Abstract
Tumor cell migration is a key step for formation of cancer metastasis. The mammalian target of rapamycin (mTOR), a highly conserved and ubiquitously expressed serine-threonine kinase, has been intensely studied for over a decade as a central regulator of cell growth, proliferation, differentiation and survival. Recent data have shown that mTOR also plays a critical role in the regulation of tumor cell motility and cancer metastasis. Here we briefly review recent advances about mTOR signaling in tumor cell motility. We also discuss recent findings about the mechanism by which rapamycin, a specific inhibitor of mTOR, inhibits cell motility in vitro and metastasis in vivo.
Keywords: mTOR, Rapamycin, S6K1, Akt, Cell motility, Metastasis
1. INTRODUCTION
Cancer metastasis, one of the hallmarks of malignancy, is the primary cause of death in most cancer patients. Cancer cells can “break away”, “leak”, or “spill” from a primary tumor, circulate through the vascular or lymphatic system, and settle down to establish a secondary tumor within normal tissues elsewhere in the body (Kedrin et al., 2007). The process of cancer metastasis consists of a series of sequential, interrelated steps involving angiogenesis, local invasion, intravasation, transit in the blood or lymph, extravasation and metastasis growth in distant organs (Woodhouse, 1997; Iiizumi et al., 2008). Many of these steps require cell migration, which allow the cells to change position within the tissues.
The migration mechanisms that tumor cells use to spread within the tissues are similar to those that occur in normal or non-neoplastic cells during physiological processes such as embryonic morphogenesis, inflammatoryimmune responses, wound healing, and angiogenesis (Friedl and Brocker, 2000). Cell migration through tissues results from a highly integrated multistep cellular events (Lauffenburger and Horwitz, 1996; Friedl and Brocker, 2000; Ridley et. al., 2003). First, responding to a migration-promoting agent, the moving cell becomes polarized and elongates, extending protrusions in the direction of migration. These protrusions, which can be spike-like filopodia, or large and broad lamellipodia, are usually driven by actin polymerization and are stabilized by adhering to the extracellular matrix or adjacent cells via transmembrane receptors linked to the actin cytoskeleton (Mitchison and Cramer, 1996). Subsequently, forward extension of a lamellipodium and retraction of the trailing edge results in the translocation of the cell body (Friedl and Wolf, 2003).
Reorganization of the actin cytoskeleton is the primary mechanism of cell motility and is essential for most types of cell migration (Schmidt and Hall, 1998). During cell migration, the actin cytoskeleton is dynamically remodeled, and this reorganization produces the force necessary for cell migration (Pollard and Borisy, 2003). In fibroblasts, a number of kinases, including phosphatidylinositol 3′kinase (PI3K), protein kinase B/Akt, mammalian target of rapamycin (mTOR), and ribosomal protein S6 kinases (S6K), interacts with the actin cytoskeleton in advancing lamellipodia (Berven et al., 2004). Here we briefly review the role of mTOR signaling pathway in cell motility and discuss the advances of the mechanism by which rapamycin inhibits cell motility in vitro and metastasis in vivo.
2. mTOR REGULATION OF CELL MOTILITY
A. A Tale of Rapamycin and mTOR
Rapamycin, a macrocyclic lactone produced by Streptomyces hygroscopicus, was first found from a soil sample of Easter Island (Rapa Nui) during a discovery program for anti-microbial agents in 1975 (Sehgal et al., 1975; Vezina et al., 1975). It was initially developed as an anti-fungal agent and subsequently discovered to have equally potent immunosuppressive and anti-tumor properties (Vezina et al., 1975; Douros and Suffness, 1981; Eng et al., 1984; Linhares et al., 2003). In 1999, rapamycin (Rapamune, Sirolimus), as an immunosuppresive drug, was approved by the Food and Drug Administration (FDA) in USA (Huang and Houghton, 2001). In addition, the results from many laboratories found that it can also act as a cytostatic agent, slowing or arresting growth of cell lines derived from rhabdomyosarcoma (Dilling et al., 1994; Hosoi et al., 1999), neuroblastoma, glioblastoma (Geoerger et al., 2001), small cell lung cancer (Seufferlein and Rozengurt, 1996), osteoscarcoma (Ogawa et al., 1998), pancreatic cancer (Grewe et al., 1999), breast cancer, prostate cancer (Pang and Faber, 2001; van der Poel et al., 2003), murine melanoma and B-cell lymphoma (Muthukkumar et al., 1995; Busca et al., 1996). So far, pharmaceutical companies have developed several rapamycin analogs (termed rapalogs) such as CCI-779 (Temsirolimus, Wyeth, Madison, NJ, USA), RAD001 (Everolimus, Novartis, Novartis, Basel, Switzerland), AP23573 (Deforolimus, ARIAD, Cambridge, MA, USA), 32 deoxy-rapamycin (SAR943) or zotarolimus (ABT-578, Abbott Laboratories, Abbott Park, IL, USA), which have improved solubility, stability and bioavailability, for malignancies (Rizzieri et al., 2008), chronic allergic inflammation (Eynott et al., 2003) or cardiovascular stent implantation (Abizaid et al., 2007).
In the early studies on rapamycin, FKBP12 (FK506-binding protein of 12 kDa) was identified as its direct cellular receptor in the budding yeast (Koltin et al., 1991). Deletion of the RBP1 gene, a homologue of mammalian FKBP12, yielded viable cells that are resistant to rapamycin, whereas expression of human FKBP in rapamycin-resistant yeast mutants restores sensitivity to rapamycin (Heitman et al., 1991; Koltin et al., 1991). In the subsequent yeast genetic screens for mutations that rescue the anti-proliferative effect of rapamycin, two rapamycin target genes named TOR1 (the target of rapamycin 1) and TOR2 were identified (Heitman et al., 1991; Cafferkey et al., 1993). Extensive studies revealed the action mechanism of rapamycin: upon entering the cells, rapamycin binds to the intracellular receptor (FKBP12), forming an inhibitory complex which binds a region in the C terminus of TOR proteins termed FRB (FKB12-rapamycin binding), thereby exerting its cell growth-inhibitory and cytotoxic effects by inhibiting the functions of TOR1 and TOR2 (Kunz and Hall, 1993; Helliwell et al., 1994; Chen et al., 1995; Choi et al., 1996). While both TOR1 and TOR2, sharing 70% homology and encoding ~280 kDa proteins, are targeted by rapamycin and mediate translation in yeast. Cells lacking TOR2 are nonviable but arrest at all points in the cell cycle. TOR1, in contrast to TOR2, is not essential, but cells lacking both TOR1 and TOR2 arrest growth within one cell cycle in G1 phase (Kunz et al., 1993). In addition, the lethality of a TOR2 disruption cannot be suppressed even by overexpression of TOR1. This suggests that TOR2 has at least two essential functions (Hall, 1996). The first is shared with TOR1, i.e. rapamycin-sensitive, and is required for translation initiation and G1 progression, whereas the second is unique to TOR2 and is involved in cell cycle-dependant organization of the actin cytoskeleton (Zheng et al., 1995; Schmidt et al., 1996). Subsequent biochemical studies extended this model to mammalian cells and led to the discovery and identification of the mammalian counterpart, mTOR (Brown et al., 1994; Chiu et al., 1994; Sabatini et al., 1994; Sabers et al., 1995).
B. The Structure of mTOR
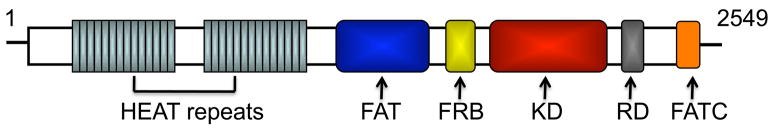
mTOR, also known as FRAP (FKBP12-rapamcyin-associated protein), RAFT1 (rapamycin and FKBP12 target), RAPT 1 (rapamycin target 1), or SEP (sirolimus effector protein), is an 289 kDa atypical serine/threonine kinase (Brown et al., 1994; Chen et al., 1994; Chiu et al., 1994; Sabatini et al., 1994). mTOR and yeast TOR proteins share >65% identity in their carboxy-terminal catalytic domains and >40% identity in overall sequence (Abraham, 1998). mTOR is considered a member of the phosphatidylinositol 3-kinase (PI3K)-kinase-related kinase (PIKK) superfamily since the C-terminus of mTOR shares strong homology to the catalytic domain of PI3K (Kunz et al., 1993; Keith and Schreiber, 1995). All the proteins in this family, including others, such as MEC1, TEL1, RAD3, MEI-41, DNA-PK, ATM, ATR, and TRRAP, contain a characteristic C-terminal PI kinase homology domain (Schmelzle and Hall, 2000). The different members of this family are associated with diverse cellular functions, such as control of cell growth, cell cycle and DNA damage checkpoints, recombination and maintenance of telomere length (Brown and Schreiber, 1996; Thomas and Hall, 1997; Canman et al., 1998). TOR comprises several domains and seems to be highly conserved from yeast to mammalian cells. At the amino-acid level, human, mouse and rat TOR proteins share a 95% identity (Hay and Sonenberg, 2004; Janus et al., 2005). mTOR contains 2549 amino acids and its domain structure is depicted in Figure 1. The N-terminus of mTOR consists of up to 20 tandemly repeated HEAT motifs including Huntingtin, elongation factor 3 (EF3), A subunit of protein phosphatase 2A (PP2A), and TOR. The C-terminus consists of mutated FRAP-ataxia-teleangiectasia (FAT), a transformation/transcription domain-associated protein domain, an FKPB12-rapamycin binding (FRB) domain, a catalytic kinase domain, a probable auto-inhibitory (repressor domain or RD domain), and a FAT carboxy-terminal (FATC) domain. Tandem HEAT repeats are present in many proteins and may form an extended superhelical structure to create multiple large interfaces for protein-protein interaction, whereas the FAT and FATC domains may modulate catalytic kinase activity of mTOR (Gingras et al., 2001; Perry and Kleckner, 2003).
