Abstract
Metastasis suppressor proteins regulate multiple steps in the metastatic cascade, including cancer cell invasion, survival in the vascular and lymphatic circulation, and colonization of distant organ sites. Understanding the biology of metastasis suppressors provides valuable mechanistic insights that may translate to therapeutic opportunities. Several reports have explored novel strategies for restoring metastasis suppressor function, including gene transfer, induction of previously suppressed gene expression and exogenous administration of gene product. Pathways activated downstream of metastasis suppressor loss can also be targeted. Although none of these strategies are yet in routine clinical use, several are being tested preclinically and in clinical trials.
Myriad oncogenes and tumour suppressor genes have been functionally implicated in the process of transformation and tumorigenesis; these genes positively and negatively regulate the subsequent development of a primary tumour1. By contrast, a growing body of literature has defined another class of genes that function, positively or negatively, in the regulation of metastasis2, the complex process through which malignant cancer cells leave a primary organ site, invade through basement membranes and connective tissue structures, journey to a distant site through the lymphatic or haematogenous circulation and finally establish a clinically detectable foothold in a distant organ3. Genes that regulate these steps in the metastatic cascade are similar to those that regulate transformation and tumorigenesis in the sense that they can be either promoters or suppressors of the phenotype. Just as tumour promoters such as oncogenic Ras or SRC and tumour suppressors such as PTEN or p53 regulate tumorigenesis, similar promoters and suppressors regulate metastasis.
For a few of these metastasis genes, loss of expression or function is requisite for the development of distant metastases, because they suppress one of the key steps of invasion, dissemination, arrest, survival and growth in a second parenchyma. Genes that inhibit metastasis but do not affect the ability of the transformed cells to produce a tumour at the primary site (which would define them as tumour suppressors) are known as metastasis suppressor genes. In this Review, we will provide perspective on these genes, discuss the rationale for targeting metastasis suppressor genes as a therapeutic modality, and review several cases in which such strategies have begun to show promise.
Perspective on metastasis suppressor genes
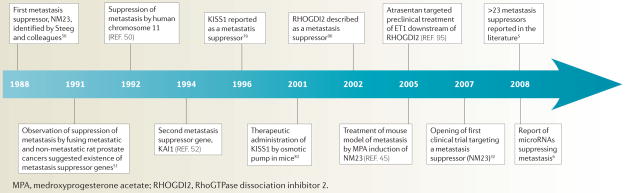
Until recently, few metastasis suppressor genes had been characterized: 5 years ago the list included only eight. In terms of broad ontology, these genes all had a similar function: regulating key cell signalling pathways, including both G-protein-coupled and tyrosine kinase receptor signalling, and small GTPase and MAPK signal transduction, reviewed recently4. However, in the ensuing few years the field has grown drastically, with one recent report reviewing more than 23 separate genes, including additional genes regulating key signalling pathways and genes regulating other functions as diverse as adhesion, migration, cell death and angiogenesis5. Further enriching the biological complexity, in 2008 an entire new class of microRNAs that suppress metastasis was described6. These molecules have been demonstrated to regulate metastasis through their ability to bind to the 3′ untranslated regions of and coordinately regulate key genes that mediate the metastatic phenotype7. Key milestones in the development of this field are summarized in the TIMELINE and recent reviews5,8,9.
Timeline.
Key advances in the metastasis suppressor field
One reason that there have been relatively few metastasis suppressor genes described until recently is that their identification and characterization not only involves a convergence of several types of data, but also requires an in vivo metastasis model for testing suppressor function that models the natural history of the specific tumour type with reasonable fidelity. This has been greatly aided by the widespread availability of several immunocompromised mouse strains for xenograft models10. Experiments to identify metastasis suppressors often include screens to identify candidate genes by comparing cells or tissues of different metastatic competence, examining the expression or mutation status of such candidates in human tumour tissues and, indispensably, showing in an in vivo metastasis assay that reconstitution of the suppressor in fact does suppress metastasis formation without abrogating proliferation or tumorigenesis (BOX 1).
Box 1 . The identification and characterization of metastasis suppressor genes.
Screens of cells of differing metastatic properties to identify differentially expressed candidate genes
Strategies have used screens such as chromosome transfer to screen for metastasis suppressor loci50, differential display and subtractive hybridization techniques30, and comparative microarray studies of cell lines exhibiting differing in vivo metastatic potentials90. Recently, workup of candidate metastasis suppressor genes and targets has been aided by available microarray expression data for many tumour histologies108, as candidates can be quickly examined across different stage tumours for expression patterns consistent with potential metastasis suppressor function40,90.
Correlation of loss of candidate gene expression or function with development of metastasis in patients
Validation of a candidate metastasis suppressor gene should include examination of its expression or function in human tumour tissues, ideally from actual metastases. However, resection of such lesions is rare for most malignancies. Instead, surrogate measures of metastatic ability are used, including whether loss of expression of the candidate in the primary tumour is associated with the development of metastasis in patient follow-up92, clinicopathological surrogates of aggression (stage, grade and so on) or patient survival.
Demonstration of in vivo suppression of metastasis
Typically, control and derivative cells exogenously re-expressing a candidate metastasis suppressor are used to prove suppression of metastasis without hindering tumorigenesis, the sine qua non for defining a metastasis suppressor protein. These experiments generally use tumour-derived cell lines introduced into rodents10. Spontaneous metastasis assays examine cells in an orthotopic xenograft109 or at subcutaneous or other sites in a heterotopic xenograft110. These evaluate the majority of the steps in the metastatic process, but not all tumour types have xenograft models amenable to this assay. Experimental metastasis assays involve injection of tumour cells directly into the arterial or venous circulation111, examining only the latter portion of the metastatic process, but can be faster and more reproducible than spontaneous metastasis assays. Either way, effects on primary tumour growth must be ruled out by subcutaneous or orthotopic xenograft assays as well.
From function to therapeutic insights
The basic rationale for development of therapies targeting metastasis suppressor genes is the same as that for targeting metastasis in general11,12. However, mechanistic insights obtained from studying these proteins have led to the identification of what we believe is a key vulnerability of metastatic cancer and perhaps the most relevant target for therapeutic intervention: metastatic colonization at the second tissue site. Interestingly, circulating cancer cells or even those found as single or small clusters in distant organs are not necessarily the harbingers of clinically overt metastatic disease13, although they are surprisingly pervasive14,15 and seem to occur early in localized disease in both patient series16,17 and animal models18,19. By contrast, colonization occurs when these disseminated cells grow from single or clusters of ectopic cancer cells (micrometastases) into clinically apparent macrometastases, a step that is extremely inefficient20–22. Even a US epidemiological survey speaks to the issue of the generality and relevance of this early seeding phenomenon in several of the most common cancer types. Steeg et al. examined the US National Cancer Institute SEER database, analysing tumour staging at diagnosis for several common cancers and found that, of cancer cases without clinically apparent metastases at diagnosis, between 29% and 37% of patients nonetheless exhibit regional lymph nodes that are positive for tumour cells23.
In short, many tumours may have already disseminated widely on a cellular basis before the time of diagnosis24. Thus, as other authors have noted25,26, in patients whose primary tumours are successfully treated, subsequent suppression of metastatic colonization is a salient therapeutic niche. This is a variation of the adjuvant therapy concept, which attempts to destroy cells in distant organs by delivering therapy following definitive treatment of the primary tumour. In the case of metastasis suppressor-targeted therapy, the same goal would be pursued, but by targeting the function of metastasis suppressor genes that work specifically at this key juncture rather than through conventional cytotoxic agents. Either way, this kind of suppression attempts to delay recurrence, which may equate to a functional cure in patients who die from non-cancer-related causes.
Though not all metastasis suppressor genes have been demonstrated to suppress this rate-limiting colonization step, a growing number of studies have demonstrated that several of them function at this point27; that is, in experimental assays modelling a setting of widely disseminated cells, their expression has been shown specifically to suppress single and small clusters of viable cells in a dormant state in the second parenchyma, whereas control cells progress to form gross macrometastatic nodules. The genes identified to date include KISS1, KAI1 (also known as CD82), NM23 (also known as NM23-H1 and NME1) and MAP2K4, which have been reviewed recently28,29, and others of the growing number of metastasis suppressors may also function at this stage. It also bears mentioning the caveat that these and other metastasis suppressor genes may also function at other steps in the metastatic cascade. Indeed, many have been shown experimentally to suppress processes such as cellular motility, invasion and survival during dissemination (FIG. 1). However, given the early, wide dissemination of cancer cells, these properties are likely to prove less adaptable to the colonization therapeutic niche.
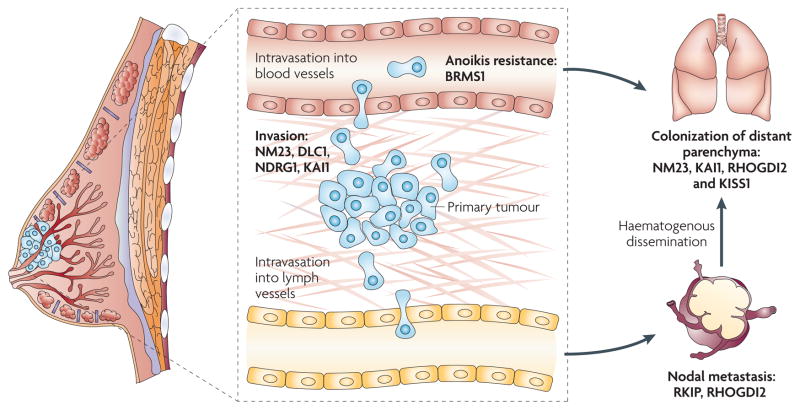
Figure 1. Metastasis suppressor genes and steps in the metastatic cascade in human cancer.
