Abstract
Mediator is the central coactivor complex required for regulated transcription by RNA polymerase (Pol) II. Mediator consists of 25 subunits arranged in the head, middle, tail and kinase modules. Structural and functional studies of Mediator are limited by the availability of protocols for the preparation of recombinant modules. Here, we describe protocols for obtaining pure endogenous and recombinant complete Mediator middle module from Saccharomyces cerevisiae that consists of seven subunits: Med1, 4, 7, 9, 10, 21 and 31. Native mass spectrometry reveals that all subunits are present in equimolar stoichiometry. Ion-mobility mass spectrometry, limited proteolysis, light scattering and small-angle X-ray scattering all indicate a high degree of intrinsic flexibility and an elongated shape of the middle module. Protein–protein interaction assays combined with previously published data suggest that the Med7 and Med4 subunits serve as a binding platform to form the three heterodimeric subcomplexes, Med7N/21, Med7C/31 and Med4/9. The subunits, Med1 and Med10, which bridge to the Mediator tail module, bind to both Med7 and Med4.
INTRODUCTION
Mediator is the essential coactivator complex required for regulated transcription by eukaryotic RNA polymerase (Pol) II (1–4). Saccharomyces cerevisiae Mediator has a molecular weight of 1.4 MDa and consists of 25 polypeptide subunits (Figure 1). The single subunits were assigned to four different structural modules: the head, middle, tail and kinase modules (5,6). Electron microscopic studies of Mediator across several species [S. cerevisiae (5,7,8), Schizosaccharomyces pombe (9), mouse (5) and human (10–12)] show a dynamic arrangement of the modules. Detailed structural information on some Mediator subunit domains, subunits and subcomplexes, was obtained by X-ray crystallography (13–16) and by nuclear magnetic resonance (NMR) (17,18) (Figure 1B). In particular, structures of two subcomplexes of the middle module are known: the Med7N/31 (15) and the Med7C/21 (13) heterodimers (Med7N and Med7C stand for the N- and C-terminal regions of Med7). Many subunit interactions have been mapped biochemically or by yeast two-hybrid analysis (16,19–21).
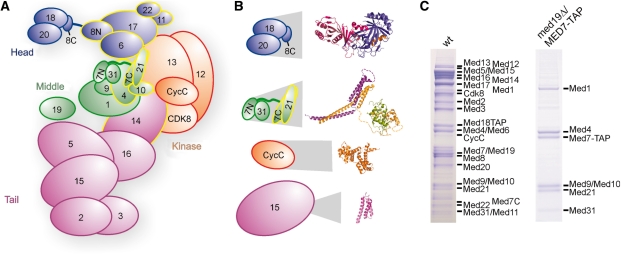
Figure 1.
Endogenous Mediator and its middle module. (A) Schematic view of Mediator subunit arrangement and modular structure, taking into account known and found (see below) subunit interactions. Subunits that are essential for yeast viability are outlined in yellow. (B) Available detailed structural information on subunits and subcomplexes of Mediator (13–17). Structures are enlarged in proportion to the full subcomplex or subunit sizes. (C) SDS-PAGE analysis of endogenous Mediator and 7-subunit Mediator middle module purified from wild-type (left) and med19Δ yeast strains containing a C-terminal TAP tag on the Med7 subunit. Copurifying proteins from 4 l yeast cells were separated on a 12% NuPAGE gel (Invitrogen), and bands were stained with Coomassie blue. The identity of all Mediator subunits could be confirmed by mass spectrometry (data not shown).
Mediator modules and submodules serve specific and overlapping functions in the regulation of subsets of genes (15,22,23). The Mediator head and middle modules together form a functional core (24), the subunits of which are conserved throughout eukaryotes (25–29). The detailed Mediator architecture and molecular mechanisms, however, remain elusive, mainly because structural information on the level of the modules is lacking. Such information would clarify the relative orientation of Mediator subunits within and between modules. Structural studies of these two core modules, however, require their preparation in large quantities and pure form. Thus far, only the head module is available in reconstituted form (20).
Here, we describe the purification of the complete 7-subunit middle module of Mediator from yeast, and as a recombinant complex after heterologous subunit coexpression in Escherichia coli. Pure middle module preparations were analyzed by various biophysical methods to determine subunit stoichiometry and module topology, and can be used for structural and functional studies in the future.
MATERIALS AND METHODS
Purification of endogenous middle module
The med19Δ yeast strain (BY4741; MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YBL093C::clonNAT) was obtained from Stephan Jellbauer (Gene Center Munich) and C-terminal TAP-tags were introduced on Med7, Med15 or Med18, respectively, using a kanMX4-marker by means of vector pYM13 (30). Yeast cultures were cultivated and the protein complexes purified using tandem affinity purification as described (15).
Preparation of recombinant middle module
Monocistronic vectors were cloned by standard procedures. Bi- or tricistronic vectors were generally created by introducing additional ribosomal binding sites by means of PCR into pET- and pCDF-vectors (Novagen) as described (13,16) and illustrated in Figure 2. A detailed listing of all vectors used and created for this study can be found in Supplementary Table S1. For expression, E. coli BL21 (DE3) RIL cells (Stratagene) were transformed with one to three plasmids and grown in LB medium at 37°C to an optical density at 600 nm (OD600) of 0.6. Expression was induced with 0.5 mM IPTG for 16 h at 18°C. Cells were lysed using a high-pressure homogenizer (Avestin) (large-scale protein purification) or by sonication (Branson) (small-scale purification).
