Arf3 associates with the TGN in a manner that is both temperature-sensitive and uniquely dependent on BIGs. TGN localization and release at 20°C are readily separated and depend on pairs of residues absolutely conserved and unique to Arf3 present at opposite ends of the protein. These results suggest that Arf3 plays a unique function at the TGN.
Abstract
It is widely assumed that class I and II Arfs function interchangeably throughout the Golgi complex. However, we report here that in vivo, Arf3 displays several unexpected properties. Unlike other Golgi-localized Arfs, Arf3 associates selectively with membranes of the trans-Golgi network (TGN) in a manner that is both temperature-sensitive and uniquely dependent on guanine nucleotide exchange factors of the BIGs family. For example, BIGs knockdown redistributed Arf3 but not Arf1 from Golgi membranes. Furthermore, shifting temperature to 20°C, a temperature known to block cargo in the TGN, selectively redistributed Arf3 from Golgi membranes. Arf3 redistribution occurred slowly, suggesting it resulted from a change in membrane composition. Arf3 knockdown and overexpression experiments suggest that redistribution is not responsible for the 20°C block. To investigate in more detail the mechanism for Arf3 recruitment and temperature-dependent release, we characterized several mutant forms of Arf3. This analysis demonstrated that those properties are readily separated and depend on pairs of residues present at opposite ends of the protein. Furthermore, phylogenetic analysis established that all four critical residues were absolutely conserved and unique to Arf3. These results suggest that Arf3 plays a unique function at the TGN that likely involves recruitment by a specific receptor.
INTRODUCTION
Imaging of live cells revealed that the Golgi complex is not static as initially assumed from its intricate structure (Rambourg and Clermont, 1990; Mogelsvang et al., 2004), but rather surprisingly dynamic and linked to several other organelles by active bidirectional transport routes (Lippincott-Schwartz et al., 1998; Glick and Nakano, 2009). This traffic involves the formation of cargo carriers that depends on the spatially and temporally regulated membrane recruitment of specific coat proteins (COPs) from the cytoplasm (Bonifacino and Glick, 2004). The COPI coat, first identified in situ at the periphery of the Golgi complex (Orci et al., 1986), has been implicated in both anterograde and retrograde traffic between the Golgi and vesiculotubular clusters (Schekman and Mellman, 1997; Duden, 2003). Packaging of endosome-targeted cargo at the trans-Golgi network (TGN) involves the coat protein clathrin and several adaptor proteins (APs), including the multimeric AP-1, -3, and -4, and monomeric gamma ear Golgi-localized Arf-binding proteins (GGAs; Braulke and Bonifacino, 2008; De Matteis and Luini, 2008). Formation of carriers from the TGN to the cell surface remains poorly understood but involves phosphate kinase D (Bard and Malhotra, 2006) and lipid transfer proteins that includes four-phosphate-adaptor proteins, or FAPPs (Godi et al., 2004; De Matteis and Luini, 2008).
The recruitment of COPI (Donaldson et al., 1992; Fischer et al., 2000), clathrin, and its adaptors (Robinson and Kreis, 1992; Ooi et al., 1998; Boehm et al., 2001; Puertollano et al., 2001; Boman et al., 2002) at the Golgi complex is controlled by ADP-ribosylation factors (Arfs) as well as their regulatory guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (D'Souza-Schorey and Chavrier, 2006; Nie and Randazzo, 2006). Sequence comparison of the six mammalian Arfs delineates three classes (Chavrier and Goud, 1999; Li et al., 2004): class I (Arfs 1, 2, and 3), class II (Arfs 4 and 5), and class III (Arf6). With the exception of Arf6 that functions at the plasma membrane (D'Souza-Schorey et al., 1995; Peters et al., 1995; Donaldson, 2003), Arfs localize primarily at the Golgi complex (Chun et al., 2008). Specific recruitment/activation of a multitude of effector proteins by a very limited number of Arfs may involve unique combinations of Arfs and their regulatory proteins Arf-GEFs and/or GTPase activating proteins. Only two of the six known Arf-GEF subfamilies, Golgi-specific brefeldin A resistance factor 1 (GBF1) and brefeldin A–inhibited guanine nucleotide exchange factors (BIG), localize to the Golgi complex (Cox et al., 2004; Mouratou et al., 2005). These GEFs are large multidomain enzymes (Mouratou et al., 2005; Bui et al., 2009) that can activate both class I and class II Arfs but localize to distinct subcompartments of the Golgi complex. GBF1 localizes to cis-compartments where it facilitates recruitment of the COPI coat, whereas BIGs regulate membrane association of clathrin adaptors on trans-compartments (Kawamoto et al., 2002; Shinotsuka et al., 2002a; Zhao et al., 2002; Garcia-Mata et al., 2003; Manolea et al., 2008).
The multiple Arfs present on Golgi membranes could also contribute to specificity if they localized to distinct compartments or interacted with unique effectors. Taylor et al. (1992) provided the first evidence for distinct biochemical properties between Arf1 (GGBF) and Arf3 (GGBF*), indicating that Arfs may play nonredundant roles. At the time, the authors speculated “that GGBF and GGBF* may direct assembly of coats onto different organelles such as the Golgi and the trans-Golgi network.” Unfortunately, several follow-up studies failed to support this hypothesis and suggested instead that Arf3 and Arf1 play redundant roles at the Golgi complex. For example, multiple Arf3 effectors were identified, including Arfaptin 1, Arfaptin 2 (Kanoh et al., 1997), mitotic kinesin-like protein 1 (MKLP1; Boman et al., 1999) and phospholipase D (PLD; Cockcroft et al., 1994), but none of these discriminated between Arf1 and Arf3. Furthermore, in vitro and in vivo studies established that Golgi-localized GEFs could activate equally Arf1 and Arf3 (Kawamoto et al., 2002; Shin et al., 2004; Islam et al., 2007). Lastly, Donaldson and colleagues identified a centrally located MXXE motif, present in all class I Arfs, that targets Arf1 to its receptor membrin on cis-Golgi membranes (Honda et al., 2005) and should target Arf3 similarly.
