the magnitude of the increase in the Ubx promoter is much smaller than that observed for 18,366–368, probably reflecting the fact that the Ubx promoter remains open in the posterior segments while the blocking of the IRER is for the whole embryo.
Figure 6. Chromatin Modification of the IRER.
(A) Schematic representation of the IRER locus, including IRER_left (red bar) and IRER_right (orange). The positions of DNA amplicons for quantification of ChIP results are shown below the IRER map relative to the DNA sequence coordinates of chromosome 3L (Dm genome release 4.3).
(B–G) ChIP assays performed on embryos at sensitive stage (red) and resistant stage (blue) using anti-H3K27Me3 (B), anti-H3K9Me3 (C), anti-Pc (D), anti-HP1 (E), anti-Psc (F), and anti-Ac-H3 (G). Precipitation of DNA fragments with antibodies was quantified by QPCR and shown in recovery rates. The coding region of Act5C was used as a background control for all the antibodies. For positive controls, Ubx promoter region was chosen for anti-H3K27Me3; H23 for anti-H3K9Me3, anti-HP1, and anti-Ac-H3; and the BXD-PRE for anti-Pc and anti-Psc. Several independent assays were performed for each antibody and a representative figure was shown.
(H and I) Timing of H3K27 and H3K9 trimethylation, respectively. ChIP results from embryos at sensitive stage (3–7 hr AEL, red), middle stage (7–9 hr AEL, yellow) and resistant stage (13–16 hr AEL, blue) were normalized to the recovery rate of the positive controls in the resistant stage. Three independent assays were performed for each stage, and the values are shown as mean ± SD.
There is also a significant increase of H3K9 trimethylation throughout the IRER (Figure 6C), especially in the center of IRER_left (18,371–382), which corresponds to the region that has the strongest resistance to DNase I (Figure 5). It is interesting to note that, in comparison, the highest level of H3K27 trimethylation is at the left boundary of the IRER (18,366–368). To test whether trimethylated H3K9 is indeed associated with the formation of heterochromatic structure in the IRER, the antibody against Heterochromatin Protein 1 (HP1) was used for ChIP assays. The distribution profile of HP1 in the tested region is quite similar with that of trimethylated H3K9 (Figure 6E), further indicating that the IRER indeed undergoes the transition from a relatively open structure to a heterochromatin-like structure.
Just as trimethylated H3K9 is often bound by HP1, trimethylated H3K27 is associated with Polycomb Repressive Complex 1 (PRC1), including Polycomb (Pc) and Posterior Sex Combs (Psc). The Bithoraxoid Polycomb Response Element (BXD-PRE) region, known to be bound by PRC1, was used as the positive control. A significant increase of specific Pc and Psc binding to the IRER was detected in the resistant embryos (Figures 6D and 6F), suggesting that PcG-mediated silencing is involved in blocking the IRER. However, instead of specifically binding to a localized PRE, we found that the binding of Pc and Psc is widespread in all of the tested loci in the IRER.
Several other types of histone modification, including di- and trimethylation of H3K4, dimethylation of H3K9 and H3K27, acetylation of H3K9, and phosphorylation of H3S10 were investigated in the same region as well (data not shown). Of those, only a moderate decrease (30%–50%) of H3 acetylation was observed in resistant-stage embryos compared to sensitive-stage ones (Figure 6G). This may also contribute to the structural transition, since acetylated H3 is considered as one of the euchromatic marks (Jenuwein and Allis, 2001). However, the magnitude of change is not comparable to that observed for trimethylated H3K27 and H3K9.
To determine the timing of histone modifications, we performed ChIP analysis in embryos between 7 and 9 hr AEL (late stage 11 and stage 12) (‘‘Middle’’; Figures 6H and 6I). Compared to the sensitive stage, both H3K27 and H3K9 trimethylation profiles changed significantly in the IRER during the transitional ‘‘middle’’ stage. Thus, it is impossible to distinguish which of the two modifications happened first. It is quite possible that the two distinct modifications happened in parallel. Interestingly, the enrichment of trimethylated H3K27 at region 366 during the middle stage is already as high as that at the resistant stage, while at other sites in the IRER there is an increase of H3K27 trimethylation between the middle stage and the late resistant stage. This suggests that this modification may be initiated from the left boundary of the IRER. Another difference between H3K9 and H3K27 trimethylation is that there is a relatively low level, but significant, increase of H3K27 trimethylation in the reaper promoter and immediate enhancer region (363 and 365), whereas H3K9 trimethylation is much more limited to the core of the IRER.
