Abstract
The sirtuin (silent information regulator 2) proteins are NAD+-dependent deacetylases that are implicated in diverse biological processes including DNA regulation, metabolism and longevity. Homologues of the prototypic yeast Sir2p have been identified in all three kingdoms of life, and while bacteria and archaea typically contain one to two sirtuins, eukaryotic organisms contain multiple members. Sirtuins are regulated in part by the cellular concentrations of the noncompetitive inhibitor, nicotinamide, and several synthetic modulators of these enzymes have been identified. The x-ray crystal structures of several sirtuin proteins in various liganded forms have been determined. This wealth of structural information, together with related biochemical studies, have provided important insights into the catalytic mechanism, substrate specificity, and inhibitory mechanism of sirtuin proteins. Implications for future structural studies to address outstanding questions in the field are also discussed.
1. Introduction
Sirtuins compose the Class III family of histone deacetylase enzymes (HDACs) that, unlike the Class I and II HDACs, require the cofactor NAD+ as a substrate. Sirtuins catalyze the removal of an acetyl moiety from the ε-amino group of lysine residues within protein targets [1, 2] to yield the deacetylated protein product, nicotinamide, and 2’-O-acetyl-ADP-ribose [3, 4]. The founding member of this protein family, Saccharomyces cerevisiae Sir2p, one of five yeast sirtuin proteins including HST1–4, is a limiting factor in yeast aging, as deletion of the SIR2 gene results in reduced replicative lifespan [5], and additional copies of SIR2 results in increased yeast replicative lifespan [6]. Furthermore, Sir2p is required for the lifespan extension that results from restricting the caloric intake of yeast cells [7]. The sirtuin protein family is broadly conserved in all three kingdoms of life [8] and increased sirtuin expression in higher eukaryotes leads to increased lifespan in worms [9], and flies [10],, and increased longevity due to a calorie restricted diet in some of these organisms is sirtuin dependent [10]. Humans have seven sirtuin proteins (SIRT1–7) [11]. Human SIRT1 is implicated to play a role in a number of age-related human diseases and biological functions including cell survival, apoptosis, stress resistance, fat storage, insulin production, glucose homeostasis, and lipid homeostasis through direct deacetylation or regulation of its many known in vivo targets including p53, Ku70/Bax, FOXO, PPARγ, PGC1α, UCP2, LXR, and NFκB (reviewed in [12, 13]). Bacteria and archaea also harbor sirtuin proteins, typically one or two, and they target DNA regulatory proteins and metabolic enzymes including the chromatin protein Alba [14] and acetyl-CoA synthetase [15].
The activity of sirtuin family members can be modulated by several known effectors. Nicotinamide, a reaction product and noncompetitive inhibitor of sirtuin proteins [2, 16], is a physiological regulator of this family of proteins [17]. Yeast cells grown in the presence of nicotinamide show a dramatic reduction in silencing, an increase in rDNA recombination, and a shortening of replicative lifespan [16]. Nicotinamide exerts its inhibitory effect on deacetylation by reacting with a reaction intermediate to reform β-NAD+ at the expense of deacetylation [18, 19], a process termed base exchange. Several compounds have also been identified and characterized as small molecule sirtuin inhibitors such as sirtinol [20], splitomycin [21], cambinol [22], tenovin [23], Ro-318220 [24], surfactin [25], suramin [26], and the most potent known sirtuin inhibitors, indole EX527 analogs [27]. It is not clear where most of these compounds bind to the sirtuin enzymes or how they exert their inhibitory effect, although the observation that several of these inhibitors have different potencies against different members of the sirtuin family suggest that they do not exclusively target the conserved enzyme active site.
In order to aid in understanding the catalytic mechanism, substrate specificity, and inhibitory mechanism of the sirtuin protein family, the three-dimensional structures of several sirtuin homologues have been determined by x-ray crystallography. This wealth of structural information together with complementary biochemical studies has provided valuable insights into these activities. Still, specific questions about the sirtuin catalytic and nicotinamide inhibition mechanism, the exact biological role of regions N- and C-terminal to the catalytic core, and the inhibition mechanism of synthetic sirtuin inhibitors remain and will require the structure determination of other sirtuin complexes such as ones containing later reaction intermediates, nicotinamide together with reaction intermediates, sirtuin homologues with other N- and C-terminal regions, and other sirtuin protein/inhibitor complexes.
2. Overall structure of the sirtuin catalytic core
Sequence alignment of the sirtuin proteins indicates that they contain an approximately 275 amino acid conserved catalytic core region (Figure 1). Additionally, the sirtuin proteins have N- and C-terminal flanking regions that are variable in length and sequence [8, 28]. Consistent with the sequence conservation among the sirtuin proteins, the catalytic core regions of known sirtuin structures show a high degree of structural superposition (Figure 2). The catalytic core region adopts an elongated shape containing a large and structurally homologous Rossmann-fold domain, characteristic of NAD+/NADH binding proteins; a more structurally diverse, smaller, zinc-binding domain; and several loops connecting the two domains. These loops form a pronounced, extended cleft between the large and small domains where the NAD+ and acetyl-lysine containing peptide substrates enter from opposite sides and bind to the enzyme (Figure 3). The amino acids involved in catalysis and the reactive groups of both bound substrate molecules are buried within a protein tunnel in the cleft between the two domains (Figure 3), the region of the enzyme that contains the highest sequence conservation within the sirtuin enzymes (Figure 1).
Figure 1.
Sequence alignments of Sirtuins for which three-dimensional structures have been determined. An alignment of S. cerevisiae Hst2 and Sir2p, A. fulgidus Sir2-Af1 and Sir2-Af2, T. maritima Sir2Tm, E. Coli cobB, and H. sapiens SIRT2 and SIRT5. The secondary structural elements for Hst2 are shown above the sequence alignment; β sheets are indicated with orange arrows, α helices with green rectangles, loops with black lines and dashed lines indicate unstructured regions. Cyan circles indicate residues that participate in NAD+ binding, purple circles indicate residues that participate in acetyl-lysine peptide binding, a green star indicates the general base residue and blue triangles indicate the residues involved in zinc binding. Solid lines beneath the sequence alignment indicate which regions of the proteins compose the Rossmann-fold domain (magenta), cofactor binding loop (black), small domain (blue), and loop regions (green).
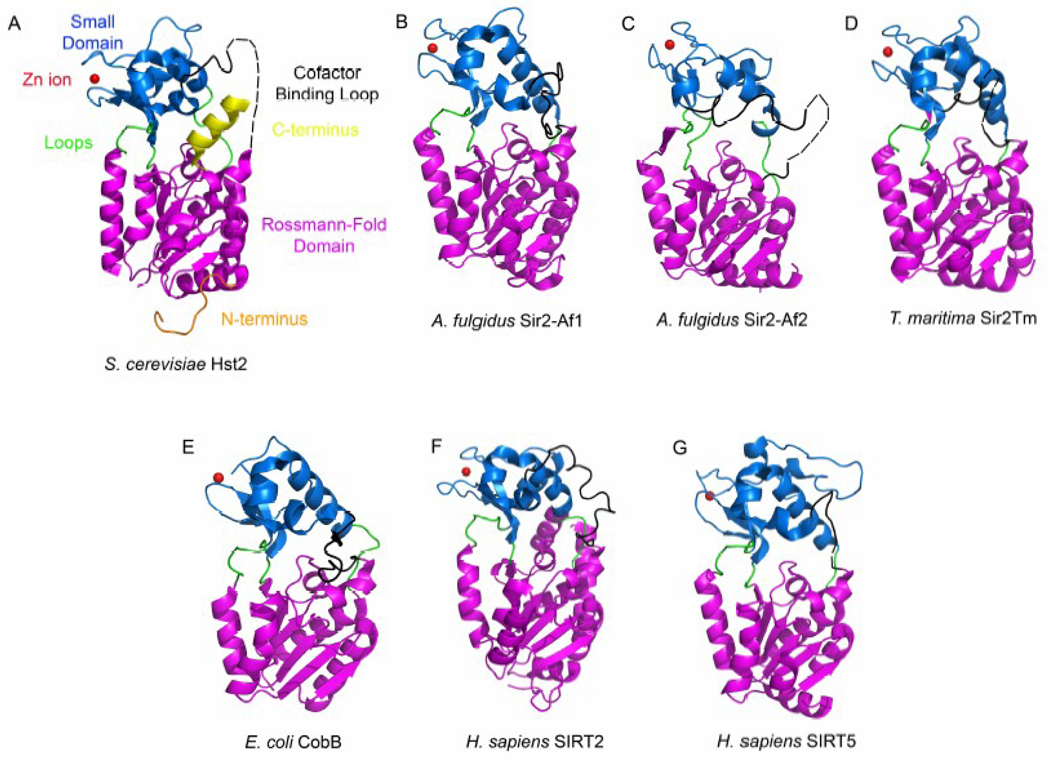
Figure 2.
Three-dimensional structure of each sirtuin protein that has been determined. The enzymes are shown in cartoon representation without bound ligands. The Rossmann-fold domain is indicated in magenta, the small domain in blue, the loops in green, the cofactor binding loop in black, the Zn ion in red, the N-terminal region in orange, and the C-terminal region in yellow. Disordered regions are indicated with dashed lines. A) S. cerevisiae Hst2 (pdb accession number 1Q14) B) A. fulgidus Sir2-Af1 (1ICI) C) A. fulgidus Sir2-Af2 (1MA3) D) T. maritima Sir2Tm (2H4J) E) E. Coli cobB (1S5P) F) H. sapiens SIRT2 (1J8F) G) H. sapiens SIRT5 (2B4Y).
