Abstract
Eukaryotic translation elongation factor 1A (eEF1A) is one of the most abundant protein synthesis factors. eEF1A is responsible for the delivery of all aminoacyl-tRNAs to the ribosome, aside from initiator and selenocysteine tRNAs. In addition to its roles in polypeptide chain elongation, unique cellular and viral activities have been attributed to eEF1A in eukaryotes from yeast to plants and mammals. From preliminary, speculative associations to well characterized biochemical and biological interactions, it is clear that eEF1A, of all the translation factors, has been ascribed the most functions outside of protein synthesis. A mechanistic understanding of these non-canonical functions of eEF1A will shed light on many important biological questions, including viral-host interaction, subcellular organization, and the integration of key cellular pathways.
Keywords: Actin, Apoptosis, Nuclear Transport, Protein Turnover, RNA Viruses, Translation Elongation Factors
Introduction
Protein synthesis can be divided into three fundamental stages: initiation, elongation, and termination. Translation elongation requires several soluble proteins called eEFs.2 During elongation, cognate aa-tRNA are delivered to the A site of the ribosome by eEF1A. Once a codon/anticodon match is detected, eEF1A deposits the aa-tRNA and is itself released from the ribosome. A peptide bond can now be made, thereby elongating the growing polypeptide. eEF2 then catalyzes the movement of the peptidyl-tRNA·mRNA complex from the A site to the P site of the ribosome, positioning the next codon in the A site and allowing the process to repeat. This minireview focuses on eEF1A, briefly discussing its canonical function in translation elongation and then describing other cellular activities of this highly abundant protein. Although there are many examples where components of the translational apparatus have been linked to a process distinct from protein synthesis (1), eEF1A provides perhaps the most examples. Our goal is to bring together the work from many different laboratories that support the hypothesis that eEF1A is a central regulator involved in the coordination of many different cellular and viral processes.
Special Delivery: The Canonical Role of eEF1A in Translation
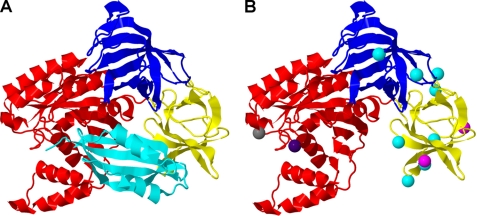
eEF1A is a GTP-binding protein and the homolog of the bacterial protein EF-Tu. The GTP-dependent binding of aa-tRNA to eEF1A, binding of the complex to the ribosome, and decoding are essential steps for both efficient and accurate gene expression (reviewed in Ref. 2). In addition, eEF1A requires the activity of a GEF, eEF1Bαγ, to promote GDP release and reactivate the protein for aa-tRNA delivery (3). A combination of kinetic, genetic, and structural information has provided a rich understanding of this canonical role of eEF1A and EF-Tu (3–5). High resolution crystal structures of Saccharomyces cerevisiae eEF1A (3, 6) and bacterial EF-Tu in multiple functional conformations (5) and bound to the ribosome (7) provide detailed information for the interpretation of the function of the protein in translation. As shown in Fig. 1A, the crystal structure of eEF1A forms three well defined domains involved in specific aspects of its function. Domain I binds GTP, domain II is implicated in aa-tRNA binding, and domains I and II interact with eEF1Bα (Fig. 1A). These studies also provide a platform for understanding the plethora of non-canonical activities ascribed to this elongation factor and how these activities compete with the canonical functions of the protein (Fig. 2).
FIGURE 1.
Structural organization of eEF1A. A, the structure of eEF1A from S. cerevisiae bound to the catalytic C terminus of eEF1Bα (cyan). eEF1A has three well defined domains. Domain I (red) binds GTP, domain II (yellow) is proposed to bind the aminoacyl end of the aa-tRNA, and domains II and III (blue) are linked to actin binding. eEF1Bα binds eEF1A in domains I and II. B, mutations affecting the non-canonical functions of eEF1A are indicated by colored spheres. Mutations in domain I include T22S (dark blue), which inhibits TBSV replication, and D156N (gray), which affects protein turnover. eEF1A mutations defective in nuclear transport, E286K and E291K (magenta), are found in domain II. Mutations that affect actin organization, N329D, N329D, Y355C, K333E, H294A, Q296R, F308L, S405P, and N305S (cyan), are shown in domains II and III. The figure was prepared with Jmol using Protein Data Bank (PDB) 1F60 (6).
