Summary
Extracellular signals regulate protein translation in many cell functions. A key advantage of control at the translational level is the opportunity to regulate protein synthesis within specific cellular subregions. However, little is known about mechanisms that may link extracellular cues to translation with spatial precision. Here we show that a transmembrane receptor, DCC, forms a binding complex containing multiple translation components, including eukaryotic initiation factors, ribosomal large and small subunits, and monosomes. In neuronal axons and dendrites DCC colocalizes in particles with translation machinery, and newly synthesized protein. The extracellular ligand netrin promoted DCC-mediated translation and disassociation of translation components. The functional and physical association of a cell surface receptor with the translation machinery leads to a generalizable model for localization and extracellular regulation of protein synthesis, based on a transmembrane translation regulation complex.
Introduction
Transmembrane receptors can provide a direct link between extracellular signals and intracellular machinery. It has long been known that receptor cytoplasmic domains can associate with two types of intracellular machinery: the cytoskeleton, and cytoplasmic signaling proteins (Flanagan and Koch, 1978; Rodbell, 1980; Ullrich and Schlessinger, 1990). From there, regulatory information can be conveyed by signaling pathways to regulate functional outputs throughout the cell, including gene expression at both transcriptional and translational levels (Pawson and Nash, 2003).
One key advantage of controlling gene expression at the translational level is that this allows protein synthesis to be spatially localized to specific subregions of the cytoplasm. This localization of protein synthesis is thought to be important in diverse biological functions, such as setting up the primary axes of the Drosophila embryo, cell migration and adhesion, axon guidance, and regulation of the synapse (St Johnston, 2005; Sutton and Schuman, 2006; Lin and Holt, 2008; Rodriguez et al., 2008). Moreover, genome scale analysis in the early Drosophila embryo indicates that most mRNAs show localization to diverse and specific subcellular sites, including at the cell membrane, implying that mechanisms to regulate localized translation participate in many, if not most, cell functions (Lecuyer et al., 2007). Regarding mechanisms for translational regulation, a great deal of progress has been made in identifying ribonucleoprotein complexes that mediate mRNA localization and translational control (St Johnston, 2005; Kiebler and Bassell, 2006; Rodriguez et al., 2008), and molecular pathways that transmit signals from cell surface receptors and regulate the translational machinery (Tee and Blenis, 2005; Sonenberg and Hinnebusch, 2007). However, it is still not well understood how extracellular signals may regulate translation with spatial precision.
Translation can be divided into three main stages: initiation, elongation and termination. The primary site for translational regulation in most systems is the initiation step, and therefore key roles in translational control are played by components involved in initiation (Sonenberg and Hinnebusch, 2007). During initiation, the ribosomal 40S and 60S subunits join the mRNA in a series of steps mediated by eukaryotic initiation factors (eIFs). The small subunit forms the 43S preinitiation complex by recruiting the initiator tRNA, bound to eIF2, a protein that is an important target of translational control. This complex then recruits mRNAs with a 5' cap through the action of eIF4E, a cap-binding protein that is a key mediator of multiple pathways of translational control. After scanning to the initiator AUG codon, the small subunit is joined by the large subunit to form the monosome, and elongation can then proceed on polysomes.
In neurons, roles for protein translation have been identified in both dendrites and axons. Developing axons can translate protein, and roles for protein synthesis have been proposed in axon growth, guidance, and regeneration. Within the axon, there can be further localization of newly synthesized proteins, preferentially within one segment along the length of an axon, or on one side of a growth cone, exposed locally to an extracellular cue (Campbell and Holt, 2001; Zhang et al., 2001; Brittis et al., 2002; Ming et al., 2002; Leung et al., 2006; Yao et al., 2006; Willis et al., 2007). In dendrites, protein synthesis is well established to have roles in synaptic plasticity, including learning and memory. Proteins can be synthesized locally within the dendrite in response to extracellular signals, and localization of translation at or near individual synapses may play a role in plasticity that is synapse-specific (Steward and Schuman, 2003; Sutton and Schuman, 2006).
To explain how protein synthesis could occur preferentially in the subcellular region of a neuron receiving an extracellular signal, one model is that transmembrane receptors would operate through signaling pathways that act at a distance, influencing translation with a range extending over part of the cell. We set out here to test an alternative, although not mutually exclusive, model based on the precedent of transmembrane receptor association with other types of machinery inside the cell. In this model, transmembrane receptors that regulate protein synthesis would colocalize with translation machinery in a particle where they would be integrated in a molecular binding complex. To test this second model, we focused on DCC, a well characterized receptor in axon growth and guidance (Keino-Masu et al., 1996; Fazeli et al., 1997), and also found postsynaptically in dendrites (Parent et al., 2005). DCC is one of the receptors for the extracellular factor netrin (Keino-Masu et al., 1996; Fazeli et al., 1997), and netrin is known to promote protein synthesis in axons (Campbell and Holt, 2001).
The results here show a physical complex of DCC with components involved in translation initiation, including eIFs, ribosomal subunits, and monosomes. Furthermore, DCC functionally mediated translational regulation in response to its ligand netrin-1. These studies lead to a generalizable model with transmembrane association of cell surface receptors and the translation machinery contributing to the specificity, efficiency, and spatially precise control of translation, based on a novel type of transmembrane complex regulated by extracellular cues.
Results
DCC colocalizes with translation machinery
In initial experiments to explore a potential association of DCC and translation machinery, we tested for colocalization in spinal commissural axon growth cones, where DCC promotes growth and chemoattraction in response to netrin (Keino-Masu et al., 1996; Fazeli et al., 1997). In particular, colocalization would be of interest at filopodial tips, where DCC is known to be located, and which are highly motile structures that explore the environment and respond to extracellular cues (Drees and Gertler, 2008). Immunofluorescence localization showed punctate labeling of both DCC and ribosomal markers, and overlap was seen in a subset of puncta, with a diameter of approximately 0.2 to 1 μm (Figures 1A-1D, S1A-S1C and S1H). Notably, colocalization was consistently seen at the tips of filopodia. Quantitation of the overlap within the growth cone indicated approximately 35% of DCC puncta overlapping with ribosomal puncta, and approximately 83% of ribosomal puncta overlapping with DCC puncta. When Pearson's correlation coefficient was calculated for individual DCC puncta that showed detectable ribosome labeling, in >90% of cases the value was between 0.5 and 0.95, confirming true colocalization of puncta rather than fortuitous overlap (see Experimental Procedures).
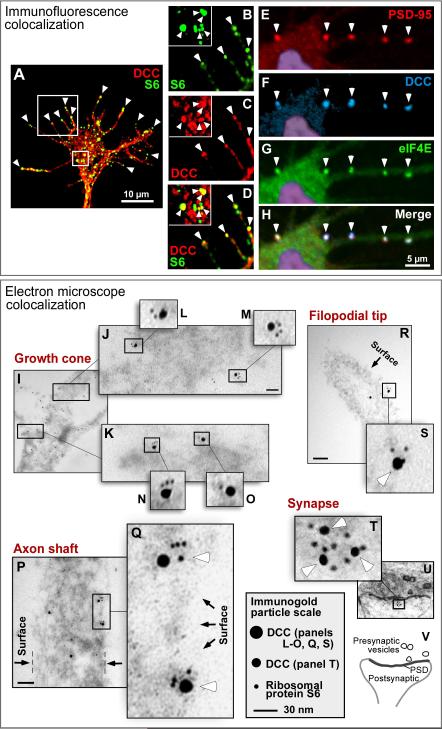
Figure 1. DCC colocalizes with translational components.