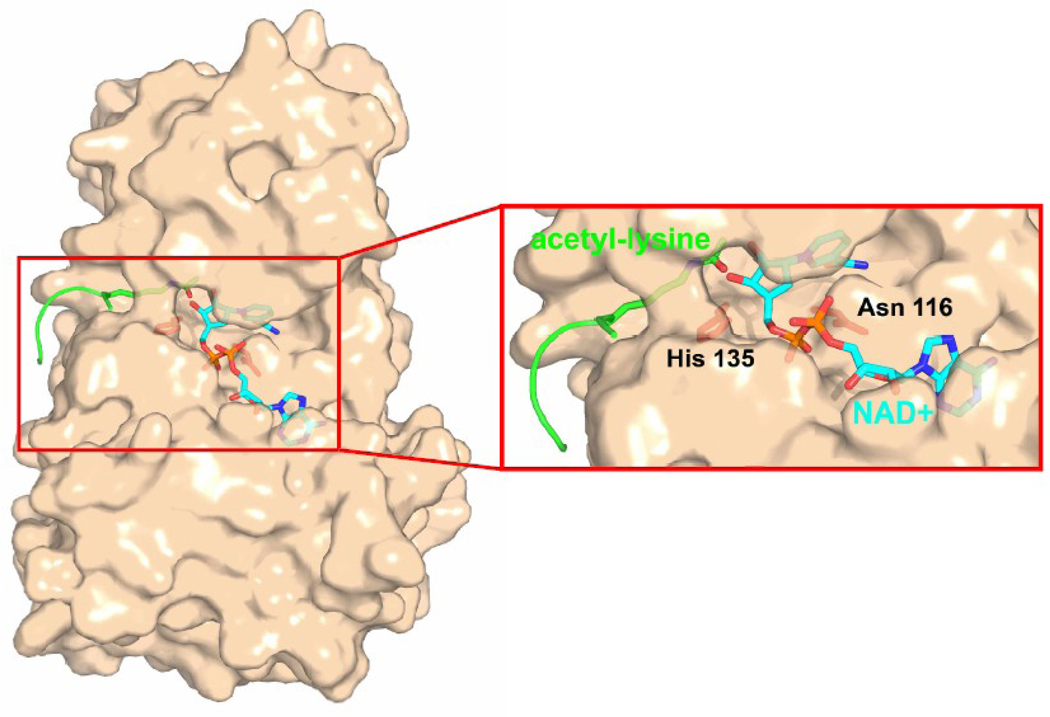
Figure 3.
Sirtuin active site binding cleft. The Hst2 protein is shown in tan surface representation. The catalytic residues H135 and N116 are shown in red stick representation in the close-up view. The peptide substrate backbone is shown in green cartoon representation, the acetyl-lysine side chain is shown in green cpk stick representation, and the NAD+ substrate molecule is shown in cyan cpk stick representation.
2.1 The Rossmann-fold domain
The large domain has a classical open α/β Rossmann-fold structure. Six parallel β strands form a central β sheet which is sandwiched between several α helices (the exact number of α helices depends upon the specific sirtuin protein) on either side of the β sheet [29] (Figure 2). The Rossmann-fold domain contains many of the hallmarks of a typical NAD-binding site such as a conserved Gly-X-Gly sequence important for phosphate binding, a pocket to accommodate an NAD+ molecule, and charged residues responsible for ribose group binding [30]. Interestingly, the Rossmann-fold domain of the sirtuin proteins is most similar to the inverted Rossmann-fold enzymes, where the adenine base of NAD+ binds to the C-terminal half and the nicotinamide group of NAD+ binds to the N-terminal half of the β sheet [31]. Structures of sirtuin enzymes bound to NAD+ (discussed below) confirm that NAD+ does bind to this protein family in an inverted conformation.
2.2 The Zn-binding domain
The small, structural Zn2+-binding domain results from two insertions in the Rossmann-fold domain and is the most diverse region between sirtuins for which structures have been determined in terms of primary sequence, three-dimensional structure and position relative to the large domain (Figure 2). The two structural modules of the small domain consist of a three-stranded antiparallel β sheet and a variable α helical region dependent on the sirtuin protein [30]. A Zn2+ molecule is bound by the sulphydryl groups of two pairs of strictly conserved cysteine residues in the β sheet module in a tetrahedral conformation [29], with the exception of CobB, which is liganded by only two visible cysteine residues and contains only three of the four expected zinc-coordinating cysteine residues according to sequence alignment (Figure 1). The three-stranded four-cysteine metal binding site resembles the Zn-ribbon structure of transcription factors TFIIS [32], TFIIB [33], and the RNA polymerase II subunit RPB9 [34]. The small domain contains the conserved sequence motif Cys-X2–4-Cys-X15–40-Cys-X2–4-Cys, a characteristic Zn-binding motif. Although the location of the zinc ion is too far from the enzyme active site to participate in catalysis, its presence is required for deacetylase activity, as mutation of all four zinc-binding cysteines to alanine or addition of the zinc chelator o-phenanthroline inhibits the in vitro deacetylase activity of sirtuin enzymes [29]. The role of the zinc ion seems to be structural, as it is postulated to be required for holding together the β strands in the small domain. One of the most interesting characteristics of the small domain is a conserved salt bridge (between R/K178 and E186, Hst2 numbering, unless otherwise indicated) [29, 35], which seems to be important for positioning the small domain with respect to the large domain.
The small domains of the archaeal and bacterial Sir2 enzymes show a similar overall topology, while the small domains of the eukaryotic Sir2 enzymes show the greatest variability in secondary structure (Figure 2). One proposal to explain the divergence in primary sequence, secondary and tertiary structures among Sir2 homologues is that divergent small domains may be involved in substrate-specific binding. Since each homologue likely deacetylates different protein targets, the greater variability of the small domain in eukaryotic proteins might reflect the greater number of sirtuin proteins that exist in eukaryotes and therefore the greater number of substrates that need to be discriminated between [36]. The position of the small domain relative to the Rossmann-fold domain is also dependent on the conformation of bound NAD+ [37] or the presence of acetyl-lysine in the enzyme active site (discussed below). Human SIRT5, for example, contains an extra loop and helix (helix α9) inserted into the small domain (Figure 2H) that is sequentially conserved in other mitochondrial sirtuins and may prove to be structurally important for mitochondrial localization [35]. Taken together, the diversity in the small domain may have a potential role in regulating protein-protein interactions, which may be important for substrate specificity, enzyme localization, and potentially for modulation of enzyme activity. Indeed, structural differences between sirtuin proteins within the small domain might be exploited as binding sites for sirtuin-selective small molecule inhibitors.
2.3 The cofactor binding loop region
The four loops that connect the small and large domains form the cleft that acts as the enzyme active site. Both the NAD+ and acetyl-lysine substrates bind in this cleft (Figure 3). This region has the most sequence homology among the protein family, and mutation of several residues in this region disrupt histone deacetylase activity, underscoring the importance of this region in sirtuin catalysis. Generally the largest of these four loops (the β1-α2 loop in Hst2) is one of the most conformationally dynamic regions of the sirtuin enzymes (Figure 4). Since this loop forms part of the cofactor NAD+ binding site and its conformation is largely dependent on the conformation and identity of the bound NAD+ molecule or intermediate, we will refer to it as the cofactor binding loop [29]. The cofactor binding loop appears to be disordered or highly flexible (as evidenced by high B-factors) when NAD+ is not bound (Figure 4A), and becomes ordered upon NAD+ binding into a relatively open conformation with residues of the loop (D43-Y52) about 7 Å away from the NAD+ binding site (Figure 4B) [38]. Residues (A33, G34, T37, F44 and D43) of the cofactor binding loop form part of the so-called “C pocket,” the region of the enzyme where the nicotinamide moiety of NAD+ binds, and they mediate almost half of the enzyme contacts to NAD+ [38, 39], which implies that this cofactor binding loop plays an important role in NAD+ binding. The relationship between the presence of bound NAD+ and the relative order and positioning of the cofactor binding loop is consistent among all homologues for which structures have been determined [29, 30, 40, 41], and can be concurrent with a rigid body rotation of the small domain relative to the large domain [42]. The cofactor binding loop region maintains the open, ordered conformation when a molecule designed to mimic the dissociative transition state of nicotinamide-dissociated NAD+, DADMe-NAD+ [43], or the oxocarbenium ion intermediate-like molecules, ADP-Ribose or ADP-HPD, are bound to the enzyme [38, 44], implying that nicotinamide cleavage does not affect the conformation of the preferred microstate of the dynamic cofactor binding loop. When the sirtuin reaction product 2’-O-acetyl-ADP-ribose is bound to the enzyme active site, the cofactor binding loop adopts a more closed conformation bringing residues (D43-Y52) of the loop ~5.5 Å closer to the nicotinamide binding pocket, partially occluding the site (Figure 4C) [38, 41, 45], suggesting that the loop may also play a role in nicotinamide release. The switch between the open and closed conformations of the cofactor binding loop seems to be dependent upon the the presence of the bound alkylamidate intermediate directly after nucleophilic attack of acetyl-lysine on an NAD+-derived intermediate and not upon nicotinamide cleavage or complete reaction product formation, as the closed conformation of the loop is also seen when an analog of the O-alkylamidate intermediate, S-alkylamidate, is bound to the enzyme [43]. When the reaction product nicotinamide is bound in the C-pocket either in complex with acetyl-lysine, NAD+ or ADP-ribose, the cofactor binding loop adopts slightly different conformations which facilitate slightly different interactions between nicotinamide and the C-pocket residues, although it is not clear whether the binding of different ligands of the ternary complex or the different nicotinamide conformations cause the differences in conformation of the cofactor binding loop [44, 46].