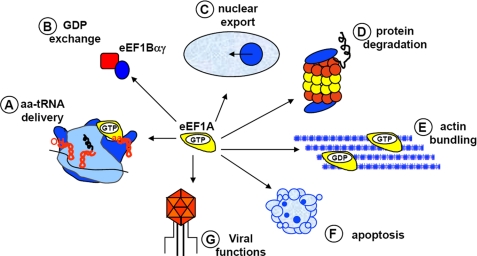
FIGURE 2.
Canonical and non-canonical functions attributed to eEF1A. The canonical function of eEF1A is to bind aa-tRNA in a GTP-dependent manner and deliver it to the ribosome (A). The exchange of GDP is promoted by the GEF eEF1Bαγ (B). The non-canonical functions include roles in nuclear export events (C), turnover of misfolded proteins (D), binding and bundling of the actin cytoskeleton as well as other cytoskeletal components (E), apoptosis (F), and the viral life cycle (G).
Movin' Out: eEF1A and the Nuclear Export Process
Although the canonical role of eEF1A in translation is associated with a cytoplasmic function, several reports have linked the protein to nuclear export. The first work isolated eEF1A in a high copy suppressor screen of a synthetic lethal nuclear transport mutant in S. cerevisiae. Budding yeast with reduced eEF1A expression or those expressing eEF1A domain II mutants, E286K or E291K (Fig. 1B), showed defects in nuclear export of aa-tRNA (reviewed in Ref. 8). These eEF1A mutants are near the proposed binding site for the aminoacyl end of the tRNA. eEF1A was also shown to exhibit aa-tRNA-dependent binding to the nuclear export machinery in mammalian cells (9, 10). Because eEF1A binds aa-tRNA in its canonical role, whether this is really a non-canonical function or part of efficient channeling of protein synthesis components within the cell can be debated. However, more recently, eEF1A was identified as a participant in the transcription-dependent nuclear export of proteins from the nucleus (11), which is clearly a distinct function from protein synthesis. In both export processes, it was postulated that this function of eEF1A was executed from the cytoplasmic side of the nuclear envelope because eEF1A appeared to be excluded from the nucleus. However, a recent study demonstrated that eEF1A could be detected in the nucleus of a specific budding yeast nuclear export mutant (msn5Δ), suggesting that eEF1A can access the nucleus (12). Therefore, a role for eEF1A on the nuclear side of the membrane cannot be excluded.
The Yin and Yang: eEF1A and Proteolysis
At first glance, the thought of eEF1A playing a role in protein degradation seems counterintuitive. However, upon further consideration, the high concentration of eEF1A in close proximity to the ribosome might facilitate a potential role for this protein in protein quality control and co-translational degradation. Initial support for this hypothesis came from in vitro experiments where eEF1A was purified as a factor required for the degradation of Nα-acetylated proteins (13). Other experiments showed that eEF1A could be UV cross-linked to a nascent polypeptide chain as well as a misfolded protein but not to a folded protein (14). eEF1A could even facilitate the folding of denatured proteins in in vitro activity assays (14). However, in vivo evidence supporting a role for eEF1A in co-translational degradation is still preliminary. eEF1A was isolated as a suppressor of a growth defect associated with deletion of genes in the proteolytic pathway (15). It was subsequently shown to interact with the 26 S proteasome and that this association increases when translation is inhibited (15). Intriguingly, the half-life of a reporter protein is significantly increased in S. cerevisiae expressing eEF1A with an D156N mutation (Fig. 1B) in the GTP-binding domain (15). This mutant was designed and determined to affect nucleotide specificity and the requirement of the GEF eEF1Bα. The combination of these data supports a model in which eEF1A plays a role in regulating protein degradation, perhaps by linking unfolded proteins with the proteasome. Future experiments will need to address the pivotal question of how the diverse roles of eEF1A in protein synthesis and quality control are integrated in the cell.
