Summary
At the onset of sporulation in Bacillus subtilis, two potential division sites are assembled at each pole, one of which will be used to synthesize the asymmetrically positioned sporulation septum. Using the vital stain FM 4-64 to label the plasma membrane of living cells, we examined the fate of these potential division sites in wild-type cells and found that, immediately after the formation of the sporulation septum, a partial septum was frequently synthesized within the mother cell at the second potential division site. Using time-lapse deconvolution microscopy, we were able to watch these partial septa first appear and then disappear during sporulation. Septal dissolution was dependent on σE activity and was partially inhibited in mutants lacking the σE-controlled proteins SpoIID, SpoIIM and SpoIIP, which may play a role in mediating the degradation of septal peptidoglycan. Our results support a model in which σE inhibits division at the second potential division site by two distinct mechanisms: inhibition of septal biogenesis and the degradation of partial septa formed before σE activation.
Introduction
Early in the Bacillus subtilis and Escherichia coli cell cycles, a complex of cell division proteins assemble into a ring-like structure at the future site of septation (recently reviewed by Bouche and Pichoff, 1998; Nanninga, 1998). This complex includes the conserved proteins FtsZ and FtsA (Bi and Lutkenhaus, 1991; Addinall and Lutkenhaus, 1996; Levin and Losick, 1996; Ma et al., 1996), as well as enzymes required to synthesize septal peptidoglycan (Weiss et al., 1997; Nanninga, 1998; Wang and Lutkenhaus, 1998). These proteins remain at the cell midpoint until late in the cell cycle, when an unknown signal triggers the onset of septal biogenesis, and the ring constricts as the septum grows inward (Addinall et al., 1996; Bi and Lutkenhaus, 1991; Pogliano et al., 1997). As septation nears completion, future division sites are assembled in both nascent daughter cells, and the key cell division protein FtsZ begins polymerizing at these new sites (Sun and Margolin, 1998). While models for the order and dependence of assembly of cell division proteins are beginning to emerge (Addinall et al., 1997; Daniel et al., 1998; Wang and Lutkenhaus, 1998), very little is known about the mechanisms by which the onset of constriction is regulated.
In B. subtilis, the onset of sporulation is marked by the relocalization of the division site from the cell midpoint to near the cell pole, producing two daughter cells of differing size: the smaller forespore and the larger mother cell (Stragier and Losick, 1997). This alteration in the site of division is mediated by the assembly of two apparently identical complexes of cell division proteins, one near each pole of the early sporangium (Levin and Losick, 1996). After the bipolar assembly of cell division complexes, one is activated, synthesizing the asymmetrically positioned sporulation septum. The second potential division complex (which is located within the larger mother cell after formation of the sporulation septum) remains inactive and is ultimately disassembled. Despite their differential regulation, both potential division sites are capable of supporting cell division. Mutants that fail to activate the mother cell transcription factor σE produce disporic sporangia containing two forespores (Piggot and Coote, 1976; Illing and Errington, 1991; Setlow et al., 1991; Lewis et al., 1994). This observation suggests that σE controls the synthesis of a protein (or proteins) that inhibits septation at the second potential division site. However, this protein remains unidentified, and the mechanism by which σE regulates division remains unclear.
The σE transcription factor controls the biogenesis of several proteins required for the striking reorganization of the sporulation septum during engulfment (Illing and Errington, 1991; Smith and Youngman, 1993; Frandsen and Stragier, 1995; reviewed by Piggot et al., 1994; Stragier and Losick, 1997). During this phagocytosis-like event, the two adjoining cells produced by asymmetric division are converted into a sporangium in which one cell (the smaller forespore) lies within the cytoplasm of the other (the mother cell). An early step in engulfment is the removal of peptidoglycan from within the sporulation septum, starting from the middle of the septum and progressing towards the edges. Mutations in three genes transcribed by σE (spoIID, spoIIM and spoIIP) inhibit this step, retaining peptidoglycan near the edges of the septum and blocking engulfment (Lopez-Diaz et al., 1986; Smith et al., 1993; Piggot et al., 1994; Frandsen and Stragier, 1995). The products of these genes are thought to mediate the hydrolysis of peptidoglycan within the septum, which may be a prerequisite for membrane migration. Consistent with this hypothesis, one of these proteins (SpoIID) is homologous to a family of proteins that regulate the activity of the cell wall hydrolase N-acetylmuramoyl-L-alanine amidase (Kuroda et al., 1992; Lazarevic et al., 1992), while the other two are membrane proteins lacking obvious homologues. After the removal of septal peptidoglycan, the mother cell membrane migrates around the forespore, apparently starting near the edge of the septum. These membranes fuse when they meet on the distal side of the forespore, producing a forespore bounded by two membranes: an inner membrane derived from the forespore cytoplasmic membrane and an outer membrane of opposite polarity derived from the mother cell. The mechanism by which the engulfing membranes move remains mysterious as, thus far, bacteria have no obvious motor proteins that may be involved in this process.
Seeking a method by which engulfment and cell division could be studied readily, we turned to the vital membrane stain N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl) pyridinium dibromide (FM 4-64; Molecular Probes), which has been used in Dictyostelium (Heuser and Clark, 1993), yeast (Vida and Emr, 1995), neurons (Miesenbock and Rothman, 1997) and E. coli (Sun and Margolin, 1998). We found that FM 4-64 was capable of revealing septal biogenesis as well as engulfment, and that neither growth nor sporulation was inhibited by its presence. When combined with optical sectioning, which allows the collection of images at different focal planes, and deconvolution microscopy, which corrects these digital images for the light scattering inherent in fluorescent microscopy, we were able to distinguish between complete and partial septa during both vegetative growth and sporulation. Using these methods, we describe an abortive cell division event during the early stages of B. subtilis sporulation.
Results
Use of FM 4-64 and deconvolution microscopy to visualize septal biogenesis during vegetative growth
FM 4-64 (Molecular Probes) is a cell-impermeant membrane stain thought to integrate into the outer leaflet of biological membranes (Haugland, 1996). To determine whether this stain was useful for the study of cell division during B. subtilis growth and sporulation, we first examined exponentially growing cultures of wild-type B. subtilis that had been grown in the presence of FM 4-64 and stained with 4,6-diaminidino-2-phenylindole (DAPI) immediately before visual examination. Optical sectioning and deconvolution microscopy were used to remove light scattered by the lens and originating from other focal planes (Hiraoka et al., 1990; Scalettar et al., 1996). Deconvolution significantly improves the effective resolution and contrast of fluorescent images; the improvement is especially dramatic for fluorescently labelled membranes, which are inherently difficult to resolve, because light from membranes in other focal planes obscures and decreases contrast within the image.