The biological process of metastasis is a complex cascade with multiple steps in which suppressor activity may prevent clinically apparent metastasis. Given the prevalence and mortality of human breast cancer, we focus here on this tissue type, although these principles apply to most solid malignancies. In the primary tumour, deregulation of oncogenes and tumour suppressor genes mediates the conversion of normal cells to a neoplastic phenotype1. By definition, metastasis suppressor genes do not prevent these steps, but must function subsequently in the cascade as shown. Invasion of the basement membrane, stroma and vasculature constitutes one key, negative prognostic turning point in the natural history of breast and other cancer types. Many metastasis suppressor genes, including NM23 (REF. 40), DLC1 (REF. 115), KAI1 (REF. 116) and NDRG1 (REF. 70) have been shown to function in in vitro and in vivo surrogates of invasion in breast cancer cells. Nodal metastasis is also a key prognostic stage for breast cancer patients117, now aggressively managed clinically. Loss of the metastasis suppressors Raf kinase inhibitor protein (RKIP)66 and RhoGTPase dissociation inhibitor 2 (RHOGDI2)93 have been shown to be associated with nodal metastasis in breast cancer. Survival during circulatory dissemination is another step of the metastatic cascade that has been studied in less detail; however, several reports have suggested that resistance to anoikis is important for metastasis118 and that the metastasis suppressor gene BRMS1 (breast cancer metastasis suppressor 1) increases anoikis114,119. Because increasing data suggest that tumours are widely disseminated on an individual cellular basis even at the time of diagnosis of localized disease, the role of metastasis suppressors in preventing outgrowth of isolated single or cellular clusters (micrometastases), known as the ‘colonization stage’ of metastasis, is compelling. NM23 (REF. 29), KAI1 (REF. 54), RHOGDI2 (REF. 94) and KISS1 (REFs 82,120) have been shown to function at this stage.
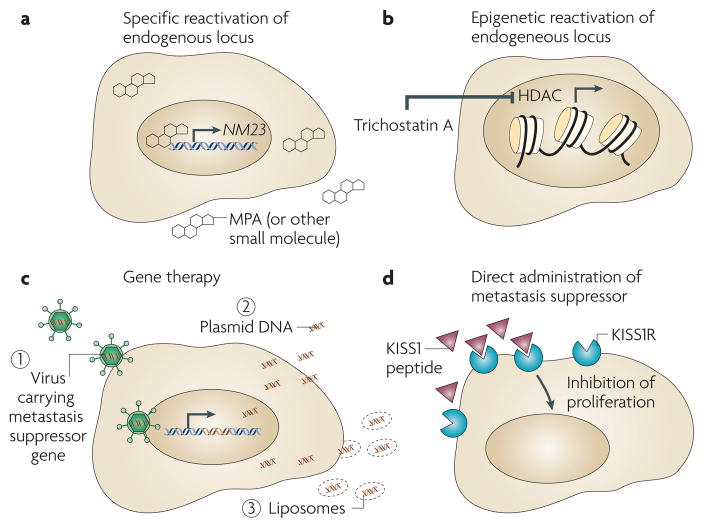
The involvement of metastasis suppressors in the formation of macrometastases suggests that functionally reconstituting these proteins could be beneficial. In the case of several metastasis suppressors, recent key reports have developed strategies for reconstituting protein function that have already proved promising in pre-clinical studies (TABLE 1). These strategies fall into three broad categories, including reconstitution of metastasis suppressor expression by induction of the endogenous locus or by gene therapy (FIG. 2a–c), direct administration of the suppressor protein itself (FIG. 2d) and targeting essential downstream pathways that are activated by loss of suppressor function (FIG. 3). Below, we discuss representative studies that highlight these approaches as well as discuss future strategies that use advanced pharmacogenomic and bioinformatic approaches aimed at discovering agents that effectively target these key players in metastatic vulnerability.
Table 1.
Metastasis suppressor genes: functions and reported targeting strategies
| Symbol | Alias(es) | Function(s) | Potential targeting strategy |
|---|---|---|---|
| BMP4 | BMP2B | Soluble cytokine | Direct therapeutic administration of suppressor protein* |
| BRMS1 | None | Chromatin and transcriptional regulation; regulation of gap junctions | None published at present |
| CTGF | CCN2, IGFBP8 | Soluble cytokine | None published at present |
| DLC1 | ARHGAP7 | Regulation of RhoGTPase signalling | Re-induction of endogenous gene through HDAC inhibition69 |
| KAI1 | CD82, kangai 1 | Inhibition of EGFR signaling; induction of senescence through interaction with DARC | Therapeutic re-induction of endogenous gene by plant extracts60; viral62 and non-viral61 gene therapy |
| KISS1 | KiSS-1, metastin | Soluble ligand for G-protein-coupled receptor | Direct therapeutic administration of suppressor protein83; possibly small molecule mimetics84 |
| MKK4 | MAP2K4 | Signal transduction | Antibody-mediated activation pathway upstream of MKK4 (REF. 122) |
| NDRG1 | CAP43, DRG1, RTP | Unknown | Induced by iron chelators123, p53 (REF. 124) and PTEN expression125 |
| NM23 | NME1, NM23-H1 | Histidine kinase activity to KSR1, decreasing Ras signalling; regulation of downstream gene expression | Re-induction of endogenous gene42,47,48; viral gene therapy49; inhibition of downstream genes40 |
| RHOGDI2 | ARHGDIB, LyGDI, GDID4 | Regulation of Rho family member activation; regulation of downstream gene expression | Inhibition of downstream genes95 |
| RKIP | PEBP1 | Binds to and inhibits Raf kinase activity and downstream signalling | Epigenetic re-induction of endogenous gene67 |
R. Anderson, unpublished data, also presented at the 2008 Meeting of the American Association for Cancer Research-Metastasis Research Society, Vancouver, Canada. BMP4, bone morphogenetic protein 4; BRMS1, breast cancer metastasis suppressor 1; DARC, Duffy chemokine receptor; EGFR, epidermal growth factor receptor; HDAC, histone deacetylase; MKK4, MAPK kinase 4; RHOGDI2, RhoGTPase dissociation inhibitor 2; RKIP, Raf kinase inhibitory protein.
Figure 2. Strategies for restoring metastasis suppressor function.
Given the key roles shown for metastasis suppressors in vivo and the association of their loss with negative outcomes in patients, re-expression of these proteins would seem a rational therapeutic strategy. This has been accomplished by strategies that are both specific — for example, re-induction of endogenous NM23 expression by medroxyprogesterone acetate (MPA)45 (a) — and general — for example, the use of chromatin-modifying drugs67,70,71 (b), including trichostatin A, which induces repressed gene expression by inhibiting histone deacetylases (HDACs). Exogenous viral gene therapy for re-expression of metastasis suppressor protein has also been reported (c) and has shown promise in preclinical models49,61,62. Non-viral vectors121 for suppressor re-expression, including naked plasmid and cationic liposomes, have also been reported61. Direct administration of the metastasis suppressor protein is another strategy (d). In at least two cases, the metastasis suppressor protein itself is a soluble, secreted factor, amenable to direct therapeutic administration, in a fashion analogous to that of insulin. In 2001, Ohtaki et al. reported the use of osmotic pumps to administer KISS1 to prevent the development of metastasis83. This may proceed through induction of dormancy through KISS1R or another receptor82 and, for KISS1, small molecule mimetics are also under development84.
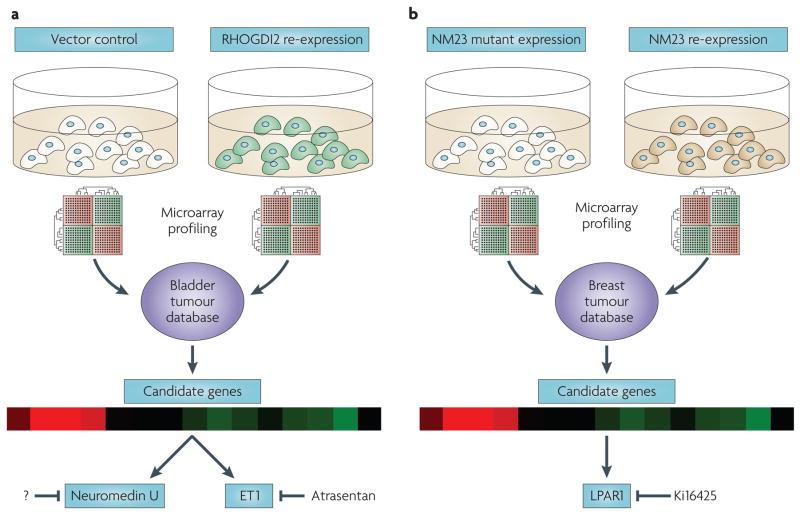
Figure 3. Targeting key genes downstream of metastasis suppressors.
In the case of two metastasis suppressor genes, NM23 (REFs 40,100) and RhoGTPase dissociation inhibitor 2 (RHOGDI2)95,98, investigators have found by microarray expression profiling that loss of metastasis suppressor expression results in transcriptional deregulation. a. By re-expressing RHOGDI2 in metastatic bladder cancer cells and comparing transcripts repressed by the suppressor to genes overexpressed in invasive bladder tumours, Titus et al. discovered both endothelin 1 (ET1), which is druggable by an inhibitor of its receptor, atrasentan, and neuromedin U, which is a potent pro-metastatic gene that cannot yet be targeted therapeutically. b. By microarray profiling breast cancer cells re-expressing NM23 and mutants lacking suppressor activity, then screening differentially expressed transcripts for appropriate correlations to NM23 expression levels in human tumours40, Horak et al. uncovered LPAR1, a lysophosphatidic acid receptor, among other genes, as a key downstream target potentiating motility and metastasis100 in breast cancer cells. An antagonist of LPAR1, Ki16425, has been reported and is being evaluated for therapeutic activity32.