Even though the majority of data to date suggest that Arf1 and Arf3 play redundant roles and localize to similar membranes, more recent observations suggest that they may perform different functions after all. In an attempt to identify specific function for each of the Arfs, Volpicelli-Daley et al. (2005) discovered that double knockdown of Arf1+Arf4 and Arf3+Arf4 caused dramatically different effects. More recently, Chun et al. (2008) reported that Arf3, unlike Arf1 or class II Arfs, did not localize to ER-Golgi intermediate compartment structures.
The present study extends our previous observations and establishes that Arf3 localizes specifically to the trans-side of the Golgi complex where BIGs regulate its membrane association. This unique localization depends on two unique and absolutely conserved residues at the C-terminus. Interestingly, temperature shift to 20°C causes Arf3 to redistribute to cytosol and this behavior can be readily separated from localization since it depends on two conserved amino acids at the N-terminus.
MATERIALS AND METHODS
Reagents and Antibodies
Media and tissue culture reagents were purchased from Invitrogen (Carlsbad, CA). Disposable plasticware and culture six-well plates were purchased from Falcon Plastics (Oxnard, CA). Brefeldin A (BFA) and all other chemicals, unless otherwise indicated, were purchased from Sigma-Aldrich (St. Louis, MO). Polyclonal anti-green fluorescent protein (GFP; G. Eitzen, University of Alberta, Edmonton, AB, Canada) and anti-Arf3 (R. Kahn, Emory University, Atlanta, GA) were used for immunoblotting at 1:1000 and 1:250, respectively. Monoclonal antibodies used for immunofluorescence (IF) were as follows: anti-Arf clone 1D9 (Abcam, Cambridge, MA) at 1:400, anti-GGA3 clone 8 (BD Biosciences Pharmingen, Oakville, ON, Canada) at 1:200, anti-p115 (clone 7D1; Waters et al., 1992; a kind gift from D. Shields, Alberta Einstein School of Medicine) at 1:50, anti-HA clone 3F10 (Roche Diagnostics, Laval, QC, Canada) at 1:100, anti-βCOP clone M3A5 (Allan and Kreis, 1986; a kind gift from Dr. T. Kreis, University of Geneva, Geneva, Switzerland) at 1:500, and anti-γ adaptin (AP-1) clone 88 (BD Biosciences Pharmingen) at 1:600. Polyclonal antibodies used for IF were as follows: anti-GBF1 (9D2) and anti-BIG1 (9D3) as previously described (Zhao et al., 2002), anti-TGN46 (AbD Serotec, Kidlington, Oxford, United Kingdom) at 1:500, anti-GFP a kind gift from Dr. Luc Berthiaume (University of Alberta) at 1:2500; anti-GM130 (a kind gift from Dr. A. DeMatteis, Department of Cell Biology and Oncology, Consorzio Mario Negri Sud, Santa Maria Imbaro, Italy) at 1:100. Secondary antibodies used were Alexa 488/594-conjugated goat anti-rabbit and Alexa 488/594/660-conjugated goat anti-mouse antibodies (Molecular Probes, Eugene, OR) at 1:600 and 18- and 12-nm conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) at 1:10 and 1:20, respectively.
Construction and Expression of Plasmids
The construction of the plasmids encoding Arf1 and Arf3 tagged with either GFP or mCherry has been previously described (Chun et al., 2008). Additional vectors encoding untagged or hemagglutinin (HA)-tagged forms of Arf3 were constructed by inserting a PCR fragment into the XhoI and KpnI sites of the pcDNA4/TO vector (Invitrogen) modified by inversion of the multiple cloning site. The hArf3 encoding fragment was generated using pET21d-Arf3 (Berger, 1998) as template. The construction of HA-tagged Arf3 involved first insertion of annealed oligonucleotides encoding the HA epitope followed by a stop codon between the KpnI and HindIII sites of pcDNA4/TO(-). In the second step, a PCR fragment encoding Arf3 was ligated between the XhoI and KpnI sites of pcDNA4/TO(-)-HA.
Arf1_3 and Arf3_1 swap chimeras were constructed using PCR and primers designed to include an XhoI site at the 5′end and a KpnI site at the 3′ end. PCR reactions were performed using a plasmid encoding hArf3 as template. The resulting PCR products were digested with XhoI and KpnI and ligated into similarly cut pEGFP-N1 vector. Plasmids encoding HA-tagged forms of those chimeras were generated by transferring the XhoI/KpnI fragment into the pcDNA4/TO(-)-HA vector described above. Similar approaches were used to generate GFP-tagged forms of Arf3 or Arf1 bearing single or double point mutations at the N- or C-termini. All mutant forms of Arf3 folded properly because they associated with the Golgi complex; more importantly, BFA readily displaced mutants from membranes indicating that Golgi association depended on activation by nucleotide exchange (data not shown).
