Abstract
Dysregulated macrophage cholesterol homoeostasis lies at the heart of early and developing atheroma, and removal of excess cholesterol from macrophage foam cells, by efficient transport mechanisms, is central to stabilization and regression of atherosclerotic lesions. The present study demonstrates that transient overexpression of STARD3 {START [StAR (steroidogenic acute regulatory protein)-related lipid transfer] domain 3; also known as MLN64 (metastatic lymph node 64)}, an endosomal cholesterol transporter and member of the ‘START’ family of lipid trafficking proteins, induces significant increases in macrophage ABCA1 (ATP-binding cassette transporter A1) mRNA and protein, enhances [3H]cholesterol efflux to apo (apolipoprotein) AI, and reduces biosynthesis of cholesterol, cholesteryl ester, fatty acids, triacylglycerol and phospholipids from [14C]acetate, compared with controls. Notably, overexpression of STARD3 prevents increases in cholesterol esterification in response to acetylated LDL (low-density lipoprotein), blocking cholesteryl ester deposition. Thus enhanced endosomal trafficking via STARD3 induces an anti-atherogenic macrophage lipid phenotype, positing a potentially therapeutic strategy.
Keywords: atherosclerosis, cholesterol efflux, lipid transport, macrophage foam cell, steroidogenic acute regulatory protein-related lipid transfer domain 3/metastatic lymph node 64 (STARD3/MLN64)
Abbreviations: ABC, ATP-binding cassette; ACAT, acyl-CoA:cholesterol acyltransferase; apo, apolipoprotein; CHO, Chinese-hamster ovary; CYP, cytochrome P450; ER, endoplasmic reticulum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HDL, high-density lipoprotein; LDL, low-density lipoprotein; AcLDL, acetylated LDL; LXR, liver X receptor; MLN64, metastatic lymph node 64; MENTAL domain, MLN64 N-terminal domain; NPC, Niemann–Pick type C; Q-PCR, quantitative real-time PCR; SREBP, sterol-regulatory-element-binding protein; StAR, steroidogenic acute regulatory protein; START, StAR-related lipid transfer; STARD, START domain
INTRODUCTION
Dysregulated macrophage cholesterol homoeostasis lies at the heart of early and developing atheroma, the principal cause of coronary heart disease. Macrophage ‘foam cells’, laden with cholesterol and cholesteryl ester, result from unregulated uptake of modified lipoproteins by macrophage scavenger receptors (CD36, CD68 and SR-AI/AII), and influence both plaque stability and progression [1]. Removal of excess cholesterol from macrophage ‘foam’ cells is central to lesion regression and stabilization, and can be orchestrated, at least in vitro, by ABC (ATP-binding cassette) lipid transporters such as ABCA1, ABCG1 and ABCG4, and apo (apolipoprotein) acceptors, such as apoAI and apoE [2]. Efficient intracellular cholesterol transport is pivotal in marshalling appropriate cholesterol homoeostasis mechanisms, regulating sterol-responsive transcription factors, such as LXRα/β (liver X receptors α/β) and SREBPs (sterol-regulatory-element-binding proteins), and controlling cholesterol content of organelles, lipid rafts and membranes, and storage as cytosolic droplets of cholesteryl ester [2,3]. Despite this, the proteins involved in non-vesicular cholesterol transport mechanisms remain poorly understood.
The START [StAR (steroidogenic acute regulatory protein)-related lipid transfer] domain is a 210 amino acid conserved ‘helix-grip’ fold, providing an adaptable binding site for lipids such as cholesterol, oxysterols, phospholipids and ceramides. In humans, START domains are found in 15 distinct proteins (STARD1–STARD15), implicated in non-vesicular lipid transport, cell signalling and lipid metabolism [4,5]. The prototypic member of this family, StAR (STARD1), delivers cholesterol from the outer to the inner mitochondrial membrane, to the CYP (cytochrome P450) side-chain-cleavage enzyme involved in steroidogenesis [6]. Overexpression of StAR can also decrease macrophage lipid content, inflammatory status and arterial cholesterol levels [7,8], and increase cholesterol efflux to apoAI via a mechanism dependent upon mitochondrial sterol 27-hydroxylase (CYP27A1), LXR activation and increases in ABCA1 mRNA and protein expression [9]. However, one ‘downside’ of StAR overexpression in macrophages is the induction of lipogenesis [9], possibly mediated by LXR-dependent increases in SREBP1c expression, a problem also associated with non-sterol LXR agonists.
The other member of the STARD1 subfamily STARD3 [also known as MLN64 (metastatic lymph node 64)] is a 54 kDa protein, co-amplified within chromosome band 17q12, a region containing the potent oncogene ERBB2 in human breast carcinoma [10–12]. STARD3 is a late endosomal protein with two distinct conserved cholesterol-binding domains: a region of four transmembrane helices with three short intervening loops, called the MENTAL domain (MLN64 N-terminal domain), and the C-terminal ‘START’ domain [13]. The MENTAL domain may maintain cholesterol at the late endosomal membrane, prior to its shuttle to cytoplasmic acceptor(s) via the START domain.