Functions of Histone Modifiers Are Required for the Sensitive-To-Resistant Transition
Trimethylation of H3K27 is carried out by Polycomb Repressive Complex 2 (PRC2), which contains three core components, Suppressor of zeste 12 [Su(z)12], Extra sexcombs (ESC), and Enhancer of zeste [E(z)]. Trimethylation of H3K9 is catalyzed by the histone methyltransferase Su(var)3-9. The histone deacetylase (Hdac1/rpd3) is involved in and required for both modifications. In searching for the key chromatin modifiers responsible for putting the inhibitory markers in the IRER, we examined γ-ray responsiveness in embryos mutated for genes involved in chromatin modulation. The list of the genes/alleles tested is presented in Table S4. In summary, we found that a significant delay of the sensitive-to-resistant transition was observed in embryos mutated for Hdac1, Su(var)3-9, Su(z)12, and Pc. The timing of the transition is monitored via in situ hybridization for reaper and hid, respectively, in irradiated embryos. There is a remarkable synchronicity between the responsiveness of reaper and hid in all of the tested mutants (Table S4), which strongly indicates that the same mechanism controls the responsiveness of both genes.
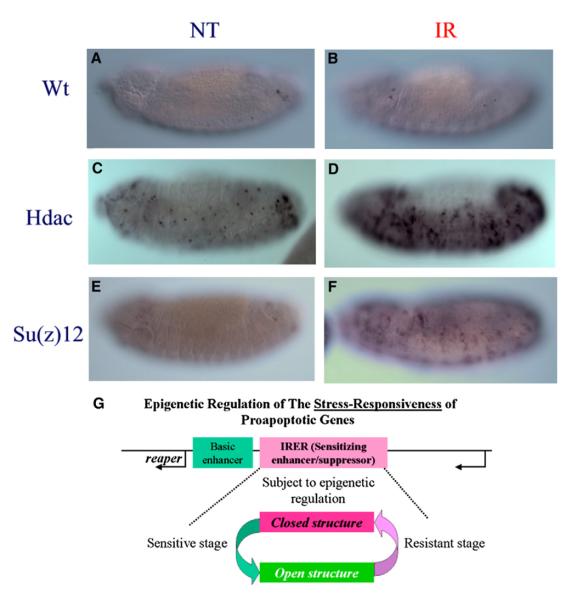
In wild-type embryos the sensitivity of reaper and hid to γ-ray irradiation is diminished once the germ band begins to retract (early or middle stage 12). In all of the Hdac, Su(var)3-9, Su(z)12, and Pc mutants, the responsiveness remained during the germ band shortening process and, in some mutant alleles, after the germ band fully retracted to the ventral side (stages 13–14) (Figures 7A–7F). There is a noticeable increase of base level reaper (and hid) expression in untreated Hdac mutant embryos (Figure 7C versus 7A), which probably reflects the general loss of suppression in these mutants. However, there is no increase of base level reaper (hid) expression in the Su(z)12 mutants, which nonetheless showed similar delay of the sensitive-to-resistant transition. All of these alleles were originally identified as dominant modifiers. For instance, the Hdac alleles were identified as dominant suppressors of position effect variegation observed for In(1)wm4 (Mottus et al., 2000). For most of the alleles tested, we noticed that the delay of transition was perceivable even in heterozygous embryos, although it was much more profound with homozygous mutants (distinguished with GFP balancer). When pooled embryos laid by heterozygous parents were tested for DNase I sensitivity, there was a detectable difference in the center of IRER_left (18,371–382) between the mutant strains and the wild-type strain at 10–13 hr AEL (stages 12–13) (Figure S4).
