Abstract
The formation of a primary endocytic vesicle is a dynamic process involving the transient organization of adaptor and scaffold proteins at the plasma membrane. Epsins and Eps15-like proteins are ubiquitin-binding proteins that act early in this process. The yeast epsins, Ent1 and Ent2, carry functional ubiquitin-interacting motifs (UIMs), whereas the yeast Eps15-like protein, Ede1, has a C-terminal ubiquitin associated (UBA) domain. Analysis of mutants lacking early endocytic adaptors reveals that the ubiquitin-binding domains (UBDs) of Ent2 and Ede1 are likely to function primarily to mediate protein-protein interactions between components of the early endocytic machinery. Cells that lack epsin and Ede1 UBDs are able to internalize activated, ubiquitinated receptors. Furthermore, under conditions in which epsin UIMs are important for receptor internalization, receptors internalized via both ubiquitin-dependent and ubiquitin-independent signals require the UIMs, indicating that UIM function is not restricted to ubiquitinated receptors. Epsin UIMs share function with non-UBD protein-protein interaction motifs and domains in Ent2 and Ede1, and the Ede1 UBA domain appears to negatively regulate interactions between endocytic proteins. Together, our results suggest that the ubiquitin-binding domains within the yeast epsin Ent2 and Ede1 are involved in the formation and regulation of the endocytic network.
Keywords: ubiquitination, endocytosis, receptor internalization, Eps15, UIM, UBA, EH domain
Introduction
Endocytosis is an essential process for regulating cell morphology and homeostasis. The formation of vesicles at the plasma membrane is highly regulated and is responsible for transporting transmembrane proteins and lipids into the cell. A dynamic network of endocytic proteins, including clathrin adaptors and clathrin, assembles at sites of vesicle formation where cargo is recruited into the nascent vesicle bud (1-3). Assembly of actin regulators and membrane-binding proteins at the endocytic network then facilitates vesicle invagination and scission (4-7). The tightly coordinated assembly and dissociation of endocytic network constituents requires specific protein-protein and protein-lipid interactions (8, 9).
Endocytic adaptors and clathrin initiate the assembly of this network. Endocytic adaptors are multisubunit complexes or monomeric proteins that localize to the plasma membrane via lipid-binding domains and recruit clathrin to the membrane to form lattices enclosing nascent buds (3, 10-12). These adaptors, including epsins, AP180 proteins and AP-2 complexes, recognize signals that target cargo for internalization. Adaptors also assemble later-acting endocytic proteins, known as scaffold proteins, that stabilize the adaptor-cargo-clathrin complex and regulate the assembly of actin required for endocytic vesicle maturation and internalization.
Endocytic adaptors are conserved from yeast to mammals, and the clathrin- and actin-dependent endocytic pathway in yeast is an excellent model for clathrin-dependent internalization that occurs in mammalian cells. In Saccharomyces cerevisiae, the epsin and AP180 adaptors are functionally important and have overlapping roles (13, 14). The epsins Ent1 and Ent2 are an essential gene pair with endocytic functions that are mostly redundant (15). They interact with phosphatidylinositol-(4,5)-bisphosphate at the plasma membrane via an Epsin N-terminal homology (ENTH) domain and with ubiquitin via tandem ubiquitin interacting motifs (UIMs). Both yeast epsins have two asparagine-proline-phenylalanine (NPF) motifs that are required for their proper localization during endocytosis and for interaction with the Eps15 homology (EH) domain-containing proteins, Ede1 and Pan1 (14, 16-19). At the C-terminus, Ent1 and Ent2 bind to clathrin through a clathrin-binding motif (15, 18). The AP180 proteins, Yap1801 and Yap1802, have an N-terminal ANTH lipid-binding domain, multiple NPF motifs and a C-terminal clathrin-binding motif (16). Previous studies suggested that different protein-protein and protein-lipid interaction domains of the yeast monomeric clathrin adaptors have partially overlapping functions during receptor internalization. Specifically, Ent1 NPF motifs function cooperatively with the lipid and clathrin binding regions (14).
Ede1 is one partner of the epsin NPF motifs and is required for normal rates of receptor internalization (14, 20, 21). Ede1 and the similar mammalian protein Eps15 are endocytic scaffold proteins that are proposed to play a role in stabilization of the adaptor-cargo complex (18, 22, 23). Both proteins have three comparably spaced EH domains at the N-terminus and each has a C-terminal ubiquitin-binding domain, which binds ubiquitin directly (21, 24). In yeast, Ede1 functionally replaces Ent1 ubiquitin binding during receptor internalization (21), and in mammalian cells, the epsins and the UIMs of Eps15 function redundantly during internalization (25). At this time, however, specific mechanistic information about the role of epsin and Ede1 UBDs in endocytosis is not available.
Here we investigate the phenotypic consequences of mutating epsin and Ede1 UBDs on receptor internalization. We find that ubiquitinated cargo can be internalized in the absence of ubiquitin-binding by epsins and Ede1. Moreover, in a genetic background in which receptor internalization depends on epsin UIMs, internalization of cargo carrying ubiquitin or linear peptide signals is severely impaired. These observations suggest that cargo binding is not the major function of the epsin UIMs or Ede1 UBA domain. Instead, we demonstrate that the UIMs of Ent2 are functionally redundant with the network-forming Ede1 EH domains. Furthermore, we provide evidence that the Ede1 UBA domain negatively regulates EH domain-mediated protein-protein interactions.
Results
Ubiquitin binding by epsins and Ede1 is not required for receptor internalization
Previous studies from our laboratory suggested that Ede1 and the epsin UIMs share a common function in the process of receptor internalization. Specifically, we demonstrated that deletion of Ent1 UIMs caused a significant internalization defect in cells lacking wildtype epsins and Ede1, but not in cells lacking only epsins (21). To determine whether the functional redundancy between Ede1 and the UIMs of the yeast epsins is due to the Ede1 ubiquitin-binding domain, we analyzed the effect of mutating the Ede1 UBA domain on receptor internalization in ent1Δ ent2Δ ede1Δ (3Δ) cells expressing Ent1 lacking functional UIMs (Ent1UIMΔ).
To measure receptor internalization in mutant yeast strains we employed an assay that measures the internalization of [35S]-labeled α-factor pheromone by its G protein-coupled cell surface receptor, Ste2. When we performed these assays on 3Δ cells expressing Ent1UIMΔ and an Ede1 clone harboring a deletion of the C-terminal UBA domain (ede1UBAΔ), we observed no defect in Ste2 internalization as compared to cells expressing Ede1WT (Figure 1A). To confirm that deletion of the UBA domain eliminated the ability of Ede1 to bind ubiquitin in this strain background, we tested whether Ede1UBAΔ expressed in ent1UIMΔ ent2Δ cells could bind ubiquitin immobilized on Sepharose beads. Although both wildtype Ede1 and Ede1UBAΔ bound ubiquitin in the presence of a wildtype epsin, Ede1UBAΔ did not bind ubiquitin in ent1UIMΔ ent2Δ cells (Figure 1B). In the presence of wildtype Ent1, Ede1UBAΔ presumably bound to ubiquitin indirectly through the formation of an Ent1-Ede1 complex. However in cells in which neither epsins nor Ede1 carry a UBD, binding of Ede1 to ubiquitin was abolished. These findings indicate that Ent1UIMΔ and Ede1UBAΔ are able to mediate the internalization of Ste2 without the ability to bind ubiquitin.
Figure 1. Ede1 and Ent2 ubiquitin binding domains are not required for Ste2 internalization.