For Arf-GEF overexpression studies (see Figure 2), HeLa cells grown on glass coverslips to ∼50% density were transfected with 1 μg of purified pCEP4 vector plasmid encoding either GBF1 (Claude et al., 1999) or BIG1 (Mansour et al., 1999), using TransiIT LT1 (Mirus Bio, Madison, WI) according to manufacturer's instructions. Twenty-four hours after transfection, cells were briefly treated with 5 μg/ml BFA, fixed, and processed for IF using the indicated antibodies. For exogenous expression of Golgi markers, 1 μg of purified plasmids encoding GalT-GFP (gift of Jennifer Lippincott-Schwartz, Cell Biology and Metabolism Branch, NIH, Bethesda, MD) or FAPP-PH-YFP (gift of Sergio Grinstein, Hospital for Sick Children, Toronto, ON, Canada), were transfected in HeLa cells for 24 h before processing as indicated.
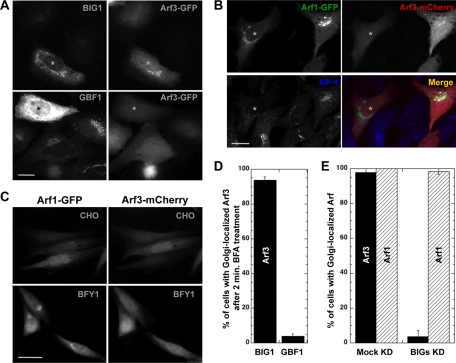
Figure 2.
Arf3 recruitment involves BIGs but not GBF1. (A) HeLa cells were cotransfected with plasmids encoding Arf3-GFP and either BIG1 or GBF1. After 24 h, cells were treated with 5 μg/ml BFA for 2 min and then fixed and stained for BIG1 or GBF1. Epifluorescence images are shown. Small asterisks label the nucleus of transfected cells. Bar, 20 μm. (B) HeLa cells were transfected with a pool of siRNA duplexes targeting BIG1 and BIG2 (BIGs KD) for 72 h. Forty-eight hours before fixation, cells were cotransfected with Arf1-GFP and Arf3-mCherry plasmids. Fixed cells were stained for AP-1, and images were acquired using a confocal microscope. Small asterisks label the nucleus in a cell with efficient BIGs KD. Bar, 20 μm. (C) CHO and BFY1 cells were cotransfected with plasmids encoding Arf1-GFP and Arf3-mCherry. After 24 h, cells were treated with 5 μg/ml BFA for 2 min and then fixed. Representative epifluorescence images selected from at least two separate experiments are shown. Bar, 20 μm. (D) Quantitative analysis of cells overexpressing BIG1/GBF1 showing the percentage of cells with Arf3 localized to the Golgi complex after BFA treatment. Error bars, mean ± SD (n ≥ 30 cells from at least four separate experiments as in A). (E) Quantitative analysis of cells with Mock KD or BIGs KD showing the percentage of cells with Arf3 or Arf1 localized to the Golgi complex. Error bars, mean ± SD (n ≥ 30 cells from at least four separate experiments as in B).
Small Interfering RNA Methods
Pools and individual small interfering RNAs (siRNAs) targeting different regions of human (h)BIG1 (MU-012207), hBIG2 (MQ-012208), and hArf3 (LQ-011581) were purchased from Dharmacon (Boulder, CO). We followed the Oligofectamine (Invitrogen) transfection protocol for HeLa cells, as described (Harborth et al., 2001). Different combinations of targeting duplexes, time points, and siRNA concentrations were assessed to optimize conditions for most effective knockdown. For BIGs knockdown, we used a pool of siRNAs targeting sequences 2 and 3 for BIG1 (75 nM each) and 1–4 for BIG2 (50 nM each). For Arf3 we used siRNAs targeting sequences either individually or as pools, at a final concentration of 200 nM. As previously reported, lower siRNA concentrations (50 nM BIG1 and 25 nM for BIG2) were sufficient for effective BIGs knockdown (Manolea et al., 2008). As control, cells were exposed to matching concentrations (200–300 nM) of a nonspecific GL2 luciferase siRNA designed as described previously (Elbashir et al., 2002).
Temperature-Shift Experiments
Most experiments were performed with cells grown on glass coverslips either in six- or 12-well plates essentially as described (Manolea et al., 2008). For the 20°C temperature shift experiments, cells were either fixed directly from the incubator for the 37°C condition or shifted to CO2-independent media (Invitrogen) prewarmed at 20°C in a water bath for the indicated time. Cells were fixed with 3% PFA at 20°C for the first 5 min and then for another 15 min at 37°C. For experiments measuring protein traffic (see Figure 5), HeLa cells were transfected with 1 μg of plasmid encoding VSVGtsO45-GFP (kind gift from Dr. John F. Presley, McGill University, Montreal, QC, Canada), shifted to nonpermissive, and then permissive temperature as described previously (Manolea et al., 2008).
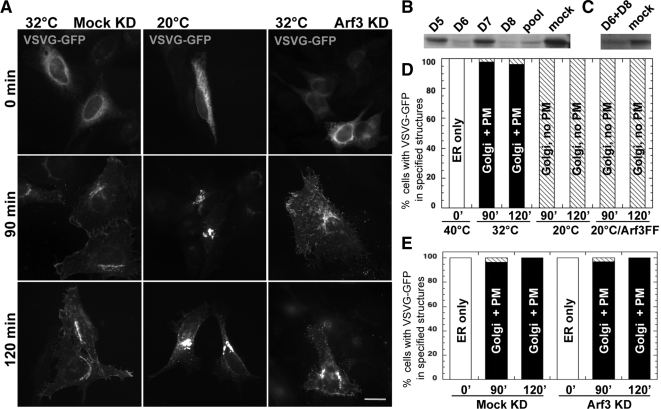
Figure 5.