Fig. 1.
Schematic structure of mTOR. Various domains discussed in the text are shown.
C. Upstream and Downstream of mTOR
In response to ligand binding, the type I insulin-like growth factor receptor (IGFR), a transmembrane tyrosine kinase, is activated via auto-phosphorylation of multiple tyrosine residues. Activated IGFR in turn phosphorylates the insulin receptor substrates 1–4 (IRS1–4) and src- and collagen-homology (SHC) adaptor proteins, which can trigger multiple downstream signal transduction pathways including the MAPK pathway (not shown) and phosphatidylinositol 3′kinase (PI3K) pathway (Figure 2) (Valentinis and Baserga, 2001). Phosphorylated IRS recruits the p85 subunit of PI3K and signals to the p110 catalytic subunit of PI3K, resulting in activation of PI3K. Activated PI3K catalyzes the conversion of phosphatidylinositol (4, 5)-bisphosphate (PIP2) to phosphatidylinositol-3, 4, 5-trisphosphate (PIP3). This pathway is negatively regulated by PTEN (phosphatase and tensin homolog on chromosome ten), also known as MMAC1 (mutated in multiple advanced cancers), a dual-specificity protein and lipid phosphatase. Increased PIP3 binds to the pleckstrin homology (PH) domain of Akt/PKB (protein kinase B) and, in combination with additional serine/threonine (S/T) phosphorylation of Akt by phosphoinositide-dependent kinase 1 (PDK1) and mTOR, results in full activation of Akt. Subsequently, activated PI3K or Akt may positively regulate mTOR, leading to increased phosphorylation of ribosomal p70 S6 kinase (S6K1) and eukaryotic initiation factor 4E (eIF4E) binding protein 1 (4E-BP1), the two best-characterized downstream effector molecules of mTOR (Huang and Houghton, 2003). Studies have placed tuberous sclerosis complex (TSC) 1/2 as a modulator between PI3K/Akt and mTOR (Gao et al., 2002; Inoki et al., 2002; Tee et al., 2002). The TSC1/2 complex acts as a repressor of mTOR function (Gao et al., 2002; Inoki et al., 2002; Tee et al., 2002). TSC2 has GTPase-activating protein (GAP) activity towards the Ras family small GTPase Rheb (Ras homolog enriched in brain), and TSC1/2 antagonizes the mTOR signaling pathway via stimulation of GTP hydrolysis of Rheb (Inoki et al., 2002; Tee et al., 2002; Garami et al., 2003; Manning and Cantley, 2003; Stocker et al., 2003; Zhang et al., 2003). Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38 (Bai et al., 2007), though this remains controversial (Wang et al., 2008). The TSC can also be activated by energy depletion through the activation of AMP-activated kinase (AMPK) (Figure 2). This, in turn, activates the TSC, which catalyzes the conversion of Rheb-GTP to Rheb-GDP and thus inhibits mTOR (Inoki et al., 2002; Tee et al., 2002; Garami et al., 2003; Manning and Cantley, 2003; Stocker et al., 2003; Zhang et al., 2003).
Fig. 2.
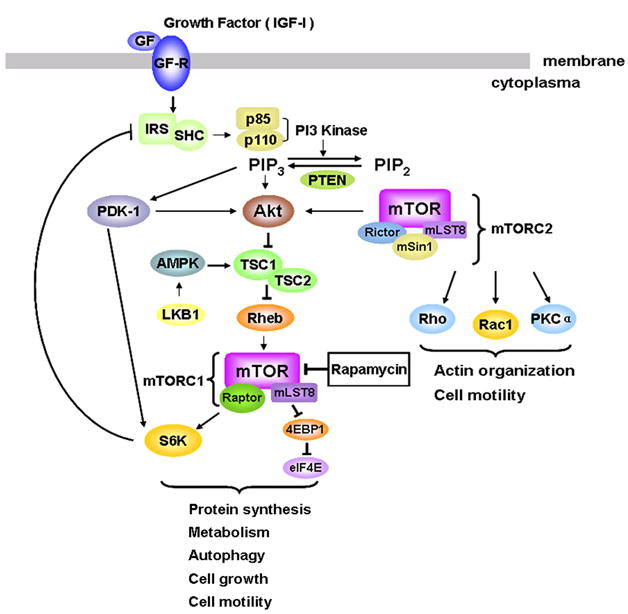
Schematic of IGF-I activation of a signaling pathway to mTOR. mTOR protein exists in two distinct complexes, mTORC1 and mTORC2. mTORC1, which receives input from several upstream pathways, including PI3K/Akt, TSC1/TSC2, and AMPK, phosphorylates downstream effectors S6K and 4EBP1 and regulates ribosome biogenesis, cell growth, autophagy, cell motility and metabolism. mTORC2 phosphorylates Akt at Ser473 and regulates the actin cytoskeleton and cell motility. Arrows represent activation, whereas bars represent inhibition.
mTOR functions as two complexes, mTORC1 and mTORC2 (Guertin and Sabatini, 2007; Polak and Hall, 2009). mTORC1 is rapamycin-sensitive, and consists of mTOR, mLST8 (also termed G-protein β-subunit-like protein, GβL, a yeast homolog of LST8), raptor (regulatory associated protein of mTOR) and PRAS40 (proline-rich Akt substrate 40 kDa), whereas mTORC2 is rapamycin-insensitive, and is composed of mTOR, mLST8, rictor (rapamycin-insensitive companion of mTOR), mSin1 (mammalian stress-activated protein kinase (SAPK)-interacting protein 1), protor (protein observed with rictor) and PRR5 (proline-rich protein 5). mTORC1 phosphorylates 4E-BP1 (eukaryotic initiation factor 4E binding protein 1) and S6K1 (ribosomal p70 S6 kinase 1), and regulates cell growth, proliferation and survival by promoting mRNA translation, ribosome biogenesis, and autophagy (Hara et al., 2002; Kim et al., 2002; Kim et al., 2003). mTORC2 phosphorylates Akt at Ser473 and regulates the actin cytoskeleton (Jacinto et al., 2004; Sarbassov et al., 2004; Sarbassov et al., 2005). Most recently, Hsp70, a heat shock protein, has been identified as a new mTORC2 component, which is required for proper assembly and activity of the kinase under basal conditions and following heat shock (Martin et al., 2008).
In mammals, the ribosomal protein S6 kinases (S6Ks) and the eukaryotic initiation factor 4E (eIF4E)-binding proteins (4E-BPs) are the best characterized downstream targets of mTOR. In mammalian, 4E-BPs, a family of translational repressor proteins, consist of three low molecular weight proteins, 4E-BP1, 4E-BP2, and 4E-BP3 (Lin et al., 1994; Pause et al., 1994; Poulin et al., 1998). 4E-BP1 (also known as PHAS-I), a repressor of the translation initiation factor eIF4E, was first identified as an adipocyte protein that underwent phosphorylation by MAP kinase in response to insulin treatment (Lin et al., 1994; Pause et al., 1994). In addition, mTOR and ATM have been identified to be involved in phosphorylation of 4E-BP1 (Brunn et al., 1997; Hara et al., 1997; Gingras et al., 1999a; Yang and Kastan, 2000). Hypophosphorylated 4E-BP1 binds with high affinity to eIF4E, and repress cap-dependent translation by blocking the binding of eIF4E to eIF4G. In response to sufficient growth factors and nutrients stimulation, six sites (Thr37, Thr46, Ser65, Thr70, Ser83 and Ser112) of 4E-BP1 can be phosphorylated (Shah et al., 2000). The phosphorylation of 4E-BP1 at multiple site induce the dissociation of the 4E-BP1 from eIF4E, allowing eIF4E to engage eIF4G (Pause et al., 1994; Marcotrigiano et al., 1999). eIF4G serves as a scaffold protein for the assembly of other initiation factors including eIF4A, which acts as an ATP-dependent RNA helicase, and further interacts with eIF3, which recruits the 40S ribosome to the 5′ end of the mRNA (Gingras et al., 1999b). Thus, 4E-BP1 phosphorylation allows several important initiation factors, as well as the 40S ribosomal subunit, to be positioned at the 5′ end of the mRNA to begin the process of scanning.