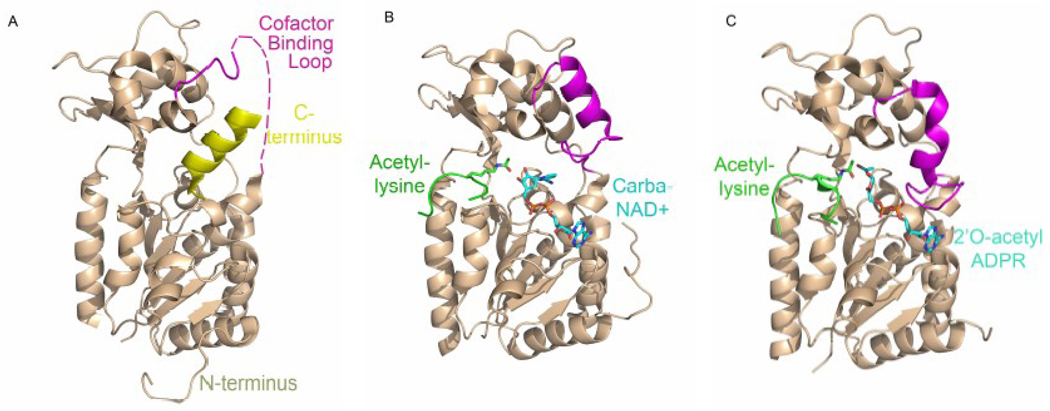
Figure 4.
Conformational changes of the cofactor binding loop upon substrate and product binding. The Hst2 protein is shown in tan cartoon representation, the C-terminus is shown in yellow, and the cofactor binding loop is shown in magenta. The peptide substrate is shown in green cartoon representation, while the acetyl-lysine residue is shown in green cpk stick representation. The substrate analog or product is shown in cyan cpk stick representation. A) The apo Hst2 enzyme, the cofactor binding loop is disordered (dashed lines). B) Hst2 with bound peptide substrate and carba-NAD+, the cofactor binding loop is in an open conformation. C) Hst2 with bound peptide substrate and 2’-O-acetyl-ADP-ribose, the cofactor binding loop is in a closed conformation.
3. Substrate binding sites
3.1 NAD+ Binding Site
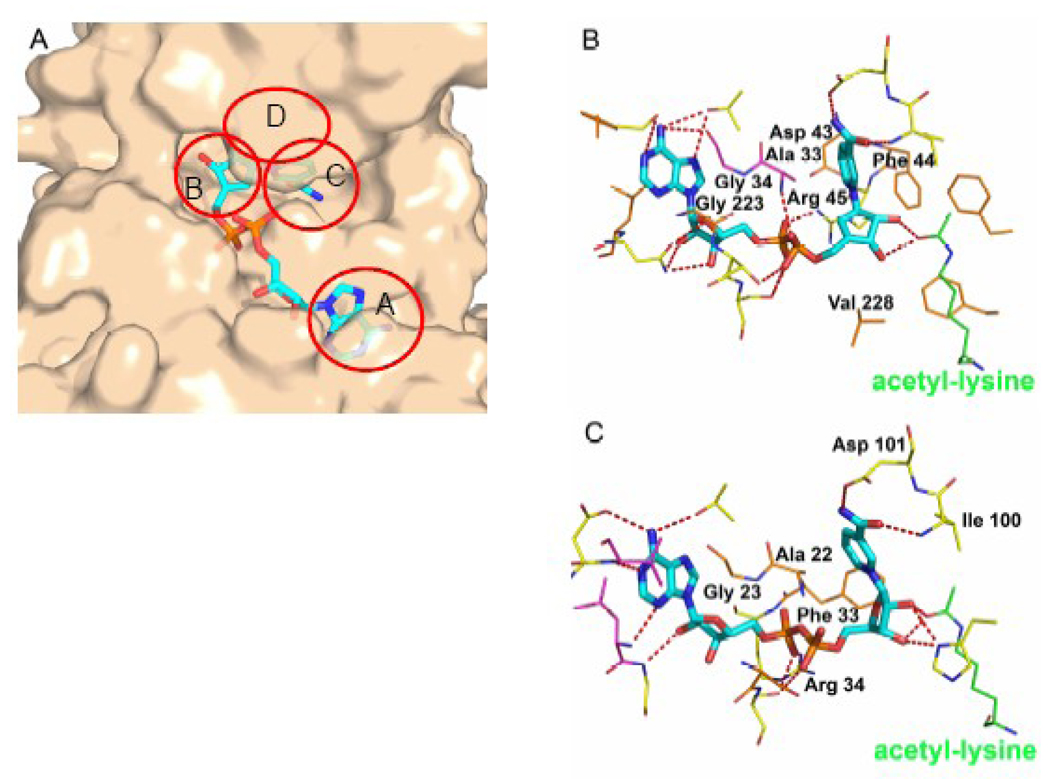
The Rossmann-fold domain forms the bottom of the NAD+ binding site while the loop regions, particularly the cofactor binding loop, forms the remainder of the pocket. When bound to a sirtuin enzyme, the NAD+ molecule adopts an extended conformation, which is commonly found when NAD+ is bound to a protein having a Rossmann-fold [29]. Unusually, the NAD+ molecule binds in an inverted direction relative to most NAD dehydrogenases [47]: the adenine base binds to the C-terminal half of the β sheet and the nicotinamide group binds to the N-terminal half. The NAD+ binding site can be divided into three regions: site A, the adenine-ribose binding site; site B, the nicotinamide-ribose binding site; and site C, the nicotinamide moiety binding site [29] (Figure 5A). The A site is shallow, mostly hydrophobic and exposed. The adenine base is surrounded by hydrophobic amino acids making van der Waals interactions, including several conserved glycines (G34 and G223) (Figure 5B). The amide nitrogen of the adenine base makes a hydrogen bond with a glutamate or serine residue in the A site. The N1 atom of the adenine base makes a hydrogen bond with a main chain amide in some sirtuin homologues. The 2’ and 3’ hydroxyls of the adenine ribose make several hydrogen bonds with asparagine residues within the A site. The oxygens of the phosphate groups make several hydrogen bonds with residues in the A site, although the exact hydrogen bonds depend upon the conformation of the cofactor binding loop [29, 41, 42]. The cofactor binding loop also makes up the ‘ceiling’ of the B site, so its conformation can also affect nicotinamide ribose binding. When the cofactor binding loop is in the closed conformation (Figure 4C), the nicotinamide ribose makes van der Waals interactions with conserved hydrophobic residues (F44, W58, and V228) (Figure 5B). Absolutely conserved residues (H135 and Q115) are located on the floor of the B site and make interactions with the 3’ oxygen and sometimes the 2’ oxygen of the nicotinamide ribose [29, 35, 41, 42] (Figure 5B). The conformation of the nicotinamide ribose is variable depending on the NAD+ analog bound (ADP-Ribose, non-productive NAD+, productive NAD+ or carba-NAD+), which other substrates are also bound to the enzyme, or even the particular sirtuin homologue [29, 35, 37, 38, 42, 45].
Figure 5.
Productive NAD+ binding interactions in several sirtuin proteins. Carba-NAD+ or NAD+ molecules are shown in cyan cpk stick representation and acetyl-lysine residues are shown in green cpk line representation. A) The Hst2 protein is shown in tan surface representation. The A, B, C and D sites are indicated by red circles. B) S. cerevisiae Hst2. Enzyme residues that make van der Waals interactions with NAD+ are shown in orange. Residues that make hydrogen bonds (red dashes) with NAD+ are shown in yellow. Residues that make both interactions are shown in magenta. C) T. maritima Sir2Tm. Residues are colored as in B.
Different conformations of the nicotinamide ribose suggest different reaction mechanisms for the dissociation of nicotinamide from the rest of the NAD+ molecule. For example, in yeast Hst2, when carba-NAD+ (a non-hydrolysable NAD+ analog) and an acetyl-lysine containing peptide are bound to the enzyme, the nicotinamide ribose ring (in this case a cyclopentane) is positioned in such a way that both the 2’ and 3’-hydroxyls make hydrogen bonds with the acetyl-lysine carbonyl oxygen, which is on the β face of the ribose group, thereby sterically occluding the acetyl-lysine carbonyl oxygen from nucleophilic attack of C1’ of the nicotinamide ribose group, suggesting that the glycosidic bond between nicotinamide and the rest of the NAD+ molecule is broken in a dissociative fashion (Figure 5B) [38]. When an oxocarbenium intermediate-like molecule, either ADP-Ribose or ADP-HPD, is bound the nicotinamide ribose ring is rotated by about 45° [38, 44] relative to the substrate analog complex. Finally, when the reaction product, 2’-O-acetyl-ADP-ribose is bound, the nicotinamide ribose ring is rotated about 90° relative to the substrate bound complex and in the opposite direction of the intermediate-like molecule [42]. This variability of the nicotinamide-ribose ring along the reaction coordinate highlights the importance of the conformation of this ring during catalysis. However, in the complex of Sir2Tm bound to an acetyl-lysine containing peptide and NAD+ in a productive conformation, the nicotinamide ribose ring is rotated 30° around the glycosidic bond relative to its position in the Hst2 structure, orienting the acetyl-lysine carbonyl toward the α face of the ribose ring, placing it within 3.2 Å from the C1’ of the ribose and positioned for nucleophilic attack (Figure 5C) [45], thus supporting a direct nucleophilic role for acetyl-lysine in nicotinamide cleavage. These differences may be explained by the use of different catalytic mechanisms for archaeal and eukaryotic sirtuins, although it seems unlikely that enzymes with conserved catalytic residues would evolve different catalytic mechanisms. The difference in conformation of the nicotinamide ribose ring may be due to the presence of a cyclopentane instead of a ribose in this position for the Hst2 structure. Alternatively, a conformational change in the ribose ring of the Sir2Tm structure may be required for enzyme catalysis, which may explain why the cognate substrates have not turned over in the time course of crystallization and why the conformation of the nicotinamide ribose is different than in other structures of this homologue with productively bound NAD+ [37].