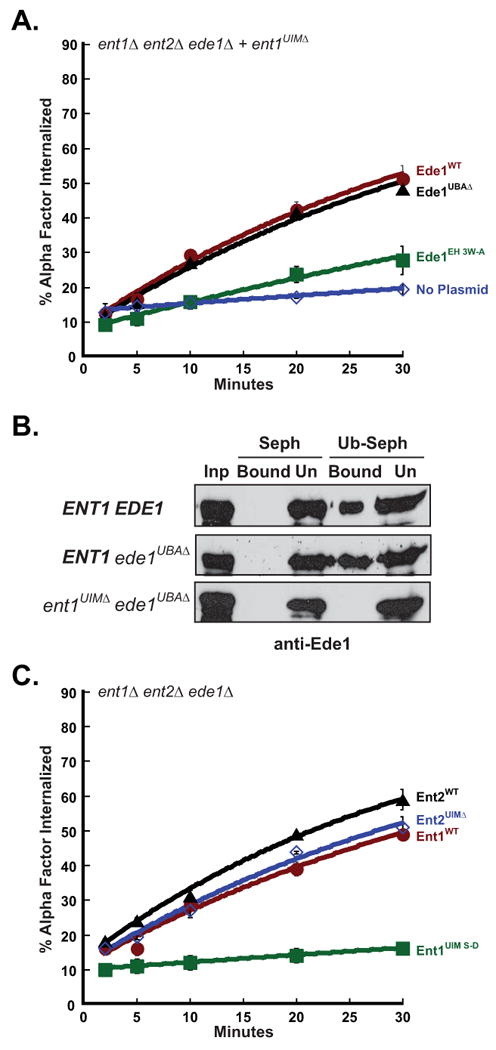
(A) The Ede1 UBA domain is not required for internalization of Ste2. Triple mutant cells, ent1Δ ent2Δ ede1Δ (3Δ), expressing Ent1UIMΔ and either Ede1WT (LHY3642), Ede1UBAΔ (LHY3643) or Ede1EH 3W-A (LHY4959) were grown at 30°C and assayed for internalization of 35S-labeled α-factor. Each point represents the average of three or more assays. Error bars represent the standard deviation. (B) An Ent1-Ede1 complex lacking ubiquitin-binding domains cannot bind monoubiquitin. Yeast lysates (Inp) prepared from 3Δ cells expressing Ent1WT and Ede1WT (LHY3638), Ent1WT and Ede1UBAΔ (LHY 3639) or Ent1UIMΔ and Ede1UBAΔ (LHY3643) were incubated with Sepharose (Seph) or ubiquitin-Sepharose (Ub-Seph) beads. The bound and unbound (Un) proteins were analyzed by SDS-PAGE and anti-Ede1 immunoblotting. (C) Ent2 UIMs are not required for Ste2 internalization. Triple mutant cells, ent1Δ ent2Δ ede1Δ (3Δ), expressing Ent1WT (LHY5718), Ent1UIM S-D (LHY5719), Ent2WT (LHY3190) or Ent2UIMΔ (LHY3191) were grown at 30°C and assayed for internalization of 35S-labeled α-factor. Each point represents the average of three or more assays. Error bars represent the standard deviation.
The results described above suggest that the UBA domain is not the region of Ede1 that functionally overlaps with epsin UIMs in receptor internalization. Further support for this idea came from the analysis of the Ede1EH 3W-A protein that carries mutations inactivating each of the three EH domains (26, 27). Unlike Ede1UBAΔ, Ede1EH 3W-A was unable to rescue Ste2 internalization in 3Δ cells expressing Ent1UIMΔ (Figure 1A). These surprising results suggest that Ede1 compensates for the function of epsin UIMs not by binding ubiquitin, but by forming protein-protein interactions mediated by EH domains.
The results described above indicate that epsin and Ede1 ubiquitin-binding is dispensable for the internalization of receptors that use ubiquitin internalization signals. To more thoroughly investigate the role of epsin UIMs in receptor internalization, we analyzed both ent1 and ent2 mutants carrying mutations that impair ubiquitin-binding by UIMs, ent1UIM S-D and ent2UIMΔ (18). As we observed with 3Δ+ent1UIMΔ, 3Δ cells expressing Ent1UIM S-D were defective for Ste2 internalization. However, when we performed these assays on 3Δ cells expressing Ent2UIMΔ, we observed only a minor defect in internalization relative to cells expressing wildtype Ent2 (Figure 1C). This finding suggests that there is a functional difference between Ent1 and Ent2 and provides further evidence that efficient receptor internalization can occur in the absence of ubiquitin-binding by Ent1, Ent2 and Ede1.
Internalization of receptors carrying ubiquitin-independent signals requires epsin UIMs
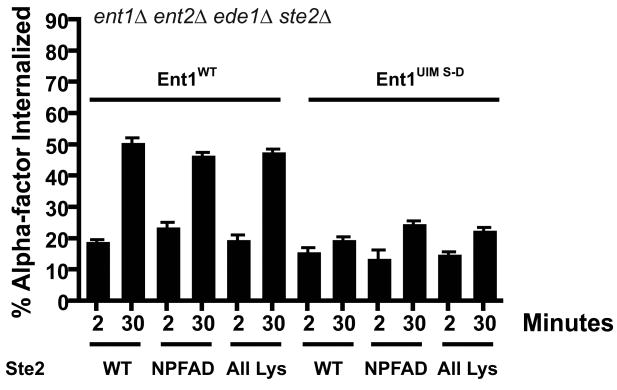
In addition to ubiquitin, yeast plasma membrane proteins can be internalized by a linear peptide signal with the consensus sequence NPFX(1,2)D, where X is any amino acid (17, 28). Wildtype Ste2 carries a weak version of this signal, GPFAD, that can be converted to a strong signal by mutation to NPFAD (17). We used an ent1Δ ent2Δ ede1Δ strain with a precise deletion of the STE2 open reading frame to test the endocytosis of variants of Ste2 internalized by different sorting signals. Ste2NPFAD contains the NPFAD linear peptide sorting signal and mutations of all the C-terminal-tail lysine residues (K→R) that serve as sites of ubiquitination. In contrast, Ste2All Lys contains an F394A point mutation that disrupts the linear peptide sorting signal, but leaves ubiquitination sites intact. In 3Δ cells expressing Ent1WT, both Ste2 variants are internalized to the same extent as exogenously expressed wildtype receptor (Ste2WT). In 3Δ+ent1UIM S-D cells, internalization of Ste2WT is defective as observed previously. Furthermore, the internalization of both Ste2All Lys and Ste2NPFAD is likewise defective in these cells (Figure 2). These results indicate that receptors carrying both ubiquitin-dependent and ubiquitin-independent internalization signals are equally reliant on the epsin UIMs. Thus mutation of the epsin UIMs affects a general step in receptor internalization required for both Ste2 sorting pathways.
Figure 2. Ent1 UIMs are required for ubiquitin-dependent and ubiquitin-independent internalization of Ste2.
Variants of Ste2, Ste2WT (LHP424), Ste2NPFAD (LHP426), which can only be internalized via the peptide sorting sequence NPFAD, or Ste2All Lys (LHP427), which can only be internalized by ubiquitination, were expressed in ent1Δ ent2Δ ede1Δ ste2Δ cells expressing either Ent1WT or Ent1UIM S-D. These strains were stimulated with radioactively labeled α-factor at 30°C as in Figure 1A, and cell aliquots taken at 2 min or 30 min were analyzed. Each bar represents the average of three assays, and error bars represent the standard deviation.
Epsin and Ede1 UBDs share an overlapping function with yeast AP180 adaptors
The previously described results suggest that epsin and Ede1 UBDs function to promote protein-protein interactions among components of the endocytic machinery, not primarily as receptors for ubiquitin internalization signals. We observed that the EH domains of Ede1, which bind to NPF motifs in several endocytic proteins (29-31), are important in the absence of epsin UIMs (Figure 1A). To identify proteins that might act with Ede1 EH domains in a protein-protein interaction that is functionally redundant with the epsin UIMs, we performed a high-throughput yeast 2-hybrid screen, using the EH domains (a.a. 1-376) of Ede1 as bait. We identified Ent1 and Ent2 in this screen, which were previously shown to bind EH domains (18). We also identified several additional yeast proteins implicated in endocytosis, including the other yeast monomeric adaptors, Yap1801 and Yap1802. As mentioned previously, the yeast AP180 adaptors function cooperatively with the epsins and harbor many of the epsin functional domains and motifs (14). A two-hybrid interaction between Ede1 EH domains and Yap1802 was previously reported (17).