Inducing expression as a therapeutic strategy
NM23
NME1 (non-metastatic cells 1), known in the field as NM23, was the first and most extensively described metastasis suppressor gene. It was discovered in 1988 by Steeg and associates using differential hybridization experiments comparing gene expression between high- and low-metastatic cell lines derived from the K-1735 murine melanoma cell line30. They identified NM23, the expression of which could abrogate spontaneous and experimental metastasis without affecting tumour cell growth in vitro or in vivo31. Subsequent studies have identified up to eight homologues in the human genome, of which NM23-H1 (herein NM23) and NM23-H2 (NME2) have been shown to function in metastasis suppression32. These findings have been confirmed by others in different experimental systems and cell types33–35.
NM23 has several interesting properties32. First, it exhibits histidine kinase activity toward several proteins, including KSR1, the kinase suppressor of Ras, resulting in increased KSR1 degradation and decreased MAPK signalling36, which has been shown in many tumour types to support survival, motility and other cellular processes that are important for metastasis37. NM23 activity may also be modulated in vivo by differential binding to endogenous and exogenous viral proteins32, including the Epstein–Barr virus (EBV) nuclear antigens 1 and 3C. This results in relief of NM23-dependent suppression of migration38 and metastasis39, suggesting that its function can be abrogated without loss of endogenous expression32. Finally, NM23 suppresses metastasis by reducing the expression of pro-metastatic genes40 (see the section ‘Targets downstream of metastasis suppressors’).
Therapeutic exploitation of NM23 function is based on studies of its promoter41, which is regulated by the glucocorticoid response pathway. Although addition of the glucocorticoid receptor agonist dexamethasone could not increase NM23 expression pharmacologically, the progesterone and atypical glucocorticoid agonist medroxyprogesterone acetate (MPA) induced expression in a glucocorticoid receptor response element-dependent manner42. MPA, commonly used at low doses for contraception, has previously been used at higher doses in breast cancer clinical trials43,44. Preclinical evaluation of MPA as an inhibitor of metastasis formation revealed promising results. Four weeks after injection of a reliably metastatic sub-line of MDA-MB-231 breast cancer cells into nude mice, lung micrometastases had formed. The animals were then randomized to either control or MPA treatment regimens that approximate the serum MPA concentrations that are achievable in human patients. Significant decreases in the proportions of animals developing clinically relevant metastases (decreased by 27% and 36% in two separate experiments) and average disease burden per animal were noted after treatment with MPA and, in animals that failed treatment, the proportion of metastases staining more strongly for NM23 by immunohistochemistry was significantly higher than that of untreated mice45. On the basis of these promising results a Phase II trial has been initiated that is examining the clinical utility of MPA with and without anti-angiogenic low-dose, frequent cyclophosphamide–methotrexate metronomic chemotherapy46 in refractory, hormone receptor–negative metastatic breast cancer (NCT00577122). NM23 has also been reported to be re-induced by treatment with retinoic acid47 and oestradiol48.
Restoration of NM23 function has also been attempted using adeno-associated virus-mediated gene therapy in a mouse model of metastatic ovarian cancer that used an aggressive variant of the SW626 cell line selected in vivo. After orthotopic injection of these cells to form a xenograft ovarian tumour, animals were treated by intraperitoneal injection of virus engineered to express NM23, which significantly decreased liver metastases (by 60%, p = 0.01) and increased the median time of survival compared with animals treated with control vector (by 35 days, p < 0.05)49.
KAI1
Re-induction of endogenous metastasis suppressor function has also been reported for the metastasis suppressor gene KAI1. The activity of KAI1 was identified initially by transferring human chromosome 11 into AT6.1 rat prostate cancer cells50,51, with subsequent cloning of the suppressing locus and observation of suppression of metastasis in vivo52. Two key suppressor functions that have been described for this membrane-bound protein are downregulation of epidermal growth factor receptor signalling, which is associated with increased receptor desensitization and endocytosis53, and a novel pathway involving KAI1 and the Duffy antigen receptor for chemokines (DARC), which is expressed on endothelial cells. Circulating KAI1-expressing cells attach to endothelium through a KAI1–DARC interaction54. This interaction is associated with cessation of proliferation and induction of senescence in KAI1-expressing disseminated cells but not in those that do not express it.
As with NM23, transcriptional regulation of KAI1 suggested several strategies for re-expression of this protein. A key early study found that p53 increased transcription of KAI1 through a p53-responsive element55. The clinical relevance of this pathway was demonstrated by immunohistochemical staining of prostate cancer tissue specimens for p53 and KAI1, showing a significant concordance of expression. Proceeding from this observation, Mashimo et al. used etoposide, an agent that induces p53, and found that it induced KAI1 expression56, suggesting that it might provide a therapeutic angle. Wu et al. treated mice with etoposide after splenic injection of gastric cancer cells and observed both induction of KAI1 expression in subsequent splenic tumours and reduced incidence of hepatic metastases57, although this anti-metastatic effect was not proved to be specific to KAI1 function. It is tempting to speculate that etoposide treatment in patients could therapeutically induce KAI1 expression and, given the ease of oral administration, might be adaptable to a long-term suppression strategy, such as low-dose metronomic chemotherapy58, a therapeutic setting in which it has been used previously59. However, given the pleiotropic effects of etoposide and p53 induction, the contribution of KAI1 induction might be minor and requires careful elucidation.
Another potential therapeutic strategy for long-term re-expression of endogenous KAI1 in prostate cancer is through the use of the soy-derived phyto-oestrogen isoflavone genistein. El Touny et al. recently reported reversal of loss of prostatic KAI1 expression in the TRAMP mouse model of prostate cancer following treatment with genistein60. In vitro, genistein treatment inhibited the invasive behaviour of tumour-derived cells from this model. This anti-invasive effect of genistein was shown to be specifically due to KAI1 induction — rather than to any of the many other genes that are probably affected by genistein treatment — by demonstration of reversion to an invasive phenotype when the induced KAI1 was depleted by small interfering RNA. However, the impact of this approach on the metastasis of the TRAMP tumours was not examined in this preclinical study, and the mechanism of KAI1 induction remains unknown.
Traditional gene therapy techniques have also been used to increase the expression of KAI1. Xu et al. compared direct injection of saline with either a KAI1 plasmid expression vector or liposomes containing the expression vector into xenografted MiaPaCa II pancreatic cancer cells established as subcutaneous tumours in nude mice. Interestingly, injection of the vector encoding KAI1 either alone or in liposomes substantially reduced spontaneous pulmonary metastasis61. Another study used a replication-deficient adenovirus to reconstitute KAI1 expression in an orthotopic non-small-cell lung cancer lymphatic metastasis mouse model62. In this study, Lewis lung carcinoma cells were directly injected into the lungs of syngeneic mice to establish tumours. Mice were treated three times by intratracheal administration of viral particles engineered to express either control lacZ or KAI1, and lung and mediastinal lymph node weights were evaluated after 3 weeks. Treatment with KAI1-expressing virus, verified by immunohistochemical evaluation of expression in tumours at necropsy, was associated with a significantly decreased weight of mediastinal metastases but not with decreased primary tumour volume.
RKIP
Finally, Raf kinase inhibitor protein (RKIP, also known as PEBP1) was recently determined to function as a suppressor of spontaneous metastasis of orthotopically implanted C4-2B human prostate cancer cells in mice63. RKIP was first identified by a yeast two-hybrid screen for proteins that bind the RAF1 kinase domain and was shown to competitively inhibit RAF1–MEK interaction and downstream signalling64. RKIP is lost in prostate and other tumour types65,66, and recent reports have suggested possible modalities for re-induction of endogenous RKIP expression.
Endogenous RKIP has been shown to be induced by treatment of cells with trichostatin A67, a histone deacetylase inhibitor, suggesting that this class of drugs, which is under evaluation for cancer treatment68, could potentially be used to induce RKIP expression. This highlights the potential use of chromatin-modifying drugs for therapeutic re-induction of silenced metastasis suppressor expression. Although relatively non-specific — that is, resulting in induction of many suppressed loci — this strategy has the potential to function for multiple metastasis suppressor genes, as has been shown for DLC1, another recently described metastasis suppressor69. Continuing with the theme of the use of drugs to de-repress metastasis suppressor genes, NDRG1 (REF. 70) has recently been reported to be induced by the DNA methyltransferase inhibitor 5-azacytidine as has NM23 (REF. 71).
Perspective
As a general therapeutic strategy, re-expression of an endogenous suppressor (FIG. 2a,b) is straightforward and appealing, especially when achieved with orally bioavailable agents. However, to date, these modalities depend on targets that have been transcriptionally downregulated instead of lost by mutagenesis or other mechanisms. Although some reports suggest that reduced transcription, rather than mutation, explains a large part of the loss of metastasis suppressor gene expression observed in different tumour types72–75, isolated reports do observe mutations76,77. In fact, the issue of restoration of endogenous expression highlights one of the potential advantages of targeting metastasis suppressors over related tumour suppressor genes: tumour suppressors are often inactivated by mutation early in the development of cancer and therefore would not be amenable to re-induction at the endogenous locus. The mechanism of loss must be established on a gene-by-gene, patient-by-patient basis in order to correctly identify and target these approaches in the clinic. As it is unlikely that most metastasis suppressor genes can be induced by agents as well tolerated as, for example, MPA, a concerted effort to discover orally bioavailable agents that can transcriptionally induce metastasis suppressors is warranted. Thus, despite the recognized difficulties of exogenous cancer gene therapy78 (FIG. 2c), such an approach can be generalized to different target genes and may prove necessary for suppressors that cannot be easily re-expressed from the endogenous locus.
Direct administration of metastasis suppressor
KISS1
Administration of the metastasis suppressor protein KISS1 has shown preclinical utility in attenuating metastasis in tumours that have lost KISS1 function. KISS1 was identified by subtractive hybridization between the human melanoma cell line C8161 and a derivative into which human chromosome 6 was transferred79. This chromosome is frequently lost in metastatic melanoma80 and can suppress metastasis in these cells. KISS1 is located on chromosome 1 but is regulated by trans-acting factors on chromosome 6. KISS1 encodes a protein that is modified into a secreted kisspeptin known as metastin, which agonizes at least one G-protein-coupled receptor known as KISS1R or GPR54 (REF. 81). Although downstream signalling events leading to metastasis suppression have yet to be elucidated, one report demonstrates that secretion of KISS1 is necessary for metastasis suppression and that secreted KISS1 maintains dormancy in disseminated cells, thus blocking metastatic colonization82.