(A-H) Immunofluorescence. (A-D) Cultured commissural axons. DCC (red) and ribosomal protein S6 (green) appeared in puncta within the growth cone, with a subset of puncta overlapping (yellow). Panels B-D are enlargement of boxes from A, showing filopodial tips, with a central growth cone area in the inset. Arrowheads: examples of colocalized puncta, notably at filopodial tips. (E-H) Cultured hippocampal dendrites. Labeling for DCC (blue), eIF4E (green), and the postsynaptic marker PSD-95 was seen in puncta at synapses (arrowheads). Nuclear DAPI staining is shown in purple. (I-V) Electron microscopy, showing double immunogold labeling for endogenous DCC (15 nm gold particles in panels I-S; 10 nm particles in T-V) and ribosomal protein S6 (5 nm particles). Some variation in particle size is caused by silver enhancement. (I-S) Cultured commissural axons. Panels I-O show distal part of growth cone near the base of the filopodia; P and Q show axon shaft, oriented vertically; R shows filopodium, with distal end toward upper left. (T-V) Cultured hippocampal neuron synapse, diagrammed in panel V. White arrowheads: examples of clustered particles. Black arrows: outer boundary of the cell. Boxes show enlargement of clustered particles. Scale bar: 70 nm in J, R and P; 15 nm in T. See also Figure S1.
Colocalization was also tested in hippocampal neuron dendrites. DCC puncta colocalized with markers of the translational machinery, including at synapses identified by triple-labeling with the postsynaptic marker PSD-95 (Figures 1E-1H, S1D-S1G and data not shown). Approximately 35% of PSD-95 puncta overlapped with DCC puncta, while most but not all DCC puncta overlapped with markers of translational machinery; for example 89% of DCC puncta overlapped with eIF4E puncta. These results appear broadly consistent with electron microscopic studies reporting that polysomes are present in approximately 10-40% of synaptic spines (Ostroff et al., 2002; Steward and Schuman, 2003).
To complement the overall view of subcellular distribution provided by fluorescence imaging, electron microscopy was used for resolution on a molecular scale. Immunogold labeling showed colocalization of DCC and ribosome markers in axonal growth cones, axon shafts, near the tips of axon filopodia, and in dendritic synaptic spines (Figures 1I-1V). As expected for ultrathin sections (80 nm), labeling density was moderate, and some particles did not show colocalization (Figure S1I), also consistent with the fluorescence results showing colocalization of some but not all puncta. While the immunolocalization procedure does not allow detailed visualization of organelles, counterstaining of the cytoplasm showed that many of the colocalized particles were near the cell surface, while others were in intracellular locations where DCC may be associated with intracellular membranes, perhaps reflecting transport to or from the cell surface. It should be noted, however, that it is not possible here to accurately quantitate the proportion of surface versus intracellular location due to the sectioning geometry; or to determine the precise location of molecules relative to one another or the cell surface, since the gold particles are on a similar scale to the molecules and are tethered via antibodies that can span up to ~30 nm. Addressing colocalization, however, distances among clustered particles were generally 0-30 nm, well within the range for immunogold detection of a direct molecular interaction.
DCC coprecipitates with translation machinery
We next tested whether DCC and translational machinery might show physical association. Initially, to provide an unbiased sampling of candidate interactors, full length DCC or vector control were transfected into 293 cells, then DCC was immunoprecipitated, and coprecipitated proteins were identified by tandem mass spectrometry. As shown in Figure 2A, translational components were among the coprecipitated proteins, including translation initiation factors (polypeptides of eIFs 1, 2, 3, 4 and 5) and proteins of the ribosomal small subunit (S10, S13 and S24) and large subunit (L5, L10a, L13, L23, L28 and L38). Most of the proteins from this list were then tested by Western blot of transfected 293 cells, confirming specific co-precipitation in every case tested (Figures 2B and S2). In addition, coprecipitation of selected proteins was confirmed with embryonic spinal cord, providing a native developmental context where the proteins are expressed at endogenous levels (Figure 3B). Importantly, however, no co-precipitation was seen with a DCC protein lacking the cytoplasmic domain (DCC-Δcyto; Figure 3C), indicating that DCC is not associated with the translation machinery merely because the DCC polypeptide is itself being synthesized there, and indicating that these associations are mediated by the DCC cytoplasmic domain.
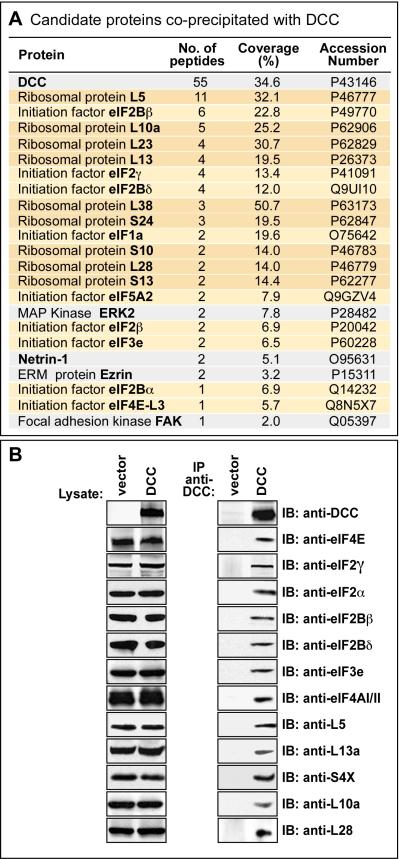
Figure 2. Tandem mass spectrometry screen identifying translational components coprecipitated with DCC.
(A) Tandem mass spectrometry identification of proteins following immunoprecipitation with anti-DCC antibody. Proteins listed showed specific coprecipitation from 293-net cells transfected with DCC, versus vector control. Ribosomal proteins are highlighted in dark orange, and initiation factors in light orange. Grey indicates proteins previously shown to interact directly with DCC (although other known interaction partners such as Nck, Src and Fyn were not seen here, presumably reflecting detection limits in this screen). Accession numbers: UniProtKB/Swiss-Prot data base. (B) Most of the candidate translational components from the mass spectrometry screen were tested by Western blot, and in every case tested were confirmed to show co-immunoprecipitation with DCC. See also Figure S2.
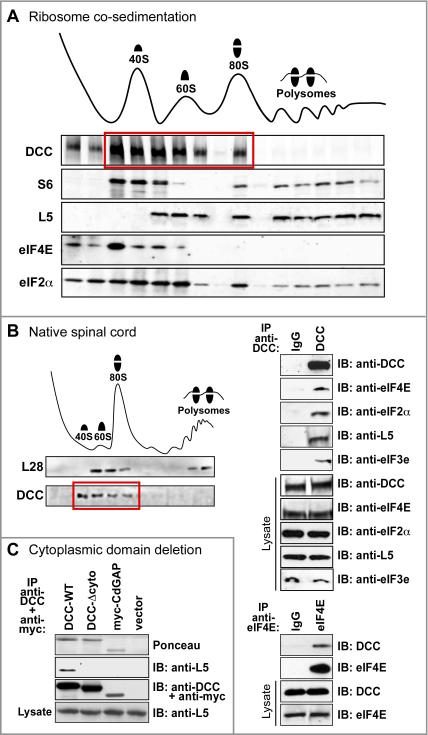
Figure 3. DCC receptor physically associates with translation machinery.