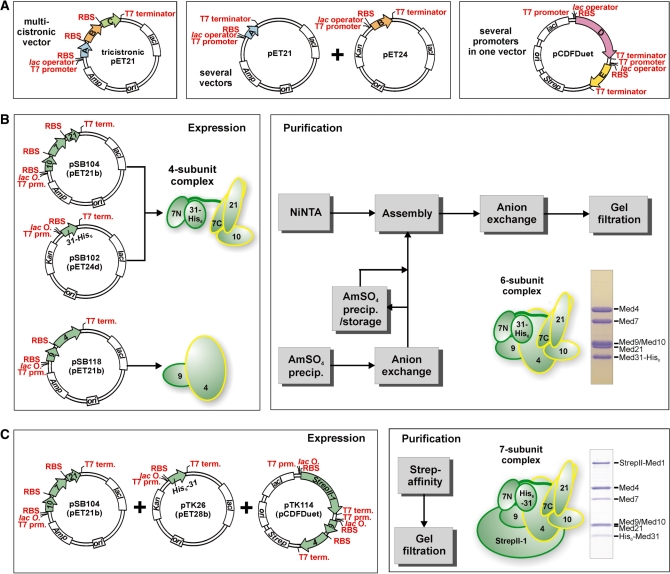
Figure 2.
Recombinant Mediator middle module. (A) Recombinant coexpression of multisubunit protein complexes in E. coli is possible by using multicistronic expression vectors, several compatible vectors cotransformed into E. coli, vectors with several promoters, and combinations of these. (B) Coexpression of 4-subunit Mediator middle complex containing Med7/10/21/31 is accomplished using a tricistronic pET21 and a monocistronic pET24 vector. The dimeric Med4/9 complex is coexpressed using a bicistronic pET21 vector. After cell-disruption and prepurification, both complexes are assembled into the 6-subunit middle complex containing Med4/7/9/10/21/31 and purified further to homogeneity. (C) Recombinant 7-subunit middle module consisting of Med1/4/7/9/10/21/31 is obtained by coexpression using a tricistronic vector encoding for Med7/10/21, a monocistronic vector encoding for His6-Med31 and a pCDFDuet vector with StrepII-tagged Med1 from multiple cloning site I (MCS I) and Med4/9 bicistronically expressed from MCS II. Purification after cell disruption is performed by StrepII-affinity purification and subsequent gel filtration.
The quaternary complex Med7/10/21/31-His6 was expressed from two plasmids encoding Med10, Med7 and Med21 in a tricistronic pET21b vector (pSB104) and Med31-His6 in a pET24d vector (pSB102). Four liters of E. coli culture were harvested and lysed in buffer A (50 mM Tris pH 8.0, 150 mM NaCl, 10 mM β-mercaptoethanol) containing protease inhibitors. After centrifugation, imidazole was added to a final concentration of 20 mM to the supernatant and loaded onto a 3 ml Ni-NTA gravity flow column (Qiagen) equilibrated with buffer A containing 20 mM imidazole. The column was washed with 20 column volumes (CVs) of buffer A containing 20 mM imidazole and eluted with buffer A containing 200 mM imidazole. The 4-subunit middle module was further purified by anion exchange and gel filtration chromatography.
The Med4/9 complex was coexpressed from a bicistronic pET21 vector (pSB118) using 2 l of E. coli culture. Cells were harvested and lysed in buffer B (20 mM Tris pH 8.0, 150 mM NaCl, 10 mM β-mercaptoethanol) containing protease inhibitors. Insoluble cell debris were removed by centrifugation and the proteins purified by ammonium sulfate precipitation and anion exchange chromatography. Ammonium sulfate precipitation was achieved by the gradual addition of saturated (20°C) ammonium sulfate solution up to 30% (v/v). After centrifugation, the pellet was resuspended and purified on a MonoQ 10/100 column (GE Healthcare) in buffer C (50 mM Tris pH 8.0, 100 mM NaCl, 10 mM β-mercaptoethanol) using a linear gradient of 20 column volumes (CVs) from 100 mM to 1 M NaCl. A six-subunit middle module comprising Med4/7/9/10/21/31 was obtained by assembling the two purified subcomplexes. Assembly was performed at 20°C on a rotating wheel with a 1.3 molar excess of Med4/9. Contaminants, excess Med4/9 and unassembled 4-subunit middle module were separated using anion exchange chromatography (buffer C, 20 CV from 100 mM to 1 M NaCl). Following concentration in a 100 kDa MWCO spin concentrator (Amicon Ultra, Millipore), the 6-subunit middle module was purified to homogeneity by gel filtration chromatography using a Superose 6 size exclusion column (GE Healthcare) with buffer A.
The complete middle module comprising Med1/4/7/9/10/21/31 was expressed from the cotransformed vectors pSB104 (encoding Med10, Med7 and Med21), pTK26 (encoding His6-Med31) and pTK114 (encoding StrepII-tagged Med1, Med9 and Med4). After cotransformation of two vectors, cells were made competent again in order to transform the third vector. Six liters of E. coli culture were harvested and lysed in buffer D [50 mM Tris pH 8.0, 150 mM NaCl, 10% (v/v) glycerol, 10 mM β-mercaptoethanol] containing protease inhibitors. After centrifugation, the complex was purified on 3 × 1 ml Strep‐Tactin MacroPrep (IBA) gravity flow columns according to the manufacturer’s instructions. Following elution by d‐Desthiobiotin (IBA) addition, the sample was concentrated in a 100-kDa MWCO spin concentrator and subjected to gel filtration using a Superose 6 size-exclusion column (GE Healthcare) with buffer A.