Shift to 20°C, but not Arf3 KD, blocks VSVG traffic at the Golgi complex. (A) HeLa cells were transfected at t = 0 with either irrelevant siRNA (Mock KD, left), no RNA (middle), or a pool of two validated Arf3 siRNA duplexes (Arf3 KD, right); some cells were cotransfected with plasmid encoding Arf3FF (not shown). Fifty hours after transfection, cells were transfected again with a plasmid encoding VSVGts045-GFP. Temperature was shifted initially at 40°C for 4 h and then to either 32 or 20°C for the length of time specified. Epifluorescence images are shown. Bar, 5 μm. (B) HeLa cells were cotransfected with the indicated Arf3 siRNAs targeting Arf3 and 1 μg of plasmid encoding for Arf3-GFP for 24 h. Immunoblots of equal amounts of detergents lysates were probed with an anti-GFP antibody. Blots shown are representative of three separate experiments. (C) HeLa cells were transfected with the indicated siRNAs for 72 h. Immunoblots of equal amounts of detergents lysates were probed with an anti-Arf3 antibody. Blots shown are representative of two separate experiments. (D) Quantitative analysis showing percent of cells with VSVG in specified structures after release at the two different temperatures and time points. (E) Quantitative analysis showing percentage of cells with VSVG in specified structures after release at indicated time points in either Mock KD or Arf3 KD cells.
Image Quantification and Analysis
Quantification of the extent of fluorescence signal overlap between various pairs of markers was performed using Metamorph software (v. 6.1, PerkinElmer Canada, Woodbridge, ON, Canada) as described previously (Manolea et al., 2008). For both markers, the areas corresponding to the Golgi complex were identified using an inclusive threshold set to contain total membrane signal. To eliminate background noise signal outside of the Golgi area, a median filter of 2 pixels was applied when necessary. The degree of colocalization was reported as a percentage of the integrated intensity of the first marker in pixels overlapping with the second marker.
To examine the protective effect of GEF overexpression on Arf3 recruitment (see Figure 2D), we scored for the presence or absence of Arf3-GFP at the Golgi after 2-min treatment with 5 μg/ml BFA. Results were expressed as the percentage of cells with Arf3 still localized to the Golgi complex in cells overexpressing either BIG1 or GBF1. To quantitate the impact of BIGs knockdown on Arf3 or Arf1 membrane recruitment (see Figure 2E), we similarly scored for the presence or absence of Arf3 or Arf1 at the Golgi complex in cells displaying BIGs knockdown or in cells treated with mock siRNAs. Efficient BIGs knockdown was measured by redistribution of AP-1 juxtanuclear staining. Results were expressed as the percentage of cells with Arf3 or Arf1 localized to the Golgi complex.
To measure the impact of temperature on the cellular distribution of WT and mutant Arf-GFP (see Figures 3 and 5), we quantified GFP signal intensity at the Golgi complex by first tracing a region of interest tightly around juxtanuclear GFP–positive structures and then obtaining the integrated signal intensity within this area. The signal intensity for the whole cell was quantified using a similar approach. Both values were corrected for background intensity obtained from a circle drawn the size of a nucleus in a region where cells were absent. Results were expressed as the fraction of total Arf3 signal present in the juxtanuclear region over total cell signal. At least 20 cells were analyzed for each time point for each experiment.
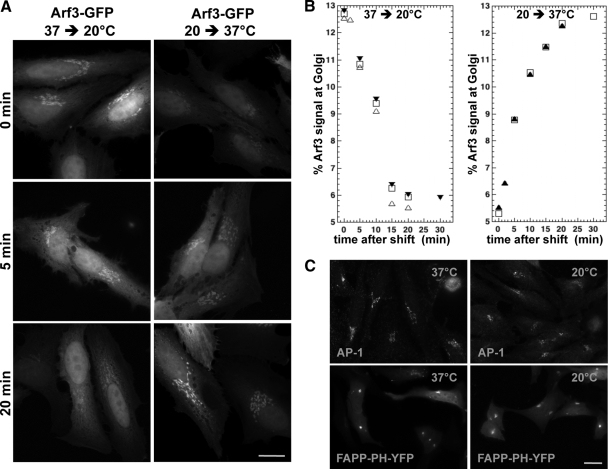
Figure 3.
Arf3 redistributes slowly between Golgi membranes and cytosol upon temperature shift to either 20 or 37°C. (A) HeLa cells were transfected with a plasmid encoding Arf3-GFP for 24 h. Cells were then shifted from 37 to 20°C (left) or kept at 20°C for 1 h and then shifted to 37°C (right). Cells were fixed at the indicated time after shift. Epifluorescence images are shown. Bar, 20 μm. (B) Quantitative analysis of Arf3 signal at the Golgi complex expressed as percent of total cell signal for temperature-shift experiments performed as in A (n ≥ 20 cells/time point from at least two separate experiments). (C) HeLa cells were transfected with a plasmid encoding FAPP-PH-YFP for 24 h. Cells were either fixed directly from 37°C (left) or shifted from 37 to 20°C (right) for either 30 min (FAPP-PH-YFP) or 2 h (AP-1) and then fixed. Cells were stained for AP-1 or not, and images were acquired using identical settings for the 37 and 20°C samples. Representative epifluorescence images selected from at least two separate experiments are shown. Bar, 20 μm.
To quantitate the impact of Arf3 knockdown and temperature shift to 20°C on VSVG (vesicular stomatitis virus glycoprotein) trafficking (see Figure 5), we scored for the presence of VSVG at 1) ER only, 2) Golgi but not plasma membrane, or 3) Golgi plus plasma membrane. Analysis was performed at each time point after release from 40°C block for both 32°C (control), 20°C-shifted cells, and cells treated with Arf3 siRNA. A minimum of 25 cells were analyzed for each condition. The fraction of cells with the indicated pattern was expressed as percentage and is shown as a function of time after shift down from 40°C.