Our previous studies have shown that cholesterol loading represses STARD3 expression, implicating this protein in dysfunctional cholesterol pathologies [14]. In the present study, we sought to establish a functional role for this endosomal cholesterol trafficking protein in macrophage lipid homoeostasis, testing the hypothesis that enhanced expression of STARD3 may be useful in the prevention of foam cell formation.
MATERIALS AND METHODS
Materials
Tissue culture reagents were purchased from Lonza; other reagents include pCMV.Script, and STARD4 and STARD5 clones (Origene), STARD3 clone (Stratagene), Amaxa monocyte/macrophage transfection reagent, NuPAGE gels and buffers (Invitrogen), antibodies (AbCAM), primers and fluorescent probes (Eurogentec), and LDL (low-density lipoprotein) (Athens Research), acetylated as described previously [14]. Peripheral human monocyte/macrophages were purchased from Lonza, and human heart aorta RNA, derived from four to seven human heart aortae, was purchased from Clontech.
Cellular studies
Human (THP-1) monocytes (ECACC 880812101) were maintained in RPMI 1640 medium containing 10% (v/v) FBS (fetal bovine serum), as described previously [14,15]. STARD3, STARD4, STARD5 and empty vector control (pCMV; 0.5 μg of DNA) were delivered to THP-1 monocytes (107 cells) using an Amaxa Human Monocyte Nucleofector® kit (VPA-1007). Transfection efficiency using protocol Y001 (78.5%) was determined using the proprietary pmaxGFP® vector provided by Amaxa and flow cytometric analysis of 50000 cells. Transfected human THP-1 monocytes (1.5×106 cells/well) were differentiated into macrophages by addition of 100 nM PMA. Cellular lipids, RNA and cell protein lysates were collected 72 h post-transfection. Macrophage foam cell prevention experiments were initiated 48 h after transfection. Macrophages were radiolabelled with [3H]oleic acid (1 μCi/ml; 10 μM) in the presence or absence of AcLDL (acetylated LDL; 50 μg/ml) and with or without an ACAT (acyl-CoA:cholesterol acyltransferase) inhibitor (447C88; 10 μM) for 24 h. Wild-type macrophages were incubated with progesterone (10 μM; 24 h) or U18666A (25 μM; 24 h) to inhibit endosomal trafficking, using ethanol vehicle (<0.1%). Cellular viability was assessed by conversion of MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide] into formazan. The caspase-Glo® 3/7 assay system (Promega) was used to detect apoptosis.
Lipid analyses
Flux of [3H]oleic acid (1 μCi/ml; 10 μM) into the cholesteryl ester pool and of [14C]acetate (1μCi/ml) into cholesterol, cholesteryl ester, fatty acid, triacylglycerol and phospholipid pools were assessed by TLC, as described previously [14,15]. Efflux of [3H]cholesterol (0.5 μCi/ml) to apoA1 (10 μg/ml) and HDL (high-density lipoprotein; 10 μg/ml) were measured as described previously [13,15]. Mass of macrophage total cholesterol, triacylglycerol and choline-containing phospholipids were measured using Infinity™ and Phospholipids-B colorimetric assays (Alpha Labs) [14,15].
Gene and protein expression
Total RNA (Tri-Reagent; Sigma–Aldrich) was isolated from macrophages and reverse-transcribed to cDNA utilizing the MMLV (Moloney-murine-leukaemia virus) reverse transcriptase (Bioline). Expression of STARD3, STARD4, STARD5, APOE, ABCA1, ABCG1, ABCG4, NR1H3 (encoding LXRα), SREBP1 and SREBP2 mRNA, relative to the housekeeping gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase), was performed by Q-PCR (quantitative real-time PCR), using DNA engine Opticon 2 (MJ Research). PCRs contained cDNA template, Q-PCR mix, molecular biology grade water and 100 nM of each forward and reverse primer, and fluorescent probe [FAM (6-carboxyfluorescein)/TAMRA (6-carboxytetramethylrhodamine)]. Thermal cycling conditions were 15 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 20 s at 60 °C, with status at 50 °C. Primers and probes are given in Table 1. The comparative 2−ΔCt method was utilized for quantification of each gene relative to GAPDH mRNA.
Table 1. Sequence of primers and probes used to quantify mRNA by PCR.