S6K1 is the other well characterized downstream of mTOR. S6K1 and 4E-BP1 function in parallel pathways that bifurcate downstream of mTOR (von Manteuffel et al., 1997). Mammalian cells contain two similar S6 kinase proteins (S6K1 and S6K2) encoded by two different genes, each of which are found as two alternatively spliced isoforms (Reinhard et al., 1992; Martin and Blenis, 2002). S6K1 is ubiquitously expressed and has been used for most of the studies on substrate phosphorylation and effects on cell growth. S6K2, which has 70% overall amino acid identity with S6K1 and harbors a potential NLS in the carboxy terminus, was discovered much later than S6K1 (Shima et al., 1998). S6K1 can be activated by a wide variety of extracellular signals. At least eight phosphorylation sites were found to be regulated by mTOR- and growth factor-dependent signaling pathways. Thr22, which is located in the activation loop, can be phosphorylated by the “loop kinase” phospholipid-dependent protein kinase 1 (PDK-1) (Alessi et al., 1998; Pullen et al., 1998). The phosphorylation of this site is a critical step of in S6K1 activation. Moreover, for S6K1 activation, mTOR can directly phosphorylates Ser371 in vitro, and this event modulates Thr389 phosphorylation by mTOR (Isotani et al., 1999; Saitoh et al., 2002). S6K1 may also be activated by TOR-insensitive signaling pathways, such as MAP kinases, which mediate phosphorylation of sites on the C-terminal autoinhititory domain (Jeno et al., 1988; Cheatham et al., 1995; Dufner and Thomas, 1999). S6K1 is known as the major ribosomal protein S6 (rpS6) kinase in mammalian cells and is pointed as a key player in the control of cell size, growth, and proliferation (Montagne et al., 1999; Avruch et al., 2001; Radimerski et al., 2002). Early studies suggest that activated S6K1 regulate protein translation through phosphorylation of the 40S ribosomal subunit, which has been suggested to increase the translational efficiency of a class of mRNA transcripts with a 5′-terminal oligopolypirymidine (5′-TOP) (Jefferies et al., 1994; Terada et al., 1994). However, this model has been recently challenged (Tang et al., 2001; Stolovich et al., 2002; Pende et al., 2004; Holz et al., 2005). New data (Holz et al., 2005) indicate that mTOR and S6K1 control on and off the eukaryotic initiation factor 3 (eIF3) translation initiation complex in a growth-factor- and rapamycin-sensitive manner. S6K1 associates with the eIF3 complex when inactive, but dissociates from the eIF3 complex upon stimulation by insulin or amino acids. Activated S6K1 then phosphorylates its translational targets, including the 40S ribosomal protein S6 and eIF4B, promoting translation initiation.
D. The Role of mTOR Signaling Pathway in Cell Motility
Early studies in the budding yeast Saccharomyces cerevisiae have demonstrated that TOR, which signals through two distinct complexes, TOR1 and TOR2, participates in the control of translational initiation and early G1 progression in response to nutrient availability (Loewith et al., 2002; Wedaman et al., 2003). Recent work has identified additional functions of TOR, including the regulation of transcription, cytoskeletal organization, cell motility and protein degradation through autophagy (Beck and Hall, 1999; Jacinto and Hall, 2003). Accumulating evidence indicates that one of the primary functions of TORC2 is actin cytoskeleton rearrangement. Deletion of TOR2 disrupts the polarized organization of the actin cytoskeleton, and the Rho GTPase pathway was required for TOR2-mediated signaling to the actin cytoskeleton (Schmidt et al., 1996; Schmidt et al., 1997). Like yeast TORC2, mTORC2 also seems to function upstream of Rho GTPases to regulate the actin cytoskeleton (Jacinto et al., 2004). In serum-starved NIH 3T3 fibroblast cells, knockdown of mTOR, rictor (mAVO3) and mLST8 (GβL) results in defective F-actin fibres (stress fibres and/or lamellipodia) formation in response to serum, whereas knockdown of raptor (mKOG) that is a component of TORC1 does not cause a detectable defect of the actin cytoskeleton (Jacinto et al., 2004). In addition, knockdown of mTORC2 prevented the phosphorylation of paxillin, which is a protein that when phosphorylated localizes and recruits other signalling molecules to focal adhesions (Schaller, 2001; Jacinto et al., 2004). In HeLa cells, lentivirus-mediated delivery of rictor leads to increased F-actin bundles concomitant with increased cytoplasmic paxillin patches that colocalize to the ends of the thick actin fibers (Sarbassov et al., 2004). Interestingly, PKCα activity is reduced in rictor or mTOR silencing cells, and in PKCα silencing cells the thick actin fibers appear more numerous, better organized and connected to the remainder of the cytoskeleton. This is consistent with the studies from yeast that demonstrate TORC2 regulate the organization of the actin cytoskeleton and that PKCα is a mediator of this function (Sarbassov et al., 2004). Molecular mechanism by which mTORC2 mediates the activation of Rho GTPases and cytoskeletal events is not well understood. However, recent studies provide a novel mechanism by which mTORC2 can control the actin cytoskeleton through the activation of Rho GTPases. Hernandez-Negrete et al. have shown that P-Rex1, which is a Rac guanine exchange factor connecting G protein-coupled receptors through GβL and PI3K signaling to Rac activation, links mTOR2 signaling to Rac activation, leading to the regulation of actin cytoskeleton and cell migration (Hernandez-Negrete et al., 2007). P-Rex family of Rac guanine exchange factors, including P-Rex1, P-Rex2a, and P-Rex2b, are able to interact directly with mTOR through their tandem DEP domains. Dominant-negative constructs and short hairpin RNA-mediated knockdown of P-Rex1 decreased leucine-induced mTOR-dependent activation of Rac and cell migration. Both P-Rex1 and mTOR are needed for Rac activation and cell migration induced by leucine. However, whether P-Rex1 and mTOR are involved in part of the same signaling route or act through independent pathways remains to be elucidated.
The initial characterization of mTORC2 led to discovery of the participation of mTORC2 signaling in cytoskeletal events and cell movement (Jacinto et al., 2004; Sarbassov et al., 2004). Recent studies have demonstrated that rapamycin-sensitive mTORC1 also plays a crucial role in the regulation of cell motility (Berven et al., 2004; Sakakibara et al., 2005; Liu et al., 2006; Liu et al., 2008). It appears that both mTORC1-mediated S6K1 and 4E-BP1 pathways are involved in the regulation of cell motility. S6K1 regulates cell motility, probably related to its regulating phosphorylation of the focal adhesion proteins, such as focal adhesion kinase (FAK), paxillin and p130Cas, and F-actin reorganization (or lamillapodia formation) (Liu et al., 2008). However, little is known about how 4E-BP1 pathway regulates cell motility.
3. RAPAMYCIN INHIBITION OF CELL MOTILITY
Multiple environmental factors such as chemokines and growth factors can promote and regulate tumor cell motility and thereby contribute to invasion and metastasis. For example, the epidermal growth factor (EGF) can promote ovarian and breast cancer cell migration, and IGF-1 can activate migration in several cancer cell lines, such as breast cancer cells, pancreatic carcinoma and melanoma cells (Brooks et al., 1997; Guvakova and Surmacz, 1999; Price et al., 1999; Alper et al., 2001). Many of these migration-enhancing factors also promote other functions, such as cell proliferation and survival. The anti-proliferative activity of rapamycin has been well established in previous studies. Recently, inhibition of motility or invasion by rapamycin has been described in several cell lines, such as porcine, murine and human aortic smooth muscle cells (Poon et al., 1996; Sun et al., 2001; Sakakibara et al., 2005), canine kidney epithelial cells (MDCKT23) and human colorectal cells (HCT-8/S11) (Attoub et al., 2000), Swiss 3T3 firoblasts (Berven et al., 2004), neutrophils (Gomez-Cambronero, 2003), ovarian cells (Wong et al., 2004), human umbilical vein endothelial cells (Kwon et al., 2005), human rhabdomyosarcoma (Rh1 and Rh30), breast carcinoma (MDA-MB-468), cervical adenocarcinoma (HeLa), and SV40 transformed African green monkey kidney firoblast (COS-1) cells (Liu et al., 2006). Diverse mechanisms may be involved in the inhibitory effect of rapamycin on cell motility or invasion. Here we briefly summarize recent findings regarding the mechanisms by which rapamycin inhibits cell migration.