Figure 7. Hdac and Su(z)12 Functions Are Required for the Sensitive-to-Resistant Transition.
Responsiveness of reaper (and hid) following irradiation was measured with ISH in stage 13 embryos. In wild-type embryos, there is no response at all (A and B). However, embryos mutated for Hdac (C and D), Su(z)12 (E and F), or Su(v)3-9, Pc, etc. (Table S4) remained responsive till stages 13–14. (G) The schematic diagram summarizes our findings. Epigenetic regulation of the sensitizing enhancer region (IRER) determines whether the proapoptotic gene(s) can be induced by cellular stresses such as DNA damage, and thus controls the sensitivity to stress-induced cell death. Such an epigenetic modification may be reversible and regulated by developmental cues.
Although the function of Su(z)12 and Pc is required for turning off the sensitivity, we were not able to observe a similar delay in mutant alleles of E(z) or Psc. This may due to the rescuing effect of the maternal deposit of E(z), which has been shown to have a longer lasting effect than that of Pc [or Su(z)12]. However, there is also little delay of transition in mutant eggs laid by homozygous E(z)S2e or transheterozygous S2e/S4e mutant females at the restrictive temperature (29°C) (Table S4). This discrepancy needs to be clarified in future studies and seems to indicate that the blocking of the IRER, although involving trimethylation of H3K27 and requiring the function of some PcG proteins, is distinct from the canonic silencing mechanism observed for PRE-mediated silencing. Furthermore, we did not observe any significant precociousness or delay of the sensitive-to-resistant transition in trithorax group mutants (Table S4).
In all of the mutants tested, eventually the sensitivity is lost after about 13 hr AEL, indicating that these mutants delayed but did not block the sensitive-to-resistant (open-to-closed chromatin) transition. The timing of transition varied among the mutant alleles; however, by developmental stage 15 (about 13–14 hr AEL), none of the mutants was responsive to irradiation as measured by reaper or hid ISH. Since the P53RE reporter construct remained responsive to irradiation till the end of embryogenesis (18–20 hr AEL) (Qi et al., 2004), the loss of responsiveness in these mutants is unlikely due to the absence of trans factor(s). Our DNase I sensitivity data also indicate that although there is a delay, eventually the IRER in the mutants becomes as inaccessible as in wild-type embryos. The blocking of the IRER in embryos mutated for the key epigenetic regulators [Hdac1, Su(var)3-9, Su(z)12, and Pc] may be mediated by other proteins that have overlapping function with the four genes. In addition, given the fact that trimethylation of H3K27 and H3K9 was initiated at about the same time, it is possible that they represent redundant mechanisms in blocking the IRER.
DISCUSSION
Irradiation responsiveness appears to be a highly conserved feature of reaper-like IAP antagonists. A recently identified functional ortholog of reaper in mosquito genomes, michelob_x (mx), was also responsive to irradiation (Zhou et al., 2005). These results highlighted that stress responsiveness is an essential aspect of functional regulation of upstream proapoptotic genes such as reaper/hid. It is also worth mentioning that several mammalian BH3 domain-only proteins, the upstream proapoptotic regulators of the Bcl-2/Ced-9 pathway, are also regulated at the transcriptional level.
In this study we showed that the irradiation responsiveness of reaper and hid is subject to epigenetic regulation during development. The epigenetic regulation of the IRER is fundamentally different from the silencing of homeotic genes in that the change of DNA accessibility is limited to the enhancer region while the promoter of the proapoptotic genes remains open. Thus, it seems more appropriate to refer this as the ‘‘blocking’’ of the enhancer region instead of the ‘‘silencing’’ of the gene. This region, containing the putative P53RE and other essential enhancer elements, is required for mediating irradiation responsiveness. Our ChIP analysis indicates that histones in this enhancer region are quickly trimethylated at both H3K9 and H3K27 at the sensitive-to-resistant transition period, accompanied by a significant decrease in DNA accessibility. DNA accessibility in the putative P53RE locus (18,368k), when measured by the DNase I sensitivity assay, did not show significant decrease until sometime after the transition period. It is possible that other enhancer elements, in the core of IRER_left, are also required for radiation responsiveness. An alternative explanation is that the strong and rapid trimethylation of H3K27 and association of PRC1 at 18,366–368 are sufficient to disrupt DmP53 binding and/or interaction with the Pol II complex even though the region remains relatively sensitive to DNase I. Eventually, the whole IRER is closed with the exception of an open island around 18,387.