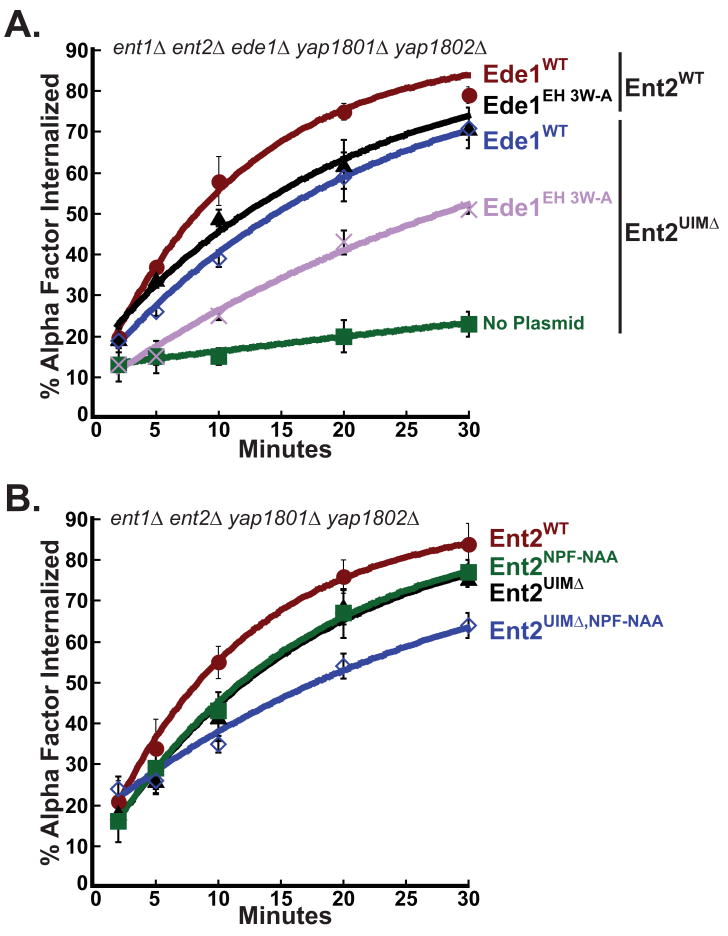
To determine whether Yap1801 and Yap1802 are Ede1-interacting proteins that might substitute for Ent2 UIM function, we expressed Ent2UIMΔ in an ent1Δ ent2Δ yap1801Δ yap1802Δ ede1Δ (5Δ) strain. We observed a ∼10 fold decrease in the rate of receptor internalization in 5Δ+ent2UIMΔ cells as compared to 5Δ+ENT2 cells (Figure 3A). In addition, we tested 5Δ strains expressing wildtype epsins or Ent2UIMΔ for growth at 30°C and 37°C on rich medium and found that the 5Δ+ent2UIMΔ strain grew poorly at 30°C and not at all at 37°C (Figure 3B). The expression of wildtype Ede1 in the 5Δ+ent2UIMΔ cells restored both receptor internalization and growth at 37°C (Figures 3A and 3B). This evidence indicates that Ede1 and the Ent2 UIMs share a common role that is crucial for receptor internalization. Furthermore, because receptor internalization mediated by Ent2UIMΔ is significantly more rapid in 3Δ cells as compared to 5Δ cells, the yeast AP180 proteins and Ent2 UIMs are likely to perform overlapping functions that are necessary for receptor internalization and cell growth at high temperature.
Figure 3. Ent2 UIMs share overlapping functions with Yap180 proteins and Ede1.
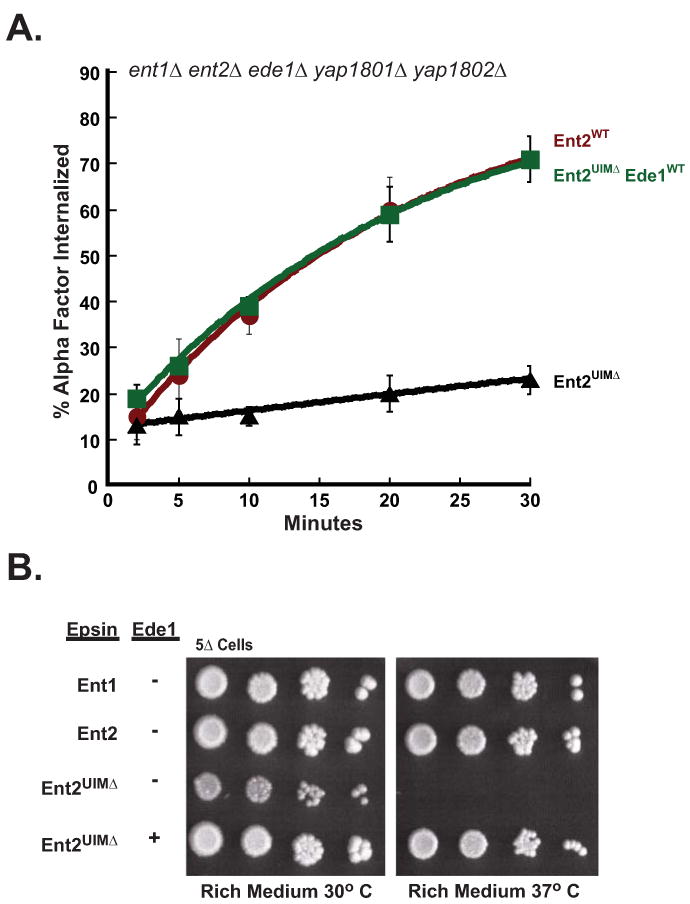
(A) Ede1 functions redundantly with Ent2 UIMs in ligand-mediated endocytosis. Quintuple deletion cells, ent1Δ ent2Δ yap1801Δ yap1802Δ ede1Δ (5Δ), expressing Ent2WT (LHY5644), Ent2UIMΔ (LHY5645), or Ent2UIMΔ together with Ede1WT (LHY5693) were assayed for internalization of radioactively labeled α-factor at 30°C. (B) Ede1 is required for growth at 37°C in cells lacking Ent2 UIMs. Quintuple mutant cells expressing Ent1WT (LHY5641), Ent2WT (LHY5644), Ent2UIMΔ (LHY5645), or co-expressing Ent2UIMΔ and Ede1WT (LHY5693) were grown on rich medium for 3 days at 30°C or 37°C.
The Ent2 UIMs and NPF motifs perform partially overlapping functions in endocytosis
Since the Ede1 EH domains and Ent1 UIMs are functionally redundant, we tested whether the Ede1 EH domains would also overlap in function with the Ent2 UIMs. We assayed Ste2 internalization in 5Δ+ent2UIMΔ cells expressing Ede1WT or Ede1EH 3W-A. The 5Δ+ent2UIMΔ ede1EH 3W-A cells exhibited a clear defect in receptor internalization (Figure 4A), indicating that the Ent2 UIMs function redundantly with the Ede1 EH domains.
Figure 4. Ent2 NPF motifs and UIM domains share a redundant function during endocytosis.
(A) The EH domains of Ede1 are required for rapid receptor internalization in the absence of Ent2 UIMs. Quintuple deletion cells co-expressing Ent2WT and Ede1WT (LHY5712), Ent2WT and Ede1EH 3W-A (LHY5648), Ent2UIMΔ and Ede1WT (LHY5693), or Ent2UIMΔ and Ede1EH 3W-A (LHY5650), were assayed for internalization of radioactively labeled α-factor at 30°C. Each point represents the average of three or more assays. Error bars represent the standard deviation. (B) The Ent2 UIMs and NPF motifs function cooperatively during receptor internalization. Quadruple deletion cells (4Δ) expressing endogenous Ede1 and either Ent2WT (LHY5627), Ent2UIMΔ (LHY5628), Ent2NPF-NAA (LHY5629) or Ent2UIMΔ,NPF-NAA (LHY5694) were assayed for internalization of radioactively labeled α-factor at 30°C. Each point represents the average of three or more assays and error bars represent the standard deviation.