Immunoelectron Microscopy
HeLa cells transfected with low levels of Arf3-GFP were prepared for immunoelectron microscopy. The cells were pelleted and fixed in a mixture of 2% glutaraldehyde and 2% paraformaldehyde in Dulbecco-PBS (D-PBS) for 20 min at 37°C. The fixed cells were rinsed in D-PBS and then dehydrated with alcohol series (30, 50, 70, and 80% ethyl-alcohol) and infiltrated with LR White (London Resin, Berkshire, United Kingdom). The infiltrated cells were embedded into gelatin capsules and polymerized under UV for 24 h at 4°C. Ultrathin sections of 60 nm were cut and loaded on a 300-mesh nickel grid without coating. A two-face double-immunolabeling technique with anti-GFP IgG and anti-GM130 IgG was performed. Both sides of the ultrathin sections on the grid were initially blocked with 5% bovine serum albumin in D-PBS for 10 min. The shiny side of the nickel grid was incubated with the first primary antibody, anti-GM130 IgG (1:100), followed by the dull side of the grid with the second primary antibody, anti-GFP IgG (1:2500), each for 2 h. The secondary antibodies, 18 nm-colloidal gold–conjugated donkey anti-rabbit IgG (1:10) and 12-nm colloidal gold–conjugated donkey anti-rabbit IgG (1:20) were also incubated sequentially for 1.5 h each. All antibodies were diluted with bovine serum albumin (1% final volume) in D-PBS, and the staining was performed at room temperature. After double-immunolabeling ultrathin sections were contrasted with 2% aqueous uranyl acetate for 15 min. The grids were examined in a Philips 410 (Mahwah, NJ) transmission electron microscope, at 80 kV equipped with a charge-coupled device camera (MegaView III, Soft Imaging System, Olympus, Melville, NY).
SDS-PAGE and Immunoblotting
Protein samples (75 μg) were analyzed by electrophoresis on 15% Tris-glycine SDS polyacrylamide gels calibrated with prestained molecular weight standards (Bio-Rad Laboratories, Hercules, CA). After separation of samples by SDS-PAGE, protein analysis by immunoblotting was carried out essentially as previously described (Harlow and Lane, 1988). After incubation with the indicated primary antibodies, the membranes were incubated with HRP-conjugated secondary antibodies (Bio-Rad Laboratories) that were detected by enhanced chemiluminescence using the ECL-plus system (GE Healthcare, Mississauga, ON, Canada) following the manufacturer's instructions.
Sequence Retrieval and Phylogenetic Analysis
Fully sequenced genomes from across the taxonomic diversity of metazoa were individually queried with Homo sapiens Arf sequences using a protein–protein Basic Local Alignment Search Tool (BLASTp). All searches were performed at the National Center for Biotechnology Information Web site (see “protein blast” at http://blast.ncbi.nlm.nih.gov/Blast.cgi/). Candidate Arf sequences with accession numbers are provided in Supplementary Table 1. The datasets were then constructed to examine the evolutionary history and distribution of Arf3. In all cases sequences were aligned using the program MUSCLE (v. 3.6; http://www.drive5.com/muscle/; Edgar, 2004) and manually adjusted such that only regions of unambiguous homology were retained for phylogenetic analysis. All alignments are available upon request. An initial set, composed of 82 taxa and 176 position contained all Arfs from the genomes examined. After elimination of class III Arfs and taxa whose sequences were highly divergent based on their representing long branches in the analysis, a second set was created containing 51 taxa and 106 positions. The final dataset focused on class I Arfs only and contained 28 taxa and 182 positions.
Datasets were analyzed by ProtTest (v. 2.4; http://darwin.uvigo.es/software/prottest.html; Abascal et al., 2005) in order to assess the optimal model of sequence evolution incorporating amino acid transition matrices, invariable sites and correction for rate-among-sites if relevant. Subsequently each dataset was subjected to three methods of phylogenetic analysis. For determination of optimal tree topology and posterior probability values MrBayes (v. 3.2.1; http://mrbayes.csit.fsu.edu/; Ronquist and Huelsenbeck, 2003) was used. Analyses were run for 2 × 106 generations and burnin values were assessed by removing trees before a graphically determined plateau. All analyses were also confirmed as having converged by having a Splits frequency below 0.1. One hundred pseudoreplicates were also analyzed by the Maximum likelihood programs PhyML (v. 2.4.4; http://www.atgc-montpellier.fr/phyml/; Guindon and Gascuel, 2003) and RAxML-VI-HPC (v. 2.2.3; http://icwww.epfl.ch/∼stamatak/index-Dateien/Page443.htm; Stamatakis, 2006).
RESULTS
Arf3 Localizes Specifically to the trans-Compartments of the Golgi Complex
A recent live cell imaging study with fluorescently tagged Arfs suggested that GFP-GBF1 and Arf3-mCherry may localize to distinct Golgi compartments (J. Chun, unpublished data). To confirm and extend this unexpected observation, we examined the relative distribution of Arf3 and several cis- and trans-Golgi markers within the Golgi complex. Most of those experiments took advantage of tagged forms of Arf3 expressed at low to moderate levels since none of the available antibodies selectively detect endogenous Arf3 in fixed cells. Some experiments used a pan-specific Arf antibody to detect overexpressed untagged Arf3.