In a preclinical study, Ohtaki et al. showed that spontaneous pulmonary metastasis of KISS1R-overexpressing B16-BL6 melanoma cells was significantly suppressed by administration of KISS1-derived peptide through an osmotic pump (by 66% (p < 0.01) compared with vehicle-treated controls)83. They hypothesized that KISS1 could be similarly administered to patients to maintain metastatic dormancy in a clinical setting. Recent reports have questioned whether the metastasis-suppressing effect of KISS1 is due to ligation of KISS1R on the cancer cell itself, a paracrine effect on neighbouring stroma or its effect on some other uncharacterized receptor82. However, this strategy seems promising. Another recent publication described development of small molecule mimetics of KISS1 (REF. 84), suggesting another potential angle for administration of KISS1. Although administration of a KISS1 peptide to patients has been undertaken and tolerated in the short term85, at least one preclinical animal model has revealed effects on the pituitary–gonadal axis, suggesting the possibility of long-term toxicity to male subjects86. Certainly any small molecule mimetic would require a similar careful preclinical and clinical trial workup. These issues must be clarified if this approach is to be successfully used in humans as it is likely that delivery of this protein would be required on a chronic basis, given the paradigm of suppression of the micrometastasis to macrometastasis transition.
BMP4
Just as KISS1 is a secreted factor, BMP4 (bone morphogenic protein 4), a member of the transforming growth factor-β superfamily of growth factors, has recently been reported to be a metastasis suppressor. Eckhardt et al. recently observed an association of reduced expression of BMP4 with increased metastatic potential in mouse mammary tumours87. They demonstrated that overexpression of BMP4 in 4T1.2 mouse breast cancer cells suppressed the formation of spontaneous metastases but not tumorigenesis, whereas stable depletion of BMP4 by RNA interference increased metastatic potential in less metastatic cell lines. Finally, they observed that intraperitoneal administration of recombinant BMP4 suppressed spontaneous metastasis and enhanced survival in a preclinical model of breast cancer metastasis (R. Anderson, unpublished data, also presented at the 2008 Meeting of the American Association for Cancer Research–Metastasis Research Society, Vancouver, Canada).
In general, it is unlikely that administration of a recombinant metastasis suppressor protein (FIG. 2d) would be applicable to many metastasis suppressor genes as most are transmembrane or intracellular proteins. Indeed, only one other secreted metastasis suppressor protein has been described: CTGF (connective tissue growth factor)88. However, these cases of KISS1 and BMP4 provide a promising, although preliminary, foundation for such a strategy.
Targets downstream of metastasis suppressors
Several metastasis suppressor genes modulate the cellular signalling of cancer cells4, which raises the question of whether the expression of downstream metastasis suppressor-regulated transcripts contribute significantly to the invasive and metastatic phenotype and whether these can be targeted therapeutically. Two recent reports suggest that this may be the case and have yielded druggable targets with clinical potential.
RHOGDI2
RHOGDI2 (RhoGTPase dissociation inhibitor 2, also known as ARHGDIB), a member of the RHOGDI family of proteins that downregulate the activation state of Rho family GTPases89, was initially identified by our group as a metastasis suppressor in human bladder cancer by comparison of transcriptional profiles of the T24 bladder cancer cell line with a metastatic derivative, T24T90,91. Candidate genes differentially expressed between these two cells were screened against a panel of microarray data from multiple human tumour types for a pattern of decreased expression as a function of stage and grade. This screen yielded RHOGDI2, which suppresses experimental pulmonary metastasis of T24T cells without affecting tumour growth rate. Moreover, we have observed that loss of expression of RHOGDI2 is associated with poor survival outcomes in bladder cancer patients92, whereas others have found that loss of RHOGDI2 is associated with nodal metastasis in breast cancer patients93 and is part of a gene expression signature of metastasis in breast tumours94.
Hypothesizing that RHOGDI2 loss leads to deregulation of signalling pathways and that resultant changes in transcription are necessary for metastasis, our group95 profiled gene expression of RHOGDI2-transfected T24T cells and control cells (FIG. 3a). Transcripts that were downregulated in RHOGDI2-transfected T24T cells, but also overexpressed as a function of stage in human bladder cancer, were selected for further study. This screen yielded ET1 (endothelin 1), a potent vasoconstrictor, as a candidate gene whose effect on the endothelin A receptor may be antagonized by atrasentan hydrochloride. It bears note that for reasons unrelated to its connection to RHOGDI2, atrasentan is currently in Phase III trials for stage IV prostate cancer (NCT00134056). Administration of atrasentan to mice injected intravenously with T24T cells reduced the proportion of animals developing metastases (53% control versus 5% atrasentan treated, p < 0.0001)95. As atrasentan is orally bioavailable and has shown activity in prostate cancer96 and pulmonary hypertension97, it would seem to be an ideal candidate for use as adjuvant therapy for patients at high risk of metastasis development after radical treatment of their primary tumour. Interestingly, RHOGDI2 also suppresses the expression of neuromedin U, a molecule with many similarities to ET1, which mediates both increased growth of metastases and increased tumour cachexia in animal models98. This again reinforces the fact that gene expression regulated by metastasis suppressors can mediate key aspects of this disease. Unfortunately, agents that block the neuromedin U receptor are not available, although development of such a product would provide another way to target key players downstream of loss of RHOGDI2.
NM23
A similar approach40 was used to discover NM23-regulated transcripts in MDA-MB-435 breast cancer cells (it is worth noting that significant controversy exists regarding whether MDA-MB-435 cells are derived from breast cancer or melanoma99). Candidate genes were identified by differential expression between control and NM23-transfected cells (FIG. 3b), and inverse correlation between candidate expression and NM23 expression in microarray data from two cohorts of human breast cancer patients was used to further winnow candidates. The lysophosphatidic acid receptor, LPAR1 (also known as EDG2), emerged from these analyses, and when exogenously expressed in breast cancer cells it rescued NM23-suppressed cellular motility40. This suggested a model in which loss of NM23 expression induces LPAR1 expression and increases cellular motility in cancer cells. As motility is only part of the metastatic process, the same group examined the in vivo metastatic properties of NM23 and LPAR1 co-expressing cells to see whether this system was relevant to lung metastasis. They found not only that LPAR1 expression enhances pulmonary retention of MDA-MB-435 cells injected intravenously in an experimental metastasis assay, but also that its co-expression with NM23 rescues NM23-suppressed pulmonary metastasis in these cells100. These findings immediately suggest a translational angle using antagonism of LPAR1 by Ki16425, an Edg family antagonist101, for which preclinical metastasis studies are ongoing102.
Future strategies and emerging technologies
The aforementioned identification of downstream targets regulated by loss of metastasis suppressor function depends heavily on microarray studies of gene expression patterns. Despite the serendipity of ET1 and LPAR1 being druggable targets, such a strategy is unlikely to work in every case. However, at a minimum every case should provide a signature of the gene expression changes induced by metastasis suppressor re-expression owing to the reversion of the cell to a non-metastatic phenotype. Intriguingly, the recent development of the Connectivity Map103 provides a novel technology for systematically targeting such a signature.
The Connectivity Map is a massive repository of gene expression data from several cell lines after treatment with >1,000 bioactive compounds, providing the signatures of gene expression induced by such compounds. Searching gene expression signatures induced by metastasis suppressor re-expression against these drug-induced signatures provides a systematic framework for the potential discovery of drugs that target an entire signature, avoiding the problem of finding individual drug-gable targets such as ET1 or LPAR1. Such a strategy might also be more potent owing to its causing the reversion of several genes that contribute to the metastatic phenotype. Advantageously, such a signature-based therapeutic approach provides a built-in, specific signature to identify cases in which such a drug is likely to work, providing a biomarker that can select patients most likely to benefit, one of the hallmarks of personalized cancer therapy.
Recently, new informatic techniques have been developed to extrapolate drug efficacy from cell line model systems to patient outcomes104,105. These techniques use microarray studies of cell line gene expression and in vitro chemotherapeutic sensitivities to generate robust and parsimonious signatures predicting drug sensitivity in cell lines, then test these signatures against independent cell lines sets or, most compellingly, outcomes of clinical trials where patient tumours were profiled with microarrays before treatment. One of these approaches, the COXEN (Coexpression Extrapolation) system developed in our group104, is adaptable to drug discovery and, in particular, to the setting of metastasis suppressor-dependent transcriptional deregulation. COXEN is an algorithm that uses data from microarray analyses and empirically determined drug sensitivities from the NCI-60 screen of cancer cell lines99 to discover genes of which expression is associated with drug sensitivity. Then a specialized algorithm compares the expression of this sensitivity-related gene set from the cell lines to the same genes from human tumours, identifying a subset of these genes whose correlated co-expression is relevant to the human tissue data and resulting in selection of a sensitivity prediction signature that works for human tumours in patients, as has been reported104.