Density for the nicotinamide moiety of NAD+ is often not visible in crystal structure solutions of sirtuin proteins crystallized in the presence of NAD+, either because it is disordered or hydrolyzed in the crystallization solution. Occasionally, the nicotinamide moiety of NAD+ can been seen in a crystal structure, but it is bound in a so-called non-productive conformation. The nicotinamide moiety of non-productively bound NAD+ is not bound in the C site and instead adopts alternate conformations which fail to account for the invariance of key residues required for catalysis located in the C site, and this conformation of NAD+ is incompatible with acetyl-lysine binding [37, 46]. When an acetyl-lysine containing peptide, a PEG ion, or the N-terminus of a symmetry-related oligomer is simultaneously bound to the enzyme active site, the nicotinamide moiety of productively bound NAD+ can bind in the C site where it makes direct contact with many invariant residues [37, 38, 45, 46]. Interestingly, the hydrophobic tunnel which acts as the acetyl-lysine binding site and not simply the peptide binding site must be occupied to facilitate the productive binding of NAD+ [37]. In this productive conformation, the bound NAD+ is compatible with both acetyl-lysine binding and enzyme catalysis. In the productive conformation of NAD+, the coplanarity of the nicotinamide moiety with the glycosidic bond is strained and the positive charge is buried in a largely hydrophobic environment, thereby destabilizing the ground state of NAD+ (Figure 5B&C) [37]. NAD+ in the productive conformation makes hydrophobic and van der Waals contacts with the invariant Gly-Ala-Gly motif, which inserts between the nicotinamide moiety and the nicotinamide ribose, and other conserved residues (I41, F40 and N116). The carbonyl oxygen of the nicotinamide moiety of NAD+ forms a hydrogen bond with the backbone nitrogen of a conserved isoleucine (117); the carbonyl amino group forms a hydrogen bond with the side chain of a conserved Asp (118) [37]. A highly conserved serine (36) is required for catalytic activity of the protein [29] and it is clear from the conformation of the nicotinamide moiety of NAD+ that the serine residue, through its network of direct and water-mediated hydrogen bonds with invariant residues involved in nicotinamide group binding, serves to maintain the protein fold or the enzyme-nicotinamide interactions it mediates [38]. The pi electron cloud of a conserved phenylalanine (44) is positioned directly above the ribose oxygen, possibly to stabilize the carbocation transition state through the formation of a cation-pi interaction, and also may act to shield the glycosidic bond and oxocarbenium intermediate from water hydrolysis [45]. The C site adopts a more closed conformation after acetyl-lysine transfer, not directly after nicotinamide cleavage [38]. The structures of sirtuin homologues with bound product (2’ or 3’-O-acetyl-ADP-ribose) reveal that residues of the cofactor binding loop (D43-R45) shift by about 5.5 Å toward the nicotinamide binding pocket and these residues along with a conserved phenylalanine (44) serve to occlude the C site and probably act to expel the product nicotinamide from this site after cleavage. Finally, when nicotinamide is absent from the C site, a series of water molecules form a network which is conserved in all sirtuin structures, mediates interactions between several residues in the nicotinamide-binding site and seems to be required to maintain the integrity of the core domain [38].
3.2 Acetyl-lysine binding site
Structures of several sirtuin homologues bound to peptides containing acetyl-lysine bearing substrates have been determined [40]. The peptide itself binds to the cleft between the small and large domains with the acetyl-lysine side chain inserting into a hydrophobic tunnel located within the cleft. The peptide backbone containing the acetyl-lysine side chain forms β sheet-like interactions with two flanking strands in the enzyme (Figure 6A) [40]. This staggered, antiparallel three-stranded β sheet is referred to as the β staple. Main chain atoms of the peptide make several hydrogen bonds with main chain atoms of catalytic residues from the flanking strands of the β staple (Figure 6B). A comparison of sirtuin enzymes with and without bound acetyl-lysine containing peptide suggest that peptide binding induces a significant shift in the position of the small domain relative to the large domain, bringing the two domains together to form the β staple motif and resulting in the correct positioning of conserved residues for formation of the acetyl-lysine binding tunnel [48]. It has been hypothesized that NAD+ and peptide binding may be cooperative, as the region surrounding the peptide binding site adopts the more closed conformation seen in the peptide bound structure when NAD+ is bound but not in the absence of NAD+ [40]. The aliphatic region of the acetyl-lysine side chain makes several van der Waals interactions with conserved hydrophobic residues (H135, V228, F184, and L188). The acetyl group appears to be positioned by a single hydrogen bond between the Nε atom of the lysine side chain and the main chain of a Val residue (182) and van der Waals interactions between the acetyl methyl group and other highly conserved residues (I117 and H135), which position the unliganded carbonyl oxygen toward the NAD+ binding B site (Figure 6C) [40]. When the peptide is bound in a ternary complex with the intermediate-like molecule ADP-ribose, or the reaction product 2’-O-acetyl-ADP-ribose, the acetyl-lysine carbonyl oxygen hydrogen bonds to the 1’-OH of the nicotinamide ribose ring [38, 42]. The β staple binding motif and the residues involved in peptide binding seem to be conserved features of acetyl-lysine peptide binding, as structures of several sirtuin proteins with bound peptides make similar interactions [36, 39, 42, 48].
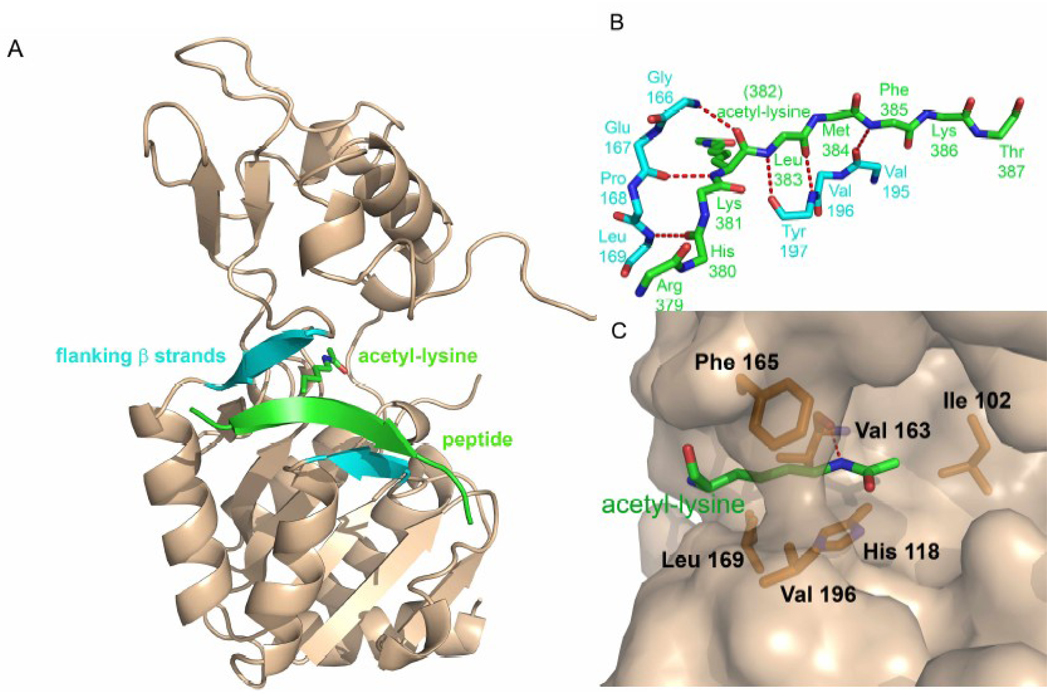
Figure 6.
Acetyl-lysine peptide binding to the sirtuin active site. A) A peptide containing an acetyl-lysine residue (green) is shown bound to the Sir2-Af2 sirtuin enzyme (tan, cartoon representation) active site forming the β staple with flanking β strands of the enzyme (cyan). B) The acetyl-lysine containing peptide (green) makes main chain β strand hydrogen bonds (red dashes) with main chain atoms of the enzyme (cyan). C) The acetyl-lysine residue (green) binds in a hydrophobic tunnel of the enzyme (tan, surface representation), making several van der Waals interactions with enzyme residues (orange) and one hydrogen bond (red dashes) with an enzyme residue.
Since only main chain atoms from both the peptide and the enzyme participate in hydrogen bonding, substrate specificity does not appear to be conferred by the primary sequence of the acetyl-lysine containing peptide, particularly in the immediate vicinity of the acetyl-lysine residue, although the enzyme residues involved in peptide binding are some of the most highly conserved in the enzyme (Figure 1). The relatively small number of protein-peptide interactions, especially outside of the acetyl-lysine residue, may reflect the fact that the peptide substrates bound to the enzymes in the crystal structures are not true in vivo targets or that substrate specificity determinants may involve other regions of the Sir2 proteins and/or regions of the substrate that are distal to the acetyl-lysine [42]. Consistent with this, comparisons of the same peptide (histone H4) bound to different sirtuin homologues or different peptides bound to different homologues show that peptide residues outside of those immediately flanking the acetyl-lysine residue diverge in direction and make variable contacts to their respective proteins. However, a study of several peptides that vary in primary sequence bound to the same sirtuin homologue, Sir2Tm, show that primary sequence in the residue immediately N-terminal (−1 position) and the residue 2 positions C-terminal (+2 position) to acetyl-lysine may govern sequence specific interactions in this homologue as these residues may hydrogen bond to the side chain of residue N165 and the main chain of G163, and make van der Waals interactions with the side chains of residues F162 and V193, respectively (Sir2Tm numbering) [48]. These van der Waals interactions have been proposed to stabilize β staple interactions when the cleft closes upon peptide binding [48]. In the same study, several peptides failed to crystallize bound to Sir2Tm, and these residues all lacked hydrophobic side chains at the +2 position and had residues other than lysine or arginine at position −1. Since residue N165 of Sir2Tm makes a hydrogen bond to the side chain of the residue in the −1 position of the peptide and residue 165 varies among sirtuin homologues, this interaction may point to one mechanism by which different sirtuins discriminate amongst substrates [48]. Interestingly, comparison of the same acetylated and deacetylated peptide bound to Sir2Tm revealed that the same lysine residue inserts into the hydrophobic tunnel in both structures despite the presence of three other lysine residues in the deacetylated peptide. This suggests that the sequence of the residues immediately N- and C- terminal to the target lysine residue do play some role in governing which lysine residue enters the hydrophobic tunnel [48].