Closet Organizer: eEF1A and the Cytoskeleton
Over the past two decades, evidence has been presented from a number of laboratories that establishes a functional link between the protein translation machinery and the cytoskeleton. For example, several components of the translational apparatus associate with the cytoskeleton, including aa-tRNA synthetases, eukaryotic initiation factors (eIFs), and eEFs (for review, see Ref. 16). In addition, efficient translation in mammalian cells has been shown to be dependent on an intact actin cytoskeleton, whereas deletion of certain actin-remodeling proteins in budding yeast has a specific effect on translation initiation (17, 18). Finally, the cytoskeleton is important for the spatially restricted translation of localized mRNAs in diverse organisms (19). At the simplest level, these observations suggest that the cytoskeleton provides a spatial concentration of the translation machinery for efficient protein synthesis. However, additional studies on the association between eEF1A and the cytoskeleton reveal that the regulation of these two processes may be reciprocal.
eEF1A was first isolated as an actin-binding protein from the slime mold, Dictyostelium discoideum (20). eEF1A can both bind and bundle actin filaments in vitro, and this activity has been evolutionarily conserved (21). Unlike its interaction with aa-tRNA, the actin bundling activity of eEF1A is not dependent on GTP (22). Interestingly, eEF1A cannot bind or bundle actin efficiently in the presence of aa-tRNA, suggesting that the binding of these two factors to eEF1A is mutually exclusive (23). These experiments raised the question of how the role of eEF1A in these diverse functions is regulated in vivo.
Genetic and biochemical experiments in budding yeast have begun to address the divergent roles of eEF1A within the cell. Overexpression of eEF1A causes a slow growth phenotype and disruption of the actin cytoskeleton in the absence of any measurable effects on protein synthesis (24). In addition, synthetic growth defects were observed when eEF1A overexpression was combined with mutations in actin, illustrating a genetic interaction between these two factors in vivo (24). A series of separation of function mutations in eEF1A has also been identified that clearly demonstrates a role for eEF1A in the organization of the actin cytoskeleton in vivo (17, 25). These mutations localize to domains II and III of eEF1A and fall into two classes (Fig. 1B). The first class of mutants is characterized by normal rates of protein synthesis, disorganization of the actin cytoskeleton, and reduced actin bundling but not binding in vitro (25). The second class of mutants displays more severe actin phenotypes combined with a slow growth phenotype and defects in translation initiation (17). The localization of the majority of these separation of function mutations within domain II, which is also implicated in aa-tRNA and eEF1Bα binding (Fig. 1B), raises important questions about how the interaction of eEF1A with these two factors affects its affinity for actin. Recent work has shown that eEF1Bα can reduce in vitro actin bundling by eEF1A, and mutations in eEF1Bα that reduce its affinity for eEF1A induce actin disorganization (26). Together, these data suggest that there exists a complex relationship in eukaryotic cells between the function of eEF1A in translation and actin organization that may be influenced by the availability of its binding factors. Finally, in addition to the direct interaction between eEF1A and actin described above, certain isoforms of eEF1A in human cells (see “To Die or Not to Die, eEF1A and Apoptosis”) may also influence the actin cytoskeleton indirectly (27, 28).
Studies using eEF1A from a number of organisms have also suggested that eEF1A may function in microtubule dynamics, although its exact role remains unclear. eEF1A or an eEF1A-like activity was first identified as a component of the mitotic apparatus in sea urchin eggs (29). In vitro experiments employing carrot eEF1A demonstrated that eEF1A could stabilize microtubules at substoichiometric amounts and bundle microtubules at approximately equimolar amounts (30, 31). In contrast, eEF1A was purified as a microtubule-severing factor from Xenopus laevis egg extracts (32). Microtubule-severing activity was also observed for mammalian forms of eEF1A both in vitro and in microinjection experiments (32). Clearly, additional experiments are required not only to address these differences in eEF1A activity but also to further understand the functional significance and regulation of these microtubule-related activities in the cell.