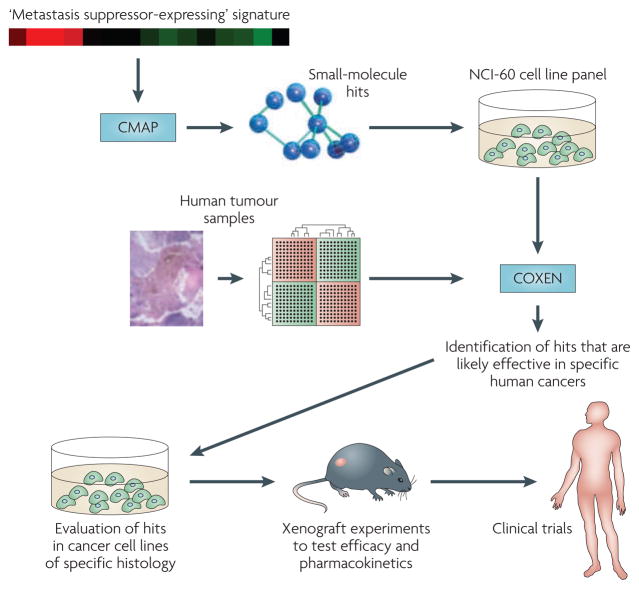
Importantly, COXEN can also facilitate computational drug discovery to identify agents or agent combinations that are highly effective in human patients based on the in vitro effects of these compounds on these cell lines. Thus, the Connectivity Map could use microarray signatures of cells with and without re-expression of metastasis suppressors to generate a list of drugs that induce the signature, and screening such drugs through COXEN and the NCI-60 screen could determine which agents both additionally show efficacy in vitro and have a high likelihood of working on patients (FIG. 4).
Figure 4. Targeting metastasis suppressor signatures through the Connectivity Map and COXen (Coexpression extrapolation).
Advanced informatic technologies including the Connectivity Map103 allow for screening an entire ‘metastasis suppressor-expressing cell’-type gene expression signature against gene expression changes induced by compounds, potentially systematically identifying molecules capable of inducing non-metastatic gene expression and phenotype. The list of hits is then further filtered using the COXEN algorithm, which provides a list of those suppressor signature-inducing agents that additionally have therapeutic efficacy in the NCI-60 cell line system99 and, most importantly, predicts function in patient tumours104. Together, this integrated approach could deliver therapeutics based on metastasis suppressor biology to the clinic with a high likelihood of efficacy.
Another recent key development in drug discovery, which is imminently applicable to targeting metastasis suppressor genes, is that of synthetic lethal screens106. Conceptually adapted from yeast biology, these screens rely upon the premise that cells lacking a specific protein (such as a metastasis suppressor) are more sensitive to specific drugs than cells that have the protein. Proof of principle of this technology has been reported for SMAD4, the pancreatic tumour suppressor gene107. Wang et al. used a pharmacological synthetic lethal strategy to screen combinatorial pharmaceutical libraries against isogenic cell lines that differ solely in exogenous expression of (and, complementarily, RNA interference depletion of) SMAD4. Such a strategy yields candidate compounds that are selectively toxic to a cell that lacks suppressor function, whether by downregulated expression, mutation or otherwise. Such approaches are probably feasible for metastasis suppressors as well.
Summary
In summary, metastasis suppressor genes define a relatively small group of proteins, of which expression or function is lost in cancer cells as metastatic competence increases. When reconstituted in model systems, they suppress in vivo metastasis without abrogating tumour growth. Experimental and epidemiological data suggest that tumours shed many cells into circulation by the time of patient diagnosis. Some of these die in the circulation or after attachment in distant organs, whereas others survive and either remain dormant as a micrometastasis or grow to colonize the organ, forming a clinical macrometastasis. Therefore, suppressing the micrometastasis-to-macrometastasis transition, which is the point at which many metastasis suppressors function, should provide a promising therapeutic window.
Although targeting a negative regulator is technically challenging, recent reports have successfully reinduced expression, administered the gene product itself or targeted druggable molecules regulated by the suppressor gene. As functional genomic and proteomic technologies increase the speed and ease of discovery of new metastasis suppressors, the field of targets is likely to swell both in number and kind, particularly given the recent description of microRNA metastasis suppressors. As the field grows, and additional novel strategies for therapeutic intervention are developed, unanswered questions will become more salient. Among others, questions of generality (the relevance of a particular metastasis suppressor to different types of cancer), specificity (the relevance of a particular metastasis suppressor to suppression of metastasis in different second sites) and toxicity to normal cells (BOX 2) will need to be addressed in a systematic manner. Future candidate metastasis suppressor genes may prove more or less tractable as targets. Nonetheless, the strategies described herein form a framework for transposing these promising findings to other genes, and novel informatic technologies might ease drug discovery and patient selection for future metastasis suppressor-based therapies.
Box 2. Key unresolved questions in metastasis suppressor research.
Generality
The relevance of most described metastasis suppressors across various types of cancer has only been superficially validated in pathological studies; rates of loss of expression and mutation, as appropriate to therapeutic strategies, must be carefully and independently validated. Given observations of early, wide cellular dissemination of tumour cells24, are metastasis suppressors that chiefly inhibit invasion the relevant targets?
Specificity
Recent reports suggest rich complexity in the genetic regulation of organ-specific tropism of metastasis112,113. It follows that metastasis suppressor function might also be highly organ specific. The ability of any given metastasis suppressor gene to suppress metastases to multiple organs has only just begun to be assessed, with two recent reports examining the role of metastasis suppressors in metastasis to multiple organs82,114.
Adaptability
As a corollary to the above questions, is it true that a strategy reconstituting a metastasis suppressor should necessarily only work in cancers where the suppressor or its function is commonly lost? Might its reconstitution in ‘irrelevant’ cancer types still target latent pathways resulting in the same goal of suppression of metastasis?
Toxicity
One of the theoretical advantages of reconstituting a lost, negative regulator such as a suppressor is that toxicity is likely to be lower than when poisoning a positive regulator that operates physiologically in normal cells. Has the observation that several metastasis suppressors impinge directly and negatively on essential cell signalling pathways4 called this postulate into question?
Integration
The integration of metastasis suppressor-targeting therapeutics into current therapeutic regimens is non-trivial: would therapy resulting in suppression of distant proliferation of micrometastases not decrease sensitivity to traditional cytotoxic chemotherapeutics?
Patient selection
Several targeted agents now available for cancer therapy have been shown to be highly efficacious in patient subsets exhibiting particular molecular lesions. What biomarkers would be used for individualized therapy targeting metastasis suppressor genes? Is a primary tumour the appropriate tissue to analyse?
Trial design
Patients who enter clinical trials often have failed standard therapies and developed gross metastatic recurrences. Could such candidates even potentially benefit from metastasis suppression therapies?
At a glance
Metastasis accounts for the preponderance of morbidity and mortality in cancer, and accumulating evidence suggests that dissemination of tumour cells may occur earlier in the development of cancer than previously appreciated.
Metastasis is regulated either positively or negatively by proteins that promote steps in the metastatic cascade or those that suppress them.
Metastasis suppressor genes encode proteins that prevent or reduce the development of metastases in vivo, without simultaneously affecting primary tumour growth. This is in contrast to tumour suppressors, which affect primary tumorigenesis. Metastasis suppressor proteins are lost primarily during cancer progression and not during transformation.
Until recently, relatively few metastasis suppressor genes had been characterized. However, expanding genomic technology has made possible in recent years the description of many metastasis suppressor genes impinging on a wide variety of cellular processes.
Although metastasis suppressor genes have been shown to work at multiple steps in the process of metastasis, several have been recently demonstrated to specifically suppress the colonization step of metastasis, the process by which solitary or small clusters of tumour cells living in a second organ site (micrometastases) grow to form clinically apparent and lethal macrometastases.
Recent work has shown techniques that restore metastasis suppressor function. These include re-expression of the gene from the endogenous locus or by exogenous gene therapy, direct administration of the protein itself and targeting important metastasis mediators downstream of the suppressor that are reciprocally induced with metastasis suppressor protein losses.
Acknowledgments
Work was supported by National Institutes of Health grant CA075115 to D.T. The authors wish to thank R. Horwitz, M. Schwartz of the University of Virginia, P. Steeg of the National Cancer Institute, and D. Welch of the University of Alabama at Birmingham for their help, suggestions and comments on the manuscript.