The finding that epigenetic regulation of the enhancer region of proapoptotic genes controls sensitivity to irradiation-induced cell death may have implications in clinical applications involving ionizing irradiation. It suggests that applying drugs that modulate epigenetic silencing may help increase the efficacy of radiation therapy. It also remains to be seen whether the hypersensitivity of some tumors to irradiation is due to the dedifferentiation and reversal of epigenetic blocking in cancer cells. On the other hand, loss of proper stress response to cellular damage is implicated in tumorigenesis (reviewed by Baylin and Ohm, 2006). The fact that the formation of heterochromatin in the sensitizing enhancer region of proapoptotic genes is sufficient to convey resistance to stress-induced cell death suggests it could contribute to tumorigenesis. In addition, it could also be the underlying mechanism of tumor cells’ evading irradiation-induced cell death. This is a likely scenario given that it has been well documented that oncogenes such as Rb and PML-RAR fusion protein cause the formation of heterochromatin through recruiting of a human ortholog of Su(v)3-9. In this regard, the reaper locus, especially the IRER, provides an excellent genetic model system for understanding the cis- and trans-acting mechanisms controlling the formation of heterochromatin associated with cellular differentiation and tumorigenesis.
Differentiation Stage-Specific Sensitivity to Irradiation-Induced Cell Death
The developmental consequence of epigenetic regulation of the IRER is the tuning down (off) of the responsiveness of the proapoptotic genes, thus decreasing cellular sensitivity to stresses such as DNA damage (Figure 7G). Epigenetic blocking of the IRER corresponds to the end of major mitotic waves when most cells begin to differentiate. Similar transitions were noticed in mammalian systems. For instance, proliferating neural precursor cells are extremely sensitive to irradiation-induced cell death while differentiating/differentiated neurons become resistant to γ-ray irradiation, even though the same level of DNA damage was inflicted by the irradiation (Nowak et al., 2006). Our findings here suggest that such a dramatic transition of radiation sensitivity could be achieved by epigenetic blocking of sensitizing enhancers.
Later in Drosophila development, around the time of pupae formation, the organism becomes sensitive to irradiation again, with LD50 values similar to what was observed for the 4–7 hr AEL embryos (Ashburner, 1989). Interestingly, it has also been found that during this period, the highly proliferative imaginal discs are sensitive to irradiation-induced apoptosis, which is mediated by the induction of reaper and hid through P53 and Chk2 (Brodsky et al., 2004). However, it remains to be studied whether the reemergence of sensitive tissue is due to reversal of the epigenetic blocking in the IRER or the proliferation of undifferentiated stem cells that have an unblocked IRER.
Silencing by a Noncanonic Mechanism?
The blocking of the IRER differs fundamentally from the silencing of homeotic genes in several aspects. First, the change of DNA accessibility and histone modification is largely limited to the enhancer region. The promoter regions of reaper (and hid) remain open, allowing the gene to be responsive to other stimuli. Indeed, there are a few cells in the central nervous system that could be detected as expressing reaper long after the sensitive-to-resistant transition. Even more cells in the late-stage embryo can be found having hid expression. Yet, the irradiation responsiveness of the two genes is completely suppressed in most if not all cells, transforming the tissues into a radiation-resistant state.
Second, the histone modification of the IRER has a mixture of features associated with pericentromeric heterochromatin formation and canonic PcG-mediated silencing. Both H3K9 and H3K27 are trimethylated in the IRER. Both HP1, the signature binding protein of the pericentromeric heterochromatin, and PRC1 are bound to the IRER. As demonstrated by genetic analysis, the functions of both Su(var)3-9 and Su(z)12/Pc are required for the silencing. Preliminary attempts to verify specific binding of PRC2 proteins to this region were unsuccessful. The fact that none of the mutants tested could completely block the transition seems to suggest that there is a redundancy of the two pathways in modifying/blocking the IRER. It is also possible that the genes we tested are not the key regulators of IRER blocking but only have participatory roles in the process.