Native mass spectrometry
The buffer exchange for the native MS analysis was performed using 10 kDa cut-off membrane spin columns (Millipore, England) by six sequential concentration/dilution cycles against 200 mM ammonium acetate pH 6.8. The concentration for the analysis was 5 µM (assuming intact protein complexes). Analysis of the intact protein complex, as well as the n-propanol and DMSO measurements were carried out on a LCT mass spectrometer (Waters, UK). Needle voltage was set to 1250 V, cone voltage varied between 100 and 150 V. Tandem mass spectrometry was performed on a modified Q-ToF; here, needle voltage was set to 1300 V and cone to 150 V. For further details see ref. 31 and references within. Ion mobility mass spectrometry (IM-MS) was carried out on a Synapt HDMS (Waters, UK) (32). The source pressure was set to 6.9 mbar, the pressure in the trap was 3.5 × 10−2 mbar, 0.7 mbar in the ion mobility separation (IMS) cell and 2 × 10−6 mbar in the ToF. The wave height in the IMS cell was fixed on 11.3 V and the wave velocity set to 250 m/s. The gas used in the trap was xenon and nitrogen in the IMS cell. Needle voltage was 1200 V and cone voltage 150 V. The bias value was set to 20 V, trap and transfer collision energy to 12 V. Cross-section calculations were done as described by ref. 33. The average volume of global proteins and protein complexes was calculated according to ref. 34.
Limited proteolysis analyses
Limited proteolysis time courses were performed to identify durations suitable for obtaining medium-size fragments. Digests were performed using 20–50 µg protein complex with 0.2 µg of chymotrypsin (Sigma C3142) in buffer A supplemented with 4 µM CaCl2 by incubation at 37°C for 1–60 min. The reactions were stopped by the addition of SDS sample buffer and were heated immediately to 95°C for 10 min. For experiments depicted in Figure 4, purified 3-, 4- and 6-subunit middle module complexes were subjected to limited proteolysis for 10 min using chymotrypsin and after stopping the reaction by addition of a protease inhibitor mixture, loaded onto a Superose 6 gel filtration column (GE Healthcare). For proteolysis, 1 µg of sample was used for 3- and 4-subunit, and 2 µg for the 6-subunit middle module. Bands of interest in individual peaks were analyzed on SDS-PAGE after TCA precipitation by MS and Edman-sequencing with a Procise 491 sequencer (Applied Biosystems) following transfer to PVDF membranes.
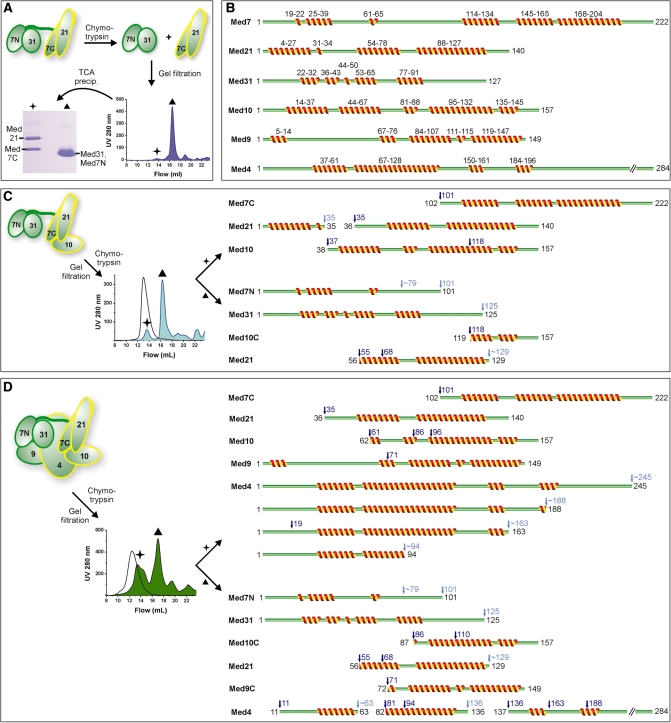
Figure 4.
Limited proteolysis analysis of Mediator middle module. (A) Recombinant complexes were subjected to limited proteolysis by chymotrypsin after identifying suitable digestion durations using time courses. The samples were subjected to gel filtration, the eluting peaks precipitated by TCA, fractions separated on SDS-PAGE and subjected to Edman sequencing. (B) Secondary structure predictions for yeast middle module subunits. Multiple sequence alignments were used whenever possible. Given are consensus predictions by HHpred (45), I-Tasser (46), PSIpred (47) and CDM (48). (C) Schematic diagram of limited proteolysis of Med7/10/21/31 using chymotrypsin, analogous to (A). The chromatogram of undigested complex is indicated by the black curve. Proteolytic cleavage sites are indicated above the protein cleavage schemes in dark blue (sequenced) and light blue (estimated from SDS-PAGE). Ambiguous C-terminal sites are marked with a tilde. In cases in which more than one N-terminus was sequenced, the alternative N-terminal cleavage sites are marked additionally. (D) Schematic diagram of limited proteolysis results for the Med4/7/9/10/21/31 complex using chymotrypsin, analogous to (C).