(A) Ribosome sedimentation from netrin-expressing 293-net cells transfected with full length DCC. DCC cosedimented in 40S, 60S and 80S fractions (red box) but not prominently in the polysome fraction. Control proteins were distributed as expected from previous studies: S6 in 40S, 80S and polysomal fractions; L5 in 60S, 80S and polysomal fractions; eIF4E primarily in the 40S fraction; and eIF2α in all four fractions. (B) DCC association with translation machinery, with all proteins expressed at endogenous levels in developing spinal cord. Left panel: DCC cosedimented in 40S, 60S and 80S fractions. Other markers such as eIF4E distributed as expected in spinal cord (not shown). Upper right panel: translational components co-immunoprecipitated with DCC. Lower right panel: DCC co-immunoprecipitated with eIF4E. (C) DCC-Δcyto mutant lacking a cytoplasmic domain did not co-precipitate ribosomal marker L5. myc-tagged mCdGAP was a negative control. See also Figure S3.
DCC cosediments with the ribosome
To investigate the interaction further, we performed sucrose gradient velocity sedimentation of ribosomes. DCC was found to co-sediment with both small and large ribosomal subunits, as well as monosomes, while little or no obvious DCC signal was seen in the polysome fraction under these conditions (Figures 3A and S3). In addition to 293 cells, a comparable distribution was obtained with native embryonic spinal cord (Figure 3B). On a per-subunit basis the association of DCC with the large and small subunits appeared approximately comparable (Figure 3A). Sedimentation profiles of the key translational regulators eIF4E and eIF2α are also shown for comparison (Figure 3A), with eIF2α cosedimenting with subunits, monosomes and polysomes (Ramaiah et al., 1992), while eIF4E cosedimented primarily with the 40S fraction as in previous studies (Kedersha et al., 2002; Napoli et al., 2008). The lack of prominent association of DCC with polysomes provided further confirmation that the association with ribosomes is not simply attributable to the DCC polypeptide itself being translated there. The results, however, appeared very consistent with a role in translational regulation, since the initiation stage involves recruitment of 40S and 60S subunits to form the 80S ribosome, and is the major site of translational control in most systems.
DCC and its cytoplasmic domain functionally affect translation
Netrin-1 promotion of axon growth and chemoattraction can be mediated not only through DCC, but also two other receptors, neogenin and DSCAM. To assess whether DCC can mediate effects of netrin-1 on translation, DCC was transfected into 293 cells, which did not contain detectable levels of endogenous netrin receptors. Translation was assessed using a dual-luciferase reporter system for cap-dependent translation with a cap-independent cistron as internal control (Figure S4A)(Kruger et al., 2001). The results indeed showed a DCC-dependent effect of netrin-1 on translation in this system (Figures 4A and S4B-S4F). Likewise, an increase in total cell protein synthesis was seen by radiolabeled amino acid incorporation (Figure S4G), although as in previous studies (for example Pfeifer et al., 2008) the signal-to-background ratio was lower than for reporter translation. The effect of netrin-1 was also tested on the association of DCC with translational components, showing that it promoted dissociation (Figures 4B and S4H). (Note, however, that an association was still detectable in 293-net cells, consistent with the results in Figure 2.) Finding that netrin promoted dissociation of DCC from translational machinery appeared very consistent with our earlier experiments which had shown an association of DCC with components involved in translation initiation, but not with polysomes (Figures 2 and 3). These results all appear consistent with a model where full length DCC associates with translation initiation machinery, and signaling by DCC within this complex promotes formation of actively elongating polysomes which are no longer associated with DCC.
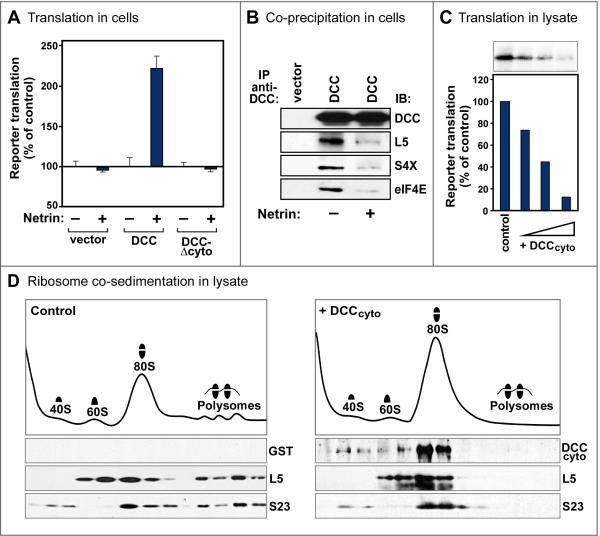
Figure 4. Effects of DCC and netrin on translation.
(A) Full length DCC mediated netrin promotion of translation. 293 cells were transfected with DCC constructs, together with a reporter plasmid for cap-dependent translation (Figure S4A). Reporter translation was normalized to a cap-independent internal control, and is compared in netrin-expressing 293-net cells (+) versus 100% in control cells (–). DCC lacking a cytoplasmic domain (DCC-Δcyto) showed no effect on translation. n=3 experiments; error bars, SEM. Without netrin, full length DCC promoted translation slightly in some experiments, although to a much lesser degree than with netrin (data not shown). (B) Netrin reduced the association of DCC with translation components. Western analysis showed reduced coprecipitation of markers of the ribosomal large subunit (L5), small subunit (S4X) and initiation factor eIF4E with exogenous DCC in netrin-expressing 293-net cells (+), compared with control 293 cells (–). An association above background remained, however, in the presence of netrin (see Figure S4H). (C) DCC cytoplasmic domain (DCCcyto) inhibited translation. Purified recombinant DCCcyto (100 ng, 200 ng, and 500 ng) or GST control protein was added to a rabbit reticulocyte lysate translation system. (D) Ribosome sedimentation profiles of reticulocyte lysates following incubation with purified recombinant DCCcyto (right) or GST control (left). DCCcyto cosedimented with 40S, 60S and 80S fractions. DCCcyto decreased the polysome fraction and increased the 80S fraction, implying that a step after 80S ribosome assembly was inhibited and became rate limiting in this assay, although earlier steps may also be affected. Ribosomal proteins distributed as expected: S23 in 40S, 80S and polysome fractions; L5 in 60S, 80S and polysome fractions. See also Figure S4.
Since our earlier results had indicated that the DCC cytoplasmic domain was required for association with translation machinery, we assessed the role of this domain in translation. The DCC-Δcyto mutant had no significant effect in mediating the effect of netrin-1, showing a requirement for the cytoplasmic domain in regulating translation (Figure 4A).
As an additional test, a truncated receptor was made containing the DCC cytoplasmic domain only (DCCcyto). Expressing a truncated cell surface receptor is a common strategy that can provide information about binding and functional properties of a specific receptor domain. Functional effects of truncated receptors are often interfering rather than activating, since they may retain the ability to bind and occupy some signaling components, while lacking other domains required for productive signaling. For DCCcyto in particular, functional effects on translation, if any, were expected to be inhibitory, since the DCC extracellular domain is required for downstream signaling (Round and Stein, 2007). When DCCcyto was tested by transfecting it into 293 cells, this caused cell death (data not shown). While this could be consistent with translation inhibition, it did not provide a good system to study translation further. When DCCcyto was instead tested in a cell-free system, the reticulocyte lysate, it was indeed found to inhibit translation, as indicated both by reduced reporter translation, and reduced polysome levels (Figures 4C and 4D). The reduction in polysomes was accompanied by an increase in the 80S fraction, indicating that a late step after 80S monosome formation became rate limiting under these conditions. Cosedimentation of the DCCcyto polypeptide in these experiments was seen with ribosomal 40S, 60S and 80S fractions (Figure 4D). Thus DCCcyto, like full length DCC, associated physically with machinery involved in translation initiation, and produced functional effects on translation, although in the case of DCCcyto these effects were inhibitory not promoting, consistent with this mutant lacking an extracellular domain. These results provided further support for an overall model where the DCC cytoplasmic domain interacts both physically and functionally with the translation initiation machinery.