Finally, within the IRER, there is a small region around 18,387 (18,386k–388k) that remains relatively open until the end of embryogenesis (Figure 5A). Interestingly, this open region is flanked by two putative noncoding RNA transcripts represented by EST sequences. If they are indeed transcribed in the embryo as suggested by the mRNA source of the cDNA library, then the ‘‘open island’’ within the closed IRER will likely be their shared enhancer/promoter region. Sequences of both cDNAs revealed that there is no intron or reputable open reading frame in either sequence. Despite repeated efforts, we were not able to confirm their expression via ISH or northern analysis. Overexpression of either cDNA using an expression construct also failed to show any effect on reaper/hid-induced cell death in S2 cells. Yet, sections of the two noncoding RNAs are strongly conserved in divergent Drosophila genomes. The potential role of these two noncoding RNAs in mediating reaper/hid expression and/or blocking of the IRER remains to be studied.
EXPERIMENTAL PROCEDURES
Additional information about the experimental procedures can be found in the Supplemental Data.
Fly Strains and Genetic Crosses
Canton S and yw Drosophila strains were used as wild-type in this study. Genetic crosses for generating defined deletions using the Exelixis insertional strains and tool kit were performed strictly as described (Thibault et al., 2004).
Embryo Staging and Irradiation
Staging of embryos and irradiation were performed as described previously (Zhou et al., 1999).
Gene Expression Analysis
Total RNA and mRNA were extracted with RNeasy Mini Kits (QIAGEN) or Poly(A) Pure (Ambion), respectively.
Real-Time PCR
Total RNA samples were treated with DNase I to remove genomic DNA. cDNA was prepared by reverse transcription of total RNA with a High-Capacity cDNA Archive Kit (Applied Biosystems). Quantitative real-time PCR (QPCR) followed protocols provided by the manufacturer. The real-time PCR step used 100 ng total cDNA/PCR well with triplicates per gene per sample. For primer sequences and detailed procedures, please refer to the Supplemental Data.
Microarray Data Analysis Identifying γ-ray-Responsive Genes
Gene expression levels in γ-ray-treated and control samples were first compared using Affymetrix Analysis Suite 5.0 by setting the untreated sample as ‘‘baseline.’’ A ‘‘change p value’’ was obtained for each probe set through this analysis. To minimize the nonsystematic error caused by random fluctuation, the ‘‘signal log ratio’’ (Log2[Exp./Control]) outputs of repeated measurements were analyzed by ‘‘one-class’’ significance analysis of microarrays (SAM) (Tusher et al., 2001). Genes (probe-sets) ranked in the top 50 based on the ‘‘relative difference’’ value by the SAM analysis (Tusher et al., 2001), and whose ‘‘change p value’’ were less than 0.001 in at least one array measurement, were selected as potential γ-ray responsive genes.
Comparison and Visualization of Array Data
For further analysis and visualization of array data, outputs from the Affymetrix Analysis Suite were loaded into the GeneSpring (Silicon Genetics) array analysis package. Genomic sequence and coordinates for each gene were extracted from datasets obtained from the Berkeley Drosophila Genome Project (http://www.fruitfly.org/). Gene lists for functional groups, such as apoptosis genes, were compiled based on functional annotation from Flybase (http:// www.flybase.org/) using the ‘‘ListG’’ program.
Functional Annotation of Gene Lists
Initial analysis of DNA array data as outlined above resulted in extensive gene (probe set) lists. To facilitate functional analysis, we annotated the lists using a Python-based ‘‘ListPro’’ program (see Table S1).
Statistical Analysis
We performed comparisons on the proportions of detectable genes to be cell death regulatory genes using the Chi-square test for paired samples. A 95% confidence interval was calculated for the proportion difference, e.g., (ps – pr) ± [1.96SE(ps – pr), where ps and pr are detectable proportions at the sensitive and resistant stages, respectively, and n equals 39. In addition, the exact p value was evaluated for the statistical significance of observing two cell death-related genes among 11 induced genes at the sensitive stage based on hypergeometric probability distributions.