Ede1 EH domains bind to NPF sequences in Ent1 and Ent2 (18). Therefore, we predicted that Ent2 NPF motif mutations might cause a defect in receptor internalization in combination with Ent2 UIM mutations in the presence of wildtype Ede1. We generated a variant of Ent2 in which both NPF motifs were mutated to NAA (ent2NPF-NAA), a mutation that eliminates binding of NPF motifs to EH domains (27). We expressed the Ent2NPF-NAA mutant in ent1Δ ent2Δ yap1801Δ yap1802Δ (4Δ) cells and found that receptor internalization was slightly defective as compared to internalization by the same strain expressing wildtype Ent2. The internalization defect in 4Δ+ent2NPF-NAA cells was the same as that in 4Δ+ent2UIMΔ cells. Mutating both UIMs and NPF sequences together (ent2UIMΔ,NPF-NAA) caused a further decrease in the rate of internalization (Figure 4B). These results suggest that epsin ubiquitin binding might serve a function similar to NPF-EH domain interactions in the organization of early endocytic machinery.
The Ede1 UBA domain has a function distinct from that of the Ent2 UIMs
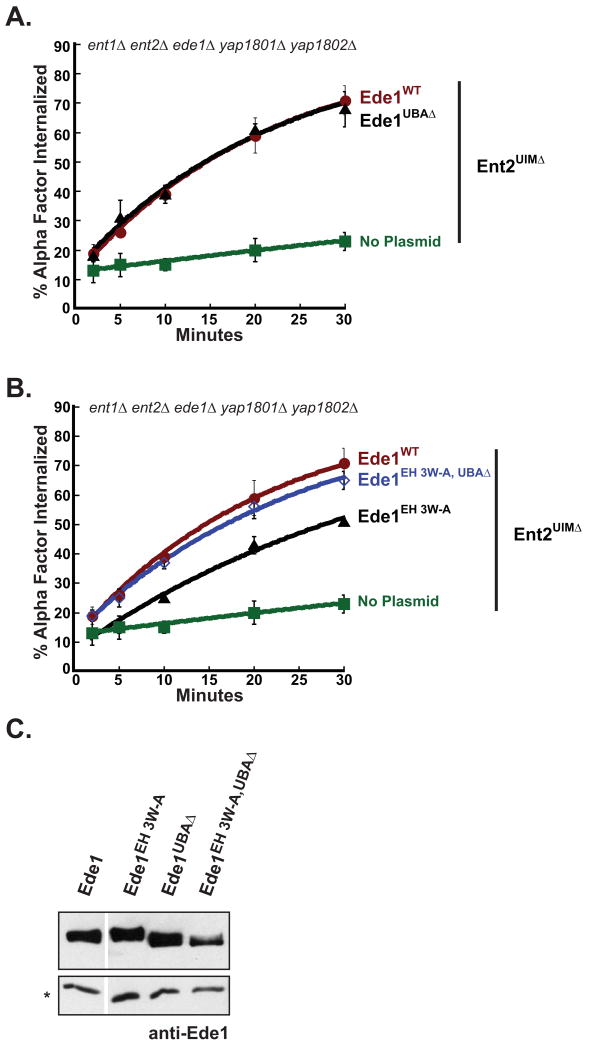
The data presented in Figure 1A suggest that the Ede1 EH domains, rather than the Ede1 UBA domain, are the part of Ede1 that is functionally redundant with the Ent1 UIMs in 3Δ cells. To test what part of Ede1 is functionally redundant with Ent2 in 5Δ cells, we compared receptor internalization in 5Δ+ent2UIMΔ cells expressing Ede1UBAΔ, Ede1EH 3W-A or Ede1WT. The 5Δ cells co-expressing Ent2UIMΔ and Ede1UBAΔ were able to internalize α-factor at the same rate as cells co-expressing Ent2UIMΔ and Ede1WT (Figure 5A). Therefore, deletion of the Ede1 UBA domain does not affect the ability of Ede1 to compensate for loss of Ent2 UIM function in 5Δ cells, consistent with the observation that the Ede1 UBA domain is not functionally redundant with the Ent1 UIMs.
Figure 5. The Ede1 UBA domain negatively regulates Ede1 function during receptor internalization.
(A) A strain lacking epsin and Ede1 ubiquitin binding domains is able to rapidly internalize ligand-bound receptors. A quintuple mutant strain co-expressing Ent2UIMΔ together with Ede1UBAΔ (LHY 5649) was assayed for internalization of radioactive α-factor at 30°C. The internalization curves for cells expressing Ede1WT (LHY5693) or no Ede1 (LHY5645) are shown for comparison. Each point represents the average of three or more assays, and error bars represent the standard deviation. (B) The UBA domain inhibits Ede1 in the absence of EH domain interactions. Quintuple deletion cells (5Δ) co-expressing Ent2UIMΔ and Ede1EH 3W-A,UBAΔ (LHY5711) were assayed for internalization of radioactively labeled α-factor at 30°C. The internalization curves for 5Δ cells expressing wildtype Ede1 (LHY5693), Ede1EH 3W-A (LHY5650), or no Ede1 (LHY5645) are provided for comparison. Each point represents the average of three or more assays. Error bars represent the standard deviation. (C) Protein levels of wildtype Ede1 and Ede1 mutants do not correlate with defects in receptor internalization. Cell lysates from the same strains used in experiments presented in Figures 3A, 4A and 4B were prepared from 2 × 108 cells and analyzed on an immunoblot probed with polyclonal anti-Ede1 antibody. The (*) denotes a cross-reacting band used as a loading control.
Also consistent with results indicating a redundant function for Ent1 UIMs and the Ede1 EH domains, the Ede1EH 3W-A mutant only partially rescued the receptor internalization defect of 5Δ cells expressing Ent2UIMΔ. We generated a mutant of Ede1 lacking functional EH domains and the UBA domain (ede1EH 3W-A,UBAΔ) to test whether deleting the UBA domain would intensify the defect caused by mutating the Ede1 EH domains. Surprisingly, the Ede1EH 3W-A,UBAΔ mutant restored receptor internalization in 5Δ+ent2UIMΔ cells to the same extent as wildtype Ede1 (Figure 5B). The effects we observed were not due to changes in mutant protein expression or stability, because the steady-state protein levels of wildtype Ede1 and Ede1 mutants did not correlate with defects in receptor internalization (Figure 5C). These observations indicate that the UBA domain does not functionally overlap with the epsin UIMs. Instead, the UBA domain appears to negatively regulate Ede1.
Ede1 is ubiquitinated
Mammalian Eps15 carries C-terminal UIMs instead of a UBA domain and is ubiquitinated. Ubiquitination of Eps15 is believed to negatively regulate the internalization of activated EGF receptor by binding to and inactivating the UIMs in an intramolecular interaction (32). Like Eps15, yeast Ede1 binds to ubiquitin via a domain at its C-terminus that appears to negatively regulate the protein's activity in receptor internalization.
To investigate whether Ede1 might also be regulated by an intramolecular interaction between its UBA domain and an attached ubiquitin, we tested whether Ede1 is ubiquitinated in yeast cells. We immuneprecipitated Ede1 from wildtype and ede1Δ cells expressing an epitope-tagged version of ubiquitin (HA-Ub) from a plasmid. Immunoblotting with anti-HA antibodies detected a precipitated protein that was specific to cells expressing Ede1 and HA-Ub, indicating that Ede1 is a ubiquitinated protein (Figure 6A). This protein migrated as a discrete band and was similar in molecular mass to Ede1, suggesting that the modification was monoubiquitin or a short oligo-ubiquitin chain. Although treatment of mammalian cells with a receptor ligand enhances Eps15 ubiquitination (33), stimulation of yeast cells with α-factor had no effect on the ubiquitination of Ede1 (Figure 6A). Ede1 immuneprecipitated from cells carrying a temperature-sensitive mutation in the catalytic domain of the Rsp5 ubiquitin ligase was not detectably modified at the non-permissive temperature (Figure 6B), providing more evidence that Ede1 is ubiquitinated. Together, these data demonstrate that Ede1, like Eps15, is modified by ubiquitin, consistent with the idea that Ede1 is negatively regulated by an intramolecular ubiquitin-UBA domain interaction.
Figure 6. Ede1 is ubiquitinated in an Rsp5-dependent manner.