NR1H3 encodes LXRα.
| Gene | Forward primer | Reverse primer | Probe sequence |
|---|---|---|---|
| STARD3 | 5′-CCTGCCCCGGTACCTCAT-3′ | 5′-GCGCTGTCGCAGGTGAA-3′ | 5′-AGAGCCTCGCGGCCACCATGT-3′ |
| STARD4 | 5′-AACATCAGACAGGCCTTCCA-3′ | 5′-GAGAGCGGTGTGAGGTGCTT-3′ | 5′-TAGCGCGCCGTAGCTTCCA-3′ |
| STARD5 | 5′-GTGGACTTGGTGCTAGTCAAGAGA-3′ | 5′-GACATAACGGATGCTCCACATG-3′ | 5′-ATGTGGACCATCAGTTCCAACGCCA-3′ |
| ABCA1 | 5′TGTCCAGTCCAGTAATGGTTCTGT-3′ | 5′-AAGCGAGATATGGTCCGGATT-3′ | 5′-ACACCTGGAGAGAAGCTTTCAACGAGACTAACC-3′ |
| ABCG1 | 5′-ACGTGCCCTTTCAGATCAATGT-3′ | 5′-GACGGCTGCGACGTCCATC-3′ | 5′-CCCAGTGGCCTACTGCAGCATCGT-3′ |
| ABCG4 | 5′-GGTTCATGTCCCACGTGGTT-3′ | 5′-GCCGGTGTTGTTGAAGACCTT-3′ | 5′-TCTACCTGCATATTGGCGACGATGCC-3′ |
| APOE | 5′-TGGGTCGCTTTTGGGATTAC-3′ | 5′-CCATCAGCGCCCTCAGTT-3′ | 5′-CTGCTCAGCTCCCAGGTCACCCA-3′ |
| NR1H3 | 5′-GAACAACTGGGCATGATCGA-3′ | 5′-AAGGAGCGCCGTTACACT-3′ | 5′-AAGCTCGTCGCTGCCCAGCAA-3′ |
| SREBP1 | 5′-GCTCCTCCATCAATGACAAAATC-3′ | 5′-GCTTGAGTTTCTGGTTGCTGTGT-3′ | 5′-AAGGCCATCGACTACATTCGCTTTCTGC-3′ |
| SREBP2 | 5′-CGGTAATGATCACGCCAACA-3′ | 5′-TGGTATATCAAAGGCTGCTGGAT-3′ | 5′-TCAGCACCACTCCGCAGACGAGG-3′ |
| GAPDH | 5′-CACATGGCCTCCAAGGAGTAA-3′ | 5′-TGAGGGTCTCTCTCTTCCTCTTGT-3′ | 5′-cTGGACCACCAGCCCCAGCAAG-3′ |
Macrophage lysates were collected in RIPA buffer plus Complete™ protease cocktail inhibitor (Roche). Protein lysates (15–30 μg/well) were separated by SDS/PAGE [NuPAGE; 10% (w/v) gels], transferred on to a PDVF membrane and probed using rabbit anti-STARD3 (1:1000 dilution) or anti-GAPDH (1:1000 dilution) polyclonal antibodies, and a murine anti-ABCA1 monoclonal antibody (1:1000 dilution). Detection was achieved using appropriate secondary antibodies (1:1000 dilution) and the ECL (enhanced chemiluminescence) detection system [14]; densitometry was performed using Scion Image software.
Statistical analyses
Values are means±S.E.M., and the number of independent experiments is indicated in the Figure legends. Significant differences (P<0.05) were determined using a Student's t test or repeated measures ANOVA, followed by Dunnett's or Tukey–Kramer post-hoc tests, as appropriate. *P<0.05, **P<0.01 and ***P<0.001 for comparisons are indicated.
RESULTS
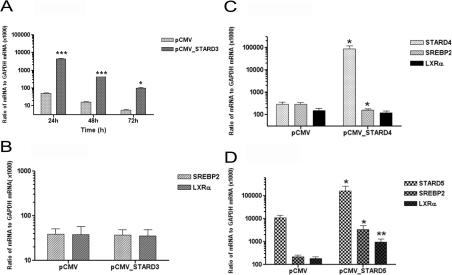
Comparison of the endogenous expression of STARD3 mRNA, relative to housekeeping gene GAPDH, in human THP-1 monocytes, THP-1 macrophages, human peripheral blood monocyte/macrophages and human heart aortae is shown in Table 2. Transient overexpression of STARD3 in human THP-1 monocyte/macrophages increased mRNA levels by 87-fold (24 h; P<0.001), 20-fold (48 h; P<0.001) and 18-fold (72 h; P<0.05) (Figure 1A), which translated into 2.2-fold (P<0.05) increases in the levels of STARD3 protein after 72 h (Figure 2B) compared with the empty vector control; all values were normalized to the level of the housekeeping protein GAPDH. Cell viability was not altered by STARD3 overexpression after 72 h (25.4±2.4 μM formazan in the empty vector control compared with 29.2±6.4 μM formazan in the STARD3-overexpressing cells; n=6 independent experiments; P=0.59), and the levels of macrophage caspase 3/7 activity did not change when STARD3 overexpression was compared with the empty vector control (6971±270 arbitrary fluorescent units in empty vector control compared with 7632±1905 in STARD3-overexpressing cells; n=3 independent experiments; P=0.509). No changes in steady-state levels of mRNA encoding sterol-responsive transcription factors SREBP1 (not detectable), SREBP2 or NR1H3 were observed, as judged by Q-PCR (Figure 1B). In comparison, overexpression of genes encoding the cytosolic cholesterol transporters STARD4 (Figure 1c) and STARD5 (Figure 1D) exerted distinct influences on gene expression of these transcription factors: STARD4 was associated with a reduced (P<0.05) expression of SREBP2, whereas STARD5 overexpression was associated with marked increases (P<0.05) in both SREBP2 and NR1H3 mRNA levels.