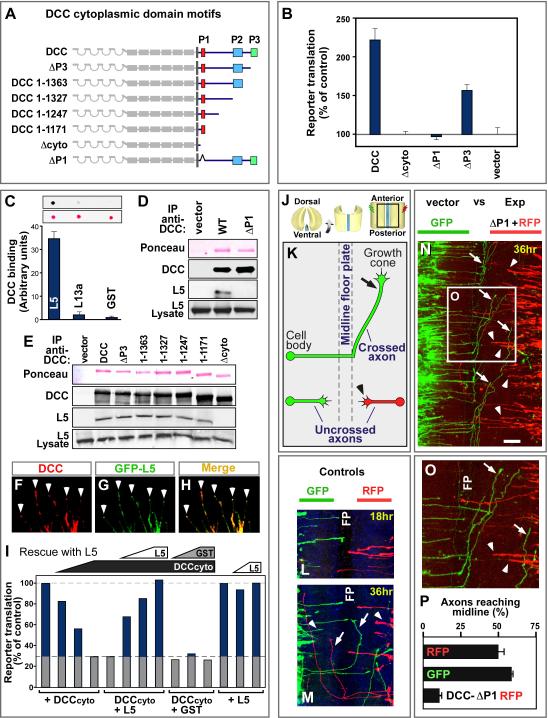
Involvement of the DCC P1 motif and ribosomal protein L5
The DCC intracellular domain contains three motifs, P1, P2 and P3, which are conserved from C. elegans to mammals, and are therefore likely to mediate downstream functions (Figure 5A)(Garbe and Bashaw, 2004; Round and Stein, 2007). When DCC mutants were tested in the reporter assay for cellular translation, P3 deletion caused some reduction of translation (DCC-ΔP3 mutant; Figure 5B). This is expected, since P3 enhances DCC dimerization, which is believed to mediate signal transduction across the membrane, and it also binds known signaling proteins (Round and Stein, 2007) which might in principle modulate translation. A more drastic effect was seen upon deletion of the P1 motif (DCC-ΔP1 mutant), which completely blocked the ability of DCC to promote translation (Figure 5B). Correspondingly, the P1 deletion reduced the association of DCC with translational components close to background levels (Figure S5A).
Figure 5. Involvement of the DCC P1 domain and ribosomal protein L5.
(A) DCC cytoplasmic domain contains three conserved motifs: P1, P2 and P3. A nested series of deletions were made, and an internal deletion of the P1 motif. (B) DCC promotion of translation was prevented by P1 deletion (DCC-ΔP1), and partially reduced by P3 deletion (DCC-ΔP3). The assay was as in Figure 4A, with cap-dependent reporter translation normalized to a cap-independent control. n=3 experiments; error bars, SEM. (C-I) Physical and functional interactions with ribosomal protein L5. (C) Far Western dot blot quantitation of direct binding of purified recombinant DCC cytoplasmic domain to purified recombinant L5. Ribosomal protein L13a and GST provide controls. Ponceau stain shows similar loading. n=4 experiments; error bars, SEM. (D, E) Western analysis of endogenous L5 co-immunopreciptation with DCC, showing L5 associated with all mutants in the nested deletion series except DCC-Δcyto, and not with DCC-ΔP1. (F-H) Immunofluorescence of DCC (red) and GFP-L5 (green) co-transfected into 293 cells, showing colocalization in neurite-like extensions, notably at the tips (arrowheads). (I) Functional interaction between ribosomal protein L5 and DCCcyto. The inhibitory effect of DCCcyto in a reticulocyte lysate (see Figure 4C) was rescued in a dose-dependent manner by recombinant L5 (100 ng, 200 ng, and 500 ng). GST control at the same concentrations did not rescue. L5 alone (200 ng and 500 ng) had no noticeable effect. Column 5 is a duplicate of column 4 to clarify control value. (J-P) Effect of DCC-ΔP1 as a dominant inhibitory mutant on commissural axon pathfinding in developing spinal cord. Chick spinal cords at E3.5 (HH 23) were electroporated and analyzed 36 hr later, compared with controls electroporated on the contralateral side of the same spinal cord. (J) Illustration of spinal cord open book preparations, with imaged area in box. (K) Diagram of axon trajectories seen in panel N: axons grow toward the midline floor plate (arrowheads), cross it, then turn and grow anteriorly (arrow). (L, M) Control plasmids expressing GFP and RFP; equivalent axon numbers, growth rates and patterns were seen for both tracers after 18 or 36 hours. (N, O) Axons electroporated with DCC-ΔP1 (right side, red) compared to GFP control (left side, green) showed greater numbers remaining at locations midway toward the floorplate (red; examples indicated by arrowheads), and fewer had reached or crossed the floor plate compared with control (green; examples indicated by arrows). Panel O shows enlargement of boxed area. (P) Quantitation of axons reaching the midline after electroporation with control or DCC-ΔP1 mutant construct. A total of 530 axons were analyzed in seven experiments; error bars, SEM. Scale bar in N, 100 μm. FP, floor plate. See also Figure S5.
Among the translational components that coprecipitated with DCC in the mass spectrometry analysis, the most prominent was ribosomal protein L5, with 11 peptides (Figure 2A). L5 also appeared interesting because it is thought to be located at or near the ribosomal P site, and L5 mutations can affect peptidyl-tRNA binding (Fabijanski and Pellegrini, 1981; Meskauskas and Dinman, 2001; Spahn et al., 2001; Takahashi et al., 2005). Purified recombinant L5 was therefore tested for binding to purified recombinant DCC cytoplasmic domain, and the results showed direct binding (Figure 5C). While this direct association of DCC with a ribosomal protein is striking, it should be emphasized that this is unlikely to be the only interaction of DCC relevant to translation (see Discussion).
A common strategy to test the biological function of protein associations is to mutate the interaction site. We were therefore interested to test whether the association of DCC with L5 could be mapped to a specific site within the DCC primary sequence. When nested deletions were examined removing successively larger regions from the C-terminus (Figure 5A), a region containing the P1 motif was found to be sufficient for L5 binding (Figure 5E). The P1 motif was found also to be necessary for L5 binding, as shown by testing the DCC-ΔP1 mutant (Figure 5D). It may be worth noting that although the P1 motif can also interact with co-receptors in the UNC5 family, they mediate repellent actions of netrin whereas commissural neurons are attracted, and they are not expressed in commissural neurons or the cell lines used in this study (Round and Stein, 2007; and unpublished data). The experiments here show that the P1 motif, which we had already found to be required to promote translation, was also the sequence necessary and sufficient for association of DCC with ribosomal protein L5.
DCC and L5 were also examined for overlapping subcellular expression. Since none of the L5 antibodies tested gave a signal in immunocytochemistry, a GFP-L5 fusion protein was co-expressed with DCC in 293 cells, where DCC promotes neurite-like extensions. Colocalization was seen in the neurite-like extensions, notably at their tips (Figures 6F-6H). Similarly, GFP-L5 expressed in commissural neurons colocalized with DCC at filopodial tips in axon growth cones (Figure S5B).
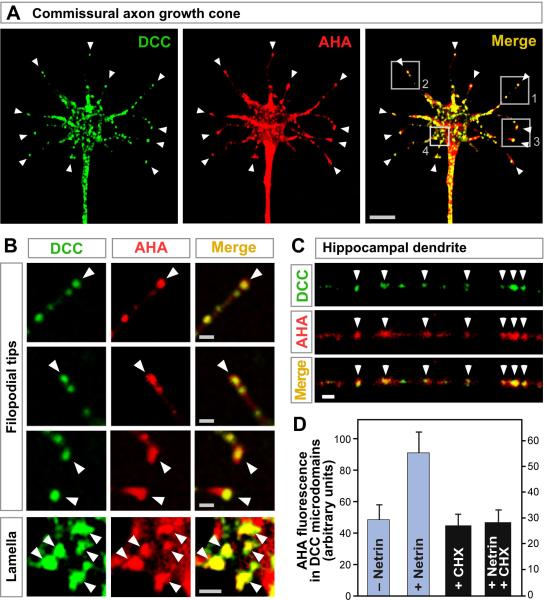
Figure 6. DCC colocalizes with newly synthesized protein.