Coexpression and copurification pull-down assays
Coexpression was performed as described above using 50 ml cultures for StrepII-affinity purifications or 7 ml cultures for Ni-NTA purifications. Cell lysates were clarified by centrifugation and copurification pull-down assays performed in batch. StrepII-affinity purifications were performed using 25 µl of Strep‐Tactin MacroPrep (IBA). The clarified lysates were incubated with the beads at 4°C on a rotating wheel for 1 h. Beads were washed with 3 × 1 ml of buffer A and the samples eluted by d-Desthiobiotin. Ni-NTA purifications were performed using 40 µl of MagneHis beads (Promega). The clarified lysates were incubated with the beads at 4°C on a rotating wheel for 15 min. Beads were washed with 2 × 1 ml of buffer A and the samples eluted by 400 mM Imidazole in buffer A. Samples were analyzed on SDS-PAGE.
Small-angle X-ray scattering
Small-angle X-ray scattering (SAXS) data were collected at the X33-Beamline (EMBL/DESY, Hamburg, Germany). Scattering patterns from 6-subunit Mediator middle module solutions comprising Med4/7/9/10/21/31 were measured in buffer A at 5 mg/ml two times in order to exclude potential radiation damage errors. The ATSAS software package (35) was used for data processing.
RESULTS
Endogenous Mediator middle module
It was previously reported that the middle module is lost from Mediator that is purified from a med19Δ strain under stringent conditions (36). To purify middle module from a med19Δ strain, we introduced a C-terminal tandem affinity purification (TAP) tag on the middle module subunit Med7. TAP purification resulted in pure middle module without the need for the previously used urea dissociation (36), indicating that the tag further destabilizes the Mediator. Fusing a TAP-tag with Med15 or Med18 did not lead to purification of individual parts of Mediator (Supplementary Figure S1). The high yield and purity of the preparation from the med19Δ/MED7-TAP strain (Figure 1C) enabled us to determine unambiguously by MS (see ‘Materials and Methods’ section) that the endogenous yeast Mediator middle module comprises the subunits Med1, Med4, Med7, Med9 (Cse2), Med10 (Nut2), Med21 (Srb7) and Med31 (Soh1), consistent with previous description (19,21,27,37).
Recombinant middle module
We previously used heterologous coexpression of physically associated Mediator subunits in E. coli to obtain milligram quantities of Mediator subcomplexes of up to three subunits (13,15,16). Here, based on commercial pET- and Duet vectors (Novagen), we created poly-cistronic expression constructs with additional ribosomal binding sites between middle module subunit open reading frames (Figure 2). For expression of a 4-subunit complex comprising Med7, 10, 21 and 31, we constructed a tricistronic pET21b vector encoding Med10, 7 and 21 (Figure 2B) and a pET24d vector encoding C-terminally hexahistidine-tagged Med31. These vectors were cotransformed into E. coli BL21(DE3) RIL cells (Stratagene) and after coexpression, the 4-subunit complex could be purified (Figure 2B, ‘Materials and Methods’ section). To obtain a 6-subunit complex that comprised also subunits Med4 and Med9, these two subunits were coexpressed separately using a bicistronic pET21b vector. A partially purified Med4/9 heterodimer was assembled with the pure 4-subunit complex and the resulting 6-subunit complex was purified to homogeneity. To obtain complete 7-subunit middle module, three vectors were cotransformed into E. coli BL21(DE3) RIL cells, the tricistronic vector encoding Med10, 7 and 21, a vector encoding N-terminally hexahistidine-tagged Med31, and a vector encoding Med4, Med9 and N-terminally StrepII-tagged Med1 (Figure 2C, ‘Materials and Methods’ section). After coexpression of the seven proteins from these three vectors, the 7-subunit module was purified in two steps, including a StrepII-affinity step (Figure 2 C, ‘Materials and Methods’ section).
Subunit stoichiometry
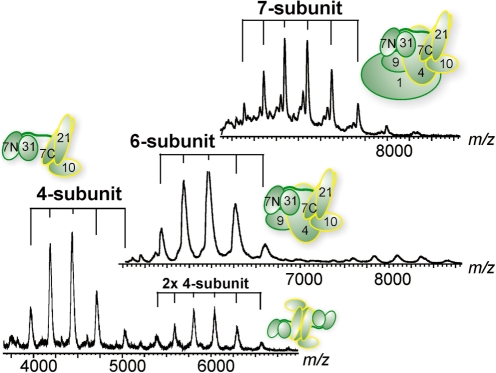
To investigate the subunit stoichiometry in the recombinant middle module and its subcomplexes, we used native MS that allows determination of molecular weights of entire protein complexes (38,39). The detected masses were 191 650 ± 60 Da for the complete 7-subunit middle module (191 335 Da expected), 124 760 ± 50 Da for the 6-subunit complex (124 471 Da expected) and 75 840 ± 30 Da for the 4-subunit complex (75 620 Da expected) (Figure 3 and Supplementary Figure S2A). We calculated expected masses taking into account that the N-terminal methionines of Med4, 7, 10, 21 and 31 were lacking (Edman sequencing data, not shown). The slightly higher experimental masses can be explained by incomplete desolvation of the complexes (40). The MS analysis also revealed that the 4- and 6-subunit complexes tend to dimerize. This is likely due to exposed hydrophobic surfaces, since addition of DMSO eliminated the dimers from the recorded MS spectra (Supplementary Figure S2B). These results reveal that only a single copy of each subunit is present in the complexes and establish the equimolar subunit stoichiometry of the middle module.
Figure 3.
Native mass spectrometry analyses of Mediator middle module. Shown are from top to bottom spectra of the 7-, 6- and 4-subunit middle module complexes. All individual subunits are present in equimolar stoichiometry. The distributions are labeled accordingly to the corresponding schematics.