A. By mTORC1 Mediated S6K1 and 4E-BP1/eIF4E Pathways
It is widely recognized that rapamycin inhibits protein synthesis, cell proliferation, growth and survival by inhibiting mTORC1-mediated S6K1 and 4E-BP1/eIF4E pathways (Ferrari et al., 1991; Brown et al., 1994; Crouch, 1997; Dufner and Thomas, 1999). Increasing evidence indicates that rapamycin inhibits cell motility and tumor metastasis also through targeting these two pathways. In Swiss 3T3 fibroblasts, thrombin stimulation induced the activity of S6K1 and the formation of more actin filaments. Pretreatment of cells with rapamycin inhibited the formation of stress fibre, showing that the inhibitory effect of rapamycin could be accounted for the action on S6K1 activity (Crouch, 1997). S6K1, Akt1, PDK1, and p85 PI3K were localized to the actin arc, a caveolin-enriched cytoskeletal structure located at the leading edge of migrating Swiss 3T3 cells (Berven et al., 2004). Activated mTOR was also enriched at the actin arc (Berven et al., 2004), suggesting that activation of the mTOR-S6K1 signaling pathway is important to cell migration. Furthermore, in EGF-stimulated fibroblasts, rapamycin treatment inhibited the formation of arcs. In transwell migration assay, rapamycin also effectively inhibited EGF-stimulated cell migration (Berven et al., 2004). The antimigratory effect of rapamycin and the critical role of mTOR-S6K1 pathway in cell motility were explored in other cell types. In either GM-CSF- or IL-8-treated neutrophil, pre-incubation with rapamycin strongly inhibited chemotaxis and chemokinesis in these cells. Consistently, rapamycin inhibited both the increase of enzymatic activitiy and the phosphorylation of S6K1 which were induced by GM-CSF. The pre-incubation of neutrophils with FK506, a structural analog of rapamycin and an FKBP12-binding compound that lacks the ability to inhibit mTOR, did not have significant effect on chemotaxis and actin ploymerization, suggesting that the effect of rapamycin was directly through the mTOR pathway (Gomez-Cambronero, 2003). Moreover, Sakakibara et al. (2005) showed that rapamycin markedly diminished chemotaxis of a human arterial smooth muscle cell line E47 cells and rat aortic vascular smooth muscle cells (SMCs) toward the matrix protein fibronectin as well as type I collagen and laminin. Fibronectin dose-dependently induced the phosphorylation of both mTOR and S6K1. Pretreatment of rapamycin completely inhibited phosphorylation of mTOR and S6K1, indicating that mTOR and S6K1 play an important role in the rapamycin-sensitive signaling pathway which is responsible for fibronection-induced migration (Sakakibara et al., 2005). In another study performed in human ovarian cancer cell lines SKOV-3 and CaOV-3, constitutively expression of active variants of S6K1 showed similar motile response of HGF and induced scattering to HGF-dependent levels even in the absence of HGF stimulation. Pretreatment with mTOR inhibitor rapamycin, MAPK inhibitor PD98059 or pan-PKC inhibitor GF109203X, only rapamycin significantly inhibited cell scattering in SKOV-3 and CaOV-3 cells, indicating that mTOR, but not ERK1/2 and PKC activity, was required for HGF to activate S6K and finally induce cell migration in these cells (Zhou and Wong, 2006). Evidence for participation of mTORC1-S6K1 signaling in cell motility is further supported by our recent studies showing that rapamycin inhibited IGF-induced motility in a panel of cell lines, such as rhabdomyosarcoma (Rh1 and Rh30), breast carcinoma (MDA-MB-468), cervical adenocarcinoma and SV40 transformed African green monkey fibroblast (COS-1) cells, and this inhibitory effect is dependent on mTOR kinase activity (Liu et al., 2006). In Rh30 cells, expression of mTORrr (rapamycin resistant mutant of mTOR), but not mTOR-SIDA ( kinase-dead mTORrr), prevented rapamycin inhibition of phosphorylation of S6K1 and 4E-BP1, and disrupted rapamycin inhibition of IGF-I-stimulated cell motility. These results indicate that rapamycin inhibition of IGF-I-stimulated cell motility is a consequence of inhibition of mTOR kinase activity. In the single cell motility assay, downregulation of S6K1 by using lentiviral shRNA effectively inhibited IGF-I-stimulated cell motility, and cells expressing constitutively active S6K1 are resistant to rapamycin inhibition of IGF-I-stimulated cell motility. Similarly, ectopic expression of constitutively hypophophorylated 4E-BP1 dramatically inhibited IGF-I-stimulated cell motility, downregulation of 4E-BP1 by shRNA attenuated rapamycin inhibition of cell motility. These data strongly suggest that the inhibition of rapamycin on IGF-induced motility may through the suppression of mTORC1-mediated S6K1 and 4E-BP1/eIF4E-signaling pathways (Liu et al., 2006). We also observed that rapamycin inhibited IGF-I-induced F-actin reorganization and phosphorylation of the focal adhesion proteins by disruption of mTOR-raptor complex. Both S6K1 and 4E-BP1 pathways, mediated by the mTOR-raptor complex, are involved in the regulation of IGF-I-stimulated F-actin reorganization. However, only S6K1 pathway controls IGF-I-stimulated phosphorylation of the focal adhesion proteins (Liu et al., 2008).
B. By the Cyclin-dependent Kinase Inhibitor p27Kip1 Pathway
Treatment with rapamycin results in accumulation of the cyclin-dependent kinase inhibitor p27Kip1 in the cells, leading to cell cycle arrest in G1 phase (Nourse et al., 1994). Partial resistance to rapamycin was found in p27−/− mouse embryo fibroblasts and p27−/−splenic T lymphocytes (Luo et al., 1996), suggesting that there are p27-dependent and independent pathways that determines rapamycin antiproliferative sensitivity. Recent findings indicate that p27Kip1 is also involved in the regulation of the antimigratory property of rapamycin. Rapamycin inhibited bFGF-induced migration of vascular smooth muscle cells in a dose-dependent manner, with IC50 of ~2 nM (for wild-type cells) and ~200 nM (for p27-null cells) (Sun et al., 2001). When exposed to 100 nM rapamycin, p27-null cells exhibited approximately 35% inhibition of cell migration, whereas wild-type cells showed nearly 80% reduction in cell migration (Sun et al., 2001). The results suggest that there exist p27Kip1-dependent and -independent mechanisms that affect rapamycin antimigratory activity as well. In multiple effects of rapamycin on cell signalling, a common step is binding to the intracellular receptor FKBP12 and forming an inhibitory complex, rapamycin-FKBP12, which also inhibits mTOR, S6K and 4E-BP1 phosphorylation (Brown et al., 1994; Chen et al., 1995; Brunn et al., 1997). Poon et al. showed that molar excesses of FK506 competed rapamycin off from FKBP12 and blocked the inhibitory effect of rapamycin on migration, suggesting that both the p27Kip1-dependent and p27Kip1-independent antimigratory effects of rapamycin are mediated through binding to FKBP12 (Poon et al., 1996). Studies have also shown that rapamycin derivative RAD001 effectively inhibited muscle cell migration in vitro and in vivo, which are partially mediated by p27Kip1 as well (Daniel et al., 2004).
Cell migration, which is a critical element for several events in cancer progression, is a highly orchestrated multi-step process, including polarization, protrusion, adhesion and de-adhesion (Ridley et al., 2003). Many proteins, such as PP2A, Rac/Cdc42, integrins, focal adhesion kinase, Src, and extracellular signal-regulated kinase 1/2 (Erk1/2) participate in this process. Activation of Erk1/2 by IGF-I is associated with mitogenesis and cell motility (O’Connor, 2003; Reddy et al., 2003). Inhibition of PP2A activity promotes motility in a number of transformed cells and cancer cell lines (Benefield et al., 1997; Jackson and Young, 2003). Recent studies have demonstrated that mTOR mediates phosphorylation of Erk1/2 (Thr202) through direct or indirect regulation of PP2A (Harwood et al., 2008). More studies are required to investigate whether rapamycin inhibits cell motility by impacting PP2A-Erk1/2 pathway.