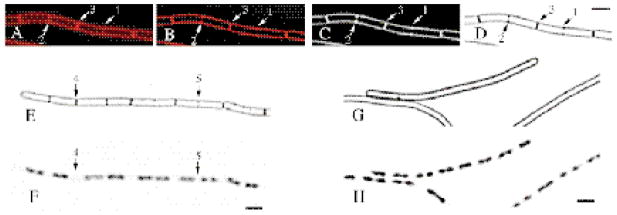
In Fig. 1, we show the effect of deconvolution on FM 4-64-stained, exponentially growing B. subtilis. Before deconvolution, the membranes appeared fuzzy, and a red haze filled the cytoplasm (Fig. 1A). After deconvolution, the FM 4-64-stained cytoplasmic membranes appeared as uniformly bright lines surrounding a nearly black cytoplasm (Fig. 1B). Deconvolution allowed identification of partial septa that were either poorly detected (Fig. 1A and B, arrow 1) or that appeared to traverse the bacterium completely (Fig. 1A and B, arrow 2) in the unprocessed image. In both cases, partial septa appeared as two dots of membrane staining oriented approximately perpendicular to the long axis of the bacterium (Fig. 1B, arrows 1 and 2), while complete septa stained more brightly and appeared to traverse the bacterium completely (Fig. 1B, arrow 3). The remaining images shown here are deconvolved medial sections presented as inverted black and white images (Fig. 1C and D). This converts the images into black membranes or DNA on a white background (Fig. 1D), thereby improving the ability of these images to be reproduced. Both complete and partial septa were normally observed only between two well-separated chromosomes (Fig. 1E and F, arrows 4 and 5). The formation of partial and complete septa was greatly reduced by depletion of the cell division protein FtsZ, demonstrating that an active division apparatus is required for their formation (Fig. 1G and H). Occasionally, single foci of increased membrane staining were also observed in wild-type cells (Fig. 1E) or after depletion of FtsZ (Fig. 1G). These could represent early stages of septal biogenesis, similar to the initial assembly of the cell division protein DivIB into foci at future division sites (Harry and Wake, 1997). However, because such membrane foci also appear in bacteria that are lysing (not shown), we do not consider them to be partial septa here; rather, we define the earliest partial septa as those with two spots of membrane staining across the long axis of the bacteria, and the latest partial septa as those whose midpoint stains less intensely than the cytoplasmic membrane (Experimental procedures).
Fig. 1.
FM 4-64 staining and deconvolution microscopy distinguish between partial and complete septa.
A–F. Exponentially growing wild type (PY79) stained with FM 4-64. A. The original medial section in colour.
B. After deconvolution, partial septa (arrows 1 and 2) are clearly distinguished from complete septa (arrow 3).
C. Conversion of image (B) to black and white; as in photographic images, fluorescence is shown in white on a black background.
D. Inversion of image (C) to black membranes on white.
E. Exponentially growing PY79 (LB, 37°C), stained with FM 4-64 and (F) DAPI to reveal DNA. Both complete (arrow 4) and partial (arrow 5) septa are located in a gap between well-separated chromosomes.
G. Strain KP444 after depletion of FtsZ as described in Experimental procedures and stained with FM 4-64 and (H) DAPI. The bar equals 2 μm.
Observation of septum formation and engulfment during sporulation
At the onset of sporulation, the cell division protein FtsZ assembles into rings at two potential division sites near each cell pole of the sporangium (Levin and Losick, 1996). One of these two potential division sites is used to synthesize the sporulation septum, giving rise to two daughter cells differing in size. The smaller of these two cells (the forespore) will become a spore, after two unusual events: the translocation of the forespore chromosome across the septum and the engulfment of the forespore by the larger mother cell.
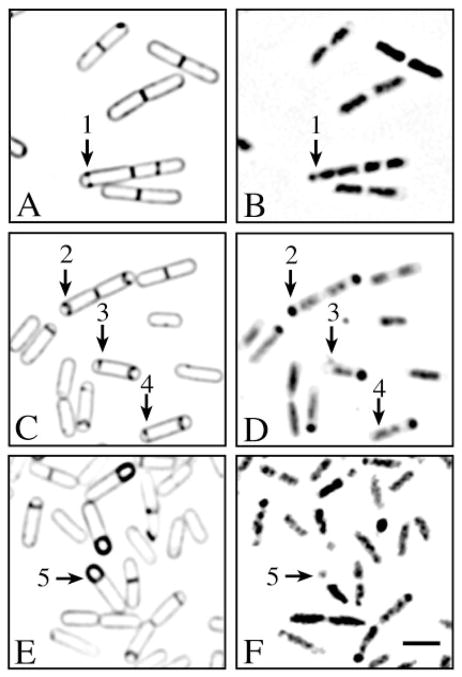
FM 4-64 was added to a sporulating culture either at the time of resuspension or 1 h before resuspension, with no detectable effect on either the rate or the efficiency of spore formation. One hour after the induction of sporulation (t = 1 h), partial septa were observed near the pole of some bacteria; these we infer to be partial sporulation septa (Fig. 2A, arrow 1). These partial septa were located between a small, condensed region of the chromosome and an elongated chromosome (Fig. 2B, arrow 1). Thirty minutes later (t = 1.5 h), complete sporulation septa were prevalent (Fig. 2C; arrow 2). About 9% of these sporangia contained a partial septum at the forespore-distal pole of the mother cell (Fig. 2C, arrows 3 and 4; Table 1), suggesting that the second potential division site had initiated septal biogenesis. Although there are two potential division sites marked by FtsZ rings in each sporangium initially (Levin and Losick, 1996), we rarely observed simultaneous partial septa at both sites, and more frequently observed sporangia with one complete and one partial septum. This suggests that, under the conditions used here, septal biogenesis initiates sequentially at these two sites. At the onset of engulfment, the septum began to curve smoothly around the forespore, without bulging notably into the mother cell (Fig. 2C, arrow 2). Sporangia that had completed engulfment, which were prevalent 30 min later (t = 2 h), lacked partial septa within the mother cell (Fig. 2E, arrow 5; Table 1), suggesting that either the partial septa had regressed or the sporangia containing them had lysed.
Fig. 2.
A time course of sporulation in the wild-type strain PY79. Sporulation was initiated by resuspension in the presence of FM 4-64. Membrane staining (A, C and E) and DNA staining are shown (B, D and F).
A and B. After 1 h of sporulation, partial sporulation septa were observed by FM 4-64 staining (arrow 1).
C and D. After 1.5 h of sporulation, complete sporulation septa were observed (arrow 2). Some sporangia had a second partial septum near the forespore-distal pole of the mother cell (arrows 3 and 4).
E and F. After 2 h of sporulation, engulfment was complete in many sporangia (arrow 5). As reported previously (Setlow et al., 1991), the DAPI staining of the forespore chromosome was reduced in many sporangia that had completed engulfment (arrow 5). This reduced staining intensity was reversed by fixation (not shown). The bar equals 2 μm.
Table 1.