Table 2. Endogenous gene expression of STARD3, relative to GAPDH, in human (THP-1) monocytes and macrophages, human peripheral blood monocyte/macrophages and human heart aorta.
Results are from three individual cDNA preparations, derived from 4×107 human peripheral blood monocyte/macrophages (Lonza), the equivalent number of THP-1 monocytes and macrophages, and from a sample (50 μg) of four to seven pooled human heart aortae (Clontech).
| Source of RNA | Expression of STARD3 mRNA to GAPDH mRNA (×1000) |
|---|---|
| Human THP-1 monocytes | 1.2±0.2 |
| Human THP-1 macrophages | 0.8±0.1 |
| Human peripheral blood monocyte/macrophages | 1.5±0.4 |
| Human heart aorta | 0.4±0.1 |
Figure 1. Overexpression studies: STARD3, STARD4 and STARD5.
(A) Transient overexpression of STARD3 compared with empty vector control (24–72 h) in human THP-1 monocyte/macrophages overexpressing STARD3. (B) Effect of STARD3 overexpression (72 h) on levels of SREBP2 and NR1H3 (LXRα) mRNA. Effect of overexpression of STARD4 (C) and STARD5 (D) on the gene expression of the same transcription factors in (B). Values are means±S.E.M. of three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 compared with the empty vector control (pCMV).
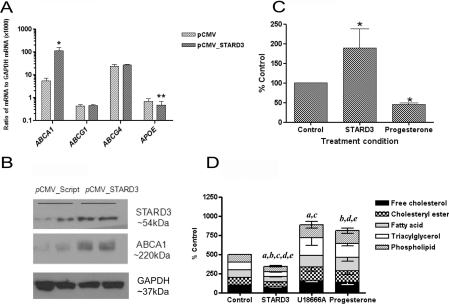
Figure 2. Effects of STARD3 overexpression on macrophage lipid homoeostasis.
(A) Levels of ABCA1, ABCG1, ABCG4 and APOE mRNA in monocyte/macrophages overexpressing STARD3 (72 h), compared with empty vector control, in three independent experiments. (B) Levels of STARD3 and ABCA1 protein, compared with the housekeeping protein GAPDH, in THP-1 monocyte/macrophages (72 h). Blots are representative of three independent experiments. The effect of STARD3 overexpression on (C) cholesterol efflux to apoAI (10 μg/ml) in five independent experiments, and (D) biosynthesis of lipids from [14C]acetate (1 μCi/ml) in four independent experiments compared with empty vector controls,. Endosomal inhibitors U18666A (25 μM) and/or progesterone (10 μM) are included as positive controls. All values are means±S.E.M. *P<0.05 and **P<0.01 compared with the empty vector control (pCMV). In (D), significant differences (P<0.05) from the control incubation for each lipid are indicated as a, free cholesterol; b, cholesteryl esters; c, fatty acids; d, triacylglycerol; and e, phospholipids.
Overexpression of STARD3 was associated with significant (P<0.05) increases (20.7-fold) in ABCA1 mRNA (Figure 2A) and protein (2.6-fold) (Figure 2B); in contrast, no changes in ABCG1 and ABCG4 mRNA (Figure 2A) were observed between STARD3-overexpressing macrophages and control cells following normalization with GAPDH. These changes in gene expression strongly predicted functional increases in cholesterol efflux to apoAI, rather than to HDL, following STARD3 overexpression, and this proved to be correct, despite a small reduction in gene expression of the endogenous acceptor APOE. [3H]Cholesterol efflux to apoAI (10 μg/ml; 24 h) was enhanced by 80% (P<0.05) (Figure 2C), whereas efflux to HDL (10 μg/ml; 24 h) did not change significantly (5.27±0.58% of HDL-specific efflux in empty vector control compared with 6.9±2.5% of HDL-specific efflux in STARD3-overpexressing cells; n=3; P=0.479). In contrast, inhibition of endosomal trafficking using progesterone (10 μM), blocked apoAI-specific cholesterol efflux by 54% (n=3; P<0.01). Decreases in the biosynthesis of cholesterol (25.5%; P<0.05), cholesteryl ester (34.1%; P<0.01), fatty acid (27.3%; P<0.05), triacylglycerol (41.9%; P<0.01) and phospholipids (27.7%; P<0.05) from [14C]acetate were observed in STARD3-overexpressing macrophages (Figure 2D) compared with the empty vector control. No change in total cholesterol (26.1±3.91 μg/mg of protein in empty vector control compared with 27.2±3.15 μg/mg of protein in STARD3-overexpressing cells), triacylglycerol (83.7±10.9 μg/mg of protein in empty vector control compared with 86.3±21.3 μg/mg of protein in STARD3-overexpressing cells) or phospholipid mass (42.3±3.96 μg/mg of protein in empty vector control compared with 47.1±8.31 μg/mg of protein in STARD3-overexpressing cells) occurred over the time scale investigated. Again, inhibition of endosomal trafficking using U18666A (25 μM) or progesterone (10 μM) induced the opposite effect, increasing the incorporation of [14C]acetate into each lipid pool (P<0.05) (Figure 2D).