4. RAPAMYCIN AND TUMOR METASTASES
In vitro, rapamycin is effective in blocking the cell growth and motility of a number of tumor cell lines. In vivo, rapamycin also potently inhibits metastases of transplanted tumors in different mouse models. Guba et al. (2002) showed that injection syngenic CT-26 adenocarcinoma cells intraportally in BALB/c mice stimulated metastasis of colon cancer to the liver. In the metastatic area, rapamycin markedly decreased hepatic tumor-replacement area. In multiple quantitative analyses on the tumors in dorsal skin-fold chambers, the measurement in rapamycin-treated mice showed a smaller vascularized tumor area and lower vascular density than in control mice, suggesting that rapamycin can suppress tumor growth by inhibiting angiogenesis. It was also suggested in this study that the antiangiogenic effect of rapamycin is related to reduced translational production of vascular endothelial growth factor (VEGF) and blockage of VEGF-induced vascular endothelial cell stimulation (Guba et al., 2002). In other studies, Luan et al. found that rapamycin prevented tumor growth and pulmonary metastasis both in the murine and the human renal cell cancer (RCC) pulmonary metastasis models (Luan et al., 2002; Luan et al., 2003). The mechanistic studies suggested that the inhibitory effect of rapamycin on tumor growth and metastasis is linked, at least in part, to the ability to block tumor cell production of tumor promoting cytokines (VEGF-A and TGF-β1) which have been implicated in the development of breast cancer metastasis to bone (Yin et al., 1999; Luan et al., 2003). In non-small cell lung cancer cell (KLN-205) model, rapamycin is also highly efficacious in preventing the formation and development of pulmonary metastases (Boffa et al., 2004). mTORC1-mediated S6K1 and 4E-BP1 pathways were found to be linked to ezrin related metastatic behavior (Wan et al., 2005). Ezrin, a substrate of tyrosine kinases, binds adhesion molecules such as CD43, CD44, and intercellular adhesion molecule-1, is believed to be involved in intracellular signal transduction which is related to cell migration and metastasis (Gould et al., 1989; Fazioli et al., 1993; Tsukita et al., 1994). Wan et al. (2005) showed that reduction of ezrin expression by RNAi or disruption of ezrin function by transfection of a dominant-negative ezrin-T567A mutant led to decreased phosphorylation of both S6K1 and 4E-BP1. Rapamycin inhibited mTORC1-mediated phosphorylation of S6K1 and 4E-BP1, leading to decreased cell migration and invasion in K7M2 cells. Furthermore, rapamycin inhibited pulmonary metastasis in tumor-inoculated SCID beige mice (Wan et al., 2005).
More recently, the effect of rapamycin on lymphangiogenesis and lymph node metastasis was investigated by using an experimental metastatic model in nude mice (Huber et al., 2007; Kobayashi et al., 2007). The results showed that rapamycin significantly decreased the number and the area of lymphatic vessels in the primary tumors in nude mice. The lymph node metastasis was significantly suppressed in rapamycin-treated nude mice. Several studies have provided evidence that the VEGF-C/VEGFR-3 signal transduction pathway plays a causal role in tumor-associated lymphangiogenesis and lymphatic metastasis (Lin et al., 2005; Roberts et al., 2006; Takizawa et al., 2006). Kobayashi et al. (2007) revealed that mTOR, p38, and JNK play important roles in VEGF-C expression. Rapamycin repressed protein and mRNA expressions of VEGF-A and VEGF-C in B13LM cells under serum-starved or normal culture condition (Kobayashi et al., 2007), implying that rapamycin inhibits lymphangiogenesis in vivo probably by inhibition of VEGF-C expression. Rapamycin also impairs downstream signaling of VEGF-A and VEGF-C through mTOR-S6K1 pathway in lymphatic endothelial cells (Huber et al., 2007). Inhibition of PI3K or mTOR suppresses phosphorylation of S6K1 and tube formation induced by fibroblast growth factor-2 in lymphatic endothelial cells (Matsuo et al., 2007), further suggesting involvement of mTORC1-mediated S6K1 pathway in lymphangiogenesis. Most recent studies have shown that IL-20 increases intracellular free calcium concentration, Akt and eNOS phosphorylations as well as perinuclear nitric oxide (NO) production, and stimulates tube formation in lymphatic endothelial cells (hTERT-HDLEC), which is PI3K and mTOR dependent (Hammer et al., 2009). Whether mTOR regulates lymphangiogenesis through calcium/NO signaling remains to be defined.
5. SUMMARY
mTOR, a master kinase, regulates cell growth, proliferation, differentiation and survival. A variety of transformed or tumor cells with dysregulated mTOR signaling have shown higher susceptibility to inhibitors of mTOR than normal cells. Therefore, mTOR becomes an attractive target for tumor-selective therapy. Increasing evidence has shed new insight into the role of mTOR in the regulation of cell motility. Early studies have demonstrated the participation of mTORC2 signaling in cytoskeletal events and cell movement. Recent studies have further shown that rapamycin-sensitive mTORC1 also plays a crucial role in the regulation of cell motility. Rapamycin inhibits cell motility through targeting S6K1 and 4E-BP1 pathways, similar to the mechamism by which it inhibits cell proliferation in mammalian cells. Cell migration is a highly orchestrated multi-step process and many proteins may participate in this process. Many questions remain obscure, including whether and how other downstream effector molecules of mTOR, such as S/T phosphatases (PP2A, PP4, PP5 and PP6), Akt, PKC, GTPases, and focal adhesion proteins, are involved in the regulation of cell motility; and how mTOR interacts with other signaling pathway or proteins, leading to the regulation of cell migration; and how mTOR regulates cell attachment/de-attachment. Addressing these questions should enhance our molecular understanding of tumor cell motility and may provide novel therapeutic strategies that would reduce metastatic spread of tumor cells, blocking metastatic progression and increasing patient survival.
Acknowledgments
This work was supported in part by NIH grant CA115414 (S.H.), and American Cancer Society Award RSG-08-135-01-CNE (S.H.).
References
- Abizaid A, Lansky AJ, Fitzgerald PJ, Tanajura LF, Feres F, Staico R, Mattos L, Abizaid A, Chaves A, Centemero M, Sousa AG, Sousa JE, Zaugg MJ, Schwartz LB. Percutaneous coronary revascularization using a trilayer metal phosphorylcholine-coated zotarolimus-eluting stent. Am J Cardiol. 2007;99:1403–1408. doi: 10.1016/j.amjcard.2006.12.064. [DOI] [PubMed] [Google Scholar]
- Abraham RT. Mammalian target of rapamycin: immunosuppressive drugs uncover a novel pathway of cytokine receptor signaling. Curr Opin Immunol. 1998;10:330–336. doi: 10.1016/s0952-7915(98)80172-6. [DOI] [PubMed] [Google Scholar]
- Alessi DR, Kozlowski MT, Weng QP, Morrice N, Avruch J. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr Biol. 1998;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- Alper O, Bergmann-Leitner ES, Bennett TA, Hacker NF, Stromberg K, Stetler-Stevenson WG. Epidermal growth factor receptor signaling and the invasive phenotype of ovarian carcinoma cells. J Natl Cancer Inst. 2001;93:1375–1384. doi: 10.1093/jnci/93.18.1375. [DOI] [PubMed] [Google Scholar]
- Attoub S, Noe V, Pirola L, Bruyneel E, Chastre E, Mareel M, Wymann MP, Gespach C. Leptin promotes invasiveness of kidney and colonic epithelial cells via phosphoinositide 3-kinase-, rho-, and rac-dependent signaling pathways. FASEB J. 2000;14:2329–2938. doi: 10.1096/fj.00-0162. [DOI] [PubMed] [Google Scholar]
- Avruch J, Belham C, Weng Q, Hara K, Yonezawa K. The p70 S6 kinase integrates nutrient and growth signals to control translational capacity. Prog Mol Subcell Biol. 2001;26:115–154. doi: 10.1007/978-3-642-56688-2_5. [DOI] [PubMed] [Google Scholar]
- Bai X, Ma D, Liu A, Shen X, Wang QJ, Liu Y, Jiang Y. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science. 2007;318:977–980. doi: 10.1126/science.1147379. [DOI] [PubMed] [Google Scholar]
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Benefield J, Meisinger J, Petruzzelli GJ, Young MR. Endothelial cell response to human head and neck squamous cell carcinomas involves downregulation of protein phosphatases-1/2A, cytoskeletal depolymerization and increased motility. Invasion Metastasis. 1997;17:210–220. [PubMed] [Google Scholar]