Double labeling experiments in normal rat kidney (NRK) cells confirmed that Arf3 localizes preferentially to a compartment containing trans-Golgi markers that is clearly separate from the cis-Golgi (Figure 1A). Quantifying the extent of overlap with various markers revealed that membrane-bound Arf3 colocalizes with the trans-Golgi markers BIG1 (∼80%) and galactosidase T (GalT)-GFP (∼84%), whereas it overlaps to a much smaller extent with the cis-Golgi markers GBF1 (∼37%) and p115 (∼30%; Figure 1B). Similar results were obtained in HeLa cells (Supplementary Figure 1). Importantly, Arf3 tagged with either GFP or the smaller HA epitope localized like untagged Arf3 to trans-compartments labeled with GalT-GFP (Figure 1, A and B). Gold immunolabeling of cells expressing Arf3-GFP confirmed that Arf3 localized to cisternae opposite of membranes containing the well-characterized (Marra et al., 2007) cis-Golgi marker GM 130 (Figure 1C). In some images Arf3-GFP localized to a tubular network (Figure 1C, inset). We conclude that Arf3 localizes specifically to the trans-side of the Golgi complex and that usage of either of the tags does not interfere with the specific localization of the protein.
Figure 1.
Arf3 localizes to a compartment distinct from the cis-Golgi, in a tag-independent manner. (A) NRK cells were transfected with plasmids encoding Arf3-GFP, Arf3-mCherry, Arf3-HA, untagged Arf3, and/or GalT-GFP, as indicated. Fixed cells were stained for the specified markers, and images were acquired using a confocal microscope. Bar, 5 μm. (B) Quantitative analysis of signal overlap between Arf3 and the specified markers. Error bars, mean ± SD (n ≥ 16 cells from two separate experiments). (C) HeLa cells transfected with Arf3-GFP were prepared and examined by immuno-EM as described in Materials and Methods. Eighteen- and 12-nm gold particles correspond to GM130 and Arf3-GFP, respectively. Bar, 200 nm.
BIGs Are Critical for Arf3 Recruitment to the trans-Side of the Golgi Complex
The specific localization for Arf3 described above and the fact that the only Arf-GEFs enriched at the trans-side of the Golgi complex are BIG1 and BIG2 (Zhao et al., 2002) prompted us examine a likely functional link between BIGs and Arf3. We tested this hypothesis by a series of three complementary experiments. We first examined if overexpression of BIG1 prevented dispersal of Arf3-GFP after short treatment with the drug BFA. BIG1 overexpression should abrogate the effect of BFA on Arf3-GFP in a manner similar to the selective effects of GBF1 overexpression on COPI (Claude et al., 1999; Kawamoto et al., 2002) or that of BIGs on clathrin adaptors (Shinotsuka et al., 2002b; Manolea et al., 2008). As predicted, Arf3-GFP fully dispersed in control cells shortly after BFA addition, but remained Golgi-localized in cells overexpressing even moderate levels of BIG1 (Figure 2A). In contrast, overexpression of GBF1 even to very high levels did not prevent BFA-induced redistribution of Arf3-GFP (Figure 2A). Quantitative analysis of cells over expressing either BIG1 or GBF1 confirmed that Arf3-GFP still localized to juxtanuclear membranes in 94 ± 2% of BIG1-overexpressing cells but in only 4 ± 1.3% of GBF1-overexpressing cells in the presence of BFA (Figure 2D).
To extend those observations and establish that BIGs are required for Arf3 activation and recruitment to trans-Golgi compartments, we performed complementary siRNA knockdown experiments. We previously established that knockdown of BIGs disperses the TGN but affects neither the GBF1/COPI system nor the maintenance of a polarized Golgi stack (Manolea et al., 2008). Because Arf1 localizes preferentially toward the cis-Golgi (Honda et al., 2005), it should depend primarily on GBF1, not BIGs for its recruitment. We therefore predicted the cis- and trans-localized Arf1 and Arf3 would be differentially affected by knockdown of BIGs. To test this possibility, we cotransfected HeLa cells with plasmids encoding Arf1-GFP and Arf3-mCherry and examined the impact of BIGs knockdown on their distribution. As predicted, effective BIGs knockdown, as measured by dispersal of juxtanuclear AP-1, caused redistribution of Arf3 but not Arf1 (Figure 2B). Quantitative analysis showed Arf3 associated with the Golgi complex in only 4 ± 4% of cells with AP-1 redistributed, whereas Arf1 was still membrane recruited in 98 ± 3% of BIGs knockdown cells (Figure 2E).
Arf3 Distribution Remains Sensitive to BFA in CHO Mutant Cells with BFA-resistant GBF1/COPI System
The unexpected dependence of Arf3 membrane recruitment on BIGs prompted us to test the functional link between the BIGs and Arf3 using one additional approach. This time we took advantage of a CHO-derived mutant cell line, BFY1 that acquired a Golgi-specific resistance to BFA so the Golgi stack remains intact under conditions where BFA disperses the TGN and endosomes (Yan et al., 1994). Current evidence suggests that BFY1 cells acquired a BFA-resistant GBF1/COPI system but retained a BIGs/clathrin system sensitive to BFA. This gave us the opportunity to test if Arf3 remained sensitive to short treatment with BFA in the BFY1 cell line, confirming that Arf3 functions with BIGs at the TGN.
We first verified the predicted BFA resistance and sensitivity of the GBF1/COPI and BIGs/clathrin systems, respectively. As expected, whereas all tested markers were sensitive to BFA in the parental CHO cell line (Supplementary Figure 2, top), the GBF1/COPI system acquired resistance to short BFA treatment while BIGs represented by AP-1 remained sensitive in BFY1 cells (Supplementary Figure 2, bottom). More importantly, Arf1 and Arf3 displayed the predicted differential sensitivity to BFA in BFY1 cells. Indeed, COPI or Arf1 remained membrane associated in the same BFY1 cells that show redistributed Arf3 or AP-1, respectively (Supplementary Figure 2, bottom).