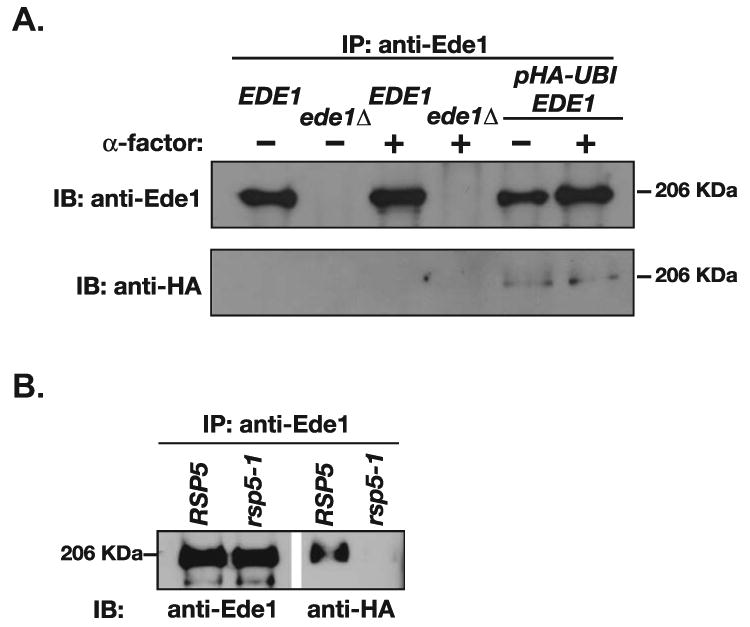
(A) Ede1 is ubiquitinated. Ede1 was precipitated from denatured lysates of ede1Δ (LHY900), EDE1 (LHY291) and EDE1 HA-UBI (LHY2432) cells prepared before and after stimulation with α-factor. The precipitates were analyzed by immunoblotting using α-Ede1 or α-HA antisera. (B) Ubiquitination of Ede1 requires the catalytic activity of the Rsp5 ubiquitin protein ligase. RSP5 HA-UBI (LHY4548) and rsp5-1 HA-UBI (LHY5056) cells were incubated for 1 h at 37°C (the non-permissive temperature for rsp5-1 cells). Cell lysates were then prepared and subjected to immuneprecipitation with Ede1 antiserum. The immuneprecipitates were analyzed by immunoblotting using α-Ede1 or α-HA antisera.
Discussion
Epsins and their yeast homologues, Ent1 and Ent2, are thought to act as adaptors for ubiquitinated endocytic cargo because they bind to specific plasma membrane phosphoinositides and clathrin, and they have putative cargo-recognition domains in the form of UIMs. In mammalian cells, epsin and Eps15 co-immuneprecipitate with activated, ubiquitinated EGF receptors in a UIM-dependent manner, prompting the conclusion that these proteins bind directly to ubiquitin internalization signals (25). Furthermore, although deletion of epsin UIMs has a small effect on receptor internalization in yeast, this defect is greatly enhanced in cells that also lack Ede1 (21), consistent with the idea that the epsin and Eps15/Ede1 UBDs might serve as redundant receptors for ubiquitinated cargo.
In this study we found that yeast cells internalized ubiquitinated receptors efficiently even in the absence of epsin and Ede1 UBDs. Expression of Ede1UBAΔ in strains that are defective for epsin ubiquitin binding (3Δ+ent1UIMΔ and 5Δ+ent2UIMΔ) restored internalization of activated Ste2. Furthermore, the internalization of receptors carrying ubiquitin-independent linear peptide signals required Ent1 UIMs to the same extent as receptors carrying ubiquitin signals. The simplest interpretation of these observations is that the primary function of epsin and Ede1 UBDs is not as receptors for ubiquitin internalization signals.
Our results are consistent with recent findings in mammalian cells demonstrating that the Eps15 UIM domains are not required for the internalization of activated, ubiquitinated receptors. The Met (hepatocyte growth factor) receptor is ubiquitinated in response to ligand binding and requires Eps15 for internalization (34-37). In contrast to EGFR, which requires the ubiquitin-binding domains of the epsins and Eps15 for internalization (25), the Met receptor does not require Eps15 UIMs. Instead, Met receptor internalization requires the coiled-coil region and the proline-rich region of Eps15 (37).
The identity of receptors for ubiquitin signals in the yeast endocytic machinery remains unknown. Perhaps multiple proteins with UBDs serve as ubiquitin internalization signal receptors, and Ent1, Ent2 and Ede1 UBDs act as receptors in addition to having other early endocytic functions. Sla1, a protein that arrives shortly after clathrin during vesicle formation (5, 38), serves as a receptor for a linear peptide internalization signal (17) and also binds to ubiquitin through its third SH3 domain (39). It is possible that Sla1 contributes to a complex of redundant cargo adaptors that bind to the ubiquitin internalization signal. Proteins that act later during internalization are less likely to recruit ubiquitinated cargo, however several of these proteins, including the endophilin homologue Rvs167, also bind to ubiquitin directly (our unpublished data).
We observed differences in the requirement for Ent1 and Ent2 UIMs during receptor internalization. Cells appear to tolerate the loss of Ent1 function in internalization more readily than that of Ent2 because Ent2UIMΔ was able to function as the sole source of epsin in the absence of Ede1 whereas Ent1UIMΔ and Ent1UIM S-D were not. An internalization defect with Ent2UIMΔ was not observed until Yap1801 and Yap1802 are also eliminated. The observed differences in phenotype between the ent1UIMΔ and ent2UIMΔ mutations are consistent with previously observed differences between Ent1 and Ent2. Ent2 is more abundant and has a higher affinity for clathrin than Ent1, and, unlike Ent1, can colocalize with clathrin in the absence of accessory proteins such as Sla2 (11, 13, 40). These findings suggest that Ent2, rather than Ent1, is the primary epsin at the internalization step of endocytosis.
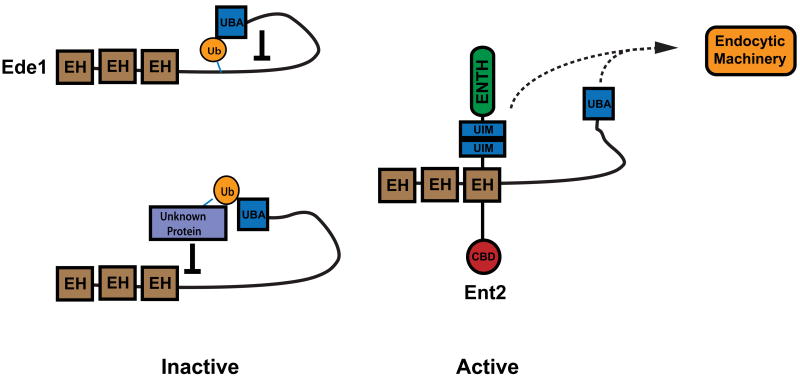
What is the primary function of the epsin and Ede1 UBDs? The Ent1 UIMs and the Ent1 clathrin-binding region share a related function (14), and the Ent1 and Ent2 UIMs overlap functionally with EH-NPF protein interactions (this study). Thus, we suggest that the yeast epsin UIMs mediate protein-protein interactions that stabilize the endocytic network or engage later-acting endocytic machinery. Mutation of the UIMs likely results in the disruption of downstream endocytic machinery required for both ubiquitin and non-ubiquitin mediated receptor internalization. Mutation of the protein-interaction domains within Ent1/2 has been shown to disrupt the spatiotemporal localization of Pan1, which in turn affects the organization of the actin cytoskeleton (14, 41). In contrast, the Ede1 UBA domain appears to negatively regulate Ede1 in the absence of EH-NPF interactions. Furthermore, Ede1 is ubiquitinated. To explain these observations, we propose a model in which the Ede1 UBA domain binds to ubiquitin conjugated to lysines within Ede1 itself or to ubiquitinated targets, as has been proposed for Eps15 (42). These interactions could hold Ede1 in an inhibited state that would be released upon binding of the Ede1 EH domains to an NPF-containing epsin or Yap180 adaptor (Figure 7). Deubiquitination of the UBA domain target would prevent re-binding and provide temporal direction to endocytic network assembly and function.
