Abstract
Models of mammalian regulatory networks controlling gene expression have been inferred from genomic data, yet have largely not been validated. We present an unbiased strategy to systematically perturb candidate regulators and monitor cellular transcriptional responses. We apply this approach to derive regulatory networks that control the transcriptional response of mouse primary dendritic cells (DCs) to pathogens. Our approach revealed the regulatory functions of 125 transcription factors, chromatin modifiers, and RNA binding proteins and constructed a network model consisting of two dozen core regulators and 76 fine-tuners that help explain how pathogen-sensing pathways achieve specificity. This study establishes a broadly-applicable, comprehensive and unbiased approach to reveal the wiring and functions of a regulatory network controlling a major transcriptional response in primary mammalian cells.
Introduction
Regulatory networks controlling gene expression serve as decision-making circuits within cells. For example, when immune dendritic cells are exposed to viruses, bacteria or fungi, they respond with transcriptional programs that are specific to each pathogen (1)and are essential for establishing appropriate immunological outcomes (2). These responses are initiated through specific receptors, such as Toll-like receptors (TLRs), that distinguish broad pathogen classes, and are propagated through well-characterized signaling cascades (2). However, how the transcriptional network is wired to produce specific outputs remains largely unknown.
Two major observational strategies have associated regulators with their putative targets on a genome scale (3): cis-regulatory models rely on the presence of predicted transcription factor binding sites in the promoters of target genes (3-5), whereas trans-regulatory models are based on correlations between regulator and target expression (3-5), (6). Since promoter binding sites and correlated expression are weak predictors of functional regulator-target linkages, such approaches are limited in their ability to produce reliable models of transcriptional networks (3). A complementary strategy is to systematically perturb every regulatory input and measure its effect on expression of gene targets. This strategy has been successfully employed in yeast (7-9) and sea urchin (10), but not in mammals.
A perturbation-based strategy for network reconstruction
We developed a perturbation strategy for reconstructing transcriptional networks in mammalian cells, and applied it to determine a network controlling the responses of DCs to pathogens (Fig. 1). First, we profiled gene expression at nine time points following stimulation with five pathogen-derived components and identified specific and shared genes that respond to each stimulus (Fig. S1A). We used these profiles to identify 144 candidate regulators whose expression changed in response to at least one stimulus (SOM) (Fig. S1B, top). We also identified a signature of 118 marker genes (Fig. S1B, bottom) that captures the complexity of the response. We generated a validated lentiviral shRNA library for 125 (of the 144) candidate regulators (Fig. S1C, top), used it to systematically perturb each of the regulators in DCs, stimulated the cells with a pathogen component, and profiled the expression of the 118 gene signature (11) (Fig. S1C, bottom). Finally, we used the measurements from the perturbed cells to derive a validated model of the regulatory network (Fig. S1D).
Fig. 1. A systematic strategy for network reconstruction.
The strategy consists of four steps, from left, (1) state measurement using arrays, (2) selection of regulators and response signatures, (3) network perturbation with shRNAs against each regulator followed by measurement of signature genes, and (4) network reconstruction from the perturbational data.
Gene expression programs in response to TLR agonists
We measured genome-wide expression profiles in DCs exposed to PAM3CSK4 (‘PAM’), a synthetic mimic of bacterial lipopeptides, polyI:C, a viral-like dsRNA, LPS, a purified component from Gram negative E. coli, gardiquimod, a small molecule agonist, and CpG, a synthetic ssDNA. These compounds are known agonists of TLR2, TLR3, TLR4, TLR7 and TLR9, respectively. PolyI:C also activates MDA-5, and LPS can also act through co-receptors such as CD14. We therefore refer to the ligands rather than their receptors for clarity. Based on pilot experiments (Fig S2, SOM), we measured mRNA expression at 0.5, 1, 2, 4, 6, 8, 12, 16, and 24 hours following stimulation with these pathogen components.