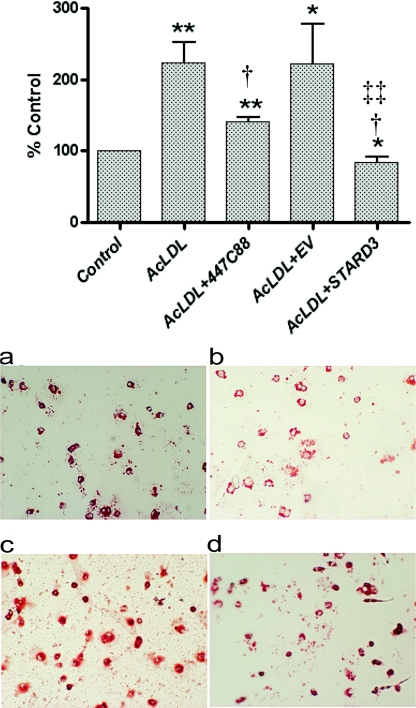
Importantly, when STARD3-overexpressing cells were challenged with AcLDL (50 μg/ml; 24 h), no significant increases in cholesterol esterification occurred, as judged by the incorporation of [3H]oleate into the cholesteryl ester pool; in fact, a small but significant (P<0.05) decrease was observed. In contrast, in untreated and empty vector control cells, cholesterol esterification increased by approx. 2.2-fold (P<0.01) when treated with AcLDL under the same conditions (Figure 3); a change blocked by the ACAT inhibitor 447C88 (10 μM). Equally, an apparent reduction in intensity of Oil Red O staining was observed in macrophages overexpressing STARD3, compared with empty vector control, after treatment with AcLDL (50 μg/ml; 24 h) (Figure 3).
Figure 3. Effect of STARD3 overexpression on macrophage foam cell formation.
Upper panel, incorporation of [3H]oleate (10 μM; 1 μCi/ml) into the cholesteryl ester pool following incubation with AcLDL (50 μg/ml; 24 h) in the presence or absence of the ACAT inhibitor 447C88 (10 μM) in wild-type cells, and in cells transfected with either empty vector (EV) or STARD3. Values are means±S.E.M. of four independent experiments. *P<0.05 and **P<0.01 compared with controls; †P<0.05 compared with AcLDL alone; and ‡‡P<0.01 compared with empty vector control treated with AcLDL. Lower panel, Oil Red O staining in empty vector control (a and c) and STARD3-overexpressing cells (b and d) incubated in the absence (a and b) or presence (c and d) of AcLDL (50 μg/ml; 24 h).
DISCUSSION
The START family of lipid trafficking may be involved in non-vesicular cholesterol transport, regulating sterol-responsive transcription factors, controlling the cholesterol content of organelles, lipid rafts and membranes, and storage of cholesterol as cytosolic droplets of cholesteryl ester, although the mechanisms remain poorly understood at present.
Notably, our previous studies have demonstrated that overexpression of distinct cholesterol and oxysterol-binding proteins within the START family of lipid trafficking proteins, STARD1 (StAR) [9], STARD3, STARD4 and STARD5, exert different effects on gene expression of the sterol-responsive transcription factors SREBP2 and LXRα. Overexpression of the mitochondrial cholesterol transporter StAR is associated with the induction of lipogenesis [9], possibly via LXR-dependent induction of SREBP1c. Transient overexpression of cytosolic STARD4 decreased SREBP2 mRNA levels by approximately half (Figure 1C), whereas overexpression of the gene encoding cytosolic STARD5 [16] protein resulted in marked increases in SREBP2 and NR1H3 mRNA levels compared with controls (Figure 1D). Overexpression of STARD4 and STARD5 have been shown previously to activate an LXR reporter plasmid in NIH-3T3 cells [17]. In contrast, STARD3 overexpression did not have an impact on either SREBP2 or NR1H3 mRNA levels (Figure 1B) and repressed lipogenesis (Figure 2D). Although some of the observed effects may be due to differences in gene expression and translation levels, despite the delivery of equivalent amounts of DNA to the same number of cells, they may also reflect the efficiency and/or directionality of the cellular cholesterol transport achieved [3,4].