- Metastatic colonization
The process by which disseminated tumour cells, present as single or small clusters of cells in a second parenchyma (micrometastasis), grow to form a clinically detectable metastatic nodule (macrometastasis)
- Epstein–Barr virus
(EBV). EBV is a herpes family virus that commonly causes infectious mononucleosis in humans. Infection with EBV has been implicated in Burkitt’s lymphoma and nasopharyngeal carcinoma, and may also be involved in the pathogenesis of other tumours
- Glucocorticoid response pathway
This nuclear hormone receptor pathway directs the regulation of genes involved in metabolism and immune function. Transcription by the glucocorticoid receptor is ligand-induced by glucocorticoid steroid hormones
- Metronomic chemotherapy
Administration of chemotherapeutic drugs at comparatively low doses on a frequent or continuous schedule, with no extended interruptions, in contrast to traditional maximum tolerated dose chemotherapy
- TRAMP
An autochthonous transgenic mouse model of prostate cancer. Various stages of progressive prostate disease can be observed in TRAMP mice, with focal adenocarcinomas developing between 10 and 20 weeks of age with 100% frequency
- Mediastinal lymph node
Lymphatic tissue occurring in a central region of the chest between the lungs and bordered by the thoracic inlet above and the diaphragm below, where lung and other cancers frequently metastasize
- Orthotopic xenograft
Establishment of a tumour by injecting human cells into the same rodent organ from which the human tumour was derived
- Heterotopic xenograft
Establishment of a tumour by injecting human cells into a different rodent organ than that from which the human tumour was derived, often for technical reasons
- Histone deacetylases
Enzymes that regulate chromatin structure and function through the removal of the acetyl group from the lysine residues of core nucleosomal histones, generally repressing transcription
- DNA methyltransferases
Enzymes that catalyse transfer of a methyl group to cytosines in DNA, generally repressing transcription
- Anoikis
Cell death in response to loss of matrix attachments
- Osmotic pump
Generally small implantable pumps that use a salt concentration gradient to draw in interstitial fluid and generate pressure to expel a drug in a regulated fashion
- Synthetic lethal
Describes a genetic phenomenon in which the combination of two otherwise non-lethal mutations or molecular lesions results in an unviable cell. In the case of drug discovery, the pharmaceutical agent, otherwise non-toxic, substitutes for one of these lesions and becomes lethal only in the presence of the second molecular lesion
Footnotes
DATABASES
ClinicalTrials.gov: http://clinicaltrials.gov/
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
SMAD4
National Cancer Institute Drug Dictionary: http://www.cancer.gov/drugdictionary/
atrasentan hydrochloride | dexamethasone | etoposide | medroxyprogesterone acetate | MPA
UniProtKB: http://www.uniprot.org
BMP4 | BRMS1 | CD82 | DARC | DLC1 | ET1 | KISS1 | KISS1R | KSR1 | LPAR1| MAP2K4 | neuromedin U | NME1 | p53 | PEBP1 | PTEN | RAF1 | SRC
FURTHER INFORMATION
Dan Theodorescu’s homepage: http://www.uvamellon.org/
US National Cancer Institute SEER database: http://seer.cancer.gov
All LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Fidler IJ, Poste G. The “seed and soil” hypothesis revisited. Lancet Oncol. 2008;9:808. doi: 10.1016/S1470-2045(08)70201-8. [DOI] [PubMed] [Google Scholar]
- 4.Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nature Rev Cancer. 2003;3:55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- 5.Stafford LJ, Vaidya KS, Welch DR. Metastasis suppressors genes in cancer. Int J Biochem Cell Biol. 2008;40:874–891. doi: 10.1016/j.biocel.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 6.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma L, Weinberg RA. MicroRNAs in malignant progression. Cell Cycle. 2008;7:570–572. doi: 10.4161/cc.7.5.5547. [DOI] [PubMed] [Google Scholar]
- 8.Rinker-Schaeffer CW, O’Keefe JP, Welch DR, Theodorescu D. Metastasis suppressor proteins: discovery, molecular mechanisms, and clinical application. Clin Cancer Res. 2006;12:3882–3889. doi: 10.1158/1078-0432.CCR-06-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iiizumi M, Liu W, Pai SK, Furuta E, Watabe K. Drug development against metastasis-related genes and their pathways: a rationale for cancer therapy. Biochim Biophys Acta. 2008;1786:87–104. doi: 10.1016/j.bbcan.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welch DR. Technical considerations for studying cancer metastasis in vivo. Clin Exp Metastasis. 1997;15:272–306. doi: 10.1023/a:1018477516367. [DOI] [PubMed] [Google Scholar]
- 11.Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steeg PS, Theodorescu D. Metastasis: a therapeutic target for cancer. Nature Clin Pract Oncol. 2008;5:206–219. doi: 10.1038/ncponc1066. References 11 and 12 provide important perspectives on the rationale for targeting metastasis in human cancer, aside from the issue of suppressor genes. As developmental therapies targeting these genes must be adapted to this specialized clinical setting, such understanding is essential. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelkey TJ, Frierson HF, Jr, Bruns DE. Molecular and immunological detection of circulating tumor cells and micrometastases from solid tumors. Clin Chem. 1996;42:1369–1381. [PubMed] [Google Scholar]
- 14.Glaves D, Huben RP, Weiss L. Haematogenous dissemination of cells from human renal adenocarcinomas. Br J Cancer. 1988;57:32–35. doi: 10.1038/bjc.1988.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fidler IJ. Metastasis: guantitative analysis of distribution and fate of tumor embolilabeled with 125I-5-iodo-2′-deoxyuridine. J Natl Cancer Inst. 1970;45:773–782. [PubMed] [Google Scholar]
- 16.Alix-Panabieres C, Riethdorf S, Pantel K. Circulating tumor cells and bone marrow micrometastasis. Clin Cancer Res. 2008;14:5013–5021. doi: 10.1158/1078-0432.CCR-07-5125. [DOI] [PubMed] [Google Scholar]
- 17.Engel J, et al. The process of metastasisation for breast cancer. Eur J Cancer. 2003;39:1794–1806. doi: 10.1016/s0959-8049(03)00422-2. [DOI] [PubMed] [Google Scholar]
- 18.Butler TP, Gullino PM. Quantitation of cell shedding into efferent blood of mammary adenocarcinoma. Cancer Res. 1975;35:512–516. [PubMed] [Google Scholar]
- 19.Husemann Y, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. Reference 19, coupled with the above reports on tumour cell dissemination, is a watershed in our understanding of how widely and early tumours disseminate. [DOI] [PubMed] [Google Scholar]
- 20.Cameron MD, et al. Temporal progression of metastasis in lung: cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer Res. 2000;60:2541–2546. [PubMed] [Google Scholar]
- 21.Wong CW, et al. Apoptosis: an early event in metastatic inefficiency. Cancer Res. 2001;61:333–338. [PubMed] [Google Scholar]
- 22.Weiss L. Metastatic inefficiency. Adv Cancer Res. 1990;54:159–211. doi: 10.1016/s0065-230x(08)60811-8. [DOI] [PubMed] [Google Scholar]
- 23.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nature Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 24.Weinberg RA. Leaving home early: reexamination of the canonical models of tumor progression. Cancer Cell. 2008;14:283–284. doi: 10.1016/j.ccr.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nature Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 26.Chambers AF, MacDonald IC, Schmidt EE, Morris VL, Groom AC. Clinical targets for anti-metastasis therapy. Adv Cancer Res. 2000;79:91–121. doi: 10.1016/s0065-230x(00)79003-8. [DOI] [PubMed] [Google Scholar]
- 27.Kauffman EC, Robinson VL, Stadler WM, Sokoloff MH, Rinker-Schaeffer CW. Metastasis suppression: the evolving role of metastasis suppressor genes for regulating cancer cell growth at the secondary site. J Urol. 2003;169:1122–1133. doi: 10.1097/01.ju.0000051580.89109.4b. [DOI] [PubMed] [Google Scholar]
- 28.Hedley BD, Allan AL, Chambers AF. Tumor dormancy and the role of metastasis suppressor genes in regulating ectopic growth. Future Oncol. 2006;2:627–641. doi: 10.2217/14796694.2.5.627. [DOI] [PubMed] [Google Scholar]
- 29.Horak CE, Lee JH, Marshall JC, Shreeve SM, Steeg PS. The role of metastasis suppressor genes in metastatic dormancy. APMIS. 2008;116:586–601. doi: 10.1111/j.1600-0463.2008.01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steeg PS, et al. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988;80:200–204. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- 31.Leone A, et al. Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell. 1991;65:25–35. doi: 10.1016/0092-8674(91)90404-m. [DOI] [PubMed] [Google Scholar]
- 32.Steeg PS, Horak CE, Miller KD. Clinical-translational approaches to the Nm23-H1 metastasis suppressor. Clin Cancer Res. 2008;14:5006–5012. doi: 10.1158/1078-0432.CCR-08-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parhar RS, et al. Effects of cytokine mediated modulation of Nm23 expression on the invasion and metastatic behavior of B16F10 melanoma cells. Int J Cancer. 1995;60:204–210. doi: 10.1002/ijc.2910600213. [DOI] [PubMed] [Google Scholar]
- 34.Leone A, Flatow U, VanHoutte K, Steeg PS. Transfection of human nm23-H1 into the human MDA-MB-435 breast carcinoma cell line: Effects on tumor metastatic potential, colonization, and enzymatic activity. Oncogene. 1993;8:2325–2333. [PubMed] [Google Scholar]
- 35.Liu F, Zhang Y, Zhang XY, Chen HL. Transfection of the nm23-H1 gene into human hepatocarcinoma cell line inhibits the expression of sialyl Lewis X, a 1,3 fucosyltransferase VII, and metastatic potential. J Cancer Res Clin Oncol. 2002;128:189–196. doi: 10.1007/s00432-001-0314-1. [DOI] [PubMed] [Google Scholar]
- 36.Hartsough MT, et al. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J Biol Chem. 2002;277:32389–32399. doi: 10.1074/jbc.M203115200. [DOI] [PubMed] [Google Scholar]