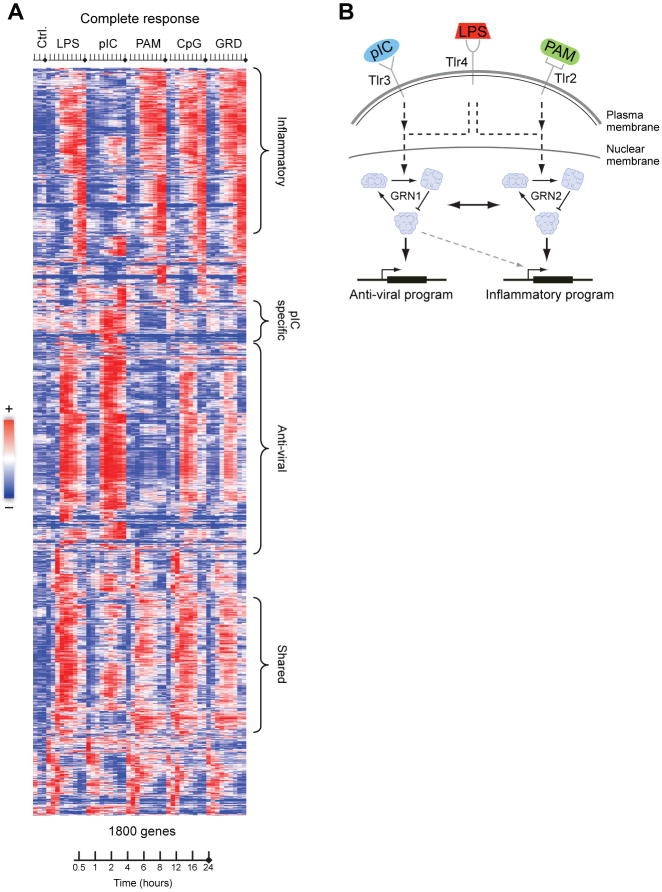
The observed transcriptional responses were classified into a ‘PAM-like’ program and a ‘polyI:C-like’ program, as well as a shared response (24.5% shared by PAM/polyI:C/LPS). The LPS response (Fig. 2A, B, Fig. S3) was largely the union of the ‘PAM-like’ and ‘polyI:C-like’ programs. This is partly explained by the known signaling pathways activated by these agonists. PAM binds TLR2 and signals through the MYD88 pathway; polyI:C binds TLR3 and MDA-5 and signals mostly through the TRIF and IPS-1 pathways, respectively; and LPS binds TLR4 and co-receptors and uses both pathways (12). It is also consistent with the known induction of an anti-viral response by polyI:C and LPS (13). The ‘PAM-like’ program is enriched for NFκB and inflammatory responsive genes (P<6.1*10-8), whereas the ‘polyI:C-like’ program is enriched for IRFs, viral- and interferon-responsive genes (ISGs, P<8.3*10-24). We thus term them the ‘inflammatory-like’ and ‘anti-viral-like’ programs. A small number of genes are specific to a single stimulus. For example, ∼250 genes are polyI:C-specific (1250 are shared with LPS), including several Type I IFNs (e.g. IFNA2, IFNA4, Fig. 2a). Surprisingly, 82% of the gardiquimod (TLR7) and CpG (TLR9) response was shared with the LPS response, but with a weaker anti-viral component (Fig. S4). This observation is unexpected given their different signaling mechanisms (14), but is highly reproducible and robust (Fig. S4, SOM).
Fig. 2. Gene expression response to pathogen components.
A, mRNA profiles of the 1800 genes whose expression was induced to at least 1.7-fold from baseline level in both duplicates of at least one time point in CD11c+ dendritic cells stimulated with the indicated pathogen component across a time course of 0, 0.5, 1, 2, 4, 6, 8, 12, 16, 24 hours (tick marks; GRD – gardiquimod). Replicates were collapsed and genes hierarchically clustered. Rows – genes, columns – experiments, red – induced from baseline, blue – repressed from baseline, white – unchanged from baseline. B, Model illustrating the differential networks controlling the anti-viral (‘polyI:C-like’) and inflammatory (‘PAM-like’) programs.