Figure 7. Models for UBA-mediated regulation of Ede1.
When not bound to Ent2, Ede1 is inhibited in a UBA domain-dependant manner. The UBA domain of Ede1 may interact with either ubiquitin conjugated directly to Ede1 or to other ubiquitinated endocytic proteins. When bound to Ent2 via EH-NPF interactions, Ede1 is active in receptor internalization. The ubiquitin-binding domains of Ent2 and Ede1 may also interact with other endocytic machinery (dashed lines), contributing to the formation of the endocytic scaffold.
Ubiquitination of the epsins may also be involved in protein-protein interactions and regulation. Our attempts to detect ubiquitinated forms of Ent1 or Ent2 have been unsuccessful, although published data suggest that they are ubiquitinated. For example, mammalian epsins are ubiquitinated (43), and Ent2 has been identified in a high-throughput screen for substrates of the E3-ligase Rsp5 (44). It is possible that epsin ubiquitination is important for the formation and regulation of the early endocytic scaffold.
Our study suggests that the Ent2 UIMs and Ede1 UBA domain are involved in the organization of the endocytic network. Understanding the role of UBDs in the formation of protein interaction networks during vesicle formation will provide clues to the role of ubiquitin-binding proteins in other membrane trafficking events and in different cellular processes.
Materials and Methods
Media and reagents
Yeast strains were grown in rich medium (2% bactopeptone, 1% yeast extract, 2% glucose supplemented with 20 mg/l adenine, uracil and tryptophan) or synthetic minimal media (YNB; US Biological, Swampscott, MA).
Monoclonal antiserum was prepared against the hemagglutinin (HA) epitope from the 12CA5 hybridoma cell line. Anti-Ede1 polyclonal antiserum was raised in rabbits against a recombinant Ede1 fragment (amino acids 1-400) expressed and isolated from E. coli. This antiserum specifically recognized a ∼200 kDa protein present in lysates prepared from wildtype but not ede1Δ∷TRP1 cells (our unpublished data).
35S-labeled α-factor was purified from yeast as described previously (45).
Strains and plasmids
Strain transformations, growth and genetic manipulations were performed using standard techniques. Strains used are listed in Table 1. The construction of the 3Δ strain used in this study was described previously (21). The 4Δ and 5Δ yeast strains used in this study and the method for exchanging ENT2 plasmids in these strains were described previously (14). Because of an essential function performed by the epsin ENTH domain, strains must carry a form of epsin with a functional ENTH domain (15). The ent1Δ ent2Δ ede1Δ ste2Δ strain was generated by the targeted deletion of STE2 in the 3Δ strain.
Table 1. Strains used in this study.
| Strain | Genotype |
|---|---|
| LHY291 | his3 trp1 lys2 ura3 leu2 bar1 |
| LHY2432 | same as LHY291 + HA-UBI [TRP1] |
| LHY4548 | rsp5∷HIS3 leu2 ura3 his3 trp1 lys2 bar1 GAL + RSP5 [TRP1] + HA-UBI [URA3] |
| LHY5056 | same as LHY4548 + rsp5-1 [TRP1] + HA-UBI [URA3] |
| LHY3190 | ede1Δ∷KAN ent2Δ∷KAN ent1Δ∷HIS3 ura3 his3 leu2Δ trp1 bar1 + ENT2 [TRP1] |
| LHY3191 | same as LHY3190 + ent2UIMΔ [TRP1] |
| LHY3638 | same as LHY3190 + ENT1[TRP1], EDE1 [URA3] |
| LHY3639 | same as LHY3190 + ENT1[TRP1], ede1UBAΔ [URA3] |
| LHY3642 | same as LHY3190 + ent1UIMΔ [TRP1], EDE1 [URA3] |
| LHY3643 | same as LHY3190 + ent1UIMΔ [TRP1], ede1UBAΔ [URA3] |
| LHY4959 | same as LHY3190 + ent1UIMΔ [TRP1], ede1EH 3W-A [URA3] |
| LHY5627 | ent1∷LEU2 ent2∷HIS3 yap1801∷HIS3 yap1802∷LEU2 ura3-52 his3-Δ200 trp1Δ 901 leu2-3, 112 lys2-801 suc2-Δ9 bar1 + ENT2 [TRP1] |
| LHY5628 | same as LHY5627 + ent2UIMΔ [TRP1] |
| LHY5629 | same as LHY5627 + ent2NPF-NAA [TRP1] |
| LHY5694 | same as LHY5627 + ent2UIMΔ,NPF-NAA [TRP1] |
| LHY5641 | same as LHY5627 + ENT1 [TRP1] |
| LHY5644 | ent1∷LEU2 ent2∷HIS3 yap1801∷HIS3 yap1802∷LEU2 ede1∷KANR ura3-52 his3-Δ200 trp1Δ 901 leu2-3,112 lys2-801 suc2-Δ9 bar1 + ENT2 [TRP1] |
| LHY5645 | same as LHY5644 + ent2UIMΔ [TRP1] |
| LHY5693 | same as LHY5644 + ent2UIMΔ [TRP1], EDE1 [URA3] |
| LHY5648 | same as LHY5644 + ENT2 [TRP1], ede1EH 3W-A,UBAΔ [URA3] |
| LHY5649 | same as LHY5644 + ent2UIMΔ [TRP1], ede1UBAΔ [URA3] |
| LHY5650 | same as LHY5644 + ent2UIMΔ [TRP1], ede1EH 3W-A [URA3] |
| LHY5711 | same as LHY5644 + ent2UIMΔ [TRP1], ede1EH 3W-A,UBAΔ [URA3] |
| LHY5712 | same as LHY5644 + ENT2 [TRP1], EDE1 [URA3] |
| LHY5718 | same as LHY3190 + ENT1 [TRP1] |
| LHY5719 | same as LHY3190 + ent1UIM S-D [TRP1] |
| LHY5723 | ent2Δ∷KANR ede1Δ∷KANR ent1Δ∷NatR ste2Δ∷leu2 his3Δ ura3Δ lys2Δ trp1Δ bar1Δ + ENT1 [TRP1] + STE2 [URA3] |
| LHY5724 | same as LHY5723 + ENT1 [TRP1], ste2NPFAD [URA3] |
| LHY5725 | same as LHY5723 + ENT1 [TRP1], ste2All Lys [URA3] |
| LHY5727 | same as LHY5723 + ent1UIM S-D [TRP1], STE2 [URA3] |
| LHY5728 | same as LHY 5727 + ent1UIM S-D [TRP1], ste2NPFAD [URA3] |
| LHY5729 | same as LHY5727 + ent1UIM S-D [TRP1], ste2All Lys [URA3] |
The plasmids used in this study are listed in Table 2. The plasmids encoding ENT2WT (LHP823), ent1UIMΔ (LHP1431), ENT1WT (LHP2703) and ent1UIM S-D (LHP2774) were described previously (14). Plasmids encoding the Ste2 variants, STE2WT (LHP424), ste2NPFAD (LHP426) and ste2All Lys (LHP427) were also described previously (46). The ent2UIMΔ (LHP1412) plasmid was generated by precise deletion of sequences encoding both UIMs (a.a. 175-225) in LHP823 using Quikchange™ mutagenesis (Stratagene, La Jolla, CA). The ent2NPF-NAA (LHP1924) and ent2UIMΔ,NPF-NAA (LHP2790) plasmids were generated by mutating Pro348, Phe349, Pro492, and Phe493 to alanine by Quikchange™ mutagenesis in LHP878 and LHP1412, respectively.