Frequency of septation at the second potential division site in wild-type and spoIID sporangia.
| Percentage of sporangia with given phenotype at the specified time after onset of sporulation |
||||
|---|---|---|---|---|
|
t = 90 mina |
t = 180 minb |
|||
| Wild type | spoIID | Wild type | spoIID | |
| Only one polar septum (partial or complete) | 91 | 87 | 99.5 | 81 |
| Partially disporicc | 9 | 13 | 0.25 | 11 |
| Completely disporicd | 0 | 0 | 0.25 | 8 |
Some 22% of the cells in a wild-type culture (592 scored) and 15% of the cells in a spoIID mutant culture (445 scored) had entered sporulation, as judged by the presence of least one partial or complete sporulation septum.
Some 45% of the cells in a wild-type culture (875 scored) and 46% of the cells in the spoIID mutant culture (444 scored) had entered sporulation, judged as described above.
These sporangia contained a complete sporulation septum and an incomplete septum within the mother cell.
These sporangia contained a complete sporulation septum and an apparently complete septum within the mother cell. The second fore-spore usually contained an incompletely translocated chromosome.
While the septa were curving, the FM 4-64 fluorescence of the septal membranes remained approximately twice as intense as the single, cytoplasmic mother cell membrane, reflecting the presence of two membranes in this region: the forespore septal membrane and the mother cell septal membrane (see Experimental procedures). In the region of the sporangium, where engulfment was complete, the FM 4-64 staining was approximately threefold more intense than the mother cell cytoplasmic membrane. This is likely to reflect the presence of three membranes in this region: the forespore membrane, the mother cell-derived engulfing membrane and the mother cell cytoplasmic membrane. Thus, although the forespore-derived and mother cell-derived membranes surrounding the forespore are not always visualized separately by electron microscopy, the FM 4-64 staining intensity suggests that they remain as distinct membranes. Our data suggest that the increase in membrane area necessary to allow the mother cell membranes to wrap around the forespore is accomplished either by increased membrane biogenesis or by utilization of intracellular lipid pools, rather than by fusing the two septal membranes, which would result in a lower FM 4-64 staining intensity.
Observation of septal regression and engulfment using time-lapse photography
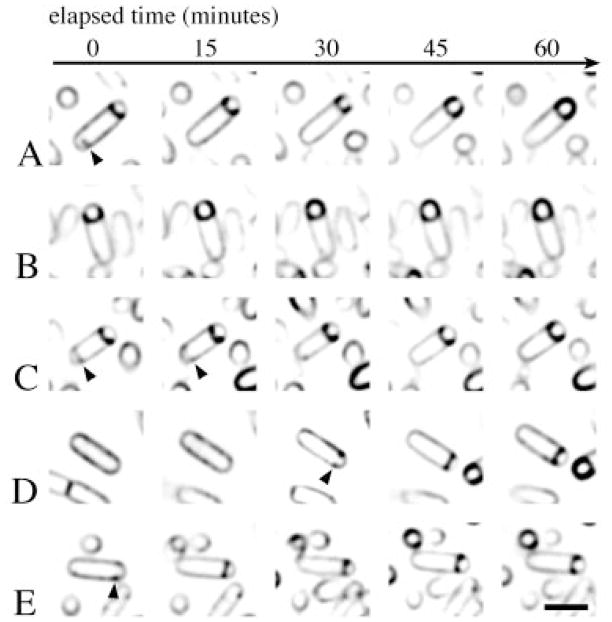
To determine whether the partial septa observed within the mother cell regressed as sporulation proceeded or if the sporangia containing them lysed, time-lapse microscopy was used to follow individual sporangia through polar septation and engulfment. About 1.5 h after the onset of sporulation in the presence of FM 4-64, the bacteria were adhered to a poly L-lysine-treated coverslip and placed on a heated chambered slide. Images were collected every 15 min for a total of 1 h. During this time, some sporangia completed engulfment (Fig. 3A–C), and others formed a sporulation septum (Fig. 3D and E). Approximately 35% of the sporangia with one complete sporulation septum contained a second, partial septum at one time point that was absent later in the time course (Fig. 3A and C, arrowheads). Such sporangia were able to complete engulfment and had not lysed by the end of the experiment. Thus, partial septa are able to be synthesized and degraded without compromising the completion of engulfment, and without causing cell lysis, at least for the duration of our experiment. While some of the partial septa at the forespore-distal end of the mother cell invaginated symmetrically (Fig. 3B), many appeared to be asymmetrical (Fig. 3A; see also Fig. 2C, arrows 3 and 4). This is in contrast to partial sporulation septa, which normally grew inwards more symmetrically (Fig. 3D and E; see also Fig. 2A, arrow 1). These results suggest that either the synthesis or the dissolution of the second partial septa occurs asymmetrically.
Fig. 3.
Observation of engulfment by time-lapse microscopy. Sporulation of the wild-type strain PY79 was induced by resuspension in the presence of FM 4-64 and affixed to a coverslip (Experimental procedures). The slide was heated to approximately 30°C, and five images were collected every 15 min (five columns, left to right). For ease of presentation, five individual sporangia (rows A–E) were selected from the field. Partial septa are visible within the mother cells of the sporangia shown in (A) and (C) (arrows). The bar equals 2 μm.
Kinetics of septation in wild-type sporangia and in sporangia lacking σE
In the absence of the mother cell-specific transcription factor σE, complete septa are synthesized at both potential division sites, producing ‘disporic’ sporangia with two forespores and a centrally positioned mother cell lacking a chromosome (Illing and Errington, 1991; Setlow et al., 1991; Lewis et al., 1994); see Fig.(4). Thus, σE (which becomes active only after the formation of the first sporulation septum) is required to prevent septation at the second potential division site within the mother cell. To understand the mechanism by which σE inhibits dispore formation further, we investigated the timing of division at the two potential division sites in a mutant (spoIIGB) that lacks σE.
Fig. 4.
A time course of sporulation in the absence of σE. Sporulation was initiated by resuspension in the presence of FM 4-64. Membrane staining (A, C and E) and DNA staining (B, D and F) are shown.
A and B. After 1.5 h of sporulation, disporic sporangia are visible (arrow 1); sometimes in sporangia in which neither chromosome has been fully translocated (arrow 1).
C and D. After 2 h of sporulation, dispores in which both chromosomes have been fully translocated are visible (arrow 2).
E and F. After 3 h of sporulation, some of the forespores appear to have collapsed or lysed (arrow 3), while others appear to be separating from the mother cell (not shown). The bar equals 2 μm.