Identification of candidate regulators and a response signature
To select potential regulators that mediate the observed transcriptional response, we focused on regulator genes whose expression changes during pathogen sensing (a reasonable assumption for many mammalian responses (15, 16), including pathogen-sensing (1, 4). First, we reconstructed an observational trans-model of gene regulation (Figs. S1B, top, S5A, SOM), that associated 80 modules of co-regulated genes with 608 predictive regulators (4, 17, 18)(SOM, Fig. S5B), automatically chosen out of a curated list of 3287 candidate regulators (SOM). Filtering identified 117 regulators above a minimal expression signal in at least one experiment (Fig. S5B). These included known regulators from the NFKB, STAT and IRF families as well as unexpected candidates such as the circadian regulator Timeless and the DNA methyltransferase Dnmt3a. Second, we added 5 constitutively expressed regulators whose cis-regulatory elements are enriched in the responsive genes (SOM). Third, to capture delayed responses or nonlinear relations, we incorporated 22 regulators with at least a 2-fold change in expression. This resulted in 144 candidate regulators, with a distribution of expression patterns similar to the general response (Figs. S6, S7, S8 and Table S1), The regulators' expression under LPS was conserved between DCs and functionally similar macrophages, (Pearson correlation r∼0.9 at 1h, Fig. S9A) as well as between human macrophages and mouse DCs (r∼0.6 at 2h, Fig. S9B) supporting the functional relevance of the regulators' transcription.
To identify highly informative reporter genes for monitoring the effects of perturbing regulators, we devised GeneSelector (Fig. S10A, Table S2, SOM). GeneSelector incrementally chooses genes (from our full expression dataset) whose expression profile improves our discrimination of stimuli given the previously chosen genes. Using this approach, we identified the optimal time point (six hours post activation, Fig. S10B) and a set of 81 genes that distinguishes the stimuli (SOM). We added 37 candidate regulators with detectable expression at the 6h time point, creating a signature of 118 genes. Finally we added 10 control genes whose expression levels were unchanged under all stimuli, but whose (constant) basal levels varied from very low to high.
Perturbations, profiling and modeling
We generated validated lentiviral shRNAs that knocked down expression of 125 of our 144 candidate regulators by at least 75% (Fig. S11, Table S3, SOM) and 32 shRNAs with no known gene targets as controls in bone marrow DCs (Fig S12, Fig S13, Table S4, SOM). To carry out our perturbational study, we selected a single treatment, LPS, that activates the majority of both the ‘inflammatory-like’ and ‘anti-viral-like’ programs. Following stimulation of shRNA-perturbed DCs with LPS for six hours, we used nCounter (11) to count transcripts of the 118 reporter and 10 control genes.
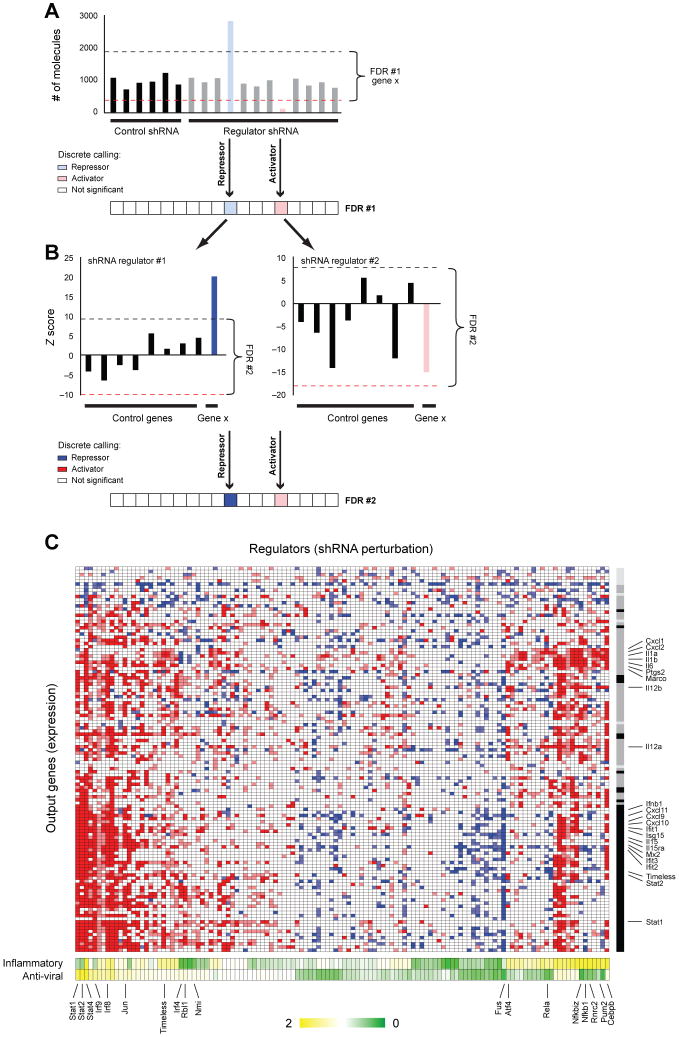
The changes in signature gene expression resulting from infection with each shRNA were used to construct a model that associated regulators to their targets. We expect increases in the transcript levels of reporter genes whose repressors are targeted by knockdown, and decreases in reporters whose activators are targeted. Our False Discovery Based (FDR) model estimates the significance of a change in transcripts in DCs infected with a given shRNA (SOM). We control for gene-specific noise by comparing to changes in the expression of each gene following perturbation with the control shRNAs (Fig. 3A), and for shRNA-specific noise by comparing to changes in the expression of the control genes following a given shRNA perturbation (Fig. 3B). We estimated the sensitivity of our calls from the 37 regulators, which are also included as target reporters (Fig. S14, SOM).