- Berven LA, Willard FS, Crouch MF. Role of the p70(S6K) pathway in regulating the actin cytoskeleton and cell migration. Exp Cell Res. 2004;296:183–195. doi: 10.1016/j.yexcr.2003.12.032. [DOI] [PubMed] [Google Scholar]
- Boffa DJ, Luan F, Thomas D, Yang H, Sharma VK, Lagman M, Suthanthiran M. Rapamycin inhibits the growth and metastatic progression of non-small cell lung cancer. Clin Cancer Res. 2004;10:293–300. doi: 10.1158/1078-0432.ccr-0629-3. [DOI] [PubMed] [Google Scholar]
- Brooks PC, Klemke RL, Schon S, Lewis JM, Schwartz MA, Cheresh DA. Insulin-like growth factor receptor cooperates with integrin alpha v beta 5 to promote tumor cell dissemination in vivo. J Clin Invest. 1997;99:390–398. doi: 10.1172/JCI119298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- Brown EJ, Schreiber SL. A signaling pathway to translational control. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr, Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- Busca R, Bertolotto C, Ortonne JP, Ballotti R. Inhibition of the phosphatidylinositol 3-kinase/p70(S6)-kinase pathway induces B16 melanoma cell differentiation. J Biol Chem. 1996;271:31824–31830. doi: 10.1074/jbc.271.50.31824. [DOI] [PubMed] [Google Scholar]
- Cafferkey R, Young PR, McLaughlin MM, Bergsma DJ, Koltin Y, Sathe GM, Faucette L, Eng WK, Johnson RK, Livi GP. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol Cell Biol. 1993;13:6012–6023. doi: 10.1128/mcb.13.10.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Cheatham L, Monfar M, Chou MM, Blenis J. Structural and functional analysis of pp70S6k. Proc Natl Acad Sci USA. 1995;92:11696–11700. doi: 10.1073/pnas.92.25.11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci USA. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chen H, Rhoad AE, Warner L, Caggiano TJ, Failli A, Zhang H, Hsiao CL, Nakanishi K, Molnar-Kimber KL. A putative sirolimus (rapamycin) effector protein. Biochem Biophys Res Commun. 1994;203:1–7. doi: 10.1006/bbrc.1994.2140. [DOI] [PubMed] [Google Scholar]
- Chiu MI, Katz H, Berlin V. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc Natl Acad Sci USA. 1994;91:12574–12578. doi: 10.1073/pnas.91.26.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- Crouch MF. Regulation of thrombin-induced stress fibre formation in Swiss 3T3 cells by the 70-kDa S6 kinase. Biochem Biophys Res Commun. 1997;233:193–199. doi: 10.1006/bbrc.1997.6419. [DOI] [PubMed] [Google Scholar]
- Daniel C, Pippin J, Shankland SJ, Hugo C. The rapamycin derivative RAD inhibits mesangial cell migration through the CDK-inhibitor p27KIP1. Lab Invest. 2004;84:588–596. doi: 10.1038/labinvest.3700078. [DOI] [PubMed] [Google Scholar]
- Dilling MB, Dias P, Shapiro DN, Germain GS, Johnson RK, Houghton PJ. Rapamycin selectively inhibits the growth of childhood rhabdomyosarcoma cells through inhibition of signaling via the type I insulin-like growth factor receptor. Cancer Res. 1994;54:903–907. [PubMed] [Google Scholar]
- Douros J, Suffness M. New antitumor substances of natural origin. Cancer Treat Rev. 1981;8:63–87. doi: 10.1016/s0305-7372(81)80006-0. [DOI] [PubMed] [Google Scholar]
- Dufner A, Thomas G. Ribosomal S6 kinase signaling and the control of translation. Exp Cell Res. 1999;253:100–109. doi: 10.1006/excr.1999.4683. [DOI] [PubMed] [Google Scholar]
- Eng CP, Sehgal SN, Vezina C. Activity of rapamycin (AY-22,989) against transplanted tumors. J Antibiot (Tokyo) 1984;37:1231–1237. doi: 10.7164/antibiotics.37.1231. [DOI] [PubMed] [Google Scholar]
- Eynott PR, Salmon M, Huang TJ, Oates T, Nicklin PL, Chung KF. Effects of cyclosporin A and a rapamycin derivative (SAR943) on chronic allergic inflammation in sensitized rats. Immunology. 2003;109:461–467. doi: 10.1046/j.1365-2567.2003.01672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazioli F, Wong WT, Ullrich SJ, Sakaguchi K, Appella E, Di Fiore PP. The ezrin-like family of tyrosine kinase substrates: receptor-specific pattern of tyrosine phosphorylation and relationship to malignant transformation. Oncogene. 1993;8:1335–1345. [PubMed] [Google Scholar]
- Ferrari S, Bandi HR, Hofsteenge J, Bussian BM, Thomas G. Mitogen-activated 70K S6 kinase. Identification of in vitro 40 S ribosomal S6 phosphorylation sites. J Biol Chem. 1991;266:22770–22775. [PubMed] [Google Scholar]
- Friedl P, Brocker EB. The biology of cell locomotion within three-dimensional extracellular matrix. Cell Mol Life Sci. 2000;57:41–64. doi: 10.1007/s000180050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS, Ru B, Pan D. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat Cell Biol. 2002;4:699–704. doi: 10.1038/ncb847. [DOI] [PubMed] [Google Scholar]
- Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- Geoerger B, Kerr K, Tang CB, Fung KM, Powell B, Sutton LN, Phillips PC, Janss AJ. Antitumor activity of the rapamycin analog CCI-779 in human primitive neuroectodermal tumor/medulloblastoma models as single agent and in combination chemotherapy. Cancer Res. 2001;61:1527–1532. [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999a;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999b;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- Gomez-Cambronero J. Rapamycin inhibits GM-CSF-induced neutrophil migration. FEBS Lett. 2003;550:94–100. doi: 10.1016/s0014-5793(03)00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL, Bretscher A, Esch FS, Hunter T. cDNA cloning and sequencing of the protein-tyrosine kinase substrate, ezrin, reveals homology to band 4.1. EMBO J. 1989;8:4133–4142. doi: 10.1002/j.1460-2075.1989.tb08598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe M, Gansauge F, Schmid RM, Adler G, Seufferlein T. Regulation of cell growth and cyclin D1 expression by the constitutively active FRAP-p70s6K pathway in human pancreatic cancer cells. Cancer Res. 1999;59:3581–3587. [PubMed] [Google Scholar]
- Guba M, von Breitenbuch P, Steinbauer M, Koehl G, Flegel S, Hornung M, Bruns CJ, Zuelke C, Farkas S, Anthuber M, Jauch KW, Geissler EK. Rapamycin inhibits primary and metastatic tumor growth by antiangiogenesis: involvement of vascular endothelial growth factor. Nat Med. 2002;8:128–135. doi: 10.1038/nm0202-128. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Guvakova MA, Surmacz E. The activated insulin-like growth factor I receptor induces depolarization in breast epithelial cells characterized by actin filament disassembly and tyrosine dephosphorylation of FAK, Cas, and paxillin. Exp Cell Res. 1999;251:244–255. doi: 10.1006/excr.1999.4566. [DOI] [PubMed] [Google Scholar]
- Hall MN. The TOR signalling pathway and growth control in yeast. Biochem Soc Trans. 1996;24:234–239. doi: 10.1042/bst0240234. [DOI] [PubMed] [Google Scholar]
- Hammer T, Tritsaris K, Hubschmann MV, Gibson J, Nisato RE, Pepper MS, Dissing S. IL-20 activates human lymphatic endothelial cells causing cell signalling and tube formation. Microvasc Res. 2009 Mar 9; doi: 10.1016/j.mvr.2009.02.007. [Epub ahead of print]. Cited in PubMed. [DOI] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem. 1997;272:26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- Harwood FC, Shu L, Houghton PJ. mTORC1 signaling can regulate growth factor activation of p44/42 mitogen-activated protein kinases through protein phosphatase 2A. J Biol Chem. 2008;283:2575–2585. doi: 10.1074/jbc.M706173200. [DOI] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- Helliwell SB, Wagner P, Kunz J, Deuter-Reinhard M, Henriquez R, Hall MN. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol Biol Cell. 1994;5:105–118. doi: 10.1091/mbc.5.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Negrete I, Carretero-Ortega J, Rosenfeldt H, Hernandez-Garcia R, Calderon-Salinas JV, Reyes-Cruz G, Gutkind JS, Vazquez-Prado J. P-Rex1 links mammalian target of rapamycin signaling to Rac activation and cell migration. J Biol Chem. 2007;282:23708–23715. doi: 10.1074/jbc.M703771200. [DOI] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Hosoi H, Dilling MB, Shikata T, Liu LN, Shu L, Ashmun RA, Germain GS, Abraham RT, Houghton PJ. Rapamycin causes poorly reversible inhibition of mTOR and induces p53-independent apoptosis in human rhabdomyosarcoma cells. Cancer Res. 1999;59:886–894. [PubMed] [Google Scholar]
- Huang S, Houghton PJ. Resistance to rapamycin: a novel anticancer drug. Cancer Metastasis Rev. 2001;20:69–78. doi: 10.1023/a:1013167315885. [DOI] [PubMed] [Google Scholar]
- Huang S, Houghton PJ. Targeting mTOR signaling for cancer therapy. Curr Opin Pharmacol. 2003;3:371–377. doi: 10.1016/s1471-4892(03)00071-7. [DOI] [PubMed] [Google Scholar]