4. N- and C-terminal regions
Outside of the catalytic core, sirtuin proteins have variable N- and C- terminal regions. These regions are not conserved among the protein family, and they vary in length, sequence and secondary structure. Most of the sirtuin proteins for which structures have been determined do not include these additional N- and C-terminal extensions in the structure. The few structures that do include the N- and C-terminal regions help to elucidate their roles in sirtuin function. The structure of the full length yeast Hst2 protein has been determined revealing a homotrimer in the crystal lattice as well as in solution [39]. The seven residue N-terminal region of the Hst2 protein binds in the active site cleft of a symmetry-related molecule in the crystal structure, forming the β staple interaction characteristic of sirtuin proteins with bound peptide substrates [39]. Notably, Met1 binds in the hydrophobic acetyl-lysine tunnel making similar van der Waals interactions as acetyl-lysine with conserved protein residues while the surrounding residues bind in the active site cleft in a conformation similar to that of the histone H4 peptide [42]. The presence of the seven N-terminal residues are required for Hst2 trimer formation in solution [39], highlighting a role for this region in oligomerization state maintenance. Further, addition of acetyl-lysine containing peptide displaces N-terminus binding in the Hst2 active site cleft, thereby disrupting trimer formation. In this way, the N-terminal region of the Hst2 protein may be involved in regulating oligomerization state until the appropriate substrate binds and displaces the N-terminus of Hst2.
The C-terminal region of the Hst2 protein contains α helical secondary structure and makes extensive interactions with residues within the cleft between the large and small domains of the catalytic core region of the protein, partially overlapping the nicotinamide and ribose moiety binding sites (Figure 2A) [39]. Because the α helix of the C-terminus makes interactions with residues that also contact NAD+ in the NAD+ bound structure (A33, G34, Y37, T224, S225, V228, and Q115), and the C-terminal deletion mutant of Hst2 has a significantly lower Km for NAD+ than the wild type protein, the C-terminal region within the NAD+-binding site has been proposed to affect NAD+ binding through an autoregulatory mechanism [39].
5. Structural insights into sirtuin catalysis
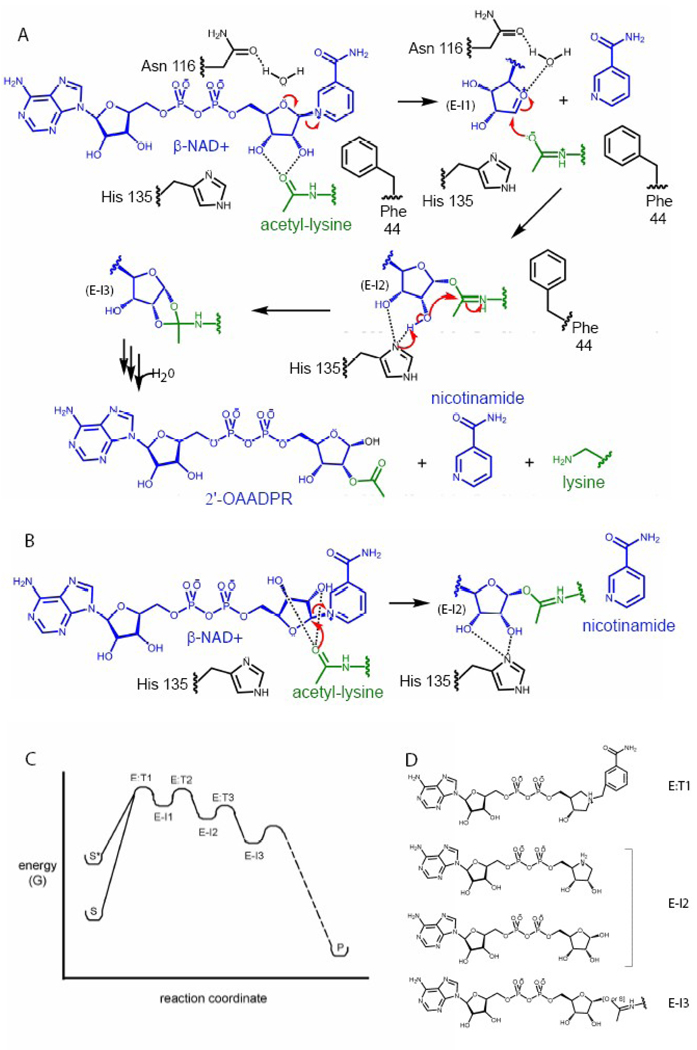
In order to obtain insights into sirtuin enzyme catalysis, several groups have determined structures of sirtuin homologues bound to proposed intermediates and transition state analogs. These structures have supported a catalytic mechanism for several steps of the sirtuin reaction (Figure 7A & B). The proposed reaction coordinate for sirtuin catalyzed acetyl transfer involves three enzyme/intermediate and three enzyme/transition state complexes (Figure 7C). The chemical structures of the intermediate and transition state analogs that have been crystallized in complex with sirtuin proteins are shown in Figure 7D. The Sir2 reaction begins when NAD+ binds to the enzyme in the productive conformation (probably induced by acetyl-lysine binding), destabilizing the ground state of the NAD+ molecule (Figure 7C, S*). It has been a matter of debate whether the glycosidic bond is broken via an SN1- or SN2-type mechanism [49], and several structural observations support one or the other mechanism. For example, the structure of Sir2Tm bound to an acetyl-lysine containing peptide and DADMe-NAD+ (Figure 7D), a moleculethat mimics the dissociative transition state of glycosidic bond cleavage (Figure 7A & C, E:T1), has been determined [43]. In the DADMe-NAD+/acetyl-lysine complex, the DADMe-NAD+ molecule binds in the NAD+ binding site in a conformation similar to that observed in the NAD+/acetyl-lysine complex of the same homologue [45]. The adenosine, phosphate and nicotinamide groups of the DADMe-NAD+ molecule superimpose with the same groups in the NAD+ structure, and the ribose group binds in a similar conformation with C1’ of the ring positioned proximal to the acetyl-lysine, except that it is closer to the amide oxygen. This movement of the C1’ of the ribose ring toward the acetyl-lysine is consistent with a dissociative mechanism for glycoside bond cleavage, where the ribosyl ribocation migrates from the nicotinamide, gains chemical reactivity at the transition state, and completes the reaction coordinate by migrating to react with the fixed carbonyl oxygen [43]. A significant caveat to the interpretation of this complex structure is that the DADMe-NAD+ molecule contains an extra carbon between the ribose ring and the nicotinamide moiety not present in the proposed transition state, and this additional length may contribute to the movement of C1’. This structure, along with the structure of carba-NAD+ bound to Hst2 showing the acetyl carbonyl oxygen inappropriately positioned for nucleophilic attack [38], supports a dissociate SN1-type mechanism for the cleavage of the glycosidic bond between nicotinamide and the rest of the NAD+ molecule (Figure 7A).
Figure 7.
The Sir2 catalytic mechanism. A) The steps of the catalytic mechanism for which there is structural support. The glycosidic bond is broken in a dissociative SN1-type manner, forming the oxocarbenium ion intermediate with an accompanying conformational change of the nicotinamide ribose ring. The acetyl-lysine is then positioned to nucleophilically attack C1’ of the oxocarbenium intermediate to form the alkylamidate intermediate, again with a conformational change in the ribose ring. F44 (Hst2 numbering) also flips to expel nicotinamide from the C pocket. The histidine general base can then deprotonate the 2’-OH directly to form the cyclic acyldioxilane intermediate. Through several steps and the addition of one water molecule the reaction products are formed. B) Nicotinamide cleavage via an SN2-type mechanism. Reaction steps after the formation of the akylamidate intermediate (E–I2) are identical as described in A). C) The reaction coordinate for the known steps of the Sir2 catalytic mechanism. S* indicates the destabilized substrate. E–I1, E–I2, and E–I3 indicate the oxocarbenium, alkylamidate and cyclic intermediates, respectively. E:T1–3 indicate the transition states required to form those intermediates, respectively. Since the remaining steps are unknown, their progress along the reaction coordinate is represented by a dashed line. D) The chemical structures of the transition state and intermediate-like molecules crystallized in complex with Sir2 enzymes.
After glycosidic bond cleavage in a dissociative manner, a positively charged, highly reactive oxocarbenium ion intermediate is proposed to form (Figure 7C, E–I1). The structure of Hst2 in complex with an acetyl-lysine containing peptide and ADP-HPD, an positively charged oxocarbenium ion intermediate-like molecule, has also been determined [44]. The ADP-HPD intermediate binds almost identically to the previously described ADP-ribose complex [38]. The positively charged ribose ring mimic of ADP-HPD is bound in a largely hydrophobic pocket and makes one water-mediated hydrogen bond with the side chain of a conserved asparagine (116). The structure of ADP-HPD bound in the sirtuin enzyme active site was significant because an initial argument against a dissociative, SN1-type reaction mechanism was the belief that a charged intermediate could not bind in such a hydrophobic environment. The fact that the positively charged ADP-HPD molecule can indeed bind in this pocket and makes a stabilizing, water-mediated hydrogen bond with the asparagines residue supports a dissociative mechanism for cleavage of nicotinamide from NAD+. However, ADP-HPD binds less tightly to the enzyme active site than the substrate NAD+, suggesting that this ligand may not be a good mimic of a reaction intermediate or transition state. This may indicate that the reaction does not proceed via an oxocarbenium ion intermediate or simply that the ADP-HPD molecule differs from the true oxocarbenium ion intermediate in ways important for intermediate binding such as bond order or pucker conformation of the ribose-like ring.