Fig. 3. Gene regulatory programs controlling the response to pathogen components.
A,B, A strategy to minimize false discovery (FDR) calls of significant changes in an output target gene resulting from knockdown of a regulator gene. A, The first FDR procedure (top) compares the expression of the gene following a perturbation with a regulator shRNA (right) to its expression upon perturbation with 32 non-targeting shRNAs (left). The dashed lines identify the gene-specific FDR-based thresholds for induction (blue line) and repression (red line). A discrete vector of significant calls (bottom) is derived from the raw data (blue – regulator represses the target gene; red – regulator induces the target gene). B, A second FDR procedure (top) compares the expression of the target gene to that of eight control (target) genes upon perturbation with the same shRNA. In the example shown, the gene's induction (left) was significant relative to the variation in expression among the control target genes resulting in a high score (bottom, dark blue), but its repression did not significantly differ from the controls, resulting in a lower score (bottom, weaker red). C. All significant (95% confidence) relations between regulators (columns) and targets (rows), colored as in B. The gray bar (right) represents the NMF-based calls for each target gene; black – anti-viral program; dark gray – inflammatory program; light gray – control genes. The bottom bar shows the degree of effect by the regulator on each program as determined by the NMF projection of the regulator's perturbation profile. (Yellow – positive effect, Green – negative effect; NMF scores are mean-normalized).
On the basis of these results we identified a densely overlapping network with 2322 significant regulatory connections, including 1728 activations and 594 repressions (Fig. 3B, red and blue, respectively, at 95% confidence Tables S5, S6 and S7). Of the 125 tested regulators, we confidently identified 100 with at least four targets. Among those were 24 hub regulators that were predicted to regulate over 25% of the 118 genes measured, and 76 specific regulators each affecting the expression of 4 to 25 genes. On average ∼14 (±8 stdv) regulators activate a target gene, and 5 (±5.8) regulators repress it. Indirect effects may account for the large number of regulators we observe for each target.
Our perturbational model captured known regulatory features of the response, but also identified novel regulators. The reporter genes partition into two main clusters based on their response to perturbations (Fig. 3B, Fig. S15A): the ‘anti-viral (polyI:C) like’ program reporters (e.g.CXCL10, ISG15, IFIT1), and the ‘inflammatory (PAM) like’ program reporters (e.g. IL1b, CXCL2, IL6, IL12b), consistent with the expression data. We also found many known regulatory relations, for example the NFκB family of transcription factors (Rel, Rela, Relb, Nfkb1, Nfkb2 and Nfkbiz) regulating their known inflammatory gene targets. Our network provided evidence for the involvement of at least 68 additional regulators in the response to pathogens, of which 11 were hubs not previously associated with this system. Interestingly, 12 regulators identified (e.g. Hhex, Fus, Bat5, Pa2g4) are in linkage disequilibrium with SNPs associated with autoimmune and related diseases in genome-wide association studies (Table S8).