- Huber S, Bruns CJ, Schmid G, Hermann PC, Conrad C, Niess H, Huss R, Graeb C, Jauch KW, Heeschen C, Guba M. Inhibition of the mammalian target of rapamycin impedes lymphangiogenesis. Kidney Int. 2007;71:771–777. doi: 10.1038/sj.ki.5002112. [DOI] [PubMed] [Google Scholar]
- Iiizumi M, Liu W, Pai SK, Furuta E, Watabe K. Drug development against metastasis-related genes and their pathways: a rationale for cancer therapy. Biochim Biophys Acta. 2008;1786:87–104. doi: 10.1016/j.bbcan.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Isotani S, Hara K, Tokunaga C, Inoue H, Avruch J, Yonezawa K. Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase alpha in vitro. J Biol Chem. 1999;274:34493–34498. doi: 10.1074/jbc.274.48.34493. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Hall MN. Tor signalling in bugs, brain and brawn. Nat Rev Mol Cell Biol. 2003;4:117–126. doi: 10.1038/nrm1018. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Jackson JL, Young MR. Protein phosphatase-2A regulates protein tyrosine phosphatase activity in Lewis lung carcinoma tumor variants. Clin Exp Metastasis. 2003;20:357–364. doi: 10.1023/a:1024012000009. [DOI] [PubMed] [Google Scholar]
- Janus A, Robak T, Smolewski P. The mammalian target of the rapamycin (mTOR) kinase pathway: its role in tumourigenesis and targeted antitumour therapy. Cell Mol Biol Lett. 2005;10:479–498. [PubMed] [Google Scholar]
- Jefferies HB, Reinhard C, Kozma SC, Thomas G. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc Natl Acad Sci USA. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeno P, Ballou LM, Novak-Hofer I, Thomas G. Identification and characterization of a mitogen-activated S6 kinase. Proc Natl Acad Sci USA. 1988;85:406–410. doi: 10.1073/pnas.85.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedrin D, van Rheenen J, Hernandez L, Condeelis J, Segall JE. Cell motility and cytoskeletal regulation in invasion and metastasis. J Mammary Gland Biol Neoplasia. 2007;12:143–152. doi: 10.1007/s10911-007-9046-4. [DOI] [PubMed] [Google Scholar]
- Keith CT, Schreiber SL. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Kishimoto T, Kamata S, Otsuka M, Miyazaki M, Ishikura H. Rapamycin, a specific inhibitor of the mammalian target of rapamycin, suppresses lymphangiogenesis and lymphatic metastasis. Cancer Sci. 2007;98:726–733. doi: 10.1111/j.1349-7006.2007.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltin Y, Faucette L, Bergsma DJ, Levy MA, Cafferkey R, Koser PL, Johnson RK, Livi GP. Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol Cell Biol. 1991;11:1718–1723. doi: 10.1128/mcb.11.3.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J, Hall MN. Cyclosporin A, FK506 and rapamycin: more than just immunosuppression. Trends Biochem Sci. 1993;18:334–338. doi: 10.1016/0968-0004(93)90069-y. [DOI] [PubMed] [Google Scholar]
- Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- Kwon YS, Hong HS, Kim JC, Shin JS, Son Y. Inhibitory effect of rapamycin on corneal neovascularization in vitro and in vivo. Invest Ophthalmol Vis Sci. 2005;46:454–460. doi: 10.1167/iovs.04-0753. [DOI] [PubMed] [Google Scholar]
- Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Lin J, Lalani AS, Harding TC, Gonzalez M, Wu WW, Luan B, Tu GH, Koprivnikar K, VanRoey MJ, He Y, Alitalo K, Jooss K. Inhibition of lymphogenous metastasis using adeno-associated virus-mediated gene transfer of a soluble VEGFR-3 decoy receptor. Cancer Res. 2005;65:6901–6909. doi: 10.1158/0008-5472.CAN-05-0408. [DOI] [PubMed] [Google Scholar]
- Lin TA, Kong X, Haystead TA, Pause A, Belsham G, Sonenberg N, Lawrence JC., Jr PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- Linhares MM, Gonzalez AM, Trivino T, Melaragno C, Moura RM, Garcez MH, Sa JR, Aguiar WF, Succi T, Barbosa CS, Pestana JO. Simultaneous pancreas-kidney transplantation initial experience. Transplant Proc. 2003;35:1109. doi: 10.1016/s0041-1345(03)00328-2. [DOI] [PubMed] [Google Scholar]
- Liu L, Chen L, Chung J, Huang S. Rapamycin inhibits F-actin reorganization and phosphorylation of focal adhesion proteins. Oncogene. 2008;27:4998–5010. doi: 10.1038/onc.2008.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Li F, Cardelli JA, Martin KA, Blenis J, Huang S. Rapamycin inhibits cell motility by suppression of mTOR-mediated S6K1 and 4E-BP1 pathways. Oncogene. 2006;25:7029–7040. doi: 10.1038/sj.onc.1209691. [DOI] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- Luan FL, Ding R, Sharma VK, Chon WJ, Lagman M, Suthanthiran M. Rapamycin is an effective inhibitor of human renal cancer metastasis. Kidney Int. 2003;63:917–926. doi: 10.1046/j.1523-1755.2003.00805.x. [DOI] [PubMed] [Google Scholar]
- Luan FL, Hojo M, Maluccio M, Yamaji K, Suthanthiran M. Rapamycin blocks tumor progression: unlinking immunosuppression from antitumor efficacy. Transplantation. 2002;73:1565–1572. doi: 10.1097/00007890-200205270-00008. [DOI] [PubMed] [Google Scholar]
- Luo Y, Marx SO, Kiyokawa H, Koff A, Massague J, Marks AR. Rapamycin resistance tied to defective regulation of p27Kip1. Mol Cell Biol. 1996;16:6744–6751. doi: 10.1128/mcb.16.12.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. Rheb fills a GAP between TSC and TOR. Trends Biochem Sci. 2003;28:573–576. doi: 10.1016/j.tibs.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell. 1999;3:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- Martin J, Masri J, Bernath A, Nishimura RN, Gera J. Hsp70 associates with Rictor and is required for mTORC2 formation and activity. Biochem Biophys Res Commun. 2008;372:578–583. doi: 10.1016/j.bbrc.2008.05.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KA, Blenis J. Coordinate regulation of translation by the PI 3-kinase and mTOR pathways. Adv Cancer Res. 2002;86:1–39. doi: 10.1016/s0065-230x(02)86001-8. [DOI] [PubMed] [Google Scholar]
- Matsuo M, Yamada S, Koizumi K, Sakurai H, Saiki I. Tumour-derived fibroblast growth factor-2 exerts lymphangiogenic effects through Akt/mTOR/p70S6kinase pathway in rat lymphatic endothelial cells. Eur J Cancer. 2007;43:1748–1754. doi: 10.1016/j.ejca.2007.04.024. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Cramer LP. Actin-based cell motility and cell locomotion. Cell. 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: a regulator of cell size. Science. 1999;285:2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- Muthukkumar S, Ramesh TM, Bondada S. Rapamycin, a potent immunosuppressive drug, causes programmed cell death in B lymphoma cells. Transplantation. 1995;60:264–270. doi: 10.1097/00007890-199508000-00010. [DOI] [PubMed] [Google Scholar]
- Nourse J, Firpo E, Flanagan WM, Coats S, Polyak K, Lee MH, Massague J, Crabtree GR, Roberts JM. Interleukin-2-mediated elimination of the p27Kip1 cyclin-dependent kinase inhibitor prevented by rapamycin. Nature. 2004;372:570–573. doi: 10.1038/372570a0. [DOI] [PubMed] [Google Scholar]
- O’Connor R. Regulation of IGF-I receptor signaling in tumor cells. Horm Metab Res. 2003;35:771–777. doi: 10.1055/s-2004-814166. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Tokuda M, Tomizawa K, Matsui H, Itano T, Konishi R, Nagahata S, Hatase O. Osteoblastic differentiation is enhanced by rapamycin in rat osteoblast-like osteosarcoma (ROS 17/2.8) cells. Biochem Biophys Res Commun. 1998;249:226–230. doi: 10.1006/bbrc.1998.9118. [DOI] [PubMed] [Google Scholar]
- Pang H, Faber LE. Estrogen and rapamycin effects on cell cycle progression in T47D breast cancer cells. Breast Cancer Res Treat. 2001;70:21–26. doi: 10.1023/a:1012570204923. [DOI] [PubMed] [Google Scholar]
- Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Kleckner N. The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell. 2003;112:151–155. doi: 10.1016/s0092-8674(03)00033-3. [DOI] [PubMed] [Google Scholar]
- Polak P, Hall MN. mTOR and the control of whole body metabolism. Curr Opin Cell Biol. 2009;21:209–218. doi: 10.1016/j.ceb.2009.01.024. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Poon M, Marx SO, Gallo R, Badimon JJ, Taubman MB, Marks AR. Rapamycin inhibits vascular smooth muscle cell migration. J Clin Invest. 1996;98:2277–2283. doi: 10.1172/JCI119038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem. 1998;273:14002–14007. doi: 10.1074/jbc.273.22.14002. [DOI] [PubMed] [Google Scholar]
- Price JT, Tiganis T, Agarwal A, Djakiew D, Thompson EW. Epidermal growth factor promotes MDA-MB-231 breast cancer cell migration through a phosphatidylinositol 3′-kinase and phospholipase C-dependent mechanism. Cancer Res. 1999;59:5475–5478. [PubMed] [Google Scholar]