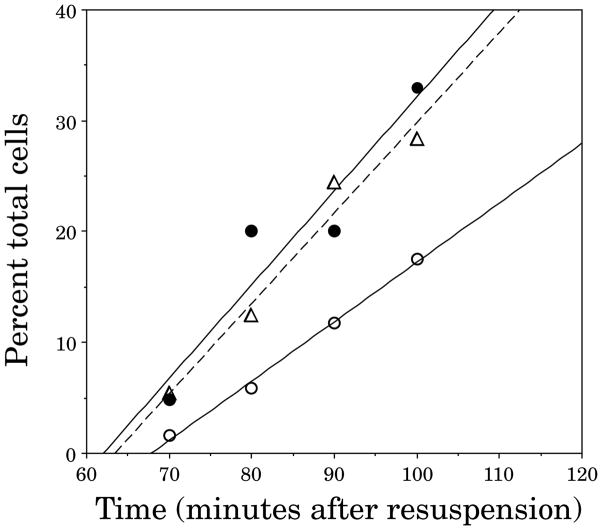
Sporulation was induced by resuspension (in the presence of FM 4-64), and images of the wild type and spoIIGB mutant cultures were collected every 10 min, deconvolved (Fig. 4) and analysed (Fig. 5). The rate of appearance of sporulation septa was approximately linear between 60 min and 100 min of sporulation, allowing extrapolation of the data to predict the time at which complete septa were first synthesized (Fig. 5). This analysis suggested that complete sporulation septa were first synthesized 63 min after resuspension in wild-type cultures and 62 min after resuspension in spoIIGB cultures, in good agreement with previous electron microscope studies, which suggested that complete septa first appear between 60 and 65 min (Partridge and Errington, 1993). As in the wild type, two partial septa were only rarely found in the same sporangium (less than 1 in 1000), suggesting that the two septa form sequentially in spoIIGB mutant sporangia. Disporic sporangia, in which the second sporulation septum appeared complete, were first observed 70 min after the onset of sporulation (Fig. 5), representing approximately 40% of all septated cells. Extrapolation of the data suggested that, in spoIIGB mutants, the second septum was complete approximately 68 min into sporulation (Fig. 5), 6 min after the completion of the first septum. After 3 h of sporulation, the forespores of some spoIIGB sporangia began to collapse (Fig. 4E and F, arrow 3), while others began to separate from the mother cell (not shown). By this time, 90% of the spoIIGB sporangia contained two polar septa, most of which contained fully translocated chromosomes in both forespores.
Fig. 5.
Kinetics of septation in wild-type and spoIIGB sporangia. Cultures of wild type (PY79) and a spoIIGB null mutant (KP161) were induced to sporulate by the resuspension method in the presence of FM 4-64 at 37°C. Beginning 1 h after the induction of sporulation, samples were taken every 10 min, stained with DAPI to visualize the chromosomes, and images collected and deconvolved. The percentage of total cells containing at least one or two complete septa at each time point was scored, plotted versus time, and a best-fit line drawn using linear regression analysis. The experiment was repeated twice with similar results; the data from one experiment are presented. Open triangles with a dashed line represent PY79 with one complete septa (at any stage of engulfment). Closed circles with a solid line represent KP161 with at least one complete septa, and open circles with a solid line represent KP161 with two complete septa. The number of cells scored at each time point is: 514 for PY79 and 311 for KP161 at t = 70 min; 121 for PY79 and 238 for KP161 at t = 80 min; 205 for PY79 and 969 for KP161 at t = 90 min; 458 for PY79 and 646 for KP161 at t = 100 min.
Three additional lines of evidence support the finding of rapid, sequential formation of septa in spoIIGB mutants. First, synthesis of the second septum appeared to commence before the completion of chromosome translocation across the first septum, because disporic sporangia in which neither chromosome was fully translocated were observed (Fig. 4A and B, arrow 1). From our data, we estimate that it takes between 10 and 15 min to translocate the chromosome across the completed sporulation septum. Hence, if formation of the second septum began more than 10 min after completion of the first, chromosome translocation across the first septum would normally be complete before the second septum was synthesized. Secondly, as septation is required for DNA translocation into the two forespores of disporic sporangia (Wu et al., 1995), the time between the appearance of sporangia with one and two completely translocated chromosomes reflects the timing between the two division events, assuming that the rate of DNA translocation is constant (as it appears to be; Lewis et al., 1994). We estimate that there is approximately 6 min between complete translocation of the first and second chromosomes, in good agreement with our estimate of the time between sequential septation based on FM 4-64 staining. Thirdly, many (25%) disporic cells are also detected by electron microscopy of spoIIGB mutants early (t = 2 h) in sporulation (Illing and Errington, 1991), supporting the idea that two complete septa are built in rapid succession.
spoIID, spoIIM and spoIIP mutants retain partial septa within the mother cell
When bacteria make septa, the cell wall grows inwards together with the invaginating membranes; thus, partial septa contain cell wall material that can be visualized in the electron microscope. Regression of partial septa is therefore likely to require hydrolysis of this peptidoglycan, which might otherwise prevent the membranes from retracting. The products of three genes in the σE regulon have been implicated in peptidoglycan hydrolysis, SpoIID, SpoIIM and SpoIIP (Piggot and Coote, 1976; Piggot et al., 1994). In wild-type sporangia, peptidoglycan is lost from the sporulation septum at the onset of engulfment, which could be necessary to make the septum more flexible so that it can wrap around the forespore. However, in sporangia lacking SpoIID, SpoIIM or SpoIIP, septal thinning is inhibited, and engulfment is blocked. As these proteins are good candidates for proteins involved in septal regression, we examined the phenotypes of null mutations in spoIID, spoIIM and spoIIP by FM 4-64 staining.
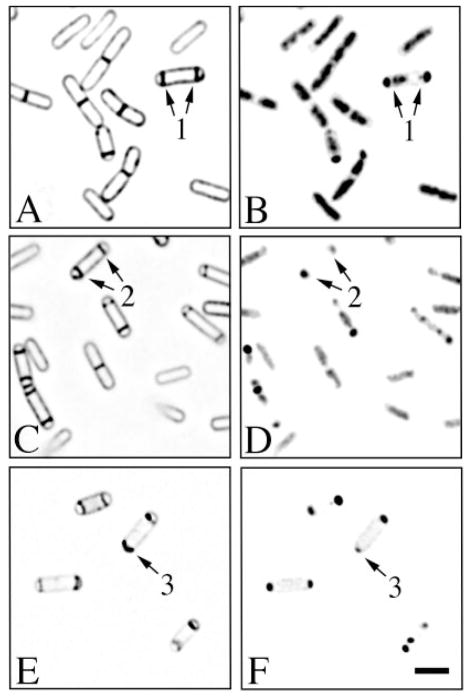
Mutants lacking either SpoIID, SpoIIM or SpoIIP had similar phenotypes. At early times of sporulation, partial septa formed at approximately the same frequency in spoIID mutant sporangia as in wild type (Fig. 6A, arrow 1; Table 1). However, many spoIID sporangia retained partial septa (Fig. 6C, arrow 2) or apparently complete septa (Fig. 6E, arrow 3) at the second division site within the mother cell until 2 and 3 h after the onset of sporulation. After 3 h of sporulation, 19% of the spoIID (444 total scored), 32% of the spoIIM (166 total scored) and 20% of the spoIIP (879 total scored) mutant sporangia had either partial or apparently complete septa within the mother cell, whereas in wild-type cultures, only 0.5% remained (875 total scored). FM 4-64 staining also readily revealed the bulging of the forespore into the mother cell (Fig. 6C, arrow 4) that has been observed previously in spoIID, spoIIM and spoIIP mutant sporangia by electron microscopy (Lopez-Diaz et al., 1986; Smith et al., 1993; Piggot et al., 1994; Frandsen and Stragier, 1995). These bulges of septal material sometimes contained portions of the forespore chromosome (Fig. 6D, arrow 4). Both partial septa (Fig. 6G) and complete septa (not shown) were readily observed by electron microscopy. The disporic sporangia formed in engulfment-defective mutants differ from those formed by spoIIGB mutants. In most spoIID sporangia, only one forespore contained a fully translocated chromosome, while the second contained either a portion of the chromosome or no chromosome (Fig. 6D and F, arrows 2 and 3); the remaining portion of the chromosome was within the mother cell. In spoIIGB sporangia, both forespores often contained a fully translocated chromosome, and the mother cell compartment often lacked DNA.