The core inflammatory and anti-viral programs
We next addressed how each regulator contributes to the generation of specific cell states. We first automatically defined the two major states induced by the five pathogen components using non-negative matrix factorization (NMF) (19)and the original array data (SOM). This procedure identified two major expression components (termed ‘metagenes’): one predominantly determined by genes from the ‘inflammatory-like’ program and the other by genes from the ‘antiviral like’ programs (Fig 2a). Next, we quantified the effects of each regulator's knockdown on these two states (Fig. 3B, Fig. S15A, Table S9), by classifying the nCounter expression measurements following a regulator's perturbation (19, 20).
Finally, we used a regulator ranking score (SOM) to assign 33 (8 known) genes as regulators of the inflammatory state and 33 (15 known) genes as regulators of the anti-viral state. This accurately classified the known activators of the inflammatory response (e.g. the NFκB factors Rela, Nfkbiz, Nfkb1, Fig. 3B, yellow in the inflammatory metagene) and of the antiviral response (e.g. Stat1, Stat2, Stat4, Irf8, Irf9 Fig. 3B, yellow in the viral metagene). Although all perturbation experiments were conducted only under LPS stimulation (a bacterial component), we correctly classified factors known to mediate the response to other stimuli. 34 additional regulators were associated with both responses, suggesting that a single regulator can control genes in either state depending on the differential timing of regulator activation, its level, or combinatorial regulation. Notably, for 12 of the transcription factors examined, we found an enriched cis-regulatory element in the appropriate metagene (SOM).
On the basis of the NMF scores (Table S9), we identified an inflammatory subnetwork (Fig. S15B), an anti-viral subnetwork (Figs. 4A, S15C), and several fine-tuning subnetworks that affect smaller numbers of genes from both responses (Figs. S15D, S16, SOM). The inflammatory subnetwork (Fig. S15B) consisted of three regulatory modes: dominant activators (Cebpb, Bcl3, Cited2) which induce more inflammatory targets than anti-viral ones; cross-inhibitors (Nfkbiz, Nfkb1, Atf4, Pnrc2) which induce inflammatory genes while repressing anti-viral ones, and specific activators (Runx1, Plagl2), that only target inflammatory genes. We observed that dominant activators mostly regulate effectors, whereas regulators are primarily controlled by cross-inhibitors.
Fig. 4. The core regulatory circuits controlling the inflammatory and anti-viral responses.
A. The antiviral sub-network shows regulatory relations between the core anti-viral regulators (blue nodes, top), their targets (boxes, bottom), each other, and inflammatory regulators (green node, top right). The two top regulators, Stat1 and 2, activate all anti-viral targets (dashed blue arrows). The second-tier regulators activate sub-sets of targets (dashed purple arrows). B, Examples of feed forward loop classes identified in the network, with fraction of each class. C. A core regulatory network of the inflammatory and anti-viral programs, consisting of the most distinct regulators, and their relation to ligands and receptors (top). Pointed arrows – induction; blunt arrows – repression; green ovals – inflammatory regulators; blue ovals – antiviral regulators. Example target genes are noted. D. nCounter expression profiles for the target genes (rows) upon perturbation with shRNAs against a subset of viral regulators (columns) and followed by stimulation with LPS (left) or polyI:C (right). All values are normalized by expression in cells infected with a control shRNA and under the same stimulus (shCtl).
Focusing on the network architecture, we found multiple feed-forward circuits in this response, where an upstream regulator controls a target gene both directly and indirectly through a secondary regulator (21)(e.g. Fig. 4B, and Tables S10, S11). The majority (76%, 4892 of 6444) of these feed-forward circuits were found to be coherent (21); having the same direct and indirect effect on the regulated gene. The vast majority (80%) are type I loops (22)with all-positive regulation (e.g. NFKBIZ activates E2F5 and both activate IL6). Such feed-forward circuits respond to persistent rather than transient stimulation, protecting the system from responding to spurious signals, as was shown for one circuit in LPS-stimulated macrophages (23). Our finding suggests that coherent feed-forward loops, especially class I (21), are a general design principle in this system and may physiologically impact this response.