(A, B) Localization of DCC (green) and AHA labeled sites of newly synthesized protein (red) in cultured commissural axon growth cone. Panel B shows enlargement of boxes in A. Arrowheads show examples of overlapping puncta (yellow) notably at filopodial tips. (C) DCC (green) and AHA (red) labeling in cultured hippocampal neuron dendrite. Arrowheads indicate overlap. (D) Quantitation of AHA labeling in DCC puncta at filopodial tips. AHA was added for 20 min; then cycloheximide (CHX) for 20 min; then 200 ng/ml netrin-1 for 10 min. Netrin induced an increase in labeling that was blocked by cycloheximide. Panels A-C were in the presence of netrin-1. Scale bars: 5 μm in panel A; 1 μm in B and C. See also Figure S6.
Having found that the P1 motif of DCC is required both for regulation of translation and for association with L5, the other side of the interaction was addressed, by testing whether perturbations of L5 might affect actions of the DCC cytoplasmic domain. Disruption of L5 within cells is problematic due to the key roles of L5 in ribosome biogenesis and function. Therefore, we used the cell-free reticulocyte system which we had already shown to be inhibited by DCCcyto. A functional interaction between L5 and DCCcyto was tested by the logic of dominant interference: if L5 provides a bridge between DCC and the ribosome, then addition of sufficient L5 should functionally occupy both its binding partners and therefore block the effects of the DCC cytoplasmic domain on translation. Accordingly, L5 addition was found to cause a dose-dependent and highly effective reversal of the DCCcyto effect on translation (Figure 5I). These experiments demonstrate a functional interaction between the DCC cytoplasmic tail and L5 in translation.
As a further test of our models, the effect of the P1 motif was tested on commissural axon behavior. Previous studies have implicated the P1 motif in axon attraction and growth promotion in C. elegans (Gitai et al., 2003; Garbe and Bashaw, 2004), and in UNC-5H dependent axon repulsion (Garbe and Bashaw, 2004; Round and Stein, 2007), and also have shown a role for the P3 motif in commissural axon guidance (Finger et al., 2002). Here, to assess P1 functionally in commissural axons, we used DCC-ΔP1 as a dominant interfering construct. The rationale for this type of dominant interfering experiment is that a mutant receptor can block the actions of endogenous wild type receptor by heterodimerization or by binding unproductively to upstream or downstream components. The DCC–ΔP1 construct was confirmed to have an interfering effect on translation, and to be expressed in neuronal growth cones (Figures S5C-S5E). To test commissural axon behavior, DCC was electroporated into chick spinal cord open book explants, using constructs which also expressed a fluorescent protein reporter, and 36 hours later axon trajectory was traced (Figures 5J-5P). Previous work has shown that DCC promotes axon growth and chemoattraction in response to floor plate derived netrin: following DCC gene disruption axons initially grow in a ventral direction but many fail to reach the floor plate (Fazeli et al., 1997). In control experiments here, full length DCC, or the use of GFP versus RFP markers, did not impair the ability of axons to reach the floor plate (Figures 6L, 6M, 6P, and data not shown). When DCC-ΔP1 was introduced, however, axons grew out in a ventral direction, but a greatly reduced proportion reached the midline (p<0.0001; Figures 6N to 6P), producing a phenotype comparable to DCC mutant mice (Fazeli et al., 1997). While the P1 motif might in principle affect other downstream processes in addition to translation, these results in spinal cord appear consistent with all our other data, and indicate that P1, a short motif that mediated translational regulation, and was necessary and sufficient for associations with ribosomal components, also affected DCC function in promoting outgrowth or attraction of spinal commissural axons toward the midline.
Newly synthesized protein colocalizes with DCC
Finding that DCC colocalizes and physically associates with the translation machinery, and regulates translation, implied that DCC should be capable of regulating protein synthesis in a localized manner within the cell. To investigate this, we used a tracer of newly synthesized protein, azidohomoalanine (AHA), which acts as an amino acid analog incorporated in place of methionine, and can be visualized after cell fixation by covalent cycloaddition with a tagged alkyne (Dieterich et al., 2007; Beatty and Tirrell, 2008). AHA labeling was found to overlap with DCC in axons of commissural neurons, notably in puncta at the tips of filopodia (Figures 6A, 6B, S6A and S6B). We also used an independent tracer of newly synthesized protein which operates by a different mechanism, Cy3-puromycin, a fluorescently tagged derivative of the aminoacyl-tRNA analog puromycin (Smith et al., 2005) and obtained comparable results (Figures S6C-S6I). In hippocampal neurons too, AHA showed punctate labeling that overlapped with DCC puncta (Figure 6C). When the effect of netrin was tested, DCC puncta in commissural axon filopodia showed increased labeling of newly synthesized protein (Figures 6D and S6G). Thus, the accumulation of newly synthesized protein within DCC microdomains was increased by netrin transmembrane signaling.
Discussion
Physical association of cellular components plays a central role in establishing both the spatial organization of the cell, and the specificity of cellular pathways. Cell surface receptors and the translation machinery are two of the basic classes of cellular components. The results here lead to a generalizable model where a transmembrane receptor interacts both extracellularly with signaling cues, and intracellularly with the translation machinery, to form a transmembrane translation regulation particle.
Interactions of DCC with protein synthesis machinery
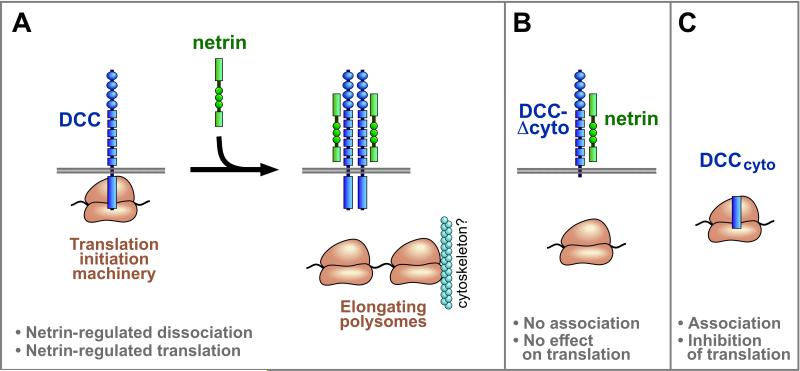
The results here showed that DCC associated with eIFs, ribosomal 40S and 60S subunits, and 80S ribosomes, though not prominently with polysomes. Taken together with finding that netrin-1 promoted translation in a DCC dependent manner, and reduced the association of DCC with translation components, these results lead to the following model. DCC forms a complex with translation initiation machinery. Signaling by DCC within this complex is enhanced by netrin, and promotes the formation of actively elongating polysomes which are no longer associated with DCC (Figure 7A). Signaling by DCC is believed to be mediated by dimerization or higher-order clustering, and in the absence of netrin, spontaneous dimerization and signaling by DCC may promote some translation, although at a lower level than with netrin. These interactions with the translation machinery are mediated via the DCC cytoplasmic domain, which was found to be necessary and sufficient for interactions with translational components (Figures 7B and 7C).
Figure 7. Models for interaction of DCC with translational machinery.