To Die or Not to Die: eEF1A and Apoptosis
eEF1A has been reported to play a role in apoptosis in metazoans. Apoptosis or programmed cell death is a highly regulated series of cellular events that lead to the elimination of damaged or unnecessary cells. Early experiments showed that the level of eEF1A expression in cultured mouse fibroblasts correlated with the rate of apoptosis upon serum withdrawal, with higher levels of eEF1A expression being associated with a faster rate of cell death (33). In another set of experiments, eEF1A was isolated in a cDNA screen to identify factors that promote survival following growth factor withdrawal of a pro-B cell line (34). An explanation for the discrepancy of these two observations may come from the fact that mammals have two isoforms of eEF1A that are ∼92% identical at the amino acid level. Although both isoforms function in translation elongation assays in vitro, they display tissue and developmentally specific expression patterns (35). eEF1A-1 is widely expressed in mouse tissues, whereas eEF1A-2 expression is found only in the heart, brain, and skeletal muscle (35, 36). Thus, the effect on apoptosis may depend on the isoform of eEF1A, a detail that was not reported in the early analyses. Recent experiments support this hypothesis. In cultured myoblasts, eEF1A-2 expression correlates with differentiation and has a protective effect against apoptosis, whereas expression of eEF1A-1 has the opposite effect (37). The reason for these different effects on apoptosis, however, is currently unclear. It is possible that the two different eEF1A isoforms may vary in their ability to express specific genes, or they may interact with different protein partners (38). It is also unknown whether the two eEF1A isoforms differ in their abilities to carry out other non-canonical functions in vivo. Further analysis of the functional differences between these two isoforms will be necessary to understand the mechanism(s) of their effects on apoptosis.
Hijacked: eEF1A and Viral Propagation
Viruses depend on host cell proteins for their replication and propagation. Therefore, it is not surprising that numerous viruses have apparently evolved to utilize eEF1A, one of the most abundant proteins in eukaryotic cells, in their life cycle. Data from a number of laboratories have implicated eEF1A in the replication of positive strand RNA viruses of both plants and animals. However, the exact role that eEF1A plays in viral replication is not clear and may vary depending on the virus. Similar to its canonical role in translation, eEF1A has been shown to bind an aa-tRNA-like element in the 3′-untranslated region of the turnip yellow mosaic virus genome and repress minus strand synthesis in vitro (39). In other viruses, eEF1A has been shown to interact with the viral RdRP, although its functional significance as a part of this complex is unknown (40–43). In still other viruses, such as the TBSV, the tobacco mosaic virus, and the West Nile virus, eEF1A binds both the viral RNA and the RdRP (44–48). Genetic analysis of both the virus and eEF1A is now illuminating the functional consequences of these interactions. Mutation of the major eEF1A-binding site in the West Nile virus genome leads to a decrease in minus strand synthesis and a corresponding decrease in viral production, suggesting that binding of eEF1A to the viral template is important for viral replication (45). Down-regulation of eEF1A in Nicotiana benthamiana inhibited tobacco mosaic virus replication and spreading, which highlights the functional significance of eEF1A in this viral life cycle (49). In TBSV, eEF1A may promote viral replication by stabilization of the p33 component of the RdRP (46). S. cerevisiae expressing a T22S mutant form of eEF1A displayed defective viral replication (Fig. 1B), suggesting that eEF1A plays a functionally significant role in TBSV replication (46). In summary, positive strand viruses may have evolved to utilize eEF1A to promote viral propagation through multiple mechanisms.
Conclusions and Future Implications
It is clear that although many functions aside from its canonical role in aa-tRNA delivery have been ascribed to eEF1A, there remains much to learn. In some cases, initial studies have advanced to convincing examples of functional interactions such as with the actin cytoskeleton. In other cases, such as recruitment as a host factor for the viral life cycle, the reassignment of function is seen in multiple different viruses. In still other cases, the results are beginning to shed light on new links to other cellular processes. These links may define important communication between cellular pathways, such as how protein synthesis links to protein turnover or cell death or how cellular organization can affect gene expression through a link to translation. The challenge remains to separate the nonspecific association of eEF1A with proteins, due to its abundance and electrostatic and RNA binding properties, from functionally significant associations. This last fact is recognized in the exclusion of the protein from the matrix-assisted laser desorption/ionization prey data set of a global mass spectroscopy-based protein complex screen (50). The examples described in this review demonstrate that eEF1A clearly has additional functions and provides unique ways to look at both secondary functions of a protein and links between different processes in the complex milieu of the cell.
Supplementary Material
Acknowledgments
We thank the members of the Kinzy laboratory for helpful comments.