The structure of of Sir2Tm bound to NAD+ and an acetyl-lysine containing p53 peptide represents a so-called “Michaelis complex” and supports an SN2-type mechanism for nicotinamide cleavage (Figure 7B) [45]. In this structure the nicotinamide-ribose ring binds in an orientation not seen in other NAD+ bound structures, that is the α-face of the ribose ring is oriented toward the acetyl-lysine carbonyl. This conformtion allows for the potential nucleophilic attack of C1’ by the carbonyl oxygen, supporting an SN2-type mechanism for nicotinamide cleavage (Figure 7B). However, the fact that the cognate reaction substrates have not turned over in the time course of crystallization (2hrs) coupled with the fact that this conformation of the ribose ring has not been seen in any other sirtuin/NAD+ complex suggests that a conformational change may be required for catalysis and these crystals may not represent a true Michaelis complex.
Following nicotinamide cleavage, the acetyl carbonyl oxygen of acetyl-lysine nucleophilically attacks the C1’ position to form the 1’-O-alkylamidate intermediate (Figure 7C, E–I2) [4, 50]. Sirtuin proteins can also de-thioacetylate thioacetyl-lysine peptides, albeit with reduced turnover most likely due to the increased carbon-sulfur bond length as compared to the carbon-oxygen bond length in acetyl-lysine [51]. Because the resulting S-alkylamidate intermediate is longer lived than the native O-alkylamidate, the S-alkylamidate intermediate was able to be trapped in complex with the Sir2Tm enzyme [43], providing the first structural evidence of the existence of this intermediate. Compared to the acetyl-lysine/NAD+ complex of the same homologue, little movement is required to form the alkylamidate intermediate. The acetyl group rotates by 30°, while the nicotinamide ribose plane tits 35°, resulting in the movement of the acetyl amide oxygen by 0.8 Å and the 2’-OH group by 1.8 Å [43]. In the S-alkylamidate complex structure, the general base (H135) is 2.5 Å from the 2’-OH and 3.5 Å from the 3’-OH, placing the 2’-OH 0.8 Å closer to the histidine than in the acetyl-lysine/NAD+ complex [43]. The proximity of the histidine to the 2’-OH suggests that the general base may deprotonate the 2’-OH directly and not through a proton shuttling mechanism involving the 3’-OH as had been previously proposed [42, 45], giving insight into a key step in the sirtuin catalytic mechanism (Figure 7A).
The nicotinamide ribose and acetyl-lysine moieties of the S-alkylamidate intermediate are also positioned by hydrogen bonds with conserved residues (V225, H135, and Q115). Several conserved phenylalanines (44, 67 and 184) serve to shield the S-alkylamidate intermediate from hydrolysis and possibly base exchange [19], the process by which nicotinamide reacts with a sirtuin reaction intermediate to reform the NAD+ and acetyl-lysine substrates (discussed below). Interestingly, the conserved phenylalanine (44) that is involved in the binding of the nicotinamide moiety of NAD+, seems to move upon cleavage of the glycosidic bond to a position that would sterically occlude nicotinamide bound to the C site. This conformational change is linked to the conformational change of the cofactor binding loop upon acetyl transfer. This movement highlights the role of this conserved phenylalanine for expulsion of nicotinamide, and shows the importance of the flexibility of the loop to correctly position this residue.
6. Product bound complexes
Structures of several sirtuin homologues have been determined with the product, O-acetyl-ADP-Ribose (OAADPR), bound to the enzyme active site. Each of these complexes have been formed by adding both substrates to the active enzymes before crystallization [41, 42, 45]. The conformation of the OAADPR is very similar to that of bound ADP-Ribose. In the Sir2-Af1 structure, the 2’-acetyl oxygen makes water mediated hydrogen bonds (with residues Y64 and R67) and the acetyl methyl group makes several van der Waals interactions (with F32, A48, W158, and F159A, Sir2-Af1 numbering) [41]. In the Hst2 structure, the position of the OAADPR is very similar to its position in the Sir2-Af1 structure, except that the carbonyl and methyl groups of the acetyl group are flipped relative to each other. In this position the acetyl oxygen makes a direct hydrogen bond with the backbone of a conserved valine (V182, Hst2 numbering), and the acetyl group makes van der Waals interactions with several conserved hydrophobic residues (F184, F67 and I117) [42].
The presence of 2’-OAADPR in the sirtuin active site after incubation of these enzymes with NAD+ and acetyl-lysine is strong support that 2’-OAADPR is indeed the catalytic product of the Sir2 reaction. The orientation of the bound OAADPR molecule shows that the majority of the NAD+ molecule binds statically throughout the catalytic reaction coordinate, with the exception of the nicotinamide ribose moiety, which appears to undergo conformational changes upon nicotinamide cleavage and more modest changes upon acetyl transfer. The sirtuin enzyme itself mimics this pattern, as most of the enzyme structure does not change depending on which substrate, intermediate-like molecule, or product is bound, with the exception of the cofactor binding loop. The cofactor binding loop becomes more ordered upon substrate binding and again changes to a more closed conformation upon acetyl transfer. These conformational changes support the two step catalytic mechanism that has been proposed for sirtuin enzymes [52].
Several structures of sirtuin enzymes bound to deacetylated lysine-containing peptides show that the presence of the acetyl group makes little conformational difference for the lysine side chain, the peptide and the enzyme [45, 48]. The relatively minor differences between the acetyl-lysine substrate and lysine product structures indicates that, while the acetyl group may play a role in positioning the NAD+ substrate for catalysis, the conformation of the lysine side chain does not seem to have much involvement in the rest of the reaction coordinate.
7. Inhibitor bound complexes
7.1 Inhibitory nicotinamide binding
Nicotinamide is a reaction product and a non-competitive inhibitor of sirtuin proteins [2, 16]. The catalytic mechanism by which Sirtuins couple NAD+ cleavage to deacetylation and the mechanism of nicotinamide inhibition have important implications for Sir2 regulation by the physiological regulators NAD+ and nicotinamide, and for development of synthetic regulators of Sirtuins. Nicotinamide exerts its inhibitory effect on deacetylation by reacting with a reaction intermediate to reform β-NAD+ at the expense of deacetylation [18, 19]. The inhibitory nicotinamide molecule may react with either the oxocarbenium or the α-1’-O-alkylamidate intermediate to reform substrate. The identity of the binding site of the inhibitory nicotinamide molecule has implications for the development of rational activators of Sirtuins that exert their effect through relief of nicotinamide inhibition. Several potential binding sites for the inhibitory nicotinamide molecule have been proposed, including the C site (the binding site of the nicotinamide moiety of NAD+) [37] or another conserved pocket in the enzyme active site, termed the D site (Figure 5A) [38].
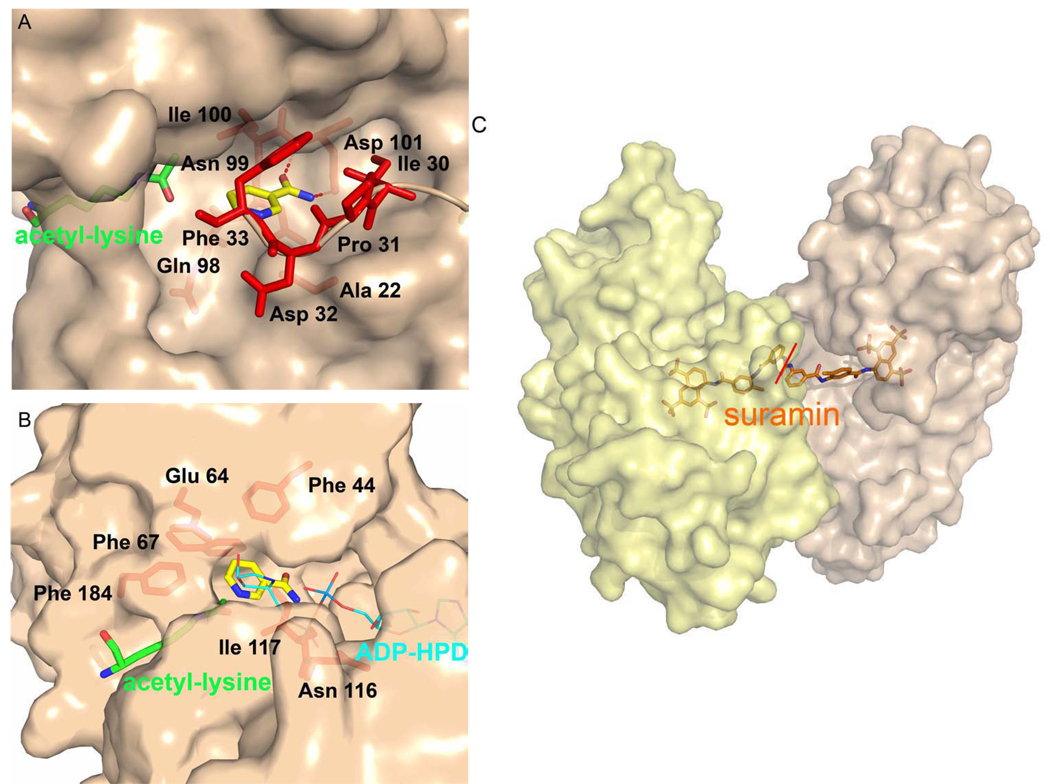
Structures of Sir2-Af2 bound to NAD+ and Sir2Tm bound to acetyl-lysine in the presence of high concentrations of nicotinamide reveal nicotinamide bound in the highly conserved C pocket [46] (Figure 5A). These structures show that nicotinamide can bind in several conformations within the C pocket, anchored by the carboxyamide group (Figure 8A). The carboxyamide amino of nicotinamide hydrogen bonds with the side chain of a conserved aspartic acid (D101) residue in the C pocket, while the carboxyamide oxygen hydrogen bonds with the backbone amino group of a conserved isoleucine (I100) and makes van der Waals interactions with several residues (N99 and I100, Sir2-Af2 numbering) [46]. The pyridine ring of nicotinamide makes more variable interactions with conserved residues (A22, I30, F33 and I100, Sir2-Af2 numbering) within the pocket, as it adopts different conformations in the structures and has correspondingly weaker electron density than the carboxyamide moiety (Figure 8A). The conformation of the cofactor binding loop also affects the interactions between the pyridine ring and residues within the C pocket. There are differences in binding between the nicotinamide moiety of NAD+ and the free nicotinamide molecule in the C site. As part of an NAD+ molecule, the carboxyamide moiety rotates approximately 150° from its most favored conformation (planar with the nicotinamide ring), which is seen in the free nicotinamide structure. Also, in the free nicotinamide structure, the pyridine ring may be flipped about the carboxyamide relative to its position in the NAD+ structure, allowing N1 of the pyridine ring to form favorable interactions with the backbones of hydrophobic residues (F44 and P42, Hst2 numbering) [46]. Although noncompetitive inhibitors like nicotinamide are expected to bind to the enzyme/substrate complex, the C site is known to bind the nicotinamide moiety of the substrate NAD+. Therefore, occupation of this binding pocket in the binary Sir2-Af2/(non-productive)NAD+ and Sir2Tm/acetyl-lysine complexes may be due to the high concentration of nicotinamide in the crystal condition. It is therefore unclear whether nicotinamide binding in this pocket is the only physiologically relevant binding site for an inhibitory nicotinamide molecule.