To unambiguously establish the difference between Arf3 and Arf1, we compared directly the response of the two Arfs in BFY1 cells cotransfected with plasmids encoding Arf3-mCherry and Arf1-GFP. As expected, Arf3-mCherry relocated to the cytoplasm after a short BFA treatment in all cells examined, whereas Arf1-GFP clearly retained its juxtanuclear localization (Figure 2C). Altogether, the three experimental approaches establish in vivo a clear functional link between the TGN-localized BIGs and the recruitment of Arf3.
Temperature Shift to and from 20°C Slowly Redistributes Arf3 between Golgi Membranes and Cytosol
It was previously demonstrated that shifting temperature to 20°C blocks cargo protein progression at trans-Golgi compartments and likely impacts TGN sorting functions (Matlin and Simons, 1983; Saraste et al., 1986; Griffiths et al., 1989). The observation that Arf3 localizes selectively to trans-Golgi compartments prompted us to examine the impact of temperature shifts on Arf3. As shown in Figure 3A (left), lowering the temperature to 20°C had a major impact on Arf3 membrane recruitment. However, redistribution of Arf3 from Golgi membranes upon temperature shift was not immediate, but proceeded with a t1/2 of ∼10 min (Figure 3B). This striking effect of temperature is fully reversible because Arf3 reassociated with Golgi membranes when the temperature was shifted back to 37°C (Figure 3A, right). As observed with downshift experiments, recruitment was delayed, with a t1/2 of ∼7.5 min (Figure 3B).
This delay in Arf3 redistribution between Golgi membranes and cytosol upon temperature shift suggests enzymatic or lipid membrane remodeling processes as key regulators of Arf3 membrane recruitment. Possible mechanisms are innumerable and potentially intractable. However, we considered three worth exploring: loss of BIGs from the TGN, loss of Golgi stack polarization, and changes in PtdIns(4)P levels. These experiments established that the reduction in Arf3 recruitment at 20°C did not result from loss of BIGs or disappearance of the TGN because the distribution of Arf1, TGN46, BIG1, or AP-1 remained unaffected (Figure 3C and Supplementary Figure 3). Furthermore, trans-Golgi markers maintained their characteristic separation from cis-compartments even after shifting temperature to 20°C (F. Manolea and D. Chen, unpublished data). To monitor the levels of phosphatidylinositol (4)-phosphate [PtdIns(4)P], we took advantage of a chimera containing YFP and two copies of the PH domain of FAPP2 (FAPP-PH-YFP) (Levine and Munro, 2002; Godi et al., 2004). To our surprise, temperature shift did not change signal intensity or pattern for FAPP-PH-YFP in the large majority of cells analyzed (Figure 3C). These experiments suggest that changes in PtdIns(4)P levels are likely not responsible for redistribution of Arf3.
Temperature Sensitivity for Membrane Recruitment of Arf3 and Arf1 Is Encrypted within Their N-terminal Helices
The sequences of human Arf1 and Arf3 differ in only two short regions at the N- and C-termini (Figure 4A). To identify which region is important for temperature sensitivity, we generated Arf3/Arf1 chimeras in which the variant regions (marked by a line in Figure 4A) were swapped. Analysis of these chimeras revealed that Arf1 constructs containing the N-terminal region of Arf3 (black region) redistributed as wild type (WT) Arf3 upon shift to 20°C (Supplementary Figure 4A). Conversely, Arf3 chimeras containing the N-terminal region of Arf1 (hatched region) appear unaffected by the temperature change, similarly to WT Arf1 (Supplementary Figure 5A). These results indicated that temperature sensitivity lies in the sequence of the N-terminal helix. Interestingly, this analysis also revealed that chimeras with the Arf1 N-terminus associated to a greater extent with the Golgi complex than those bearing the Arf3 N-terminus.
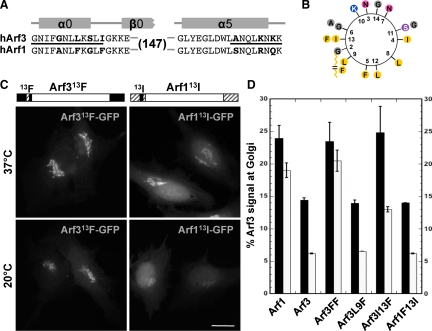
Figure 4.
The residue at position 13 in the N-terminal helix dictates the 20°C temperature sensitivity for membrane recruitment of Arf3 and Arf1. (A) Sequence alignment of N- and C-termini of Arf3 and Arf1 showing swapped regions (black line) and unique residues (bold). (B) Helical wheel representation of the N-terminal α-helix of Arf3 was obtained using DNA Strider 1.4f5. Variant amino acids present in Arf1 are shown on the outside at their respective positions. The N-terminal bound myristate is represented as a broken chain. The amino acid color code: yellow, hydrophobic; purple, serine; blue, basic; pink, asparagine; gray, alanine and glycine. (C) HeLa cells were transfected with either Arf313F-GFP or Arf113I-GFP constructs as indicated. After 24 h, cells were either kept at 37°C or shifted to 20°C for 30 min and then fixed. Epifluorescence images are shown. Schematic representation of each construct shown above each set of panels. Bar, 20 μm. (D) Quantitative analysis of Arf3-mutant signal at the Golgi complex expressed as percent of total cell signal for temperature-shift experiments performed as in C (n ≥ 6 cells/time point from at least two separate experiments).