Module topology
We next investigated the topology of the middle module by MS analysis. We observed spontaneous dissociation of subunits and subcomplexes in the buffer used for the native MS experiments. This effect could be enhanced by addition of DMSO or n-propanol (Supplementary Figure S2). Dissociation of the 7-subunit module revealed that Med1 could detach from the module separately, to result in the 6-subunit complex. Dissociation of the 6-subunit complex revealed a dimer of Med4/9, confirming the interaction between those two subunits. Dissociation of the 4-subunit complex resulted in the trimers Med7/10/31 and Med7/21/31, and in the dimers Med7/31 and Med7/21.Overall, Med7 never dissociated from the middle module, showing it is an architectural subunit, whereas Med9 and Med10 were least stably attached. To further analyze the module topology, we investigated fragmentation of the 4-, 6- and 7-subunit complexes by tandem MS. Generally consistent with solution dissociation experiments, Med9 and Med10 readily dissociated from the complexes, whereas Med7 never did, except to a low extent when the complete middle module was used. Taken together, the dissociation and fragmentation experiments are consistent with earlier structural and interaction analysis of the middle module and the existence of heterodimers Med7N/31, Med7C/21 and Med4/9 within intact middle module (13,15,19).
Exposed regions
To detect exposed and flexible protein regions, middle module complexes were subjected to limited proteolysis using chymotrypsin (see ‘Materials and Methods’ section). In an attempt to separate stable fragments, we stopped proteolysis by addition of protease inhibitor at certain time points, and subjected the samples to gel filtration. Peak fractions were TCA-precipitated and analyzed by SDS-PAGE and Edman sequencing (Figure 4). This approach could identify stable subcomplexes suitable for crystallization, such as Med7N/31 and Med7C/21, which result from proteolysis of a Med7/21/31 trimer (Figure 4A) (13,15).
Limited proteolysis of the 4-subunit complex revealed not only protease cleavage within regions predicted to form flexible loops, but also cleavage in some helical regions of Med21 (known structure) and Med10 (predicted) (Figure 4B, and C). Two subcomplexes were detected based on Med7N/31 and Med7C/21 that both contain N-terminally truncated Med10 (Figure 4C). Proteolysis of the 6-subunit complex also revealed Med7C/21 and Med7N/31 in different gel filtration fractions, but now associated with different truncated variants of Med10 (Figure 4D). Consistently, native MS analysis of an early 7-subunit complex preparation revealed an intact middle module with a truncated form of Med10 that lacked 66 N-terminal residues (data not shown). Med9 showed only one chymotrypsin cleavage site and was detected either as full-length subunit (star in Figure 4D) or as an N-terminally shortened variant (star and triangle peak in Figure 4D). Med4 was cleaved at multiple sites, but preferentially from the C-terminus.
This analysis supports a submodular architecture of the middle module based on the Med7N/31 and Med7C/21 core subcomplexes connected via a flexible Med7 linker. Med4, 9 and 10 are associated with both core subcomplexes, but their proteolytic fragments were not sufficient to hold these subcomplexes together. The C-terminal region of Med4, the N-terminal region of Med9, and the N-terminal region of Med10 are not stably bound to the core subcomplexes and may be involved in contacts with other Mediator modules or external factors. Consistently, yeast complementation assays revealed that the flexible Med4 C-terminal residues 194–250 are required for viability, whereas the N-terminal residues 1–66 are not (Supplementary Figure S3).
Intra-module subunit interactions
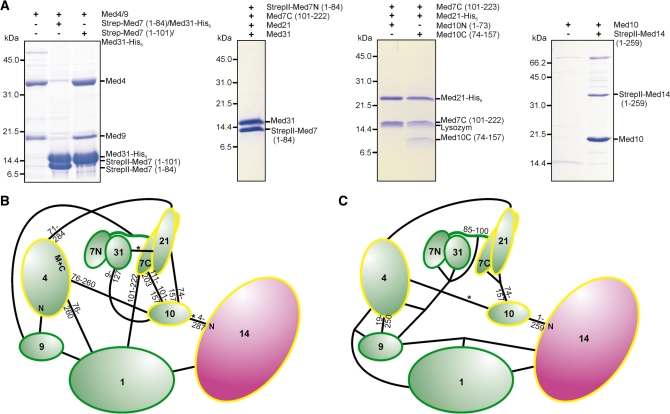
To further elucidate the subunit interactions within the middle module, we used coexpression pull-down experiments (Figure 5A and Supplementary Table S2). Our biochemical analysis generally confirmed previous subunit interactions based on yeast two hybrid (Y2H) assays and immunoprecipitation (19), but additionally allowed us to distinguish weak and strong subunit interactions, and to detect interaction domain boundaries (Figure 5B and C). Important findings from this analysis were the following. First, Med7N/31 neither bound Med7C/21 nor Med4/9. For interaction of Med4/9 to any Med7-containing complex, the Med7 linker between Med7N and Med7C was required. Second, the Med4/9 heterodimer binds to Med1. Med4/9 heterodimer formation neither required the 18 N-terminal residues of Med4 nor the predicted loop in Med9 comprising residues 19–63, consistent with the prior observation that Med9 contains two domains (compare 41). Third, Med10 and Med4 interacted weakly and Med21 stabilized this interaction. The C-terminal part of Med10 was sufficient for binding the Med7C/21 subcomplex. Med10 was also found to stably bind to Med14, confirming previous data (19) and showing that Med14 (1-259) is sufficient to establish a connection of middle module towards Mediator tail module. Finally, reported Y2H interactions between Med21 and Med31 were not confirmed. Stable expression of Med9 always required coexpression with Med4, but was not possible with Med1 or Med7, suggesting that Med9 has no direct interaction with these. Together with published data, this analysis establishes the middle module subunit interaction network (Figure 5C).