- 37.Reddy KB, Nabha SM, Atanaskova N. Role of MAP kinase in tumor progression and invasion. Cancer Metastasis Rev. 2003;22:395–403. doi: 10.1023/a:1023781114568. [DOI] [PubMed] [Google Scholar]
- 38.Subramanian C, Cotter M, Robertson E. Epstein–Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: A molecular link to cancer metastasis. Nature Med. 2001;7:350–355. doi: 10.1038/85499. [DOI] [PubMed] [Google Scholar]
- 39.Kaul R, Murakami M, Choudhuri T, Robertson ES. Epstein–Barr virus latent nuclear antigens can induce metastasis in a nude mouse model. J Virol. 2007;81:10352–10361. doi: 10.1128/JVI.00886-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horak CE, et al. Nm23-H1 suppresses tumor cell motility by down-regulating the lysophosphatidic acid receptor EDG2. Cancer Res. 2007;67:7238–7246. doi: 10.1158/0008-5472.CAN-07-0962. [DOI] [PubMed] [Google Scholar]
- 41.Ouatas T, Clare S, Hartsough M, DeLaRosa A, Steeg P. MMTV-associated transcription factor binding sites increase nm23-H1 metastasis suppressor gene expression in human breast carcinoma cell lines. Clin Exp Metast. 2002;19:35–42. doi: 10.1023/a:1013897022827. [DOI] [PubMed] [Google Scholar]
- 42.Ouatas T, Halverson D, Steeg PS. Dexamethasone and medroxyprogesterone acetate elevate Nm23-H1 metastasis suppressor gene expression in metastatic human breast carcinoma cells: new uses for old compounds. Clin Cancer Res. 2003;9:3763–3772. [PubMed] [Google Scholar]
- 43.Focan C, et al. Adjuvant high dose medroxyprogesterone acetate for early breast cancer: 13 years update in a multicentre randomized trial. Br J Cancer. 2001;85:1–8. doi: 10.1054/bjoc.2001.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hupperets P, et al. Adjuvant chemo-hormonal therapy with chclophosphamide, doxorubicin and 5-fluroruacil (CAF) with or without medroxyprogesterone acetate (MPA) for node-positive cancer patients, update at 12 years follow up. The Breast. 2001;10:35–37. doi: 10.1054/brst.2000.0180. [DOI] [PubMed] [Google Scholar]
- 45.Palmieri D, et al. Medroxyprogesterone acetate elevation of Nm23-H1 metastasis suppressor expression in hormone receptor-negative breast cancer. J Natl Cancer Inst. 2005;97:632–642. doi: 10.1093/jnci/dji111. Reference 45 provides an excellent example of design, execution and internal controls for preclinical studies of metastasis suppressor-inducing therapies. [DOI] [PubMed] [Google Scholar]
- 46.Orlando L, et al. Prolonged clinical benefit with metronomic chemotherapy in patients with metastatic breast cancer. Anticancer Drugs. 2006;17:961–967. doi: 10.1097/01.cad.0000224454.46824.fc. [DOI] [PubMed] [Google Scholar]
- 47.Liu F, Qi HL, Chen HL. Effects of all-trans retinoic acid and epidermal growth factor on the expression of nm23-H1 in human hepatocarcinoma cells. J Cancer Res Clin Oncol. 2000;126:85–90. [PubMed] [Google Scholar]
- 48.Lin KH, Wang WJ, Wu YH, Cheng SY. Activation of antimetastatic Nm23-H1 gene expression by estrogen and its alpha-receptor. Endocrinology. 2002;143:467–475. doi: 10.1210/endo.143.2.8620. [DOI] [PubMed] [Google Scholar]
- 49.Li J, et al. Inhibition of ovarian cancer metastasis by adeno-associated virus-mediated gene transfer of nm23-H1 in an orthotopic transplantation model. Cancer Gene Ther. 2006;13:266–270. doi: 10.1038/sj.cgt.7700899. [DOI] [PubMed] [Google Scholar]
- 50.Ichikawa T, et al. Localization of metastasis suppressor gene(s) for prostatic cancer to the short arm of human chromosome 11. Cancer Res. 1992;52:3486–3490. [PubMed] [Google Scholar]
- 51.Ichikawa T, Ichikawa Y, Isaacs JT. Genetic factors and suppression of metastatic ability of prostatic cancer. Cancer Res. 1991;51:3788–3792. [PubMed] [Google Scholar]
- 52.Dong JT, et al. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science. 1995;268:884–886. doi: 10.1126/science.7754374. [DOI] [PubMed] [Google Scholar]
- 53.Odintsova E, Sugiura T, Berditchevski F. Attenuation of EGF receptor signaling by a metastasis suppressor, the tetraspanin CD82/KAI-1. Curr Biol. 2000;10:1009–1012. doi: 10.1016/s0960-9822(00)00652-7. [DOI] [PubMed] [Google Scholar]
- 54.Bandyopadhyay S, et al. Interaction of KAI1 on tumor cells with DARC on vascular endothelium leads to metastasis suppression. Nature Med. 2006;12:933–938. doi: 10.1038/nm1444. [DOI] [PubMed] [Google Scholar]
- 55.Mashimo T, et al. The expression of the KAI1 gene, a tumor metastasis suppressor, is directly activated by p53. Proc Natl Acad Sci USA. 1998;95:11307–11311. doi: 10.1073/pnas.95.19.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mashimo T, et al. Activation of the tumor metastasis suppressor gene, KAI1, by etoposide is mediated by p53 and c-Jun genes. Biochem Biophys Res Commun. 2000;274:370–376. doi: 10.1006/bbrc.2000.3139. [DOI] [PubMed] [Google Scholar]
- 57.Wu Q, et al. Role of tumor metastasis suppressor gene KAI1 in digestive tract carcinomas and cancer cells. Cell Tissue Res. 2003;314:237–249. doi: 10.1007/s00441-003-0781-6. [DOI] [PubMed] [Google Scholar]
- 58.Kerbel RS, Kamen BA. The anti-angiogenic basis of metronomic chemotherapy. Nature Rev Cancer. 2004;4:423–436. doi: 10.1038/nrc1369. Reference 58 reviews metronomic chemotherapy, a novel therapeutic strategy that is adaptable to simultaneous therapy with agents targeting metastasis suppressor function. [DOI] [PubMed] [Google Scholar]
- 59.Correale P, et al. A novel metronomic chemotherapy regimen of weekly platinum and daily oral etoposide in high-risk non-small cell lung cancer patients. Oncol Rep. 2006;16:133–140. doi: 10.3892/or.16.1.133. [DOI] [PubMed] [Google Scholar]
- 60.El Touny LH, Banerjee PP. Genistein induces the metastasis suppressor kangai-1 which mediates its anti-invasive effects in TRAMP cancer cells. Biochem Biophys Res Commun. 2007;361:169–175. doi: 10.1016/j.bbrc.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu JH, Guo XZ, Ren LN, Shao LC, Liu MP. KAI1 is a potential target for anti-metastasis in pancreatic cancer cells. World J Gastroenterol. 2008;14:1126–1132. doi: 10.3748/wjg.14.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takeda T, et al. Adenoviral transduction of MRP-1/CD9 and KAI1/CD82 inhibits lymph node metastasis in orthotopic lung cancer model. Cancer Res. 2007;67:1744–1749. doi: 10.1158/0008-5472.CAN-06-3090. References 61 and 62 are two examples of the use of viral and non-viral gene therapy strategies for reconstitution of metastasis suppressor expression. [DOI] [PubMed] [Google Scholar]
- 63.Fu Z, et al. Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J Natl Cancer Inst. 2003;95:878–889. doi: 10.1093/jnci/95.12.878. [DOI] [PubMed] [Google Scholar]
- 64.Yeung K, et al. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999;401:173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- 65.Fu Z, et al. Metastasis suppressor gene Raf kinase inhibitor protein (RKIP) is a novel prognostic marker in prostate cancer. Prostate. 2006;66:248–256. doi: 10.1002/pros.20319. [DOI] [PubMed] [Google Scholar]
- 66.Hagan S, et al. Reduction of Raf-1 kinase inhibitor protein expression correlates with breast cancer metastasis. Clin Cancer Res. 2005;11:7392–7397. doi: 10.1158/1078-0432.CCR-05-0283. [DOI] [PubMed] [Google Scholar]
- 67.Beach S, et al. Snail is a repressor of RKIP transcription in metastatic prostate cancer cells. Oncogene. 2008;27:2243–2248. doi: 10.1038/sj.onc.1210860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mai A. The therapeutic uses of chromatin-modifying agents. Expert Opin Ther Targets. 2007;11:835–851. doi: 10.1517/14728222.11.6.835. [DOI] [PubMed] [Google Scholar]
- 69.Kim TY, et al. Transcriptional induction of DLC-1 gene through Sp1 sites by histone deacetylase inhibitors in gastric cancer cells. Exp Mol Med. 2008;40:639–646. doi: 10.3858/emm.2008.40.6.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bandyopadhyay S, et al. Role of the putative tumor metastasis suppressor gene Drg-1 in breast cancer progression. Oncogene. 2004;23:5675–5681. doi: 10.1038/sj.onc.1207734. [DOI] [PubMed] [Google Scholar]
- 71.Hartsough M, et al. Elevation of breast carcinoma nm23-H1 metastasis suppressor gene expression and reduced motility by DNA methylation inhibition. Cancer Res. 2001;61:2320–2327. [PubMed] [Google Scholar]
- 72.Miyazaki T, et al. Mutation and expression of the metastasis suppressor gene KAI1 in esophageal squamous cell carcinoma. Cancer. 2000;89:955–962. doi: 10.1002/1097-0142(20000901)89:5<955::aid-cncr3>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 73.Kim HL, et al. Mitogen-activated protein kinase kinase 4 metastasis suppressor gene expression is inversely related to histological pattern in advancing human prostatic cancers. Cancer Res. 2001;61:2833–2837. [PubMed] [Google Scholar]
- 74.Cropp C, et al. NME1 protein expression and loss of heterozygosity mutations in primary human breast tumors. J Natl Cancer Inst. 1994;86:1167–1169. doi: 10.1093/jnci/86.15.1167. [DOI] [PubMed] [Google Scholar]
- 75.Dong JT, et al. Down-regulation of the KAI1 metastasis suppressor gene during the progression of human prostatic cancer infrequently involves gene mutation or allelic loss. Cancer Res. 1996;56:4387–4390. [PubMed] [Google Scholar]
- 76.Becker KF, et al. E-cadherin gene mutations provide clues to diffuse type gastric carcinomas. Cancer Res. 1994;54:3845–3852. [PubMed] [Google Scholar]
- 77.Wang L, Patel U, Ghosh L, Chen HC, Banerjee S. Mutation in the nm23 gene is associated with metastasis in colorectal cancer. Cancer Res. 1993;53:717–720. [PubMed] [Google Scholar]
- 78.Roth JA, Cristiano RJ. Gene therapy for cancer: what have we done and where are we going? J Natl Cancer Inst. 1997;89:21–39. doi: 10.1093/jnci/89.1.21. [DOI] [PubMed] [Google Scholar]
- 79.Lee JH, et al. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst. 1996;88:1731–1737. doi: 10.1093/jnci/88.23.1731. [DOI] [PubMed] [Google Scholar]