In the anti-viral sub-network, we identified a two-tiered regulatory circuit combining feed-forward and feed-back loops (Fig. 4A, Table S11). This circuit has at the top the anti-viral regulators Stat1 and Stat2, which regulate a full complement of anti-viral reporters. The second-tier regulators Timeless, Rbl1 and Hhex are controlled by Stat1 and 2 and most likely form coherent feed-forward loops that target specific sub-sets of genes. Timeless, Rbl1 and Hhex also feed-back and promote the expression of the Stat regulators. This circuit is repressed through the cell cycle regulator and RNA binding protein Fus (24), acting as a single dominant inhibitor of 43 viral genes.
Finally, we derived a core network incorporating the regulators with the most substantial impact on each response, on the basis of the number, magnitude, and logic of targets that each regulator affects (SOM). The core network (Fig. 4C) has 24 regulators, 13 of which have previously been identified as key factors regulating the inflammatory or anti-viral responses, while 11 have not been previously implicated in either response. Of these, 19 are transcription factors, three are chromatin modifiers, and two are RNA binding proteins. The regulators apparently distinguish the two programs through cross-inhibition (Fig. 4C, gray lines) or dominant activation (Fig. 4C). The core network also explains how differential expression of secreted factors is specified, leading to the activation and migration of appropriate cell types for different pathogens (25) (Fig. S17, SOM).
Embedded within the many known regulators of the anti-viral response (Figs. 4C, S15C), we found a large set of regulators not previously associated with this response. These included several known regulators of the cell cycle and the circadian rhythm, including Rbl1, Jun, RB, E2F5, E2F8, Nmi, Fus, and Timeless, several of which were placed in our core network. This suggests that a cell cycle regulatory circuit was co-opted to function in the anti-viral response in DCs (with no observable effect on cell cycle progression, Fig S18). Since we identified these anti-viral regulatory relations in perturbation experiments using DCs stimulated with the bacterial component LPS, we silenced four regulators (TIMELESS, RBL1, JUN and NMI) following exposure to the viral component polyI:C. Each of the four regulators strongly impacted the antiviral program, more than was observed under LPS stimulation (Fig. 4D), and affected genes (e.g. Type I IFNs) whose expression is polyI:C-specific. Nmi affected a smaller set of genes, consistent with the model's prediction. These results demonstrate our ability to correctly predict function in unobserved conditions.
Although most anti-viral genes are induced following stimulation with the bacterial component LPS, a few critical ones are expressed specifically in polyI:C stimulation, or follow distinct patterns in each stimulus. In response to viral infection cells induce the production of interferon beta1 (IFNB1), a crucial mediator of the antiviral response. Because high levels of IFNB1 may be deleterious to the host if infected by specific bacteria (26), we predicted that specific mechanisms insulate IFNB1's regulation from the response to LPS. Indeed, although IFNB1 expression was induced in the first two hours of stimulation with LPS, this expression declined at subsequent time points, in contrast to its sustained induction following polyI:C treatment (Fig. 5A). Our model suggested that three regulators known to affect chromatin remodeling (24, 27, 28) are IFNB1 repressors in LPS (Fig. 5B): the Polycomb complex subunit Cbx4 (27), Fus (24), and the DNA methyltransferase Dnmt3a (28). Cbx4 appeared to confer antiviral specificity to IFNB1 induction as it is induced within the first two hours of PAM and LPS treatment but not by polyI:C (Fig. 5C), and Cbx4 knockdown caused induction of IFNB1 mRNA and protein during LPS treatment (Fig. 5D, Fig. S19A), but had no effect on the induction of the chemokine Cxcl10, a polyI:C and LPS-induced gene (Fig. S19B). Cbx4 knockdown did not affect IFNB1 during PAM activation (Fig. 5E), when the anti-viral response is not induced. Combined with evidence for chromatin changes around the Ifnb1 locus and its closest neighbor gene, Ptplad2 (Fig. S20A), which has a similar dependence on Cbx4, these data are consistent with an effect by Cbx4 on local chromatin organization (Figs. S20B, C). Cbx4 knockdown affected few genes (∼120 up-regulated and ∼120 down-regulated genome-wide, Table S12). Because most up-regulated genes show a precise temporal pattern in unperturbed cells akin to that of Cbx4– they are induced quickly and return to basal level by 2-4 hours (Fig. S21 A-F), we conclude that a chromatin modifier can act like a transcription factor controlling the precise expression of specific genes in the regulatory program.