(A) In the model illustrated, full length DCC associates with translation initiation machinery, including 40S, 60S and 80S ribosomes and eIFs. Signaling by DCC within this complex is enhanced by netrin, and promotes formation of actively elongating polysomes which are no longer associated with DCC. Polysomes may nevertheless tend to remain locally within micron scale structures such as a filopodial tip or synaptic spine, and this may be facilitated by their known interaction with the cytoskeleton. Transduction of a signal across the membrane by DCC and other receptors is believed to be mediated by dimerization or higher-order clustering. In the absence of netrin, spontaneous dimerization and signaling by DCC may promote some translation, although at a lower level than with netrin. Signaling by some DCC molecules in the cell would not preclude the coexistence of other DCC molecules that remain in a complex with translation initiation machinery. In addition to DCC and translation initiation machinery, the complex is proposed also to include signal transduction proteins, and DCC may regulate more than one step of initiation. (B) No prominent association with translation components or functional effect on translation was seen with DCC mutants lacking the cytoplasmic domain (DCC-Δcyto) or lacking the 20 amino acid P1 motif (DCC-ΔP1), showing a requirement of these regions for physical and functional interactions with translation machinery. (C) The DCC cytoplasmic domain by itself, DCCcyto, cosedimented with 40S, 60S and 80S ribosomes, and inhibited protein synthesis in a cell-free system, showing that this region is sufficient for physical and functional interactions with translational machinery. DCCcyto is proposed to act here as an interfering mutant, occupying downstream components but lacking an extracellular domain necessary for positive signaling. Although the diagrams are not to a precise scale, receptor length and ribosome diameter are approximately in proportion: the eukaryotic ribosome is ~20-30 nm across (Spahn et al., 2001); each Ig domain (oval) and FNIII domain (small square) is ~4 nm long; the intracellular domain of DCC (rectangle) is approximately 25% of the protein sequence as illustrated here in proportion with the extracellular length to account for potential folding, or in more extended conformation it could be up to 100 nm long.
The association of DCC with translation initiation machinery, but not prominently with the polysome fraction, is reminiscent of the key translational regulator eIF4E, which cosedimented mainly with the ribosomal small subunit and not prominently with polysomes, as confirmed under the conditions here and seen in previous results (Kedersha et al., 2002; Napoli et al., 2008). Why DCC, or eIF4E, would not show more prominent association with the polysome fraction is an interesting question, since polysomes undergo repeated rounds of initiation as successive ribosomes are added. One potential explanation is suggested by observations that the initial recruitment of translation machinery to an mRNA involves formation of specialized ribonucleoprotein structures where the mRNA is looped to place the 5’ cap next to the 3’ polyA tail, whereas subsequently added ribosomes join an mRNA loop that is already undergoing active translation and may no longer require all the same components (Amrani et al., 2008). Although eIF4E and DCC shared the property of promoting translation yet not cosedimenting prominently with polysomes, they are very different in other respects. Whereas eIF4E initially recruits mRNA to the small subunit, we found that the truncated DCCcyto fragment affected a late stage after 80S ribosome assembly. While it is possible that this late step is the primary site of regulation by full length DCC, additional stages of translation initiation are likely to be regulated by full length DCC, as discussed further below.
Regarding protein interactions linking DCC to translational control, our studies implicated the conserved cytoplasmic DCC P1 motif, and ribosomal protein L5. Previous studies of eukaryotic L5 indicate that it is located at or near the ribosomal P site, and showed that mutations in L5 can influence binding of the peptidyl-tRNA to the ribosome (Fabijanski and Pellegrini, 1981; Meskauskas and Dinman, 2001; Spahn et al., 2001; Takahashi et al., 2005). Therefore, one model could be that effects of DCC on translation might be mediated via structural changes in L5. An alternative model would be that L5 serves more passively as a tether helping link DCC to the ribosome, while translational regulation would be mediated via other DCC interactors. The second of these models may be favored by our finding that DCC preferentially affected cap-dependent rather than cap-independent translation, since functional effects transmitted by a general translation component such as L5 might be expected to influence both types of translation equally. In either model, the location of L5 is notable because it could provide a mechanism to position DCC, together with other associated proteins, near the ribosomal P site, where the ribosomal large and small subunits, the initiator tRNA, and the initiator AUG codon all come together during translation initiation.
Another protein that may be involved in linking DCC to translation is Nck-1, which has been found to bind directly to the DCC cytoplasmic domain (Li et al., 2002). In a separate study, Nck-1 was shown to bind directly to eIF2β, and to regulate translation (Kebache et al., 2002). Since eIF2β is part of the 43S pre-initiation complex, Nck-1 therefore has the necessary binding specificities to act as a bridge between DCC and the ribosomal small subunit. Thus, together, L5 and Nck-1 can provide a model for DCC to be linked to both the ribosomal large and small subunits.
DCC associates with several other signaling proteins, some of which have been implicated in translational control in other systems (Garbe and Bashaw, 2004; Round and Stein, 2007). Also, eIF4E and its upstream regulator 4E-BP1 are thought to mediate netrin signaling (Campbell and Holt, 2003; Leung et al., 2006), and we find here that eIF4E associates in a complex with DCC. We therefore favor an overall model where DCC would act as a platform for the assembly of a large multicomponent complex, containing not only ribosomes and eIFs, but also multiple signal transduction components, providing more than one pathway structurally and functionally linking DCC to translational regulation within an integrated molecular apparatus.
In relation to the lack of prominent DCC association with polysomes, it is interesting that ribosomes have long been known to associate with the cytoskeleton, and in contrast to DCC, this cytoskeletal association has been reported to involve few or no monosomes or subunits, but most or all polysomes (Lenk et al., 1977; Hesketh and Pryme, 1991). This suggests a model where DCC forms a complex with translation initiation components, whereas actively translating polysomes would instead become associated with the cytoskeleton. Cytoskeletal attachment of polysomes could provide mechanisms either for directed movement of translating polysomes, or to retain them within specific subcellular sites close to the receptor (Figure 7A).
The identification of a complex of a transmembrane receptor with translation machinery opens up many possibilities for functions of this complex in ribonucleoprotein transport, localization, metabolism and regulation. In this regard, it would be interesting to know whether the DCC-containing particles identified here have any relationship with previously described ribonucleoprotein particles (Kiebler and Bassell, 2006) or mRNA binding proteins (St Johnston, 2005; Kiebler and Bassell, 2006; Lin and Holt, 2008; Rodriguez et al., 2008). Also, while DCC may regulate general translation of all mRNAs that are present locally, an appealing model is that its association with translation components may provide a mechanism to preferentially localize and regulate a specific subset of mRNAs.
Translation in axons and dendrites
Our observations on the association of DCC with translational machinery in neurons appear consistent with previous work showing functional roles for local protein translation in chemoattractant responses to netrin (Campbell and Holt, 2001; Ming et al., 2002). Meanwhile, guidance receptors also interact directly with the cytoskeleton, and accordingly translation inhibitors do not block all axon morphological responses, as reported for example by Roche et al. (2009) for cues other than netrin, although their study like others confirmed an involvement of translation in chemoattraction. The association of DCC with sites of protein synthesis within micron scale subcellular structures such as the filopodial tip and synaptic spine, as seen here, can provide a mechanism to help regulate translation with spatial precision in processes such as axon guidance and synaptic plasticity.