Figure 5.
Intra-module subunit interactions within the middle module. (A) Middle module subunits and subcomplexes were tested for interaction with other middle module subunits or subcomplexes by coexpression and subsequent copurification pull-down assays. (B) Mediator middle module interaction map based on previously published (19) yeast-two-hybrid (Y2H) assays and structural data (13,15). The length of truncated subunit variants that gave interactions with its partner, are indicated closely to the molecules. Interactions based only on single Y2H clones are indicated by a star. (C) Mediator middle module interaction map based on coexpression and copurifications. The map integrates published data (13,15) with the findings depicted in Figures 2B, C, 5A and Supplementary Table S2. As coexpression and copurification was required to obtain stable complexes, connections to more than one partner are indicated for some proteins. Weak interactions are indicated by a star.
Module shape
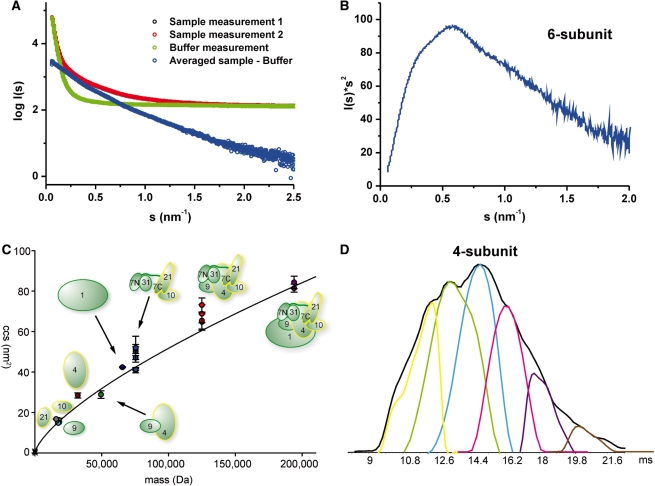
To investigate the overall shape of the middle module in solution we used light scattering and SAXS. Static light scattering analysis of the 6-subunit complex revealed a hydrodynamic radius of 66 Å, 34% larger than that of Med7C/21, which exists as a heterotetramer in solution (compare Supplementary Table S3). SAXS analysis showed that the 6-subunit middle module was partially aggregated at the high concentrations required for the analysis (Figure 6A and Supplementary Table S3). Thus, we could not reliably determine the radius of gyration and could not calculate low-resolution structural models. Nevertheless, the scattering curve shows clearly that the middle module exhibits an elongated shape (compare 42, Figure 1, e.g. model body 10). The Kratky-plot further indicates that the protein complex is folded and consists of a few globular areas (Figure 6B). Unfortunately, analysis of the 7-subunit middle module by SAXS was hampered by susceptibility of the Med1 subunit to degradation and difficulties in complex concentration.
Figure 6.
Shape of the middle module. (A) The experimental SAXS curve of 6-subunit Mediator middle module indicates an elongated protein complex. I0 signal intensity in comparison with the BSA standard measurement suggested a molecular weight of the complex far above the theoretical weight. Therefore, the Guinier radius could not be reliably determined and useful models could not be calculated. (B) The corresponding Kratky-plot shows no classical bell shape, but is stretched toward higher scattering angles. The complex is nevertheless folded and exhibits also few globular areas. (C) Ion mobility MS experiments. Plotted are the calibrated collision cross sections (CCSs) of the single subunits and (sub-)complexes (for detailed information see Supplementary Table S4). The graph indicates a general trend of CCS versus mass for globular proteins [determined both experimentally (34) and derived from globular structures deposited in the PDB http://www.rcsb.org]. The graph extends up to a mass of 801 kDa and a CCS of 229 nm2. Only the section relevant for the Mediator complexes is shown. The color-coding is: grey: Med21, cyan: Med9, orange: Med10, brown: Med4, dark green: Med4/7, dark blue: Med1, blue: 4-subunit middle, red: 6-subunit middle, purple: 7-subunit middle. (D) Flexibility of the Mediator middle module. From IM-MS measurements, we generated the averaged drift time plot (in ms) for the 4-subunit Mediator middle (charge 16+) (shown in black). Using the Drift Scope software (Waters, UK), we extracted the different conformations that contribute to the averaged drift time plot. We can detect up to six conformations that make up this charge state of the 4-subunit Mediator middle in the gas phase, thereby reflecting the flexibility of the Mediator middle module.
To add support to the results obtained in solution, we used ion mobility (IM) MS analysis. IM-MS can provide the collisional cross section (CCS), which corresponds to an averaged lateral cut through the protein (43). We applied IM-MS to the three recombinant Mediator middle module complexes (Figure 6C and Supplementary Table S4). Calculation of calibrated CCS revealed conformational flexibility in all complexes, consistent with our inability to crystallize any of these complexes and published in silico analysis data (44). This becomes more clear even when looking at an averaged drift time analysis (Figure 6D and Supplementary Figure S2C). Compared to standard proteins of similar mass (34), the most abundant conformations for the 4- and 6-subunit complexes represented extended structures. The conformation of the complete 7-subunit middle module was, however, more compact (Figure 6C). The average calculated densities for the 4-, 6- and 7-subunit complexes were 0.48, 0.50 and 0.56 g/cm3 respectively. Thus, three different experimental methods are consistent with an elongated shape of the middle module, which is further consistent with published cryo-electron microscopic data (7,9,10).