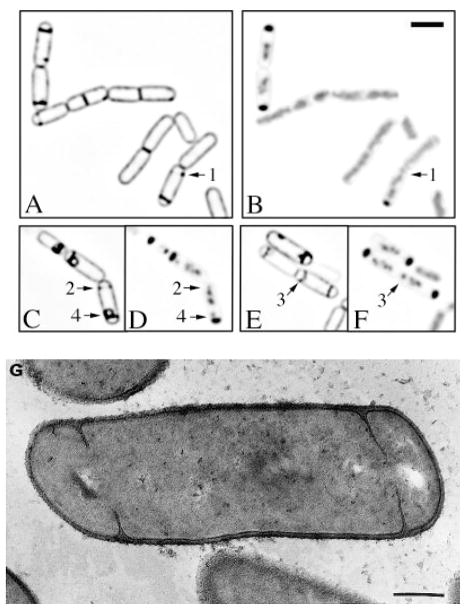
Fig. 6.
Partially disporic sporangia persist in spoIID mutant sporangia. Sporulation of the spoIID mutant strain KP522 was induced by resuspension in the presence of FM 4-64. Membrane staining (A, C and E) and DNA staining (B, D and F) are shown.
A and B. 1.5 h after the onset of sporulation, about 13% of the sporangia are partially disporic (arrow 1), compared with 9% of wild type.
C–F. 2 h after the onset of sporulation, sporangia with partial septa (C and D, arrow 2) or apparently complete septa (E and F, arrow 3) within the mother cell together comprise about 20% of the population. Many forespores have bulged into the mother cell (C and D, arrow 4). The bar equals 2 μm.
G. Electron micrograph of a spoIID mutant sporangium containing one partial septum (left side of sporangium) and a complete sporulation septum that has bulged into the mother cell (right side of sporangium). The bar is 300 nm.
Discussion
Here, we have used the membrane stain FM 4-64 to demonstrate that the two potential division sites assembled early in the spore formation pathway of B. subtilis are able to synthesize two septa rapidly and sequentially. In mutants lacking σE, which normally prevents septation at the second site, the two septa form just 6 min apart. Because septation takes several minutes and was never observed to occur simultaneously, the second division site must rapidly become active after the first septum is complete. These results suggest that, upon completion of the sporulation septum, a protein or diffusible signal molecule is released that triggers the onset of division at the second potential division site (Lewis et al., 1994; Fig. 7). A similar signal may trigger the onset of division during vegetative growth, during which cell division proteins assemble at the nascent division site well in advance of septal biogenesis (Addinall et al., 1996; Levin and Losick, 1996; Pogliano et al., 1997).
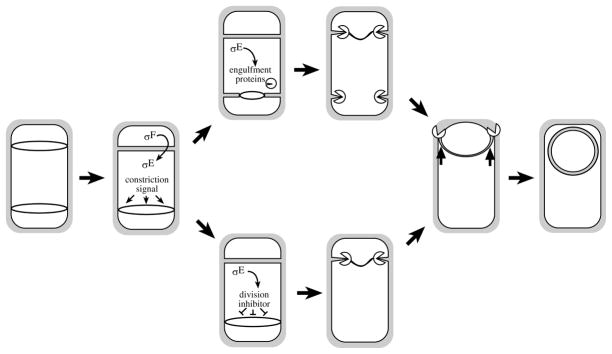
Fig. 7.
Model for cell cycle events during sporulation in B. subtilis. Entry into sporulation triggers a switch in the assembly of the cell division machinery (shown as rings) from a single point in the cell centre to sites near each cell pole (first cell). Completion of the sporulation septum at one of these two sites triggers two events: (i) activation of the second division complex for septal biogenesis; and (ii) activation of σF in the smaller cell (the forespore), which activates σE in the larger cell (the mother cell). Because both events occur shortly after the formation of the sporulation septum, there is a race between the σE-dependent inhibition of dispore formation and the cell division machinery. In some sporangia (bottom pathway), σE may become active in time to produce a putative inhibitor of division such that no visible constriction is seen. In other sporangia (top pathway), σE may become active after constriction has begun, and the signal to inhibit constriction may therefore arrive after the formation of partial septa. Dissolution of partial septa requires engulfment proteins (shown as ‘packmen’), which function to digest the cell wall from these septa, as well as from the sporulation septum.
The rapid and sequential onset of division at the two potential sites of septation suggests that the intercellular signal transduction cascade necessary for the activation of σE occurs extremely rapidly (as was also suggested by the studies of Partridge and Errington, 1993). Activation of σE is coupled to the activation of σF, which normally requires completion of the sporulation septum (recently reviewed by Stragier and Losick, 1997). We have found that, in the absence of σE, septation at the second site is complete approximately 6 min after the formation of the sporulation septum. Previous studies in which chromosome translocation was used as a measure of septation (Lewis et al., 1994) suggested that, in the absence of σE, the two division events were separated by 15–20 min. Our estimate of 6 min is strikingly shorter, suggesting that, in contrast to the proposal of Lewis et al. (1994), the window of opportunity for σE to repress the second cell division site is extremely narrow. We therefore suggest that the second potential division site at the forespore-distal pole of the mother cell initiates septal biogenesis immediately after completion of the sporulation septum and before the completion of chromosome translocation into the forespore.