Previously, we have speculated that STARD3 may traffic endosomally derived cholesterol to the ER (endoplasmic reticulum) and/or the plasma membrane [14]; even in the absence of lipoprotein-derived cholesterol, cholesterol cycles between the plasma membrane and endosomes [18]. Thus STARD3 could help to increase the cholesterol content of the ER membrane, and retention of SREBPs by the SCAP (SREBP2 cleavage-activating protein)/Insig (insulin-induced gene)-1(-2) complex, in agreement with the generalized repression of cholesterol, fatty acid and triacylglycerol biosynthesis observed in the present study. Cholesterol delivery to the ER might also be expected to expand the substrate pool available for esterification by ACAT1 (SOAT1). Instead, expansion of the cholesteryl [3H]oleate pool following exposure to AcLDL was effectively blocked in cells overexpressing STARD3. One explanation for this finding is that delivery of sterol to the ER was just sufficient to sequester SREBPs, but does not reach the threshold required to activate ACAT. In turn, this must imply that the bulk of the endosomal cholesterol trafficked by STARD3 may be efficiently directed elsewhere, perhaps to the plasma membrane to facilitate cholesterol efflux, possibly via vesicular transport facilitated by the Rab family of small GTPases. Dissociation between cholesterol transport to membrane-bound SREBP transcription factors and the substrate pool available for cholesterol esterification has been reported previously [19,20]. Kristiana et al. [20] observed clear differences in kinetics between endosomal delivery of LDL-derived cholesterol to SREBPs and ACAT in mutant CHO (Chinese-hamster ovary) cells with cholesterol trafficking defects [including NPC (Niemann–Pick type C)], contending that different cholesterol pools and/or transport pathways supply SREBPs and ACAT within the ER.
Our present findings also agree well with deletion studies of the START domain of STARD3 in mice in vivo [21]. Although relatively modest changes in lipid phenotype were observed, probably due to functional redundancies within the START family of cholesterol transfer proteins [21,22], significant increases in hepatic sterol ester were observed after feeding a high-fat diet, together with the reduced conversion of cholesterol into steroid hormones [21]. Use of a dominant-negative mutant of STARD3 (ΔSTART-STARD3) caused extensive cholesterol accumulation in CHO and COS-7 cells, accompanied by inhibition of late endosomal trafficking, similar to the phenotype caused by functional loss of NPC1/2 proteins [23]. Moreover, in cholesterol-laden cells, STARD3 becomes trapped at the periphery of cholesterol-laden lysosomes, reflecting a loss of dynamic cholesterol movement [23], and deletion of STARD3 is linked with disrupted actin-mediated dynamics of late endocytic organelles, suggesting that cholesterol binding or sensing by STARD3 in late endosomal membranes may govern actin-dependent fusion and degradative activity of that compartment [24]. Hepatic overexpression of STARD3 in vivo is associated with increased conversion into bile acids [25], although, in a separate study, apoptosis and hepatic toxicity were also reported, probably resulting from the grossly elevated and highly unphysiological levels of STARD3 utilized [26]; such changes were not observed in the present study.
Overexpression of STARD3 in the present study was also associated with increased expression of ABCA1 mRNA and protein. The latter suggests that STARD3, like NPC1, may facilitate trafficking of endosomal cholesterol and possibly ABCA1 protein through this compartment to the plasma membrane, increasing pools of membrane cholesterol and/or transporter available for efflux to apoAI [27]. The mechanism(s) by which STARD3 increases ABCA1 mRNA levels are less obvious, as no induction of LXRα was observed in our present experiments, and Npc1 inactivation reduces Abca1 protein levels, but does not alter Abca1 mRNA levels, in murine macrophages [28]. However, SREBPs (1 and 2) can exert repressive effects on ABCA1 expression, decreasing cholesterol efflux [29,30]. Thus it is possible that sequestration of SREBPs at the ER, as suggested by the co-ordinated loss of lipid biosynthesis observed in the present study, may relieve inhibition of ABCA1 gene expression and increase cholesterol efflux to apoAI. Alternatively, STARD3 expression could alter levels of ABCA1 mRNA by changing the ratios of saturated to unsaturated fatty acids [31], or perhaps via its involvement in actin-mediated dynamics of late endosomes [32] that trigger changes in actin-dependent gene expression [33].
In conclusion, STARD3 overexpression may be useful in limiting atherogenesis by up-regulating cholesterol efflux mechanisms, reducing cholesterol synthesis and inhibiting cholesterol esterification. In vivo studies are now needed to establish this contention, using murine models of atheroma.
AUTHOR CONTRIBUTION
Faye Borthwick initiated and performed the majority of the laboratory work, which was designed and supervised by Annette Graham; Anne-Marie Allen and Janice Taylor carried out additional experiments, including cell transfections, viability and caspase assays, and immunohistochemistry.
FUNDING
This work was supported by the Heart Research UK [grant number 2515/07/09].