DISCUSSION
Understanding the Mediator complex on a molecular level requires a detailed analysis of its four multisubunit modules, for which no structures are available. Here, we first confirmed the subunit composition of the complete endogenous S. cerevisiae middle module after its purification from a med19Δ strain of yeast. We then established a protocol for the expression and purification of the complete 7-subunit middle module and its 4- and 6-subunit subcomplexes. We have used the defined middle complex preparations in various biophysical assays to determine the subunit stoichiometry, subunit interactions and the elongated shape of the complex.
Light and X-ray scattering as well as IM-MS suggest that the 6-subunit middle module has an elongated shape, whereas the complete 7-subunit middle module is more compact. As revealed by interaction experiments, Med7 and Med4 subunits interact via the linker between Med7N and Med7C. Our proteolysis, interaction experiments, and MS data all suggest that they serve as a binding platform to form three stable heterodimers, Med7N/31, Med7C/21 and Med4/9. Med1 and Med10 both bridge between the heterodimers Med7C/21 and Med4/9. These middle module subcomplexes are apparently connected in a flexible manner. The Med4 C-terminus and the Med10 N-terminus are flexible and exposed, with the latter involved in binding Med14. These intrinsic flexibilities have thus far impeded crystallization of the middle module. Our interaction data not only validate most subunit contacts mapped by yeast-two hybrid analysis (19) but also identify a few false positives and reveal new details about subunit interactions.
In order to understand how Mediator integrates regulatory signals and how it enables activated transcription, future work will require elucidating the activity of the essential Mediator middle module and its parts. Despite extensive efforts, we were however unable to test the functionality of the middle module in vitro. Establishing an in vitro assay for middle module-dependent activated transcription will thus remain a future goal. In addition, the results presented here form the basis for future structural studies. The coexpression strategies established here may also be valuable for the preparation of other large and flexible multiprotein complexes by coexpression in E. coli.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Deutsche Forschungsgemeinschaft; the Sonderforschungsbereich SFB646; the Transregio 5; the EU 3D Repertoire (contract no. LSHG-CT-2005-512028); the European Molecular Biology Organization (EMBO); The Netherlands Proteomics Centre, which is part of the Netherlands Genomics Initiative; and a LMU-excellent research professorship to P.C. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stefan Benkert and other members of the Cramer laboratory for help and discussions. We thank Stephan Jellbauer (Gene Center Munich) for providing the med19Δ yeast strain, members of the Hopfner laboratory for SAXS measurement and Gregor Witte (Gene Center Munich) for discussion of SAXS data. We thank Georg Arnold and Thomas Fröhlich (LAFUGA at the Gene Center Munich) as well as Axel Imhof and members of the Zentrallabor für Proteinanalytik (Adolf-Butenandt-Institute Munich) for mass spectrometry analysis in the endogeneous and limited proteolysis experiments. Part of this study was performed at the EMBL/DESY (Hamburg, Germany).
REFERENCES
- 1.Bjorklund S, Gustafsson CM. The yeast Mediator complex and its regulation. Trends Biochem Sci. 2005;30:240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Malik S, Roeder RG. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- 4.Naar AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu. Rev. Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- 5.Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD. Conserved structures of mediator and RNA polymerase II holoenzyme. Science. 1999;283:985–987. doi: 10.1126/science.283.5404.985. [DOI] [PubMed] [Google Scholar]
- 6.Kang JS, Kim SH, Hwang MS, Han SJ, Lee YC, Kim YJ. The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 2001;276:42003–42010. doi: 10.1074/jbc.M105961200. [DOI] [PubMed] [Google Scholar]
- 7.Cai G, Imasaki T, Takagi Y, Asturias FJ. Mediator structural conservation and implications for the regulation mechanism. Structure. 2009;17:559–567. doi: 10.1016/j.str.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis JA, Takagi Y, Kornberg RD, Asturias FA. Structure of the yeast RNA polymerase II holoenzyme: mediator conformation and polymerase interaction. Mol. Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 9.Elmlund H, Baraznenok V, Lindahl M, Samuelsen CO, Koeck PJ, Holmberg S, Hebert H, Gustafsson CM. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc. Natl Acad. Sci. USA. 2006;103:15788–15793. doi: 10.1073/pnas.0607483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 2009;23:439–451. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taatjes DJ, Naar AM, Andel F., III, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- 12.Taatjes DJ, Schneider-Poetsch T, Tjian R. Distinct conformational states of nuclear receptor-bound CRSP-Med complexes. Nat. Struct. Mol. Biol. 2004;11:664–671. doi: 10.1038/nsmb789. [DOI] [PubMed] [Google Scholar]
- 13.Baumli S, Hoeppner S, Cramer P. A conserved mediator hinge revealed in the structure of the MED7.MED21 (Med7.Srb7) heterodimer. J. Biol. Chem. 2005;280:18171–18178. doi: 10.1074/jbc.M413466200. [DOI] [PubMed] [Google Scholar]
- 14.Hoeppner S, Baumli S, Cramer P. Structure of the mediator subunit cyclin C and its implications for CDK8 function. J. Mol. Biol. 2005;350:833–842. doi: 10.1016/j.jmb.2005.05.041. [DOI] [PubMed] [Google Scholar]