- 80.Welch DR, et al. Microcell-mediated transfer of chromosome 6 into metastatic human C8161 melanoma cells suppresses metastasis but does not inhibit tumorigenicity. Oncogene. 1994;9:255–262. [PubMed] [Google Scholar]
- 81.Nash KT, Welch DR. The KISS1 metastasis suppressor: mechanistic insights and clinical utility. Front Biosci. 2006;11:647–659. doi: 10.2741/1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nash KT, et al. Requirement of KISS1 secretion for multiple organ metastasis suppression and maintenance of tumor dormancy. J Natl Cancer Inst. 2007;99:309–321. doi: 10.1093/jnci/djk053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ohtaki T, et al. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411:613–617. doi: 10.1038/35079135. Reference 83 is the first report of a therapeutic strategy targeting metastasis suppressor function. [DOI] [PubMed] [Google Scholar]
- 84.Orsini MJ, et al. Metastin (KiSS-1) mimetics identified from peptide structure-activity relationship-derived pharmacophores and directed small molecule database screening. J Med Chem. 2007;50:462–471. doi: 10.1021/jm0609824. [DOI] [PubMed] [Google Scholar]
- 85.Dhillo WS, et al. Kisspeptin-54 stimulates the hypothalamic-pituitary gonadal axis in human males. J Clin Endocrinol Metab. 2005;90:6609–6615. doi: 10.1210/jc.2005-1468. [DOI] [PubMed] [Google Scholar]
- 86.Thompson EL, et al. Chronic subcutaneous administration of kisspeptin-54 causes testicular degeneration in adult male rats. Am J Physiol Endocrinol Metab. 2006;291:E1074–E1082. doi: 10.1152/ajpendo.00040.2006. [DOI] [PubMed] [Google Scholar]
- 87.Eckhardt BL, et al. Genomic analysis of a spontaneous model of breast cancer metastasis to bone reveals a role for the extracellular matrix. Mol Cancer Res. 2005;3:1–13. [PubMed] [Google Scholar]
- 88.Lin BR, et al. Connective tissue growth factor inhibits metastasis and acts as an independent prognostic marker in colorectal cancer. Gastroenterology. 2005;128:9–23. doi: 10.1053/j.gastro.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 89.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 90.Gildea JJ, et al. Cancer Res. Vol. 62. 2002. RhoGDI2 is an invasion and metastasis suppressor gene in human cancer; pp. 6418–6423. Reference 90 provides an example of how novel high-throughput technologies have enabled identification of new metastasis suppressors. [PubMed] [Google Scholar]
- 91.Harding MA, et al. Functional genomic comparison of lineage-related human bladder cancer cell lines with differing tumorigenic and metastatic potentials by spectral karyotyping, comparative genomic hybridization, and a novel method of positional expression profiling. Cancer Res. 2002;62:6981–6989. [PubMed] [Google Scholar]
- 92.Theodorescu D, et al. Reduced expression of metastasis suppressor RhoGDI2 is associated with decreased survival for patients with bladder cancer. Clin Cancer Res. 2004;10:3800–3806. doi: 10.1158/1078-0432.CCR-03-0653. [DOI] [PubMed] [Google Scholar]
- 93.Hu LD, Zou HF, Zhan SX, Cao KM. Biphasic expression of RhoGDI2 in the progression of breast cancer and its negative relation with lymph node metastasis. Oncol Rep. 2007;17:1383–1389. [PubMed] [Google Scholar]
- 94.Wang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 95.Titus B, et al. Endothelin axis is a target of the lung metastasis suppressor gene RhoGDI2. Cancer Res. 2005;65:7320–7327. doi: 10.1158/0008-5472.CAN-05-1403. [DOI] [PubMed] [Google Scholar]
- 96.Lalich M, McNeel DG, Wilding G, Liu G. Endothelin receptor antagonists in cancer therapy. Cancer Invest. 2007;25:785–794. doi: 10.1080/07357900701522588. [DOI] [PubMed] [Google Scholar]
- 97.Battistini B, Berthiaume N, Kelland NF, Webb DJ, Kohan DE. Profile of past and current clinical trials involving endothelin receptor antagonists: the novel “-sentan” class of drug. Exp Biol Med (Maywood) 2006;231:653–695. [PubMed] [Google Scholar]
- 98.Wu Y, et al. Neuromedin U is regulated by the metastasis suppressor RhoGDI2 and is a novel promoter of tumor formation, lung metastasis and cancer cachexia. Oncogene. 2007;26:765–773. doi: 10.1038/sj.onc.1209835. [DOI] [PubMed] [Google Scholar]
- 99.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nature Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 100.Horak CE, et al. Nm23-H1 suppresses metastasis by inhibiting expression of the lysophosphatidic acid receptor EDG2. Cancer Res. 2007;67:11751–11759. doi: 10.1158/0008-5472.CAN-07-3175. Reference 100 illustrates the paradigm of using gene expression profiling of cells re-expressing metastasis suppressors to determine, and hopefully target, downstream genes that mediate the metastatic phenotype. [DOI] [PubMed] [Google Scholar]
- 101.Ohta H, et al. Ki16425, a subtype-selective antagonist for EDG-family lysophosphatidic acid receptors. Mol Pharmacol. 2003;64:994–1005. doi: 10.1124/mol.64.4.994. [DOI] [PubMed] [Google Scholar]
- 102.Boucharaba A, et al. The type 1 lysophosphatidic acid receptor is a target for therapy in bone metastases. Proc Natl Acad Sci USA. 2006;103:9643–9648. doi: 10.1073/pnas.0600979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 104.Lee JK, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci USA. 2007;104:13086–13091. doi: 10.1073/pnas.0610292104. References 103 and 104 provide excellent examples of the types of novel bioinformatic technologies that could enable investigators to target metastasis suppressor signatures, facilitate in silico drug discovery and ease clinical translation through patient selection and prediction of in vivo efficacy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Potti A, et al. Genomic signatures to guide the use of chemotherapeutics. Nature Med. 2006;12:1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 106.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nature Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 107.Wang H, Han H, Von Hoff DD. Identification of an agent selectively targeting DPC4 (deleted in pancreatic cancer locus 4)-deficient pancreatic cancer cells. Cancer Res. 2006;66:9722–9730. doi: 10.1158/0008-5472.CAN-05-4602. [DOI] [PubMed] [Google Scholar]
- 108.Rhodes DR, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fidler IJ. Orthotopic implantation of human colon carcinomas into nude mice provides a valuable model for the biology and therapy of metastasis. Cancer Metastasis Rev. 1991;10:229–243. doi: 10.1007/BF00050794. [DOI] [PubMed] [Google Scholar]
- 110.Ware JL, Paulson DF, Mickey GH, Webb KS. Spontaneous metastasis of cells of the human prostate carcinoma cell line PC-3 in athymic nude mice. J Urol. 1982;128:1064–1067. doi: 10.1016/s0022-5347(17)53345-5. [DOI] [PubMed] [Google Scholar]
- 111.Fidler IJ. Selection of successive tumour lines for metastasis. Nat New Biol. 1973;242:148–149. doi: 10.1038/newbio242148a0. [DOI] [PubMed] [Google Scholar]
- 112.Minn AJ, et al. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest. 2005;115:44–55. doi: 10.1172/JCI22320. Reference 112 is a landmark study that demonstrates empirically the exquisite tissue specificity of genes regulating metastasis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smith SC, et al. Profiling bladder cancer organ site-specific metastasis identifies LAMC2 as a novel biomarker of hematogenous dissemination. Am J Pathol. 2009;174:371–379. doi: 10.2353/ajpath.2009.080538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hedley BD, et al. BRMS1 suppresses breast cancer metastasis in multiple experimental models of metastasis by reducing solitary cell survival and inhibiting growth initiation. Clin Exp Metastasis. 2008;25:727–740. doi: 10.1007/s10585-008-9184-0. [DOI] [PubMed] [Google Scholar]
- 115.Goodison S, et al. The RhoGAP protein DLC-1 functions as a metastasis suppressor in breast cancer cells. Cancer Res. 2005;65:6042–6053. doi: 10.1158/0008-5472.CAN-04-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang X, et al. Overexpression of KAI1 suppresses in vitro invasiveness and in vivo metastasis in breast cancer cells. Cancer Res. 2001;61:5284–5288. [PubMed] [Google Scholar]
- 117.Chen SL, Hoehne FM, Giuliano AE. The prognostic significance of micrometastases in breast cancer: a SEER population-based analysis. Ann Surg Oncol. 2007;14:3378–3384. doi: 10.1245/s10434-007-9513-6. [DOI] [PubMed] [Google Scholar]
- 118.Liotta LA, Kohn E. Anoikis: cancer and the homeless cell. Nature. 2004;430:973–974. doi: 10.1038/430973a. [DOI] [PubMed] [Google Scholar]
- 119.Phadke PA, Vaidya KS, Nash KT, Hurst DR, Welch DR. BRMS1 suppresses breast cancer experimental metastasis to multiple organs by inhibiting several steps of the metastatic process. Am J Pathol. 2008;172:809–817. doi: 10.2353/ajpath.2008.070772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee JH, Welch DR. Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Res. 1997;57:2384–2387. [PubMed] [Google Scholar]
- 121.Wang JT, Peng DY, Chen M, Ye JS. Gene delivery for lung cancer using nonviral gene vectors. Pharmazie. 2007;62:723–726. [PubMed] [Google Scholar]
- 122.Ohtsuka T, et al. Synergistic induction of tumor cell apoptosis by death receptor antibody and chemotherapy agent through JNK/p38 and mitochondrial death pathway. Oncogene. 2003;22:2034–2044. doi: 10.1038/sj.onc.1206290. [DOI] [PubMed] [Google Scholar]
- 123.Le NT, Richardson DR. Iron chelators with high antiproliferative activity up-regulate the expression of a growth inhibitory and metastasis suppressor gene: a link between iron metabolism and proliferation. Blood. 2004;104:2967–2975. doi: 10.1182/blood-2004-05-1866. [DOI] [PubMed] [Google Scholar]
- 124.Kurdistani SK, et al. Inhibition of tumor cell growth by RTP/rit42 and its responsiveness to p53 and DNA damage. Cancer Res. 1998;58:4439–4444. [PubMed] [Google Scholar]
- 125.Bandyopadhyay S, et al. PTEN up-regulates the tumor metastasis suppressor gene Drg-1 in prostate and breast cancer. Cancer Res. 2004;64:7655–7660. doi: 10.1158/0008-5472.CAN-04-1623. [DOI] [PubMed] [Google Scholar]