Given the rapid initiation of division at the second division site in the absence of σE, it is not surprising to find that, in wild-type sporangia, division at this site initiates frequently, producing partially disporic sporangia. However, in wild-type bacteria, this second division event did not result in the formation of a complete septum. Rather, septal biogenesis stopped, and these partial septa disappeared rapidly (Fig. 7). Thus, in a wild-type sporulating culture, partially disporic sporangia are present transiently. The partial septa we observe are likely to be identical to the ‘cloisons’ visualized by Ryter (1965) in the electron microscope. These partial septa probably result from active septal biogenesis, rather than from the presence of an inactive complex of division proteins within the mother cell, because a high percentage of sporangia have bipolar division complexes early in sporulation (63% at t = 1.5; Levin and Losick, 1996), yet we never observed two partial septa in wild-type sporangia in numerous experiments representing more than 1000 sporangia. Thus, the partial septa we observe are likely to result from active septal biogenesis; our studies demonstrate that they disappear as engulfment commences.
Our studies do not address the proportion of sporangia that undergo this abortive division event, but two factors make it likely that the proportion is significantly higher than observed in our time-lapse experiments. First, we estimate that it takes less than 6 min to synthesize an apparently complete sporulation septum, starting from a cell lacking a visible constriction. Biogenesis of a partial septum would, therefore, be expected to take significantly less than 6 min. Secondly, when partial septa were observed in our time-lapse experiments, septal dissolution was often complete by the next time point, 15 min later. Thus, partial septa may be synthesized rapidly and lost, and many partial division events may have been missed in our time-lapse experiments. Unfortunately, collecting images more frequently in the hope of estimating the prevalence of these events better is difficult given the toxicity of fluorescence to biological specimens.
Single mutants lacking one of the mother cell-produced engulfment proteins (SpoIID, SpoIIM and SpoIIP) retain about 25% partially disporic sporangia at late times in sporulation. These proteins have been suggested to play a role in peptidoglycan hydrolysis as, in the absence of any one, peptidoglycan is retained in the sporulation septum, and engulfment is blocked (Piggot et al., 1994; Stragier and Losick, 1997). We suggest that these proteins mediate dissolution of partial septa by promoting the hydrolysis of peptidoglycan, which would be a necessary step in the regression of septal membranes. This hypothesis predicts that any mutant that retains peptidoglycan within the sporulation septum might produce partially disporic sporangia. Indeed, we find that spoIIB spoVG double mutants, which retain peptidoglycan within their sporulation septum (Margolis et al., 1993), produce partially disporic sporangia (not shown). Thus, one mechanism by which σE prevents dispore formation is by mediating dissolution of partial septa formed as a result of the close timing between formation of the sporulation septum and the onset of septation at the second potential division site.
Thus far, we have been unable to simulate a mutant lacking σE (100% disporic sporangia, complete translocation of the forespore chromosomes into both forespores) by constructing multiply mutant strains lacking the known engulfment proteins (although the spoIID, spoIIM, spolIP triple mutant strain produces about twice as many dispores as the single mutants). This may suggest that there are additional unidentified engulfment proteins (Londoño-Vallejo et al., 1997). Alternatively, in addition to promoting septal dissolution, σE may also directly inhibit septal biogenesis at the second potential division site by controlling the synthesis of proteins that inhibit the division machinery before or after septation has commenced. It remains to be determined whether septal regression or inhibition of septal biogenesis is primarily responsible for preventing dispore formation. However, in the race between activation of σE and the production of abortive disporic sporangia, a positive outcome may best be ensured by using both strategies.
Previously, septal biogenesis could only be visualized reproducibly in the electron microscope; experiments based on fluorescence microscopy lacked a direct measure of septation, which was therefore assessed indirectly. FM 4-64 staining can readily distinguish between partial and complete septa, is not dependent on specialized optics and can be used together with the DNA stain DAPI and also with proteins localized by either immunofluorescence microscopy (Hofmeister, 1998) or fusions to the green fluorescent protein (Sun and Margolin, 1998). Together with quantitative image analysis and deconvolution microscopy, fluorescent membrane stains such as FM 4-64 may be able to detect invaginations and vesicles smaller than the resolution of the light microscope, because the increased membrane contained in such structures will retain more FM 4-64 and be more brightly fluorescent. This stain will be useful in studies of B. subtilis sporulation, with its dramatic reorganization of the cell envelope, and also in studies of bacterial and Archael cell cycles.
Experimental procedures
Bacterial strains and genetic manipulations
The wild-type strain used in this study is PY79 (Youngman et al., 1984). The mutations were introduced into PY79 by transformation (Dubnau and Davidoff-Abelson, 1971). KP161 is spoIIGB::erm (Kenney and Moran, 1987); KP522 is spoIID::cm; KP519 is spoIIM::Tn917 (Smith et al., 1993); KP520 is spoIIP::tet (Frandsen and Stragier, 1995); KP444 is Pspac::ftsZ (Beall and Lutkenhaus, 1991). KP521 is PY79 cotE::gfpΩkan (Webb et al., 1995). Strains KP519, KP520 and KP522 also contain the fusion cotE::gfpΩkan, which provided an indicator of σE activity, subcellular compartmentalization of σE gene expression and septation (Webb et al., 1995). Identical results were obtained in all experiments using strains lacking this fusion (not shown).
Growth conditions and FM 4-64 staining
Sporulation was induced by the resuspension method of Sterlini and Mandelstam (1969) or by nutrient exhaustion in DS medium (Sandman et al., 1988). FM 4-64 (Molecular Probes) was added at the time of resuspension in sporulation salts (t = 0) to a final concentration of 1 μg ml−1. At the appropriate time of sporulation, 0.5 ml aliquots of the sporulating culture were taken, centrifuged briefly and resuspended in 0.15 ml of the original culture supernatant. This concentrated cell suspension (2–3 μl) was placed on a microscope slide, and the DNA stained with DAPI (to a final concentration of 0.2 μg ml−1). A freshly prepared poly L-lysine-treated coverslip was used to immobilize the cells for microscopic analysis. When sporulation was induced by nutrient exhaustion, FM 4-64 was either added to the culture near the onset of sporulation or added together with DAPI at the time of slide preparation. At the concentrations used here, FM 4-64 had no detectable effect on growth rate (doubling time in LB at 37°C is 23 min with or without FM 4-64) or sporulation efficiency (1.7 × 108 spores ml−1 with FM 4-64 and 1.3 × 108 spores ml−1 without FM 4-64).
The strain KP444 contains the ftsZ gene fused to the IPTG-regulated Pspac promoter and was grown in the presence of 1 mM IPTG. To deplete cells of FtsZ, cultures of KP444 growing exponentially at 37°C in LB supplemented with 1 mM IPTG and 1 μg ml−1 FM 4-64 were centrifuged and resuspended in prewarmed LB media with 1 μg ml−1 FM 4-64 and either containing or lacking 1 mM IPTG (Beall and Lutkenhaus, 1991). After one, two and three generations of growth in the absence of IPTG, samples were stained with DAPI, affixed to a slide and images collected as described below.