- 15.Koschubs T, Seizl M, Lariviere L, Kurth F, Baumli S, Martin DE, Cramer P. Identification, structure, and functional requirement of the Mediator submodule Med7N/31. EMBO J. 2009;28:69–80. doi: 10.1038/emboj.2008.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larivière L, Geiger S, Hoeppner S, Röther S, Strässer K, Cramer P. Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat. Struct. Mol. Biol. 2006;13:895–901. doi: 10.1038/nsmb1143. [DOI] [PubMed] [Google Scholar]
- 17.Thakur JK, Arthanari H, Yang F, Pan S.-J, Fan X, Breger J, Frueh DP, Gulshan K, Li DK, Mylonakis E, et al. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature. 2008;452:604–609. doi: 10.1038/nature06836. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang S, et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 19.Guglielmi B, van Berkum NL, Klapholz B, Bijma T, Boube M, Boschiero C, Bourbon H.-M, Holstege F.CP, Werner M. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 2004;32:5379–5391. doi: 10.1093/nar/gkh878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takagi Y, Calero G, Komori H, Brown JA, Ehrensberger AH, Hudmon A, Asturias F, Kornberg RD. Head module control of mediator interactions. Mol. Cell. 2006;23:355–364. doi: 10.1016/j.molcel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Beve J, Hu GZ, Myers LC, Balciunas D, Werngren O, Hultenby K, Wibom R, Ronne H, Gustafsson CM. The structural and functional role of Med5 in the yeast Mediator tail module. J. Biol. Chem. 2005;280:41366–41372. doi: 10.1074/jbc.M511181200. [DOI] [PubMed] [Google Scholar]
- 22.Larivière L, Seizl M, van Wageningen S, Röther S, van de Pasch L, Feldmann H, Sträßer K, Hahn S, Holstege F.CP, Cramer P. Structure-system correlation identifies a gene regulatory Mediator submodule. Genes Dev. 2008;22:872–877. doi: 10.1101/gad.465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van de Peppel J, Kettelarij N, van Bakel H, Kockelkorn T.T.JP, van Leenen D, Holstege F.CP. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell. 2005;19:511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Ranish JA, Aebersold R, Hahn S. Yeast nuclear extract contains two major forms of RNA polymerase II mediator complexes. J. Biol. Chem. 2001;276:7169–7175. [PubMed] [Google Scholar]
- 25.Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S. Purification of a plant Mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell. 2007;26:717–729. doi: 10.1016/j.molcel.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Boube M, Joulia L, Cribbs DL, Bourbon H.-M. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell. 2002;110:143–151. doi: 10.1016/s0092-8674(02)00830-9. [DOI] [PubMed] [Google Scholar]
- 27.Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flanagan PM, Kelleher R.J., III, Sayre MH, Tschochner H, Kornberg RD. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 29.Kelleher RJ, III, Flanagan PM, Kornberg RD. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990;61:1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 30.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 31.Lorenzen K, Vannini A, Cramer P, Heck AJ. Structural biology of RNA polymerase III: mass spectrometry elucidates subcomplex architecture. Structure. 2007;15:1237–1245. doi: 10.1016/j.str.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 32.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-ToF instrument. Int. J. Mass Spectrom. 2007;261:1–12. [Google Scholar]
- 33.Ruotolo BT, Benesch JL, Sandercock AM, Hyung SJ, Robinson CV. Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 34.Uetrecht C, Versluis C, Watts NR, Wingfield PT, Steven AC, Heck AJ. Stability and shape of hepatitis B virus capsids in vacuo. Angew. Chem. Int. Ed. Engl. 2008;47:6247–6251. doi: 10.1002/anie.200802410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baidoobonso SM, Guidi BW, Myers LC. Med19(Rox3) regulates intermodule interactions in the Saccharomyces cerevisiae mediator complex. J. Biol. Chem. 2007;282:5551–5559. doi: 10.1074/jbc.M609484200. [DOI] [PubMed] [Google Scholar]
- 37.Linder T, Zhu X, Baraznenok V, Gustafsson CM. The classical srb4-138 mutant allele causes dissociation of yeast Mediator. Biochem. Biophys. Res. Commun. 2006;349:948–953. doi: 10.1016/j.bbrc.2006.08.099. [DOI] [PubMed] [Google Scholar]
- 38.Heck AJ. Native mass spectrometry: a bridge between interactomics and structural biology. Nat. Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 39.Sharon M, Robinson CV. The role of mass spectrometry in structure elucidation of dynamic protein complexes. Annu. Rev. Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 40.McKay AR, Ruotolo BT, Ilag LL, Robinson CV. Mass measurements of increased accuracy resolve heterogeneous populations of intact ribosomes. J. Am. Chem. Soc. 2006;128:11433–11442. doi: 10.1021/ja061468q. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi H, Kasahara K, Kokubo T. Saccharomyces cerevisiae Med9 comprises two functionally distinct domains that play different roles in transcriptional regulation. Genes Cells. 2009;14:53–67. doi: 10.1111/j.1365-2443.2008.01250.x. [DOI] [PubMed] [Google Scholar]
- 42.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uetrecht C, Rose RJ, Duijn Ev, Lorenzen K, Heck A.JR. Ion mobility mass spectrometry of proteins and protein assemblies. Chem. Soc. Rev. 2010 doi: 10.1039/b914002f. doi:10.1039/B914002F. [DOI] [PubMed] [Google Scholar]
- 44.Toth-Petroczy A, Oldfield CJ, Simon I, Takagi Y, Dunker AK, Uversky VN, Fuxreiter M. Malleable machines in transcription regulation: the mediator complex. PLoS Comput. Biol. 2008;4:e1000243. doi: 10.1371/journal.pcbi.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 48.Sen TZ, Cheng H, Kloczkowski A, Jernigan RL. A Consensus Data Mining secondary structure prediction by combining GOR V and Fragment Database Mining. Protein Sci. 2006;15:2499–2506. doi: 10.1110/ps.062125306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.