Fig 5. The polycomb component Cbx4 selectively restricts IFNB1 production under bacterial perturbations.
A,C LPS (red), polyI:C (blue) and PAM (green)-induced expression of ifnb1 (A) and cbx4 (C) derived from data in Fig. 2A. B. by nCounter) in response to LPS in DCs perturbed by control shRNAs or shRNAs targeting each of 125 regulators (format as in Fig. 3B). D. Ifnb1 mRNA levels (by qRT-PCT) following LPS treatment in unsorted mouse bone marrow DCs perturbed with an shRNA against Cbx4 (black) or a control shRNA (grey); signals are relative to t=0. E. ifnb1 mRNA levels (by nCounter) at 6h post-LPS or PAM in bone marrow DCs perturbed with an shRNA against Cbx4 or one of three control shRNAs. F. Model for bacterial-specific repression of ifnb1 by Cbx4: both polyI:C and LPS induce ifnb1 expression early (left), but only LPS induces Cbx4, which then represses the ifnb1 locus at a later time (right, top).
Taken together, our results suggest a model of a transcriptional negative feedback loop, controlling IFNB1 expression in LPS stimulation, wherein the induced pro-inflammatory regulator and chromatin modifier Cbx4 represses transcription by modifying the chromatin in the Ifnb1 locus, generating the specificity needed to drive inflammatory versus the anti-viral response (Fig. 5F). The Type I coherent feedforward loop formed by Cbx4 and Dnmt3a (Fig. 4B) is consistent with a delayed repression of IFNB1. Since neither regulator carries a sequence-specific DNA binding domain, the factors responsible for their guidance to the Ifnb1 locus remain unknown.
Discussion
A central goal of our study was to address the mechanistic basis for pathogen-specific responses. Consistent with previous studies (13), we distinguished two key programs, a PAM (TLR2)-like inflammatory response and a polyI:C (TLR3/MDA-5)-like anti-viral response, which are together induced by LPS, a gram-negative bacterial component and a TLR4-ligand. These programs reflect both qualitative and quantitative differences between the required functional responses, and are consistent with the cross-protection between certain bacteria and virus infections (13). The broad effect of LPS allowed us to focus on a single stimulus and timepoint, but screens with other stimuli may identify additional unique regulators.
We found that these two responses are controlled by two corresponding regulatory arms, uncovering a mechanistic basis for the observed transcriptional responses. These two arms are integrated into a core network of two dozen regulators which balances specific and shared responses through dominant activation and cross-inhibition. In the inflammatory response, we found several feed-forward loops, which may ensure response to only persistent, and not sporadic, signals. In the anti-viral response, we discovered a two-tiered circuit involving feedback and feed-forward loops, implicating a module of cell cycle regulators (Jun, Rbl1, Timeless and Nmi), which we directly validated. Over 75 additional genes work to further fine-tune the regulation of gene targets. This perturbational model identifies many regulatory relations that would have been missed by non-systematic approaches.
While we have benefited from the specific features of the DC system, our work establishes an unbiased, straightforward and general framework for network reconstruction in mammalian cells (SOM). In particular, we develop several strategies to leverage shRNA for the study of gene regulation. This approach can be executed at substantial scale and reasonable cost, and is compatible with the challenge of deciphering the multiple regulatory systems that operate in mammals. It can be expanded to derive increasingly detailed models, and distinguish direct from indirect targets.
Our study will facilitate the development of new computational approaches to infer regulatory models. While many computational approaches have attempted to derive observational models, their quality has been difficult to evaluate (3). The data generated here includes both expression profiles for training a model, and a perturbational unbiased screen for testing its quality (Web portal; ftp://ftp.broadinstitute.org/pub/papers/dc_network/). When we compared the perturbational model to our observational model, we found that many candidate regulators were correctly identified in both (Fig. S5, S22). However, there were also numerous false positive relations in the observational model, attributable to the fact that both the correct regulator and many others have indistinguishable expression (Figs. S22, S23).