Table 2. Plasmids used in this study.
| Plasmid | Description | Details |
|---|---|---|
| LHP472 | RSP5 | pRS416∷RSP5 [TRP1, CEN] |
| LHP1615 | rsp5-1 | pRS416∷rsp5-1 (L733S) [TRP1, CEN] |
| LHP326 | HA-UBI | YEp112∷HA-UBI [TRP1, 2μm] |
| LHP462 | HA-UBI | pES7∷HA-UBI [URA3, 2μm] |
| LHP823 | ENT2 | YCplac22∷ENT2 [TRP1, CEN] |
| LHP424 | STE2 | YCplac33∷STE2 [URA3, CEN] |
| LHP426 | ste2NPFAD | YCplac33∷ste2NPFAD (K337R, K352R, K358R, K374R, K387R, K400R, K422R, G392N) [URA3, CEN] |
| LHP427 | ste2All Lys | YCplac33∷ste2All Lys (F394A). [URA3, CEN] |
| LHP1412 | ent2UIMΔ | YCplac22∷ent2UIM- (Δaa175-225) [TRP1, CEN] |
| LHP1431 | ent1UIMΔ | YCplac22∷ent1UIM- (Δaa165-209 and Δaa 217-251) [TRP1, CEN] |
| LHP1924 | ent2NPF-NAA | YCplac22∷ent2NPF- (P348A, F349A, P492A, F493A) [TRP1, CEN] |
| LHP2799 | ent2UIMΔ,NPF-NAA | YCplac22∷ent2UIM-,NPF- (Δaa175-225, P348A, F349A, P492A, F493A) [TRP1, CEN] |
| LHP439 | EDE1 | pRS426∷EDE1 [URA3, 2μm] |
| LHP1531 | EDE1 | pRS316∷EDE1 [URA3, CEN] |
| LHP1532 | ede1UBAΔ | pRS316∷ede1 (aa1-1334) [URA3, CEN] |
| LHP1770 | ede1EH 3W-A | pRS316∷ede1 (W56A, W176A, W319A) [URA3, CEN] |
| LHP1992 | ede1EH 3W-A,UBAΔ | pRS316∷ede1 (aa1-1334, W56A, W176A, W319A) [URA3, CEN] |
| pBW0768 | ENT1 | YCplac22∷ENT1 [TRP1, CEN] |
| pBW1375 | ent1UIM S-D | YCplac22∷ent1 (S177D,S201D) [TRP1, CEN] |
LHP1531 carries a genomic fragment containing EDE1, including 485 base pairs of the promoter and 80 base pairs of the 3′ region, cloned into pRS426 (47). Truncation mutants of EDE1 were generated using Quikchange™ mutagenesis to introduce tandem stop codons at the indicated amino acid. Similarly, ede1UBAΔ (LHP1532) was generated by the addition of tandem stop codons at amino acid 1334 to precisely delete the C-terminal UBA domain. EH domain point mutants in ede1EH 3W-A (LHP1770) and ede1EH 3W-A,UBAΔ (LHP1992) were generated by mutating Trp56, Trp176, and Trp319 to alanine using Quikchange™ mutagenesis. All mutations were verified by automated DNA sequencing. The expression of mutant proteins was checked by immunoblot analysis (see below).
α-factor internalization assays
35S-labeled α-factor internalization assays were performed at 30°C using the continuous presence method, as described previously (48). Cells were grown to mid-log phase, harvested and pre-incubated at 30°C for 15 min prior to addition of 35S-α-factor. Aliquots of cells (1 × 108) were taken at the indicated time and incubated in either pH6 buffer (40 mM KH2PO4, 6.0 mM K2HPO4) or pH1 buffer (50 mM Na3C6H5O7). Samples were collected on glass-fiber filters (VWR, Batavia, IL) and analyzed using a Beckman LS6500 scintillation counter. The percent internalized α-factor for each time point was calculated by dividing the pH1 sample value by the pH6 sample value. Each data point represents an average of three or more assays and error bars represent the standard deviation.
Ubiquitin-binding experiments
Binding of proteins from yeast lysates to ubiquitin-Sepharose was performed as described previously (21) with the following modification: total yeast lysate proteins (2.0 mg) were incubated with 80 μl 25% ubiquitin-Sepharose slurry (Boston Biochem, Cambridge, MA) or Sepharose CL-4B (Pharmacia, Peapack, NJ) in 1.5 ml MES buffer at 4°C for 4 h.
Denaturing immuneprecipitations
Lysates and immuneprecipitations were carried out as described previously with minor changes (45). All buffers were supplemented with 5 mM NEM and 1 mM 1,10-orthophenanthroline to inhibit deubiquitinating enzymes. Following lysis by mechanical agitation using glass beads, SDS was added to 1% final weight by volume and lysates were incubated at 37°C for 10 min. After clarification, lysates were diluted to 2 ml. Immuneprecipitations were carried out at 4°C for 1 h, followed by incubation with Protein A Sepharose CL-4B (Amersham Biosciences, Piscataway, NJ) at 4°C for 30-60 min. Immuneprecipitates were washed as described, eluted by heating to 80-90°C in urea sample buffer and resolved by 6% SDS-PAGE. Immunoblotting was performed as described previously using α-Ede1 and α-HA antisera (39).
Acknowledgments
We thank Miroslava Protic for her work in preparing yeast strains. This work was done with the help of the Cancer Diagnostic and Therapeutics Screening, Keck Biophysics, and Monoclonal Antibody Facilities of the Robert H. Lurie Cancer Center, Northwestern University, Evanston IL. Funding was provided by grants from the National Institutes of Health (DK53257 and DK61299 to L.H., and GM60979 to B.W.).
Abbreviations
- EGF
epidermal growth factor
- EH
Eps15 homology domain
- HA
hemagglutinin
- UBA
ubiquitin associated domain
- UBD
ubiquitin binding domain
- UIM
ubiquitin interacting motif
References
- 1.Traub LM. Sorting it out: AP-2 and alternate clathrin adaptors in endocytic cargo selection. J Cell Biol. 2003;163(2):203–208. doi: 10.1083/jcb.200309175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorkin A. Cargo recognition during clathrin-mediated endocytosis: a team effort. Curr Opin Cell Biol. 2004;16(4):392–399. doi: 10.1016/j.ceb.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Maldonado-Baez L, Wendland B. Endocytic adaptors: recruiters, coordinators and regulators. Trends in Cell Biology. 2006;16(10):505–513. doi: 10.1016/j.tcb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida Y, Kinuta M, Abe T, Liang S, Araki K, Cremona O, Di Paolo G, Moriyama Y, Yasuda T, De Camilli P, Takei K. The stimulatory action of amphiphysin on dynamin function is dependent on lipid bilayer curvature. Embo J. 2004;23(17):3483–3491. doi: 10.1038/sj.emboj.7600355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123(2):305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 6.Yarar D, Waterman-Storer CM, Schmid SL. A Dynamic Actin Cytoskeleton Functions at Multiple Stages of Clathrin-mediated Endocytosis. Mol Biol Cell. 2005;16(2):964–975. doi: 10.1091/mbc.E04-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girao H, Geli MI, Idrissi FZ. Actin in the endocytic pathway: From yeast to mammals. FEBS Letters. 2008;582(14):2112–2119. doi: 10.1016/j.febslet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 9.Perrais D, Merrifield CJ. Dynamics of endocytic vesicle creation. Dev Cell. 2005;9(5):581–592. doi: 10.1016/j.devcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Meyerholz A, H L, Groos S, Esk PC, Brandes G, Ungewickell EJ. Effect of Clathrin Assembly Lymphoid Myeloid Leukemia Protein Depletion on Clathrin Coat Formation. Traffic. 2005;6(12):1225–1234. doi: 10.1111/j.1600-0854.2005.00355.x. [DOI] [PubMed] [Google Scholar]