References
- 1.Gerrity R. G. The role of the monocyte in atherogenesis. Am. J. Pathol. 1981;103:181–190. [PMC free article] [PubMed] [Google Scholar]
- 2.Schmitz G., Grandl M. The molecular mechanisms of HDL and associated vesicular trafficking mechanisms to mediate cellular lipid homeostasis. Arterioscler. Thromb. Vasc. Biol. 2009;29:1718–1722. doi: 10.1161/ATVBAHA.108.179507. [DOI] [PubMed] [Google Scholar]
- 3.Soccio R. E., Breslow J. L. Intracellular cholesterol transport. Arterioscler. Thromb. Vasc. Biol. 2007;2:1150–1160. doi: 10.1161/01.ATV.0000131264.66417.d5. [DOI] [PubMed] [Google Scholar]
- 4.Soccio R. E., Breslow J. L. StAR-related lipid Transfer (START) proteins: mediators of intracellular lipid metabolism. J. Biol. Chem. 2003;278:22183–22186. doi: 10.1074/jbc.R300003200. [DOI] [PubMed] [Google Scholar]
- 5.Alpy F., Tomasetto C. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J. Cell Sci. 2005;188:2791–2801. doi: 10.1242/jcs.02485. [DOI] [PubMed] [Google Scholar]
- 6.Miller W. L. Steroidogenic acute regulatory protein (StAR), a novel mitochondrial cholesterol transporter. Biochim. Biophys. Acta. 2007;1771:663–676. doi: 10.1016/j.bbalip.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 7.Ning Y., Bai Q., Lu H., Li X., Pandak W. M., Zhao F., Chen S., Ren S., Yin L. Overexpression of mitochondrial cholesterol delivery protein, StAR, decreases intracellular lipids and inflammatory factors secretion in macrophages. Atherosclerosis. 2009;204:114–120. doi: 10.1016/j.atherosclerosis.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ning Y., Xu L., Ren S., Pandak W. M., Chen S., Yin L. StAR overexpression decreases serum and tissue lipids in apolipoprotein E-deficient mice. Lipids. 2009;44:511–519. doi: 10.1007/s11745-009-3299-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor J. M., Borthwick F., Bartholomew C., Graham A. Overexpression of steroidogenic acute regulatory protein increases macrophage cholesterol efflux to apolipoprotein AI. Cardiovasc. Res. 2010;86:526–535. doi: 10.1093/cvr/cvq015. [DOI] [PubMed] [Google Scholar]
- 10.Alpy F., Stoeckel M. E., Dierich A., Escola J. M., Wendling C., Chenard M. P., Vanier M. T., Gruenberg J., Tomasetto C., Rio M. C. The steroidogenic acute regulatory protein homolog MLN64, a late endosomal cholesterol binding protein. J. Biol. Chem. 2001;276:4261–4269. doi: 10.1074/jbc.M006279200. [DOI] [PubMed] [Google Scholar]
- 11.Dressman M. A., Baras A., Malinowski R., Alvis L. B., Kwon I., Walz T. M., Polymeropoulos M. H. Gene expression profiling detects gene amplification and differentiates tumour types in breast cancer. Cancer Res. 2003;63:2194–2199. [PubMed] [Google Scholar]
- 12.Benusiglio P. R., Pharoah P. D., Smith P. L., Lesueur F., Conroy D., Luben R. N., Dew G., Jordan C., Dunning A., Easton D. F., Ponder B. A. HapMap-based study of the 17Q21 ERBB2 amplicon in susceptibility to breast cancer. Br. J. Cancer. 2006;95:1689–1695. doi: 10.1038/sj.bjc.6603473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alpy F., Latchumanan V. K., Kedinger V., Janoshazi A., Thiele C., Wendling C., Rio M. C., Tomasetto C. Functional characterisation of the MENTAL domain. J. Biol. Chem. 2005;280:17945–17952. doi: 10.1074/jbc.M500723200. [DOI] [PubMed] [Google Scholar]
- 14.Borthwick F., Taylor J. M., Bartholomew C., Graham A. Differential regulation of the STARD1 subfamily of START lipid trafficking proteins in human macrophages. FEBS Lett. 2009;583:1147–1153. doi: 10.1016/j.febslet.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 15.Palmer A. M., Murphy N., Graham A. Triglyceride-rich lipoproteins inhibit cholesterol efflux to apolipoprotein (apo) AI from human macrophage foam cells. Atherosclerosis. 2004;173:27–38. doi: 10.1016/j.atherosclerosis.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Agudo D., Ren S., Hylemon P. B., Montanez R., Redford K., Natarajan R., Medina M. A., Gil G., Pandak W. M. Localisation of the StarD5 cholesterol binding protein. J. Lipid Res. 2006;47:1168–1175. doi: 10.1194/jlr.M500447-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Soccio R. E., Adams R. M., Maxwell K. N., Breslow J. N. Differential gene regulation of StarD4 and StarD5 cholesterol transfer proteins. J. Biol. Chem. 2005;280:19410–19418. doi: 10.1074/jbc.M501778200. [DOI] [PubMed] [Google Scholar]
- 18.Holtta-Vuori M., Ikonen E. Endosomal cholesterol traffic: vesicular and non-vesicular mechanisms meet. Biochem. Soc. Trans. 2006;34:392–394. doi: 10.1042/BST0340392. [DOI] [PubMed] [Google Scholar]