DNase I Sensitivity Assay
Using the Apollo Genome Annotation and Curation Tool, sequences and annotations were input covering the 75C1-2 locus (18,060k–18,460k) from the Drosophila melanogaster Genome Annotation 4.0 (http://www.fruitfly.org/). For each selected 1,000 bp interval, the sequence was used as input to Primer3 for designing/selecting a set of primers that are 150–400 bp apart. The DNase I sensitivity assay was performed based on a modification of published methods (Carr and Biggin, 2000; Kalmykova et al., 2005).
Chromatin Immunoprecipitation Assay
ChIP analyses of staged embryos were performed essentially by using a protocol provided to us by Ian Birch-Machin and Shan Gao (Birch-Machin et al., 2005).
Histology
The procedures for TUNEL, in situ hybridization (ISH), and immunocytochemistry (ICC) were performed as described (Zhou and Steller, 2003). For distinguishing homozygous mutant embryos, embryos collected from Df(3L18,365–399)/TM3Ubi-GFP were first subjected to ICC with anti-GFP (Santa Cruz, 1:2000) and then subjected to ISH or TUNEL procedures.
Supplementary Material
ACKNOWLEDGMENTS
Fly strains were kindly provided by the Bloomington Stock Center, The Drosophila Stock Collection at Harvard Medical School (Dr. Artavanis-Tsakonas), Dr. Arnold Levine, Dr. Jurg Muller, Dr. Rick S. Jones, and Drs. Mottus and Grigliatti. We are grateful to Dr. Thomas Jenuwein, Dr. Rick S. Jones, and the Developmental Studies Hybridoma Bank (University of Iowa) for providing us with antibodies. Sincere appreciation is extended to Drs. He Jin, Lung-Ji Chang, Thomas Yang, Jorg Bungert, Ian Birch-Machin, Shan Gao, and Steve Russell for providing with us protocols and/or helping us to set up the DNase I sensitivity and ChIP assays. The authors also want to thank Ms. Mary Wall for proofreading of this manuscript and Drs. Jianrong Lu and Shuming Huang for helpful comments. This work is supported by NIH CA95542 to L.Z.
Footnotes
SUPPLEMENTAL DATA Supplemental Data include four figures, four tables, Supplemental Discussion, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://www.developmentalcell.com/cgi/content/full/14/4/481/DC1/.
REFERENCES
- Ashburner M. A Laboratory Handbook. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. Drosophila. [Google Scholar]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer– a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Birch-Machin I, Gao S, Huen D, McGirr R, White RA, Russell S. Genomic analysis of heat-shock factor targets in Drosophila. Genome Biol. 2005;6:R63. doi: 10.1186/gb-2005-6-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, Rio DC, Rubin GM. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell. Biol. 2004;24:1219–1231. doi: 10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Springer-Verlag; Berlin: 1985. [Google Scholar]
- Carr A, Biggin MD. Accessibility of transcriptionally inactive genes is specifically reduced at homeoprotein-DNA binding sites in Drosophila. Nucleic Acids Res. 2000;28:2839–2846. doi: 10.1093/nar/28.14.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christich A, Kauppila S, Chen P, Sogame N, Ho SI, Abrams JM. The damage-responsive Drosophila gene sickle encodes a novel IAP binding protein similar to but distinct from reaper, grim, and hid. Curr. Biol. 2002;12:137–140. doi: 10.1016/s0960-9822(01)00658-3. [DOI] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jin S, Martinek S, Joo WS, Wortman JR, Mirkovic N, Sali A, Yandell MD, Pavletich NP, Young MW, et al. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2000;97:7301–7306. doi: 10.1073/pnas.97.13.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova AI, Nurminsky DI, Ryzhov DV, Shevelyov YY. Regulated chromatin domain comprising cluster of co-expressed genes in Drosophila melanogaster. Nucleic Acids Res. 2005;33:1435–1444. doi: 10.1093/nar/gki281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Lee E, Park J, Kim E, Kim J, Chung J. In vivo p53 function is indispensable for DNA damage-induced apoptotic signaling in Drosophila. FEBS Lett. 2003;550:5–10. doi: 10.1016/s0014-5793(03)00771-3. [DOI] [PubMed] [Google Scholar]