This work was supported, in whole or in part, by National Institutes of Health Grant GM057483 (to T. G. K.). This is the sixth article in the Thematic Minireview Series on Protein Synthesis. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- eEF
- eukaryotic elongation factor
- eEF1A
- eukaryotic elongation factor 1A
- aa-tRNA
- aminoacyl-tRNA
- GEF
- guanine nucleotide exchange factor
- TBSV
- tomato bushy stunt virus
- RdRP
- RNA-dependent RNA polymerase.
REFERENCES
- 1.Kinzy T. G., Goldman E. (2000) in Translational Control of Gene Expression (Hershey J. W. B., Mathews M. B., Sonenberg N. eds) pp. 973–997, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 2.Taylor D. R., Frank J., Kinzy T. G. (2006) in Translational Control in Biology and Medicine (Sonenberg N., Hershey J. W. B., Mathews M. B. eds) pp. 59–85, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 3.Andersen G. R., Valente L., Pedersen L., Kinzy T. G., Nyborg J. (2001) Nat. Struct. Biol. 8, 531–534 [DOI] [PubMed] [Google Scholar]
- 4.Rodnina M. V., Wintermeyer W. (2009) Curr. Opin. Cell Biol. 21, 435–443 [DOI] [PubMed] [Google Scholar]
- 5.Andersen G. R., Nissen P., Nyborg J. (2003) Trends Biochem. Sci. 28, 434–441 [DOI] [PubMed] [Google Scholar]
- 6.Andersen G. R., Pedersen L., Valente L., Chatterjee I., Kinzy T. G., Kjeldgaard M., Nyborg J. (2000) Mol. Cell 6, 1261–1266 [DOI] [PubMed] [Google Scholar]
- 7.Schmeing T. M., Voorhees R. M., Kelley A. C., Gao Y. G., Murphy F. V., 4th, Weir J. R., Ramakrishnan V. (2009) Science 326, 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grosshans H., Simos G., Hurt E. (2000) J. Struct. Biol. 129, 288–294 [DOI] [PubMed] [Google Scholar]
- 9.Calado A., Treichel N., Müller E. C., Otto A., Kutay U. (2002) EMBO J. 21, 6216–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohnsack M. T., Regener K., Schwappach B., Saffrich R., Paraskeva E., Hartmann E., Görlich D. (2002) EMBO J. 21, 6205–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khacho M., Mekhail K., Pilon-Larose K., Pause A., Côté J., Lee S. (2008) Mol. Biol. Cell 19, 5296–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murthi A., Shaheen H. H., Huang H. Y., Preston M. A., Lai T. P., Phizicky E. M., Hopper A. K. (2010) Mol. Biol. Cell 21, 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonen H., Smith C. E., Siegel N. R., Kahana C., Merrick W. C., Chakraburtty K., Schwartz A. L., Ciechanover A. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7648–7652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hotokezaka Y., Tobben U., Hotokezaka H., Van Leyen K., Beatrix B., Smith D. H., Nakamura T., Wiedmann M. (2002) J. Biol. Chem. 277, 18545–18551 [DOI] [PubMed] [Google Scholar]
- 15.Chuang S. M., Chen L., Lambertson D., Anand M., Kinzy T. G., Madura K. (2005) Mol. Cell. Biol. 25, 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S., Coulombe P. A. (2010) Nat. Rev. Mol. Cell Biol. 11, 75–81 [DOI] [PubMed] [Google Scholar]
- 17.Gross S. R., Kinzy T. G. (2007) Mol. Cell. Biol. 27, 1974–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stapulionis R., Kolli S., Deutscher M. P. (1997) J. Biol. Chem. 272, 24980–24986 [DOI] [PubMed] [Google Scholar]
- 19.Martin K. C., Ephrussi A. (2009) Cell 136, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang F., Demma M., Warren V., Dharmawardhane S., Condeelis J. (1990) Nature 347, 494–496 [DOI] [PubMed] [Google Scholar]
- 21.Demma M., Warren V., Hock R., Dharmawardhane S., Condeelis J. (1990) J. Biol. Chem. 265, 2286–2291 [PubMed] [Google Scholar]
- 22.Edmonds B. T., Bell A., Wyckoff J., Condeelis J., Leyh T. S. (1998) J. Biol. Chem. 273, 10288–10295 [DOI] [PubMed] [Google Scholar]