Figure 8.
The inhibitor binding sites. Sirtuin enzymes are shown in tan surface representation. Acetyl-lysine residues are shown in green cpk stick representation, the intermediate-like molecule, ADP-HPD, is shown in cyan cpk line representation, the nicotinamide molecules are shown in yellow cpk stick representation, and the suramin molecule is shown in orange cpk stick representation. Residues involved in van der Waals and hydrogen bonding (red dashes) interactions with nicotinamide are also shown in red stick representation. A) T. maritima Sir2Tm with nicotinamide bound in the C pocket. The cofactor binding loop is shown in cartoon instead of surface representation for clarity. B) S. cerevisiae Hst2 with nicotinamide bound in the D pocket. C) H. sapiens SIRT5 dimer with bound suramin. The red line indicates the 2-fold symmetry of the suramin molecule.
The structure of Hst2 bound to carba-NAD+ and an acetyl-lysine containing peptide reveals that the acetyl group of the acetyl-lysine substrate hydrogen bonds to the 2’ and 3’ hydroxyl groups of the cyclopentane ring, presumably to help position the nicotinamide group in the highly conserved C pocket for hydrolysis, leaving the acetyl group inappropriately positioned for nucleophilic attack of the 1’ carbon. A comparison of this structure with a ternary complex in which ADP-ribose replaces carba-NAD+ reveals that the ribose ring is rotated by about 90° relative to its corresponding position in the carba-NAD+ structure with the 1’-hydroxyl group of the ADP-ribose ring pointing into another highly conserved, hydrophobic D pocket that could accommodate an incoming nicotinamide group for a base exchange reaction to reform β-NAD+ [38]. The Hst2/acetyl-lysine/ADP-HPD (Figure 7D) complex crystals were soaked with nicotinamide in order to trap a free inhibitory nicotinamide molecule bound to a sirtuin enzyme in complex with an oxocarbenium ion intermediate-like molecule (ADP-HPD). The structure revealed the presence of one nicotinamide molecule bound to the D pocket, adjacent to the β face of the ADP-HPD molecule (Figure 8B) [44]. The D pocket is formed by several residues around the pyridine ring (E64, F67, N116 and F184) and proximal to the carboxyamide moiety (F44 and F117). The density for the carboxyamide is better defined than for the pyridine ring, suggesting greater flexibility of the pyridine ring. Both the carboxyamide nitrogen and oxygen make water mediated hydrogen bonds with the backbone of a highly conserved isoleucine residue (117), while the carboxyamide nitrogen also makes water mediated hydrogen bonds with the side chain of a conserved asparagine (116) and a phosphate oxygen. The pyridine ring makes van der Waals interactions with two phenylalanine residues (F44 and F67) (Figure 8B). Nicotinamide binding in this conformation is consistent with exclusive reformation of β-NAD+ upon base exchange and the noncompetitive nature of nicotinamide inhibition. Also, a nicotinamide molecule in the D pocket can be accommodated in the S-alkylamidate structure, while a nicotinamide molecule in the C pocket cannot because of the occupancy of the cofactor binding loop, specifically a conserved phenylalanine residue (44), in this pocket. Since this intermediate may be the one involved in base exchange [50], it is a requirement that nicotinamide would bind simultaneously to this complex. Taken together, these results seem to support the D pocket as a secondary nicotinamide binding site involved in nicotinamide inhibition.
7.2 Synthetic inhibitor complex
There has also been a reported structure of a human sirtuin homologue, SIRT5, in complex with a synthetic sirtuin inhibitor, suramin [35] (Figure 8C). This structure reveals the binding mode of the inhibitor and helps explain the inhibition mechanism. Monomeric SIRT5 dimerizes in solution upon addition of suramin, and in the crystal structure one suramin molecule links two monomers of SIRT5. The buried surface area of the dimer is very limited and the interface between the two monomers is mediated by only non-polar interactions (Figure 8C). The conformation of the cofactor binding loop of SIRT5 changes and becomes more ordered in the suramin structure relative to the SIRT5/ADP-ribose complex structure due to the presence of part of the suramin molecule in the C site. Further, in the suramin bound structure of SIRT5, structural elements in the A site become disordered because this pocket is not occupied. One sulphonyl and the naphthyl groups of suramin bind in the C and B NAD+ binding sites, respectively. Several of the benzene rings of suramin bind in the peptide, but not the acetyl-lysine, binding site. This is consistent with suramin acting as a competitive inhibitor of both substrates. The sulphonyl groups make several hydrogen bonds within the enzyme active site, while many residues in the catalytic cleft make van der Waals interactions with nonpolar groups of the suramin molecule [35]. Since suramin is a large, flexible, chemically multifunctional molecule and is not specific for sirtuin enzymes, suramin itself may not be an attractive sirtuin inhibitor candidate for further development. Nonetheless, the structure of suramin bound in the SIRT5 NAD+ and acetyl-lysine binding site does suggest that the trisulfonyl naphthyl group and adjacent moieties of the suramin molecule may serve as minimal inhibitor moieties that may be used as lead scaffolds for developing new sirtuin inhibitors with increased specificity for this class of enzymes.
8. Conclusions and future prospects
The compilation of all known sirtuin structures allows several observations to be made about structural features that are important for substrate specificity and binding, enzyme catalysis and sirtuin inhibition. The sirtuin catalytic core is composed of a small Zn2+-binding domain, a larger Rossmann-fold domain and four loops that form the catalytic cleft. Both substrate molecules bind in a hydrophobic tunnel of the cleft. Several intermediates and transition states along the reaction mechanism are structurally supported, however, the exact catalytic mechanism for many of the steps that ultimately result in the formation of the 2’-OAADPR reaction product are structurally and biochemically unknown.
Structural diversity in the small domain, particularly in the non-Zn2+ binding module, suggests that this region of the enzyme plays a role in substrate discrimination, and may also play a role in protein localization, which appears to be the case for the sirtuins that are located in the mitochondria. In support of this, the position of the small domain relative to the large domain also appears to be varied among sirtuin homologues and is dependent upon substrate/intermediate/product binding. These conformational changes may be important for abrogating protein-protein interactions after initial substrate recognition and catalysis. Other factors also likely play a role in substrate specificity and mediation of protein-protein interactions. Importantly, acetyl-lysine substrate specificity appears to be mediated by sites distal to the enzyme active site, although the residue immediately N-terminal and the residue 2 positions C-terminal to acetyl-lysine may play a role in governing sequence specific interactions in the Sir2Tm homologue.
Highly conserved residues of the catalytic core domain make up the cofactor binding loop, the most dynamic region of the enzyme. The conformation of this loop seems to be dependent upon the substrate/intermediate/product bound in the NAD+ binding site, implicating it to play an important role in NAD+ binding, acetyl-lysine transfer and nicotinamide release. Conversely, the conformation of the nicotinamide ribose is the most structurally variable among all substrate/intermediate/product moieties. The position of the group along the reaction coordinate suggests both a mechanism with dissociative character for glycosidic bond cleavage and a base exchange mechanism with an oxocarbenium or alkylamidate intermediate for nicotinamide inhibition. Also in support of this catalytic mechanism, the simultaneous binding of the acetyl-lysine and NAD+ molecules serve to position the nicotinamide moiety of NAD+ in the C pocket, thereby destabilizing the ground state of NAD+ and promoting dissociative cleavage of the glycosidic bond. Interestingly, sirtuin enzymes also appear to contain a secondary binding site (D pocket) for an inhibitory nicotinamide molecule distinct from the binding site of the nicotinamide moiety of NAD+ that likely acts as an inhibitory base exchange site for nicotinamide binding.
There are several functional aspects of sirtuin enzymes that remain unresolved that could be addressed with additional structural information. With regard to the catalytic mechanism, the nature of the proposed 1’-O-alkylamidate reaction state intermediate and the precise mode of nicotinamide capture for base exchange could be resolved by cocrystallizing an appropriate 1’-O-alkylamidate reaction state mimic in the presence of nicotinamide. Several sirtuins such as Sir2 from Plasmodium Falciparum have been proposed to catalyze ADP ribosylation [53] in addition to deacetylation, presumably going through a similar 1’-O-alkylamidate intermediate and structure determinations of some of these sirtuin enzymes might provide additional key information on the catalytic mechanism.