Microscopy and image analysis
A Delta Vision optical sectioning microscope (Applied Precision) was used to collect digitally between 10 and 20 images spaced at 0.1–0.2 μm through the specimen, using standard filter sets for DAPI (360–400 nm excitation and 457–507 nm emission) and rhodamine (for FM 4-64 visualization; 555–583 nm excitation and 617–690 nm emission). These images were deconvolved using 15 iterations of the Delta Vision deconvolution software (Applied Precision), which is a constrained iterative deconvolution program. Medial sections from the deconvolved images were saved in TIFF format and imported into Adobe Photoshop. The images (consisting of white fluorescent membranes or DNA on a black background) were inverted to yield an image of black membranes (or DNA) on a white background to improve contrast and reproducibility (Fig. 1). After deconvolution and inversion, contrast was increased slightly, but the images were not processed further.
The intensity of fluorescence of FM 4-64-labelled membranes was quantified using the Delta Vision software, starting from the medial focal plane of experiments within the linear range of the CCD camera. The sum of the pixel values from a selected segment of membrane was obtained using the ‘Polygon Statistics’ command of Delta Vision (giving a measure of the total fluorescence from that segment of membrane). This value was divided by the length of the membrane in microns (obtained using the ‘Measure Distance’ command of Delta Vision; normally, a segment between 0.2 and 0.4 microns was measured) to give an average amount of fluorescence per micron (or counts per micron). Essentially the same results were obtained before and after deconvolution, except that the resolution of nearby membranes was improved after deconvolution. After this process, partial septa were conservatively defined as septa in which the minimum value of fluorescence per micron was less than that from the cytoplasmic membrane in the same cell.
Because FM 4-64 incorporates into bilayers and stains these bilayers approximately uniformly, under the conditions reported here, the relative fluorescence intensities of different regions of the membrane should reflect either the number or topology of lipid bilayers in that region. For example, septa contain two parallel membranes and so should stain twice as intensely as the cytoplasmic membrane, which is a single membrane. To test this hypothesis, the fluorescence intensity of different regions of sporangia was quantified to give a measure of the fluorescence intensity per length of membrane. In the sporangium with the flat sporulation septum at the lower right of Fig. 2E, the middle of the septal membranes had a fluorescence intensity of 61 849 counts μm−1, while the cytoplasmic membrane had a fluorescence intensity of 31 552 counts μm−1. Thus, the septal membranes stained 1.96-fold more intensely than the cytoplasmic membrane in this sporangium. In the three sporangia in Fig. 2E that had completed engulfment, the values were: upper sporangium (forespore pole, 91 468 counts μm−1; cytoplasmic membrane, 29 147 counts μm−1) middle sporangium (forespore pole, 115 991 counts μm−1; cytoplasmic membrane, 36 811 counts μm−1), bottom sporangium (forespore pole, 140 228 counts μm−1; cytoplasmic membrane, 37 682 counts μm−1). The relative fluorescence of these three forespores were 3.1-fold, 3.1-fold and 3.7-fold more intense than the cytoplasmic membranes.
Time-lapse observation of engulfment
For time-lapse observation of engulfment, cultures were stained with FM 4-64 as described above, but using a lower concentration of FM 4-64 (0.5 μg ml−1) to prolong survival of the bacteria during observation. The stained bacteria were applied to a 24 × 60 mm coverslip (poly L-lysine treated), washed (in conditioned sporulation medium) to remove weakly adherent bacteria and covered in a 50 μl drop of sporulation medium. A chambered slide was inverted over the drop, and the chamber was sealed with 50% glycerol to reduce evaporation. A heating device was constructed for chambered slides. Briefly, a radiant heating pad designed for use in reptile terrarria (ReptiTherm) was affixed to a 8 × 14 cm piece of 4-mm-thick glass. A layer of aluminium foil was used to cover the ReptiTherm, which was insulated further with a layer of blue foam pad (Recreational Equipment). A second layer of blue foam pad was cut to fit around the chambered slide and affixed to the glass. Before use, the entire device was prewarmed for at least 1 h. Using this device, the bacteria were able to sporulate in the chambered slide, completing engulfment after 45 or 60 min (Fig. 3), and producing phase bright spores after prolonged incubation. The room containing the microscope was heated to approximately 30°C, and six optical sections were collected every 15 min. Minimizing exposure of the bacteria to fluorescence was essential for assuring their survival during observation, so short exposure times were used.
Electron microscopy
Samples of sporulating cultures of PY79 and KP522 (spoIID:: cat) were fixed 2 h after the initiation of sporulation by resuspension and processed for electron microscopy, essentially as described previously (Cutting et al., 1991), but with the following modifications. The cells were fixed in 4% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.0, overnight at 4°C. Cells were post-fixed overnight at 4°C with 1% osmium tetroxide in phosphate buffer. Cell pellets were resuspended in 0.5 M ammonium chloride, dehydrated in increasing concentrations of ethanol, then embedded in Spurr’s resin. Ultra-thin sections were post-stained with 0.5% uranyl acetate and Reynold’s lead.
Acknowledgments
We thank Scott Emr for providing our first sample of FM 4-64, Patrick Stragier for providing strains, and Raffi Aroian for bringing to our attention the advantages of deconvolution microscopy. We also thank Adam Driks for many helpful suggestions on electron microscopy, and members of Richard Losick’s laboratory for their comments on this manuscript. This work was supported by the Arnold and Mabel Beckman Foundation, the Searle Scholars Program/The Chicago Community Trust and the Public Health Service (GM-57045).