Other receptors
While the study here focuses on DCC, the model of receptor association with translation machinery may be more broadly applicable. Many imaging studies have found ribosomes, or newly synthesized protein, in the same vicinity as cell surface receptors, for example in fibroblast lamellipodia, near synapses or other sites of cell-cell contact, and under the surface membrane of axons (Koenig and Martin, 1996; Steward and Schuman, 2003; Rodriguez et al., 2006; Sutton and Schuman, 2006). Although it is not clear whether this localization places ribosomes in direct association with the membrane, it is worth noting that the electron microscopic approaches that are generally used identify polysomes but not subunits or monosomes (Sutton and Schuman, 2006), whereas we found here an association of DCC with subunits and monosomes. Other potentially consistent observations include the presence of a Wnt receptor in particles that sediment with approximately the velocity of a ribosome (Bilic et al., 2007), and the association of focal adhesion proteins with initiation factors and ribosomal proteins (de Hoog et al., 2004). We find that a subset of receptors other than DCC show physical and functional interactions with the translation machinery (unpublished data). While specific molecular mechanisms are likely to vary with different receptors, the concept of a transmembrane translation regulation particle, where cell surface receptors form a physical complex with protein synthesis machinery and regulate translation, could be widespread in cell biology.
General conclusions
Regulated protein synthesis is implicated in a wide variety of cellular functions controlled by extracellular cues in both normal biology and pathology (St Johnston, 2005; Tee and Blenis, 2005; Sutton and Schuman, 2006; Lecuyer et al., 2007; Lin and Holt, 2008; Rodriguez et al., 2008). Linking a cell surface receptor in a complex together with the translation machinery could have several potential advantages. One may be to enhance the efficiency or speed of regulation by bringing components together in an integrated molecular machine. Another is the potential for each receptor to associate with a unique repertoire of mRNAs, providing a novel layer of specificity in biological regulation. A third advantage may be to allow spatial precision of translation, which may be crucial for cellular functions such as motility, adhesion, guidance, and synaptic plasticity. Association of DCC or other cell surface receptors with translation machinery in a transmembrane translation regulation particle could contribute to varied developmental, physiological and pathological processes.
Experimental Procedures
Primary neuron cultures
Commissural neuron cultures were prepared from E10.5-11.5 mouse spinal cords by trituration and plated on polylysine; >95% of cells were positive for commissural markers TAG-1 and DCC. Hippocampal neuron cultures from E18-19 rat brain were cultured for three weeks on laminin polylysine.
Coprecipitation, Western analysis, and tandem mass spectrometry
Cell lysates were prepared in Triton-X100 buffer. For spinal cord lysates, approximately 15 mouse E11.5 cords were homogenized. Immunoprecipitation used protein G–Sepharose beads washed with RIPA buffer. Far Western binding analysis used affinity purified recombinant GST fusion proteins probed with recombinant purified DCC cytoplasmic domain, followed by anti-DCC antibody and detection by chemiluminescence. For tandem mass spectrometry, after gel purification and trypsin digestion, peptides were analyzed by LC-MS/MS. All major peaks in the spectrum were assigned to predicted peptide ions, peptide representation of 138 proteins was present in immunoprecipitates from cells with DCC and not control cells, and peptides for proteins with two or less peptide hits were individually analyzed to confirm tryptic peptide identity. The entire immunoprecipitation and mass spectrometry analysis was performed twice; L5 and other ribosomal proteins and initiation factors were identified in both experimental series, and aggregate peptide counts are shown in Figure 2A.
Sucrose gradient velocity sedimentation of ribosomes
293-net cells transiently transfected with DCC were treated with cycloheximide, and lysed in 1% Triton X-100. For each spinal cord gradient, twelve cords were homogenized in low salt buffer with digitonin. Lysates were precleared, centrifuged on a sucrose velocity gradient, and eluted on a fractionator monitored by 254 nm UV.
Functional assays of translation and newly synthesized protein
Cellular cap-dependent translation assays used bicistronic reporter plasmid pRL-5'-IRES-FL (Kruger et al., 2001), which directs cap-dependent translation of Renilla luciferase (RL), and also cap-independent IRES-mediated translation of firefly luciferase (FL) which was used as an internal control. In vitro translation experiments used Retic Lysate IVT Kit (Ambion), with Xef-1 RNA template, adding purified recombinant GST-DCCcyto, GST-L5 or GST control. Translated Xef-1 protein was resolved on SDS-PAGE, transferred to nitrocellulose and quantitated by phosphoimager. For AHA labeling, cells were incubated in methionine-free medium without serum containing 100 μg/ml AHA for the times indicated, then fixed and permeabilized with Triton-X100 and incubated in Click-iT tetramethylrhodamine (Invitrogen).
Image analysis
For electron microscopy, cultured neurons were permeabilized in saponin or Triton-X100, glutaraldehyde post-fixed, embedded in resin, and 80 nm sectioned. Non-specific distribution of particles in areas devoid of cells was negligible, and controls showed no gold particle clustering. Fluorescence imaging used an inverted laser confocal at 0.25-2.0 micron per z-series slice. All fluorescence quantitation used original unprocessed image data, with no pixels at zero intensity or saturated. In panels displayed in the Figures, for consistent visibility across the intensity range, contrast and brightness were adjusted uniformly within each experimental series. To confirm that individual yellow puncta reflected true colocalization, rather than fortuitous overlap, ROIs for individual puncta were selected by automatic thresholding in one channel, then Pearson's correlation coefficient (r) was measured for the two channels. This tests all pixels within an ROI for correlation over the full range of intensity values: chance overlap corresponds to r=0; identical patterns produce r=1, which would suggest an artifact; and co-localization of two molecules in a common structure should produce an intermediate value. For the growth cone as a whole, r values up to 0.8 were obtained. As a control, a circular ROI was selected containing a central part of the growth cone, and one channel was gradually rotated; although the density of yellow pixels remained similar, the r value for correlation of intensities was confirmed to fall to approximately zero. To quantify numbers of overlapping puncta, ROIs were selected in each channel independently by automatic thresholding to select 0.2-1 μm puncta, and the number of puncta that overlapped was then counted. To quantify AHA intensity, each ROI for quantitation was selected automatically using the green channel, with a constant threshold above background, to select 0.2-1 μm puncta from twenty different filopodia in twenty different growth cones, chosen at random and blind to the red channel. Mean pixel intensity was then measured within each ROI in the red channel.