- 23.Liu G., Tang J., Edmonds B. T., Murray J., Levin S., Condeelis J. (1996) J. Cell Biol. 135, 953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munshi R., Kandl K. A., Carr-Schmid A., Whitacre J. L., Adams A. E., Kinzy T. G. (2001) Genetics 157, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gross S. R., Kinzy T. G. (2005) Nat. Struct. Mol. Biol. 12, 772–778 [DOI] [PubMed] [Google Scholar]
- 26.Pittman Y. R., Kandl K., Lewis M., Valente L., Kinzy T. G. (2009) J. Biol. Chem. 284, 4739–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amiri A., Noei F., Jeganathan S., Kulkarni G., Pinke D. E., Lee J. M. (2007) Oncogene 26, 3027–3040 [DOI] [PubMed] [Google Scholar]
- 28.Jeganathan S., Morrow A., Amiri A., Lee J. M. (2008) Mol. Cell. Biol. 28, 4549–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohta K., Toriyama M., Miyazaki M., Murofushi H., Hosoda S., Endo S., Sakai H. (1990) J. Biol. Chem. 265, 3240–3247 [PubMed] [Google Scholar]
- 30.Durso N. A., Cyr R. J. (1994) Plant Cell 6, 893–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore R. C., Durso N. A., Cyr R. J. (1998) Cell Motil. Cytoskeleton 41, 168–180 [DOI] [PubMed] [Google Scholar]
- 32.Shiina N., Gotoh Y., Kubomura N., Iwamatsu A., Nishida E. (1994) Science 266, 282–285 [DOI] [PubMed] [Google Scholar]
- 33.Duttaroy A., Bourbeau D., Wang X. L., Wang E. (1998) Exp. Cell Res. 238, 168–176 [DOI] [PubMed] [Google Scholar]
- 34.Talapatra S., Wagner J. D., Thompson C. B. (2002) Cell Death Differ. 9, 856–861 [DOI] [PubMed] [Google Scholar]
- 35.Kahns S., Lund A., Kristensen P., Knudsen C. R., Clark B. F., Cavallius J., Merrick W. C. (1998) Nucleic Acids Res. 26, 1884–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S., Francoeur A. M., Liu S., Wang E. (1992) J. Biol. Chem. 267, 24064–24068 [PubMed] [Google Scholar]
- 37.Ruest L. B., Marcotte R., Wang E. (2002) J. Biol. Chem. 277, 5418–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang R., Wang E. (2007) J. Cell. Biochem. 100, 267–278 [DOI] [PubMed] [Google Scholar]
- 39.Matsuda D., Yoshinari S., Dreher T. W. (2004) Virology 321, 47–56 [DOI] [PubMed] [Google Scholar]
- 40.Qanungo K. R., Shaji D., Mathur M., Banerjee A. K. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5952–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson C. M., Perez D. R., French R., Merrick W. C., Donis R. O. (2001) J Gen. Virol. 82, 2935–2943 [DOI] [PubMed] [Google Scholar]
- 42.Nishikiori M., Dohi K., Mori M., Meshi T., Naito S., Ishikawa M. (2006) J. Virol. 80, 8459–8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thivierge K., Cotton S., Dufresne P. J., Mathieu I., Beauchemin C., Ide C., Fortin M. G., Laliberté J. F. (2008) Virology 377, 216–225 [DOI] [PubMed] [Google Scholar]
- 44.Blackwell J. L., Brinton M. A. (1997) J. Virol. 71, 6433–6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis W. G., Blackwell J. L., Shi P. Y., Brinton M. A. (2007) J. Virol. 81, 10172–10187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z., Pogany J., Panavas T., Xu K., Esposito A. M., Kinzy T. G., Nagy P. D. (2009) Virology 385, 245–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamaji Y., Kobayashi T., Hamada K., Sakurai K., Yoshii A., Suzuki M., Namba S., Hibi T. (2006) Virology 347, 100–108 [DOI] [PubMed] [Google Scholar]
- 48.Zeenko V. V., Ryabova L. A., Spirin A. S., Rothnie H. M., Hess D., Browning K. S., Hohn T. (2002) J. Virol. 76, 5678–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamaji Y., Sakurai K., Hamada K., Komatsu K., Ozeki J., Yoshida A., Yoshii A., Shimizu T., Namba S., Hibi T. (2010) Arch. Virol. 155, 263–268 [DOI] [PubMed] [Google Scholar]
- 50.Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St. Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.