In addition to suramin, several other sirtuin effectors have also been identified, yet their molecular basis for inhibition has not been established due to the absence of additional sirtuin/effector structures. Structures of such sirtuin/effector complexes would not only provide molecular details underlying sirtuin inhibition and/or activation but might provide important information for the structure-based design of small molecule sirtuin modulators with therapeutic potential.
We have only begun to scratch the surface with regards to how the sirtuin segments N- and C- terminal to the catalytic core domain affect sirtuin biology. In the case of yeast Hst2, these regions appear to autoregulate sirtuin catalytic activity [39]. Given the large number of sirtuins in eukaryotic species and the high degree of conservation with the catalytic core domain, it is tempting to speculate that these N- and C- terminal domains of other sirtuins contribute in a significant way to sirtuin substrate specificity. Indeed, the fact that structures of peptide complexes bound to sirtuin catalytic core domains have provided relatively little information on this front suggests that the N- and C-terminal segments might play a more significant role in substrate selectivity. Therefore, structures of full-length sirtuin proteins might be useful for this purpose. Structures of sirtuin proteins bound to full-length protein substrates as opposed to peptide segments may also provide important information in this regard. Related to this issue, there is emerging evidence that sirtuin function may be modulated by other protein binding partners, such as the AROS-mediated activation of SIRT1 for p53 deacetylation [54]. Therefore, structures of relevant sirtuin/protein complexes might be particularly insightful for understanding how this fascinating family of enzymes mediates such a wide array of biological processes. Of course, structure without biochemistry and biology is just a pretty picture so such structural information will have to be appropriately complemented with functional studies to uncover the true mysteries of sirtuin enzymes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 2.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. Proc Natl Acad Sci U S A. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson MD, Denu JM. J Biol Chem. 2002;277:18535–18544. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- 4.Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL. Biochemistry. 2001;40:15456–15463. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- 5.Kim S, Benguria A, Lai CY, Jazwinski SM. Mol Biol Cell. 1999;10:3125–3136. doi: 10.1091/mbc.10.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaeberlein M, McVey M, Guarente L. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin SJ, Defossez PA, Guarente L. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 8.Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 9.Tissenbaum HA, Guarente L. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 10.Rogina B, Helfand SL. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frye RA. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 12.Kim EJ, Um SJ. BMB Rep. 2008;41:751–756. doi: 10.5483/bmbrep.2008.41.11.751. [DOI] [PubMed] [Google Scholar]
- 13.Guarente L. Cold Spring Harb Symp Quant Biol. 2007;72:483–488. doi: 10.1101/sqb.2007.72.024. [DOI] [PubMed] [Google Scholar]
- 14.Bell SD, Botting CH, Wardleworth BN, Jackson SP, White MF. Science. 2002;296:148–151. doi: 10.1126/science.1070506. [DOI] [PubMed] [Google Scholar]
- 15.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 16.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt MT, Smith BC, Jackson MD, Denu JM. J Biol Chem. 2004;279:40122–40129. doi: 10.1074/jbc.M407484200. [DOI] [PubMed] [Google Scholar]
- 18.Jackson MD, Schmidt MT, Oppenheimer NJ, Denu JM. J Biol Chem. 2003;278:50985–50998. doi: 10.1074/jbc.M306552200. [DOI] [PubMed] [Google Scholar]
- 19.Sauve AA, Schramm VL. Biochemistry. 2003;42:9249–9256. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- 20.Grozinger CM, Chao ED, Blackwell HE, Moazed D, Schreiber SL. J Biol Chem. 2001;276:38837–38843. doi: 10.1074/jbc.M106779200. [DOI] [PubMed] [Google Scholar]
- 21.Bedalov A, Gatbonton T, Irvine WP, Gottschling DE, Simon JA. Proc Natl Acad Sci U S A. 2001;98:15113–15118. doi: 10.1073/pnas.261574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, Goehle S, Kollipara R, Depinho RA, Gu Y, Simon JA, Bedalov A. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 23.Lain S, Hollick JJ, Campbell J, Staples OD, Higgins M, Aoubala M, McCarthy A, Appleyard V, Murray KE, Baker L, Thompson A, Mathers J, Holland SJ, Stark MJ, Pass G, Woods J, Lane DP, Westwood NJ. Cancer Cell. 2008;13:454–463. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapp J, Jochum A, Meier R, Saunders L, Marshall B, Kunick C, Verdin E, Goekjian P, Sippl W, Jung M. J Med Chem. 2006;49:7307–7316. doi: 10.1021/jm060118b. [DOI] [PubMed] [Google Scholar]
- 25.Chakrabarty SP, Saikumari YK, Bopanna MP, Balaram H. Mol Biochem Parasitol. 2008;158:139–151. doi: 10.1016/j.molbiopara.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 27.Napper AD, Hixon J, McDonagh T, Keavey K, Pons JF, Barker J, Yau WT, Amouzegh P, Flegg A, Hamelin E, Thomas RJ, Kates M, Jones S, Navia MA, Saunders JO, DiStefano PS, Curtis R. J Med Chem. 2005;48:8045–8054. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- 28.Frye RA. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 29.Min J, Landry J, Sternglanz R, Xu RM. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- 30.Finnin MS, Donigian JR, Pavletich NP. Nat Struct Biol. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- 31.Prasad GS, Sridhar V, Yamaguchi M, Hatefi Y, Stout CD. Nat Struct Biol. 1999;6:1126–1131. doi: 10.1038/70067. [DOI] [PubMed] [Google Scholar]
- 32.Qian X, Jeon C, Yoon H, Agarwal K, Weiss MA. Nature. 1993;365:277–279. doi: 10.1038/365277a0. [DOI] [PubMed] [Google Scholar]
- 33.Zhu W, Zeng Q, Colangelo CM, Lewis M, Summers MF, Scott RA. Nat Struct Biol. 1996;3:122–124. doi: 10.1038/nsb0296-122. [DOI] [PubMed] [Google Scholar]
- 34.Wang B, Jones DN, Kaine BP, Weiss MA. Structure. 1998;6:555–569. doi: 10.1016/s0969-2126(98)00058-6. [DOI] [PubMed] [Google Scholar]
- 35.Schuetz A, Min J, Antoshenko T, Wang CL, Allali-Hassani A, Dong A, Loppnau P, Vedadi M, Bochkarev A, Sternglanz R, Plotnikov AN. Structure. 2007;15:377–389. doi: 10.1016/j.str.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Zhao K, Chai X, Marmorstein R. J Mol Biol. 2004;337:731–741. doi: 10.1016/j.jmb.2004.01.060. [DOI] [PubMed] [Google Scholar]
- 37.Avalos JL, Boeke JD, Wolberger C. Mol Cell. 2004;13:639–648. doi: 10.1016/s1097-2765(04)00082-6. [DOI] [PubMed] [Google Scholar]
- 38.Zhao K, Harshaw R, Chai X, Marmorstein R. Proc Natl Acad Sci U S A. 2004;101:8563–8568. doi: 10.1073/pnas.0401057101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao K, Chai X, Clements A, Marmorstein R. Nat Struct Biol. 2003;10:864–871. doi: 10.1038/nsb978. [DOI] [PubMed] [Google Scholar]
- 40.Avalos JL, Celic I, Muhammad S, Cosgrove MS, Boeke JD, Wolberger C. Mol Cell. 2002;10:523–535. doi: 10.1016/s1097-2765(02)00628-7. [DOI] [PubMed] [Google Scholar]
- 41.Chang JH, Kim HC, Hwang KY, Lee JW, Jackson SP, Bell SD, Cho Y. J Biol Chem. 2002;277:34489–34498. doi: 10.1074/jbc.M205460200. [DOI] [PubMed] [Google Scholar]
- 42.Zhao K, Chai X, Marmorstein R. Structure. 2003;11:1403–1411. doi: 10.1016/j.str.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 43.Hawse WF, Hoff KG, Fatkins DG, Daines A, Zubkova OV, Schramm VL, Zheng W, Wolberger C. Structure. 2008;16:1368–1377. doi: 10.1016/j.str.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanders BD, Zhao K, Slama JT, Marmorstein R. Mol Cell. 2007;25:463–472. doi: 10.1016/j.molcel.2006.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoff KG, Avalos JL, Sens K, Wolberger C. Structure. 2006;14:1231–1240. doi: 10.1016/j.str.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 46.Avalos JL, Bever KM, Wolberger C. Mol Cell. 2005;17:855–868. doi: 10.1016/j.molcel.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 47.Rossmann MG, Liljas A, Branden CI, Banaszak LJ. Evolutionary ans structural relationships among dehydrogenases. 3rd edition ed. Vol. XI. New York: Academic Press; 1975. [Google Scholar]
- 48.Cosgrove MS, Bever K, Avalos JL, Muhammad S, Zhang X, Wolberger C. Biochemistry. 2006;45:7511–7521. doi: 10.1021/bi0526332. [DOI] [PubMed] [Google Scholar]
- 49.Smith BC, Denu JM. J Am Chem Soc. 2007;129:5802–5803. doi: 10.1021/ja070162w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith BC, Denu JM. Biochemistry. 2006;45:272–282. doi: 10.1021/bi052014t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith BC, Denu JM. Biochemistry. 2007;46:14478–14486. doi: 10.1021/bi7013294. [DOI] [PubMed] [Google Scholar]
- 52.Borra MT, Langer MR, Slama JT, Denu JM. Biochemistry. 2004;43:9877–9887. doi: 10.1021/bi049592e. [DOI] [PubMed] [Google Scholar]
- 53.Merrick CJ, Duraisingh MT. Eukaryot Cell. 2007;6:2081–2091. doi: 10.1128/EC.00114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim EJ, Kho JH, Kang MR, Um SJ. Mol Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]