- Pullen N, Dennis PB, Andjelkovic M, Dufner A, Kozma SC, Hemmings BA, Thomas G. Phosphorylation and activation of p70s6k by PDK1. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- Radimerski T, Montagne J, Rintelen F, Stocker H, van der Kaay J, Downes CP, Hafen E, Thomas G. dS6K-regulated cell growth is dPKB/dPI(3)K-independent, but requires dPDK1. Nat Cell Biol. 2002;4:251–255. doi: 10.1038/ncb763. [DOI] [PubMed] [Google Scholar]
- Reddy KB, Nabha SM, Atanaskova N. Role of MAP kinase in tumor progression and invasion. Cancer Metastasis Rev. 2003;22:395–403. doi: 10.1023/a:1023781114568. [DOI] [PubMed] [Google Scholar]
- Reinhard C, Thomas G, Kozma SC. A single gene encodes two isoforms of the p70 S6 kinase: activation upon mitogenic stimulation. Proc Natl Acad Sci USA. 1992;89:4052–4056. doi: 10.1073/pnas.89.9.4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Rizzieri DA, Feldman E, Dipersio JF, Gabrail N, Stock W, Strair R, Rivera VM, Albitar M, Bedrosian CL, Giles FJ. A phase 2 clinical trial of deforolimus (AP23573, MK-8669), a novel mammalian target of rapamycin inhibitor, in patients with relapsed or refractory hematologic malignancies. Clin Cancer Res. 2008;14:2756–2762. doi: 10.1158/1078-0432.CCR-07-1372. [DOI] [PubMed] [Google Scholar]
- Roberts N, Kloos B, Cassella M, Podgrabinska S, Persaud K, Wu Y, Pytowski B, Skobe M. Inhibition of VEGFR-3 activation with the antagonistic antibody more potently suppresses lymph node and distant metastases than inactivation of VEGFR-2. Cancer Res. 2006;66:2650–2657. doi: 10.1158/0008-5472.CAN-05-1843. [DOI] [PubMed] [Google Scholar]
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1, Amammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, Abraham RT. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- Saitoh M, Pullen N, Brennan P, Cantrell D, Dennis PB, Thomas G. Regulation of an activated S6 kinase 1 variant reveals a novel mammalian target of rapamycin phosphorylation site. J Biol Chem. 2002;277:20104–20112. doi: 10.1074/jbc.M201745200. [DOI] [PubMed] [Google Scholar]
- Sakakibara K, Liu B, Hollenbeck S, Kent KC. Rapamycin inhibits fibronectin-induced migration of the human arterial smooth muscle line (E47) through the mammalian target of rapamycin. Am J Physiol Heart Circ Physiol. 2005;288:H2861–2868. doi: 10.1152/ajpheart.00561.2004. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Schaller MD. Paxillin: a focal adhesion-associated adaptor protein. Oncogene. 2001;20:6459–6472. doi: 10.1038/sj.onc.1204786. [DOI] [PubMed] [Google Scholar]
- Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Bickle M, Beck T, Hall MN. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell. 1997;88:531–542. doi: 10.1016/s0092-8674(00)81893-0. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Hall MN. Signaling to the actin cytoskeleton. Annu Rev Cell Dev Biol. 1998;14:305–338. doi: 10.1146/annurev.cellbio.14.1.305. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Kunz J, Hall MN. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc Natl Acad Sci USA. 1996;93:13780–13785. doi: 10.1073/pnas.93.24.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal SN, Baker H, Vezina C. Rapamycin (AY-22,989), a new antifungal antibiotic. II. Fermentation, isolation and characterization. J Antibiot (Tokyo) 1975;28:727–232. doi: 10.7164/antibiotics.28.727. [DOI] [PubMed] [Google Scholar]
- Seufferlein T, Rozengurt E. Rapamycin inhibits constitutive p70s6k phosphorylation, cell proliferation, and colony formation in small cell lung cancer cells. Cancer Res. 1996;56:3895–3897. [PubMed] [Google Scholar]
- Shah OJ, Anthony JC, Kimball SR, Jefferson LS. 4E-BP1 and S6K1: translational integration sites for nutritional and hormonal information in muscle. Am J Physiol Endocrinol Metab. 2000;279:E715–E729. doi: 10.1152/ajpendo.2000.279.4.E715. [DOI] [PubMed] [Google Scholar]
- Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- Stolovich M, Tang H, Hornstein E, Levy G, Cohen R, Bae SS, Birnbaum MJ, Meyuhas O. Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol Cell Biol. 2002;22:8101–8113. doi: 10.1128/MCB.22.23.8101-8113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Marx SO, Chen HJ, Poon M, Marks AR, Rabbani LE. Role for p27(Kip1) in Vascular Smooth Muscle Cell Migration. Circulation. 2001;103:2967–2972. doi: 10.1161/01.cir.103.24.2967. [DOI] [PubMed] [Google Scholar]
- Takizawa H, Kondo K, Fujino H, Kenzaki K, Miyoshi T, Sakiyama S, Tangoku A. The balance of VEGF-C and VEGFR-3 mRNA is a predictor of lymph node metastasis in non-small cell lung cancer. Br J Cancer. 2006;95:75–79. doi: 10.1038/sj.bjc.6603209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Hornstein E, Stolovich M, Levy G, Livingstone M, Templeton D, Avruch J, Meyuhas O. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol Cell Biol. 2001;21:8671–8683. doi: 10.1128/MCB.21.24.8671-8683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci USA. 2002;99:13571–13576. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada N, Patel HR, Takase K, Kohno K, Nairn AC, Gelfand EW. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc Natl Acad Sci USA. 1994;91:11477–11481. doi: 10.1073/pnas.91.24.11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Hall MN. TOR signalling and control of cell growth. Curr Opin Cell Biol. 1997;9:782–787. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- Tsukita S, Oishi K, Sato N, Sagara J, Kawai A, Tsukita S. ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J Cell Biol. 1994;126:391–401. doi: 10.1083/jcb.126.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentinis B, Baserga R. IGF-I receptor signalling in transformation and differentiation. Mol Pathol. 2001;54:133–137. doi: 10.1136/mp.54.3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Poel HG, Hanrahan C, Zhong H, Simons JW. Rapamycin induces Smad activity in prostate cancer cell lines. Urol Res. 2003;30:380–386. doi: 10.1007/s00240-002-0282-1. [DOI] [PubMed] [Google Scholar]
- Vezina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- von Manteuffel SR, Dennis PB, Pullen N, Gingras AC, Sonenberg N, Thomas G. The insulin-induced signalling pathway leading to S6 and initiation factor 4E binding protein 1 phosphorylation bifurcates at a rapamycin-sensitive point immediately upstream of p70s6k. Mol Cell Biol. 1997;17:5426–5436. doi: 10.1128/mcb.17.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, Mendoza A, Khanna C, Helman LJ. Rapamycin inhibits ezrin-mediated metastatic behavior in a murine model of osteosarcoma. Cancer Res. 2005;65:2406–2411. doi: 10.1158/0008-5472.CAN-04-3135. [DOI] [PubMed] [Google Scholar]
- Wang Y, Huang BP, Luciani DS, Wang X, Johnson JD, Proud CG. Rheb activates protein synthesis and growth in adult rat ventricular cardiomyocytes. J Mol Cell Cardiol. 2008;45:812–820. doi: 10.1016/j.yjmcc.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Wedaman KP, Reinke A, Anderson S, Yates J, 3rd, McCaffery JM, Powers T. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:1204–1220. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AS, Roskelley CD, Pelech S, Miller D, Leung PC, Auersperg N. Progressive changes in Met-dependent signaling in a human ovarian surface epithelial model of malignant transformation. Exp Cell Res. 2004;299:248–256. doi: 10.1016/j.yexcr.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Woodhouse EC, Chuaqui RF, Liotta LA. General mechanisms of metastasis. Cancer. 1997;80:1529–1537. doi: 10.1002/(sici)1097-0142(19971015)80:8+<1529::aid-cncr2>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- Yang DQ, Kastan MB. Participation of ATM in insulin signalling through phosphorylation of eIF-4E-binding protein 1. Nat Cell Biol. 2000;2:893–898. doi: 10.1038/35046542. [DOI] [PubMed] [Google Scholar]
- Yin JJ, Selander K, Chirgwin JM, Dallas M, Grubbs BG, Wieser R, Massague J, Mundy GR, Guise TA. TGF-beta signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J Clin Invest. 1999;103:197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- Zheng XF, Florentino D, Chen J, Crabtree GR, Schreiber SL. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]
- Zhou HY, Wong AS. Activation of p70S6K induces expression of matrix metalloproteinase 9 associated with hepatocyte growth factor-mediated invasion in human ovarian cancer cells. Endocrinology. 2006;147:2557–2566. doi: 10.1210/en.2005-1404. [DOI] [PubMed] [Google Scholar]