- Lu BY, Ma J, Eissenberg JC. Developmental regulation of heterochromatin-mediated gene silencing in Drosophila. Development. 1998;125:2223–2234. doi: 10.1242/dev.125.12.2223. [DOI] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- Mizumatsu S, Monje ML, Morhardt DR, Rola R, Palmer TD, Fike JR. Extreme sensitivity of adult neurogenesis to low doses of X-irradiation. Cancer Res. 2003;63:4021–4027. [PubMed] [Google Scholar]
- Mottus R, Sobel RE, Grigliatti TA. Mutational analysis of a histone deacetylase in Drosophila melanogaster: missense mutations suppress gene silencing associated with position effect variegation. Genetics. 2000;154:657–668. doi: 10.1093/genetics/154.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom W, Chen P, Steller H, Abrams JM. Activation of the reaper gene during ectopic cell killing in Drosophila. Dev. Biol. 1996;180:213–226. doi: 10.1006/dbio.1996.0296. [DOI] [PubMed] [Google Scholar]
- Nowak E, Etienne O, Millet P, Lages CS, Mathieu C, Mouthon MA, Boussin FD. Radiation-induced H2AX phosphorylation and neural precursor apoptosis in the developing brain of mice. Radiat. Res. 2006;165:155–164. doi: 10.1667/rr3496.1. [DOI] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Peissner W, Kocher M, Treuer H, Gillardon F. Ionizing radiation-induced apoptosis of proliferating stem cells in the dentate gyrus of the adult rat hippocampus. Brain Res. Mol. Brain Res. 1999;71:61–68. doi: 10.1016/s0169-328x(99)00170-9. [DOI] [PubMed] [Google Scholar]
- Peterson C, Carney GE, Taylor BJ, White K. reaper is required for neuroblast apoptosis during Drosophila development. Development. 2002;129:1467–1476. doi: 10.1242/dev.129.6.1467. [DOI] [PubMed] [Google Scholar]
- Qi D, Larsson J, Mannervik M. Drosophila Ada2b is required for viability and normal histone H3 acetylation. Mol. Cell. Biol. 2004;24:8080–8089. doi: 10.1128/MCB.24.18.8080-8089.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogame N, Kim M, Abrams JM. Drosophila p53 preserves genomic stability by regulating cell death. Proc. Natl. Acad. Sci. USA. 2003;100:4696–4701. doi: 10.1073/pnas.0736384100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of micro-arrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- Wurgler FE, Ullrich H. Radiaosensitivity of embryonic stages. In: Ashburner M, Novitski E, editors. The Genetics and Biology of Drosophila. Academic Press; London; New York: 1976. pp. 1269–1298. [Google Scholar]
- Zhou L, Schnitzler A, Agapite J, Schwartz LM, Steller H, Nambu JR. Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system mid-line cells. Proc. Natl. Acad. Sci. USA. 1997;94:5131–5136. doi: 10.1073/pnas.94.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Song Z, Tittel J, Steller H. HAC-1, a Drosophila homolog of APAF-1 and CED-4 functions in developmental and radiation-induced apoptosis. Mol. Cell. 1999;4:745–755. doi: 10.1016/s1097-2765(00)80385-8. [DOI] [PubMed] [Google Scholar]
- Zhou L, Steller H. Distinct pathways mediate UV-induced apoptosis in Drosophila embryos. Dev. Cell. 2003;4:599–605. doi: 10.1016/s1534-5807(03)00085-6. [DOI] [PubMed] [Google Scholar]
- Zhou L, Jiang G, Chan G, Santos CP, Severson DW, Xiao L. Michelob_x is the missing inhibitor of apoptosis protein antagonist in mosquito genomes. EMBO Rep. 2005;6:769–774. doi: 10.1038/sj.embor.7400473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.