Supplementary Material
Acknowledgments
We thank Marc Kirschner, Dietmar Schmucker, Melissa Hancock, Marc Tessier-Lavigne, Nancy Kadersha, Steven Gygi, Ross Tomaino, Tom Walz, Maria Ericsson, Natalie Farny, Pam Silver, Joachim Hauber, Taka-Aki Sato, Nathalie Lamarche-Vane, John Blenis, Davie Van Vactor, Judy Glaven, Yao Chen, Dan Nowakowski, Miriam Osterfield, and Nicolas Preitner, for help, reagents, advice, and comments on the manuscript. This work was supported by grants from the NIH (J.G.F.) and NCIC (P.P.R.), an HHMI fellowship (P.A.B.), a CIHR fellowship (J.T.), and a Canada Research Chair and HFSP Career Development Award (P.P.R.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty KE, Tirrell DA. Two-color labeling of temporally defined protein populations in mammalian cells. Bioorg Med Chem Lett. 2008;18:5995–5999. doi: 10.1016/j.bmcl.2008.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilic J, Huang YL, Davidson G, Zimmermann T, Cruciat CM, Bienz M, Niehrs C. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316:1619–1622. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- Brittis PA, Lu Q, Flanagan JG. Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell. 2002;110:223–235. doi: 10.1016/s0092-8674(02)00813-9. [DOI] [PubMed] [Google Scholar]
- Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32:1013–1026. doi: 10.1016/s0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- de Hoog CL, Foster LJ, Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell. 2004;117:649–662. doi: 10.1016/s0092-8674(04)00456-8. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, Schuman EM. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc. 2007;2:532–540. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- Drees F, Gertler FB. Ena/VASP: proteins at the tip of the nervous system. Curr Opin Neurobiol. 2008;18:53–59. doi: 10.1016/j.conb.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabijanski S, Pellegrini M. Identification of proteins at the peptidyl-tRNA binding site of rat liver ribosomes. Mol Gen Genet. 1981;184:551–556. doi: 10.1007/BF00352539. [DOI] [PubMed] [Google Scholar]
- Fazeli A, Dickinson SL, Hermiston ML, Tighe RV, Steen RG, Small CG, Stoeckli ET, Keino-Masu K, Masu M, Rayburn H, et al. Phenotype of mice lacking functional Deleted in colorectal cancer (Dcc) gene. Nature. 1997;386:796–804. doi: 10.1038/386796a0. [DOI] [PubMed] [Google Scholar]
- Finger JH, Bronson RT, Harris B, Johnson K, Przyborski SA, Ackerman SL. The netrin 1 receptors Unc5h3 and Dcc are necessary at multiple choice points for the guidance of corticospinal tract axons. J Neurosci. 2002;22:10346–10356. doi: 10.1523/JNEUROSCI.22-23-10346.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan J, Koch GLE. Cross-linked surface Ig attaches to actin. Nature. 1978;273:278–281. doi: 10.1038/273278a0. [DOI] [PubMed] [Google Scholar]
- Garbe DS, Bashaw GJ. Axon guidance at the midline: from mutants to mechanisms. Crit Rev Biochem Mol Biol. 2004;39:319–341. doi: 10.1080/10409230490906797. [DOI] [PubMed] [Google Scholar]
- Gitai Z, Yu TW, Lundquist EA, Tessier-Lavigne M, Bargmann CI. The netrin receptor UNC-40/DCC stimulates axon attraction and outgrowth through enabled and, in parallel, Rac and UNC-115/AbLIM. Neuron. 2003;37:53–65. doi: 10.1016/s0896-6273(02)01149-2. [DOI] [PubMed] [Google Scholar]
- Hesketh JE, Pryme IF. Interaction between mRNA, ribosomes and the cytoskeleton. Biochem J. 1991;277(Pt 1):1–10. doi: 10.1042/bj2770001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebache S, Zuo D, Chevet E, Larose L. Modulation of protein translation by Nck-1. Proc Natl Acad Sci U S A. 2002;99:5406–5411. doi: 10.1073/pnas.082483399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keino-Masu K, Masu M, Hinck L, Leonardo ED, Chan SS, Culotti JG, Tessier-Lavigne M. Deleted in Colorectal Cancer (DCC) encodes a netrin receptor. Cell. 1996;87:175–185. doi: 10.1016/s0092-8674(00)81336-7. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Koenig E, Martin R. Cortical plaque-like structures identify ribosome-containing domains in the Mauthner cell axon. J Neurosci. 1996;16:1400–1411. doi: 10.1523/JNEUROSCI.16-04-01400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger M, Beger C, Welch PJ, Barber JR, Manns MP, Wong-Staal F. Involvement of proteasome alpha-subunit PSMA7 in hepatitis C virus internal ribosome entry site-mediated translation. Mol Cell Biol. 2001;21:8357–8364. doi: 10.1128/MCB.21.24.8357-8364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lenk R, Ransom L, Kaufmann Y, Penman S. A cytoskeletal structure with associated polyribosomes obtained from HeLa cells. Cell. 1977;10:67–78. doi: 10.1016/0092-8674(77)90141-6. [DOI] [PubMed] [Google Scholar]
- Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Meriane M, Triki I, Shekarabi M, Kennedy TE, Larose L, Lamarche-Vane N. The adaptor protein Nck-1 couples the netrin-1 receptor DCC (deleted in colorectal cancer) to the activation of the small GTPase Rac1 through an atypical mechanism. J Biol Chem. 2002;277:37788–37797. doi: 10.1074/jbc.M205428200. [DOI] [PubMed] [Google Scholar]
- Lin AC, Holt CE. Function and regulation of local axonal translation. Curr Opin Neurobiol. 2008;18:60–68. doi: 10.1016/j.conb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskas A, Dinman JD. Ribosomal protein L5 helps anchor peptidyl-tRNA to the P-site in Saccharomyces cerevisiae. RNA. 2001;7:1084–1096. doi: 10.1017/S1355838201001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming GL, Wong ST, Henley J, Yuan XB, Song HJ, Spitzer NC, Poo MM. Adaptation in the chemotactic guidance of nerve growth cones. Nature. 2002;417:411–418. doi: 10.1038/nature745. [DOI] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Ostroff LE, Fiala JC, Allwardt B, Harris KM. Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat hippocampal slices. Neuron. 2002;35:535–545. doi: 10.1016/s0896-6273(02)00785-7. [DOI] [PubMed] [Google Scholar]
- Parent AT, Barnes NY, Taniguchi Y, Thinakaran G, Sisodia SS. Presenilin attenuates receptor-mediated signaling and synaptic function. J Neurosci. 2005;25:1540–1549. doi: 10.1523/JNEUROSCI.3850-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- Pfeifer I, Elsby R, Fernandez M, Faria PA, Nussenzveig DR, Lossos IS, Fontoura BM, Martin WD, Barber GN. NFAR-1 and -2 modulate translation and are required for efficient host defense. Proc Natl Acad Sci U S A. 2008;105:4173–4178. doi: 10.1073/pnas.0711222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiah KV, Dhindsa RS, Chen JJ, London IM, Levin D. Recycling and phosphorylation of eukaryotic initiation factor 2 on 60S subunits of 80S initiation complexes and polysomes. Proc Natl Acad Sci U S A. 1992;89:12063–12067. doi: 10.1073/pnas.89.24.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche FK, Marsick BM, Letourneau PC. Protein synthesis in distal axons is not required for growth cone responses to guidance cues. J Neurosci. 2009;29:638–652. doi: 10.1523/JNEUROSCI.3845-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodbell M. The role of hormone receptors and GTP-regulatory proteins in membrane transduction. Nature. 1980;284:17–22. doi: 10.1038/284017a0. [DOI] [PubMed] [Google Scholar]
- Rodriguez AJ, Czaplinski K, Condeelis JS, Singer RH. Mechanisms and cellular roles of local protein synthesis in mammalian cells. Curr Opin Cell Biol. 2008;20:144–149. doi: 10.1016/j.ceb.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AJ, Shenoy SM, Singer RH, Condeelis J. Visualization of mRNA translation in living cells. J Cell Biol. 2006;175:67–76. doi: 10.1083/jcb.200512137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J, Stein E. Netrin signaling leading to directed growth cone steering. Curr Opin Neurobiol. 2007;17:15–21. doi: 10.1016/j.conb.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Smith WB, Starck SR, Roberts RW, Schuman EM. Dopaminergic stimulation of local protein synthesis enhances surface expression of GluR1 and synaptic transmission in hippocampal neurons. Neuron. 2005;45:765–779. doi: 10.1016/j.neuron.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. New modes of translational control in development, behavior, and disease. Mol Cell. 2007;28:721–729. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae--tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Steward O, Schuman EM. Compartmentalized synthesis and degradation of proteins in neurons. Neuron. 2003;40:347–359. doi: 10.1016/s0896-6273(03)00635-4. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Hirayama S, Odani S. Ribosomal proteins cross-linked to the initiator AUG codon of a mRNA in the translation initiation complex by UV-irradiation. J Biochem. 2005;138:41–46. doi: 10.1093/jb/mvi096. [DOI] [PubMed] [Google Scholar]
- Tee AR, Blenis J. mTOR, translational control and human disease. Semin Cell Dev Biol. 2005;16:29–37. doi: 10.1016/j.semcdb.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178:965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci. 2006;9:1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.