The high-resolution map we constructed has important biomedical implications. By identifying regulators that mediate the differential control of specific gene pairs (e.g. IL-23 vs. IL-12, Fig. S17) and entire regulatory arms (e.g. viral vs. inflammatory), it opens the way for therapeutic targeting of specific pathways to control disease or enhance vaccine efficacy. Furthermore, 12 of our regulators reside in genetic loci that were in linkage disequilibrium with SNPs associated with autoimmune and related diseases. The identified genes and their impact on DCs provide hypotheses to help explain how alleles of genes in a cascade may alter susceptibility to specific infections or immune disorders in humans.
Supplementary Material
Acknowledgments
We thank E. Lander, I. Wapinski, D. Pe'er, N. Friedman, J. Kagan, A. Luster, and V. Kuchroo for discussions and comments, L. Gaffney for assistance with artwork, S. Gupta and the Broad Genetic Analysis Platform for microarray processing, and T. Mikkelsen and the Broad Sequencing Platform for help with the ChIP-seq experiments. Supported by the Human Frontier Science Program Organization and he APF Fellowship's Claire & Emanuel G. Rosenblatt Award (IA); NIH R21 AI71060 and the NIH New Innovator Award (NH); a Career Award at the Scientific Interface from the Burroughs Wellcome Fund, the NIH Pioneer Award, and the Sloan Foundation (AR). Complete microarray data sets available at the Gene Expression Omnibus (GSEXXX).
References
- 1.Huang Q, et al. Science. 2001 Oct 26;294:870. doi: 10.1126/science.294.5543.870. [DOI] [PubMed] [Google Scholar]
- 2.Kawai T, Akira S. Int Immunol. 2009 Apr;21:317. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim HD, Shay T, O'Shea EK, Regev A. Accepted to Science. 2009 doi: 10.1126/science.1171347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramsey SA, et al. PLoS Comput Biol. 2008 Mar;4:e1000021. doi: 10.1371/journal.pcbi.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki H, et al. Nat Genet. 2009 May;41:553. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segal E, et al. Nat Genet. 2003 Jun;34:166. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 7.Capaldi AP, et al. Nat Genet. 2008 Nov;40:1300. doi: 10.1038/ng.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Z, Killion PJ, Iyer VR. Nat Genet. 2007 May;39:683. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- 9.Workman CT, et al. Science. 2006 May 19;312:1054. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erwin DH, Davidson EH. Nat Rev Genet. 2009 Feb;10:141. doi: 10.1038/nrg2499. [DOI] [PubMed] [Google Scholar]
- 11.Geiss GK, et al. Nat Biotechnol. 2008 Mar;26:317. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 12.Kagan JC, et al. Nat Immunol. 2008 Apr;9:361. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doyle S, et al. Immunity. 2002 Sep;17:251. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- 14.Kawai T, et al. Nat Immunol. 2004 Oct;5:1061. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- 15.Amit I, et al. Nat Genet. 2007 Apr;39:503. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- 16.Pe'er D, Regev A, Tanay A. Bioinformatics. 2002;18 1:S258. doi: 10.1093/bioinformatics/18.suppl_1.s258. [DOI] [PubMed] [Google Scholar]
- 17.Lee SI, et al. PLoS Genet. 2009 Jan;5:e1000358. doi: 10.1371/journal.pgen.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou H, Hastie T. Journal of the Royal Statistical Society B. 2005;67:301. [Google Scholar]
- 19.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Proc Natl Acad Sci U S A. 2004 Mar 23;101:4164. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daily JP, et al. Nature. 2007 Dec 13;450:1091. doi: 10.1038/nature06311. [DOI] [PubMed] [Google Scholar]
- 21.Mangan S, Alon U. Proc Natl Acad Sci U S A. 2003 Oct 14;100:11980. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alon U. Nat Rev Genet. 2007 Jun;8:450. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 23.Litvak V, et al. Nat Immunol. 2009 Apr;10:437. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, et al. Nature. 2008 Jul 3;454:126. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luster AD. Curr Opin Immunol. 2002 Feb;14:129. doi: 10.1016/s0952-7915(01)00308-9. [DOI] [PubMed] [Google Scholar]
- 26.Decker T, Muller M, Stockinger S. Nat Rev Immunol. 2005 Sep;5:675. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- 27.Bernstein E, et al. Mol Cell Biol. 2006 Apr;26:2560. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, et al. Biochem J. 2007 Jul 15;405:369. doi: 10.1042/BJ20061873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.