References
- Addinall SG, Lutkenhaus J. FtsA is localized to the septum in an FtsZ-dependent manner. J Bacteriol. 1996;178:7167–7172. doi: 10.1128/jb.178.24.7167-7172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinall SG, Bi E, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinall SG, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in E. coli. Mol Microbiol. 1997;25:303–310. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- Beall B, Lutkenhaus J. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev. 1991;5:447–455. doi: 10.1101/gad.5.3.447. [DOI] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Bouche JP, Pichoff S. On the birth and fate of bacterial division sites. Mol Microbiol. 1998;29:19–26. doi: 10.1046/j.1365-2958.1998.00874.x. [DOI] [PubMed] [Google Scholar]
- Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-σK processing in Bacillus subtilis. Genes Dev. 1991;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- Daniel RA, Harry EJ, Katis VL, Wake RG, Errington J. Characterization of the essential cell division gene ftsL (yllD) of Bacillus subtilis and its role in the assembly of the division complex. Mol Microbiol. 1998;29:593–604. doi: 10.1046/j.1365-2958.1998.00954.x. [DOI] [PubMed] [Google Scholar]
- Dubnau D, Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. J Mol Biol. 1971;56:209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- Frandsen N, Stragier P. Identification and characterization of the Bacillus subtilis spoIIP locus. J Bacteriol. 1995;177:716–722. doi: 10.1128/jb.177.3.716-722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry EJ, Wake RG. The membrane-bound cell division protein DivIB is localized to the division site in Bacillus subtilis. Mol Microbiol. 1997;25:275–283. doi: 10.1046/j.1365-2958.1997.4581822.x. [DOI] [PubMed] [Google Scholar]
- Haugland RP. Membrane markers of endocytosis and exocytosis. In: Spence MTZ, editor. Handbook of Fluorescent Probes and Research Chemicals. 6. Eugene, OR: Molecular Probes; 1996. pp. 406–407. [Google Scholar]
- Heuser JSQ, Clark M. Proton pumps populate the contractile vacuoles of Dictyostelium amoebae. J Cell Biol. 1993;121:311–1327. doi: 10.1083/jcb.121.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Sedat JW, Agard DA. Determination of three-dimensional imaging properties of a light microscope system. Partial confocal behavior in epifluorescence microscopy. Biophys J. 1990;57:325–333. doi: 10.1016/S0006-3495(90)82534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeister A. Activation of the proprotein transcription factor pro-sigmaE is associated with its progression through three patterns of subcellular localization during sporulation in Bacillus subtilis. J Bacteriol. 1998;180:2426–2433. doi: 10.1128/jb.180.9.2426-2433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illing N, Errington J. Genetic regulation of morphogenesis in Bacillus subtilis: roles of σE and σF in pre-spore engulfment. J Bacteriol. 1991;173:3159–3169. doi: 10.1128/jb.173.10.3159-3169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney TJ, Moran CP., Jr Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J Bacteriol. 1987;169:3329–3339. doi: 10.1128/jb.169.7.3329-3339.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda A, Rashid MH, Sekiguchi J. Molecular cloning and sequencing of the upstream region of the major Bacillus subtilis autolysis gene: a modifier protein exhibiting sequence homology to the major autolysin and the spoIID product. J Gen Microbiol. 1992;138:1064–1076. doi: 10.1099/00221287-138-6-1067. [DOI] [PubMed] [Google Scholar]
- Lazarevic V, Margot P, Soldo B, Karamata D. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-L-alanine amidase and its modifier. J Gen Microbiol. 1992;138:1949–1961. doi: 10.1099/00221287-138-9-1949. [DOI] [PubMed] [Google Scholar]
- Levin PA, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- Lewis PJ, Partridge SR, Errington J. σ factors, asymmetry and the determination of cell fate in Bacillus subtilis. Proc Natl Acad Sci USA. 1994;91:3849–3853. doi: 10.1073/pnas.91.9.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londoño-Vallejo JA, Fréhel C, Stragier P. spoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol. 1997;24:29–39. doi: 10.1046/j.1365-2958.1997.3181680.x. [DOI] [PubMed] [Google Scholar]
- Lopez-Diaz I, Clarke S, Mandelstam J. spoIID operon of Bacillus subtilis: cloning and sequence. J Gen Microbiol. 1986;132:341–354. doi: 10.1099/00221287-132-2-341. [DOI] [PubMed] [Google Scholar]
- Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis PS, Driks A, Losick R. Sporulation gene spoIIB from Bacillus subtilis. J Bacteriol. 1993;175:528–540. doi: 10.1128/jb.175.2.528-540.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesenbock G, Rothman JE. Patterns of synaptic activity in neural networks recorded by light emission from synaptolucins. Proc Natl Acad Sci USA. 1997;94:3402–3407. doi: 10.1073/pnas.94.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanninga N. Morphogenesis of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:110–129. doi: 10.1128/mmbr.62.1.110-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge SR, Errington J. Importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol Microbiol. 1993;8:945–955. doi: 10.1111/j.1365-2958.1993.tb01639.x. [DOI] [PubMed] [Google Scholar]
- Piggot PJ, Coote JG. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot PJ, Bylund JE, Higgins ML. Morphogenesis and gene expression during sporulation. In: Piggot PJ, Moran CP Jr, Youngman P, editors. Regulation of Bacterial Differentiation. Washington DC: American Society for Microbiology Press; 1994. pp. 113–137. [Google Scholar]
- Pogliano J, Pogliano K, Weiss D, Losick R, Beckwith J. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc Natl Acad Sci USA. 1997;94:559–564. doi: 10.1073/pnas.94.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter A. Etude morphologie de la sporulation de Bacillus subtilis. Ann Inst Pasteur (Paris) 1965;108:40–60. [PubMed] [Google Scholar]
- Sandman K, Kroos L, Cutting S, Youngman P, Losick R. Identification of the promoter for a spore coat protein gene in Bacillus subtilis and studies on the regulation of its induction at a late stage of sporulation. J Mol Biol. 1988;200:461–473. doi: 10.1016/0022-2836(88)90536-0. [DOI] [PubMed] [Google Scholar]
- Scalettar BA, Swedlow JR, Sedat JW, Agard DA. Dispersion, aberration and deconvolution in multi-wavelength fluorescence images. J Microsc. 1996;182:50–60. doi: 10.1046/j.1365-2818.1996.122402.x. [DOI] [PubMed] [Google Scholar]
- Setlow B, Magill N, Febbroriello P, Nakhimousky L, Koppel DE, Setlow P. Condensation of the forespore nucleoid early in sporulation of Bacillus species. J Bacteriol. 1991;173:6270–6278. doi: 10.1128/jb.173.19.6270-6278.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Youngman P. Evidence that the spoIIM gene of Bacillus subtilis is transcribed by RNA polymerase associated with σE. J Bacteriol. 1993;175:3618–3627. doi: 10.1128/jb.175.11.3618-3627.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Bayer ME, Youngman P. Physical and functional characterization of the Bacillus subtilis spoIIM gene. J Bacteriol. 1993;175:3607–3617. doi: 10.1128/jb.175.11.3607-3617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlini JM, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1997;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- Sun Q, Margolin W. FtsZ dynamics during the division cycle of live Escherichia coli cells. J Bacteriol. 1998;180:2050–2056. doi: 10.1128/jb.180.8.2050-2056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;129:321–334. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lutkenhaus J. FtsI and FtsW are localized to the septum in Escherichia coli. J Bacteriol. 1998;180:2810–2816. doi: 10.1128/jb.180.11.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CD, Decatur A, Teleman A, Losick R. Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J Bacteriol. 1995;177:5906–5911. doi: 10.1128/jb.177.20.5906-5911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Pogliano K, Carson M, Guzman LM, Fraipont C, Nguyen-Disteche M, et al. Localization of the Escherichia coli cell division protein FtsI (PBP3) to the division site and cell pole. Mol Microbiol. 1997;25:671–681. doi: 10.1046/j.1365-2958.1997.5041869.x. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Lewis PJ, Allmansberger R, Hauser PM, Errington J. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev. 1995;9:1316–1326. doi: 10.1101/gad.9.11.1316. [DOI] [PubMed] [Google Scholar]
- Youngman P, Perkins JB, Losick R. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol Gen Genet. 1984;195:424–433. doi: 10.1007/BF00341443. [DOI] [PubMed] [Google Scholar]