- 19.Du X., Pham Y. H., Brown A. J. Effects of 25-hydroxycholesterol on cholesterol esterification and SREBP processing are dissociable: implications for cholesterol movement to the regulatory pool in the endoplasmic reticulum. J. Biol. Chem. 2004;279:47010–47016. doi: 10.1074/jbc.M408690200. [DOI] [PubMed] [Google Scholar]
- 20.Kristiana I., Yang H., Brown A. J. Different kinetics of cholesterol delivery to components of the cholesterol homeostatic machinery: implications for cholesterol trafficking to the endoplasmic reticulum. Biochim. Biophys. Acta. 2008;1781:724–730. doi: 10.1016/j.bbalip.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Kishida T., Kostetskii I., Zhang Z., Martinez F., Liu P., Walkley S. U., Dwyer N. K., Blanchette-Mackie E. J., Radice G. L., Strauss J. F., III Targeted mutation of the MLN64 START domain causes only modest alterations in cellular sterol metabolism. J. Biol. Chem. 2004;279:19276–19285. doi: 10.1074/jbc.M400717200. [DOI] [PubMed] [Google Scholar]
- 22.Riegelhaupt J. J., Waase M. P., Garbarino J., Cruz D. E., Breslow J. L. Targeted disruption of StARD4 leads to modest weight reduction and minor alterations in lipid metabolism. J. Lipid Res. 2010;51:1134–1143. doi: 10.1194/jlr.M003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang M., Liu P., Dwyer N., Christenson L. K., Fujimoto T., Martinez F., Comly M., Hanover J. A., Blanchette-Mackie E. J., Strauss J. F. MLN64 mediates mobilisation of lysosomal cholesterol to steroidogenic mitochondria. J. Biol. Chem. 2002;277:33300–33310. doi: 10.1074/jbc.M200003200. [DOI] [PubMed] [Google Scholar]
- 24.Holtta-Vuori M., Alpy F., Tanhuanpaa K., Jokitalo E., Mutka A. L., Ikonen E. MLN64 is involved in actin-mediated dynamics of late endocytic organelles. Mol. Biol. Cell. 2005;16:3873–3886. doi: 10.1091/mbc.E04-12-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren S., Hylemon P., Marques D., Hall E., Redford K., Gil G., Pandak W. M. Effect of increasing the expression of cholesterol transporters (StAR, MLN64 and SCP-2) on bile acid synthesis. J. Lipid Res. 2004;45:2123–2131. doi: 10.1194/jlr.M400233-JLR200. [DOI] [PubMed] [Google Scholar]
- 26.Tichauer J. E., Morales M. G., Amigo L., Galdames L., Klein A., Quinones V., Ferrada C., Alvarez A. R., Rio M.-C., Miquel J. F., et al. Overexpression of the cholesterol-binding protein MLN64 induces liver damage in the mouse. World J. Gastroenterol. 2007;13:3071–3079. doi: 10.3748/wjg.v13.i22.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rigamonti E., Helin L., Lestavel S., Mutka A. L., Lepore M., Fontaine C., Bouhlel M. A., Bultel S., Fruchart J. C., Ikonen E., et al. Liver X receptor activation controls intracellular cholesterol trafficking and esterification in human macrophages. Circ. Res. 2005;97:682–689. doi: 10.1161/01.RES.0000184678.43488.9f. [DOI] [PubMed] [Google Scholar]
- 28.Wang M.-D., Franklin V., Sudaram M., Kiss R. S., Ho K., Gallant M., Marcel Y. L. Differentiation regulation of ATP binding cassette protein A1 expression and ApoA-I lipidation by Niemann-Pick Type C1 in murine hepatocytes and macrophages. J. Biol. Chem. 2007;282:22525–22553. doi: 10.1074/jbc.M700326200. [DOI] [PubMed] [Google Scholar]
- 29.Zhou X., He W., Huang Z., Gotto A. M., Jr, Hajjar D. P., Han J. Genetic deletion of low density lipoprotein receptor impairs sterol-induced mouse macrophage ABCA1 expression: a new SREBP1-dependent mechanism. J. Biol. Chem. 2008;283:2129–2138. doi: 10.1074/jbc.M706636200. [DOI] [PubMed] [Google Scholar]
- 30.Zeng L., Liao J., Lee T. S., Zhu M., Wang X., Stemerman M. B., Zhu Y., Shyy J. Y. Sterol responsive element binding protein (SREBP) 2 down-regulates ATP binding cassette transporter A1 in vascular endothelial cells: a novel role of SREBP in regulating cholesterol metabolism. J. Biol. Chem. 2004;279:48801–48807. doi: 10.1074/jbc.M407817200. [DOI] [PubMed] [Google Scholar]
- 31.Schmitz G., Langmann T. Transcriptional regulatory networks in lipid metabolism control ABCA1 expression. Biochim. Biophys. Acta. 2005;1735:1–19. doi: 10.1016/j.bbalip.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Holtta-Vuori M., Alpy F., Tanuanpaa K., Jokitalo E., Mutka A-L, Ikonen E. MLN64 is involved in actin-mediated dynamics of late endocytic organelles. Mol Biol. Cell. 2005;16:3873–3886. doi: 10.1091/mbc.E04-12-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castano E., Philimonenko V. V., Kahle M., Fukalova J., Kalendova A., Yildirim S., Dzijak R., Dingova-Krasna H., Hozak P. Actin complexes in the cell nucleus: new stones in an old field. Histochem. Cell Biol. 2010;133:607–626. doi: 10.1007/s00418-010-0701-2. [DOI] [PubMed] [Google Scholar]