- 11.Newpher TM, Smith RP, Lemmon V, Lemmon SK. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev Cell. 2005;9(1):87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Edeling MA, Mishra SK, Keyel PA, Steinhauser AL, Collins BM, Roth R, Heuser JE, Owen DJ, Traub LM. Molecular Switches Involving the AP-2 [beta]2 Appendage Regulate Endocytic Cargo Selection and Clathrin Coat Assembly. Developmental Cell. 2006;10(3):329–342. doi: 10.1016/j.devcel.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 13.Baggett JJ, D'Aquino KE, Wendland B. The Sla2p talin domain plays a role in endocytosis in Saccharomyces cerevisiae. Genetics. 2003;165(4):1661–1674. doi: 10.1093/genetics/165.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maldonado-Baez L, Dores MR, Perkins EM, Drivas TG, Hicke L, Wendland B. Interaction between Epsin/Yap180 Adaptors and the Scaffolds Ede1/Pan1 Is Required for Endocytosis. Mol Biol Cell. 2008;19(7):2936–2948. doi: 10.1091/mbc.E07-10-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wendland B, Steece KE, Emr SD. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 1999;18(16):4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wendland B, Emr SD. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J Cell Biol. 1998;141(1):71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howard JP, Hutton JL, Olson JM, Payne GS. Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J Cell Biol. 2002;157(2):315–326. doi: 10.1083/jcb.200110027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguilar RC, Watson HA, Wendland B. The yeast epsin Ent1 is recruited to membranes through multiple independent interactions. J Biol Chem. 2003;278:10737–10743. doi: 10.1074/jbc.M211622200. [DOI] [PubMed] [Google Scholar]
- 19.Miliaras NB, Wendland B. EH proteins: multivalent regulators of endocytosis (and other pathways) Cell Biochem Biophys. 2004;41(2):295–318. doi: 10.1385/CBB:41:2:295. [DOI] [PubMed] [Google Scholar]
- 20.Gagny B, Wiederkehr A, Dumoulin P, Winsor B, Riezman H, Haguenauer-Tsapis R. A novel EH domain protein of Saccharomyces cerevisiae, Ede1p, involved in endocytosis. J Cell Sci. 2000;113(Pt 18):3309–3319. doi: 10.1242/jcs.113.18.3309. [DOI] [PubMed] [Google Scholar]
- 21.Shih SC, Katzmann KJ, Schnell JD, Sutanto M, Emr SC, Hicke LH. Epsins and Vps27/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat Cell Biol. 2002;4:389–393. doi: 10.1038/ncb790. [DOI] [PubMed] [Google Scholar]
- 22.Torrisi MR, Lotti LV, Belleudi F, Gradini R, Salcini AE, Confalonieri S, Pelicci PG, Di Fiore PP. Eps15 is recruited to the plasma membrane upon epidermal growth factor receptor activation and localizes to components of the endocytic pathway during receptor internalization. Mol Biol Cell. 1999;10:417–434. doi: 10.1091/mbc.10.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stang E, Blystad FD, Kazazic M, Bertelsen V, Brodahl T, Raiborg C, Stenmark H, Madshus IH. Cbl-dependent ubiquitination is required for progression of EGF receptors into clathrin-coated pits. Mol Biol Cell. 2004;15(8):3591–3604. doi: 10.1091/mbc.E04-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416(6879):451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 25.Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, Transidico P, Di Fiore PP, Polo S. Clathrin-independent endocytosis of ubiquitinated cargos. Proc Natl Acad Sci USA. 2005;102(8):2760–2765. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paoluzi S, Castagnoli L, Lauro I, Salcini AE, Coda L, Fre S, Confalonieri S, Pelicci PG, Di Fiore PP, Cesareni G. Recognition specificity of individual EH domains of mammals and yeast. EMBO J. 1998;17(22):6541–6550. doi: 10.1093/emboj/17.22.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Beer T, Hoofnagle AN, Enmon JL, Bowers RC, Yamabhai M, Kay BK, Overduin M. Molecular mechanism of NPF recognition by EH domains. Nat Struct Biol. 2000;7(11):1018–1022. doi: 10.1038/80924. [DOI] [PubMed] [Google Scholar]
- 28.Tan PK, Howard JP, Payne GS. The sequence NPFXD defines a new class of endocytosis signal in Saccharomyces cerevisiae. J Cell Biol. 1996;135:1789–1800. doi: 10.1083/jcb.135.6.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polo S, Confalonieri S, Salcini AE, Di Fiore PP. EH and UIM: endocytosis and more. Sci STKE. 2003;213:re17. doi: 10.1126/stke.2132003re17. [DOI] [PubMed] [Google Scholar]
- 30.Naslavsky N, Boehm M, Backlund PS, Jr, Caplan S. Rabenosyn-5 and EHD1 interact and sequentially regulate protein recycling to the plasma membrane. Mol Biol Cell. 2004;15(5):2410–2422. doi: 10.1091/mbc.E03-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braun A, Pinyol R, Dahlhaus R, Koch D, Fonarev P, Grant BD, Kessels MM, Qualmann B. EHD Proteins Associate with Syndapin I and II and Such Interactions Play a Crucial Role in Endosomal Recycling. Mol Biol Cell. 2005;16(8):3642–3658. doi: 10.1091/mbc.E05-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fallon L, Belanger CML, Corera AT, Kontogiannea M, Regan-Klaspisz E, Moreau F, Voortman J, Haber M, Rouleau G, Thorarinsdottir T, Brice A, van Bergen en Henegouwen PMP, Fon EA. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat Cell Biol. 2006;8(8):834–842. doi: 10.1038/ncb1441. [DOI] [PubMed] [Google Scholar]
- 33.van Delft S, Govers R, Strous G, Verkleij A, Van Bergen en Henegouwen P. Epidermal growth factor induces ubiquitination of Eps15. J Biol Chem. 1997;272:14013–14016. doi: 10.1074/jbc.272.22.14013. [DOI] [PubMed] [Google Scholar]
- 34.Hammond DE, Urbe S, Vande Woude GF, Clague MJ. Down-regulation of MET, the receptor for hepatocyte growth factor. Oncogene. 2001;20(22):2761–2770. doi: 10.1038/sj.onc.1204475. [DOI] [PubMed] [Google Scholar]
- 35.Peschard P, Fournier TM, Lamorte L, Naujokas MA, Band H, Langdon WY, Park M. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol Cell. 2001;8(5):995–1004. doi: 10.1016/s1097-2765(01)00378-1. [DOI] [PubMed] [Google Scholar]
- 36.Peschard P, Park M. Escape from Cbl-mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell. 2003;3(6):519–523. doi: 10.1016/s1535-6108(03)00136-3. [DOI] [PubMed] [Google Scholar]
- 37.Parachoniak CA, Park M. Distinct Recruitment of Eps15 via Its Coiled-coil Domain Is Required For Efficient Down-regulation of the Met Receptor Tyrosine Kinase. J Biol Chem. 2009;284(13):8382–8394. doi: 10.1074/jbc.M807607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115(4):475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 39.Stamenova SD, French ME, He Y, Francis SA, Kramer ZB, Hicke L. Ubiquitin binds to and regulates a subset of SH3 domains. Mol Cell. 2007;25(2):273–284. doi: 10.1016/j.molcel.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 41.Barker SL, Lee L, Pierce BD, Maldonado-Baez L, Drubin DG, Wendland B. Interaction of the Endocytic Scaffold Protein Pan1 with the Type I Myosins Contributes to the Late Stages of Endocytosis. Mol Biol Cell. 2007;18(8):2893–2903. doi: 10.1091/mbc.E07-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, Wagner S, Kowanetz K, Breitling R, Mann M, Stenmark H, Dikic I. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat Cell Biol. 2006;8(2):163–169. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- 43.Oldham CE, Mohney RP, Miller SL, Hanes RN, O'Bryan JP. The ubiquitin-interacting motifs target the endocytic adaptor protein epsin for ubiquitination. Curr Biol. 2002;12(13):1112–1116. doi: 10.1016/s0960-9822(02)00900-4. [DOI] [PubMed] [Google Scholar]
- 44.Gupta R, Kus B, Fladd C, Wasmuth J, Tonikian R, Sidhu S, Krogan NJ, Parkinson J, Rotin D. Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol Syst Biol. 2007;3 doi: 10.1038/msb4100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 46.Dunn R, Hicke L. Multiple roles for Rsp5p-dependent ubiquitination at the internalization step of endocytosis. J Biol Chem. 2001;276(28):25974–25981. doi: 10.1074/jbc.M104113200. [DOI] [PubMed] [Google Scholar]
- 47.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110(1):119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 48.Dulic V, Egerton M, Elguindi I, Raths S, Singer B, Riezman H. Yeast endocytosis assays. Meth Enzymol. 1991;194:697–710. doi: 10.1016/0076-6879(91)94051-d. [DOI] [PubMed] [Google Scholar]