Aromatic residues in the amphipathic N-terminal helix of Arf1 have been implicated in membrane association (Antonny et al., 1997), and the two variant F residues located on the hydrophobic side of the helix (Figure 4B) were therefore selected for further analysis. We first swapped both F residues from Arf1 (F9 and F13) with the corresponding residues from Arf3 (L9 and I13) resulting in two chimeras: Arf3FF (L9F, I13F) and Arf1LI (F9L, F13I). Imaging of transfectants expressing the double mutants suggested that these two residues are indeed critical to temperature sensitivity because Arf3FF-GFP behaved as WT Arf1 and Arf1LI-GFP behaved as WT Arf3 (Supplementary Figure 4B). To determine if both residues were required, we constructed and analyzed the recruitment of single mutants before and after temperature shift (Figure 4C). Those experiments revealed that changes at position 13 had greatest impact on Golgi recruitment.
To better assess the relative contributions to Golgi complex recruitment of residues at positions 9 and 13, we quantitated the fraction of GFP signal associated with juxta-nuclear Golgi structures for both WT as well as single and double Arf mutants (Figure 4D). Quantitative analysis first revealed that a greater fraction of WT Arf1 associated with the Golgi relative to WT Arf3. Second, the majority of Arf1 remained membrane bound at 20°C, whereas temperature shift reduced Arf3 levels dramatically by ∼60%. As suggested by Antonny et al. (1997), the two F residues at positions 9 and 13 had a dramatic impact on membrane association and temperature sensitivity because Arf3FF displayed properties identical to WT Arf1. Analysis of single mutants revealed that introduction of a F residue at position 9 in Arf3 had no impact on membrane association or temperature sensitivity. In contrast, this change at position 13 dramatically increased membrane association. Analysis of the complementary Arf1F13I mutant confirmed the importance of this position because substitution of I at position 13 dramatically reduced the extent of Arf1 binding at 37°C, and this interaction now displayed temperature sensitivity similar to that of WT Arf3. These results suggest that the nature of hydrophobic residues at both positions 9 and 13 not only affects the extent of membrane association of Arf3 and Arf1 N-terminal helices, but may also allow sensing of changes in bilayer properties that occur upon shift to 20°C.
Arf3 Knockdown Does Not Disperse the TGN and Does Not Block VSVG Traffic at the Golgi Complex
Protein traffic through the secretory pathway is sensitive to shifts in temperature with two well-characterized blocks at 15 and 20°C. For example, electron microscopy (EM) studies revealed that a shift to 20°C blocks traffic of VSVG to the plasma membrane and causes its accumulation at the TGN (Griffiths et al., 1989). The observation that Arf3 localizes to the trans-elements of the Golgi complex from which it is dramatically displaced after the downshift to 20°C (Figures 1, 3, and 4) suggested a potential connection between Arf3 and the temperature-dependent block of VSVG at TGN.
To test this hypothesis, we first analyzed cells treated with a pool of two validated siRNAs that effectively block both exogenous Arf3-GFP expression from cotransfected plasmids (Figure 5B), as well as endogenous Arf3 (Figure 5C). We first confirmed that an effective 20°C block could be observed using epifluorescence. On shift to the permissive temperature, a thermosensitive form of VSVG reaches the plasma membrane in control cells in <90 min (Figure 5A, left panels). A shift to 20°C effectively prevented VSVG appearance at the plasma membrane and caused its arrest at the Golgi complex (Figure 5A, middle panels) in the vast majority of cells examined (Figure 5D). Interestingly, VSVG was not blocked at the Golgi complex, but reached the plasma membrane in all cells transfected with VSVG-GFP plasmid and Arf3 siRNAs (Figure 5, A and E). To further rule out a role for Arf3 dissociation in the 20°C block, we performed a complementary experiment using Arf3FF, a mutant that recruits onto TGN membranes better that Arf3 and is temperature insensitive (Figure 4D). As summarized in Figure 5D, overexpression of Arf3FF did not abrogate the 20°C block because VSVG did not reach the plasma membrane in all cells examined. Similar results were obtained after overexpression of the temperature-insensitive chimera Arf1_3 (data not shown). Parallel experiments established that treatment with Arf3 siRNAs had no impact on the TGN as judged by staining for TGN46 and BIG1 or the clathrin adaptors AP-1 and GGA3 (Supplementary Figure 5). These observations are consistent with previously published data, suggesting that knockdown of BIGs did not prevent VSVG trafficking (Manolea et al., 2008) and that knockdown of Arf3 by itself did not significantly impact TGN architecture, clathrin adaptors recruitment, or VSVG trafficking (Volpicelli-Daley et al., 2005). Note that even though Arf3 may not be required for VSVG exit from the Golgi complex, its redistribution at 20°C may be induced by the same mechanism that ultimately blocks VSVG in the TGN.
The C-Terminal Helix Targets Arf3 to trans-Golgi Compartments
Arf3 and Arf1 not only behave differently in response to temperature-shift but they also differ in localization within the Golgi complex; Arf3 concentrates on trans-elements of the Golgi complex (Figure 1 and Supplementary Figure 1), whereas Arf1 appears preferentially recruited on cis-elements (Honda et al., 2005). The availability of Arf3/Arf1 chimeras allowed us to determine which region directs the specific localization of Arf3. We initially expected that the same N-terminal region important for membrane binding and temperature sensitivity might also target Arf3 to the TGN. To our surprise, the C-terminal rather than the N-terminal helix in Arf3 appeared to dictate concentration of chimeras on trans-Golgi compartments (Figure 6A, top panels). Quantitative analysis of signal overlap between the Arf3/Arf1 swap chimeras and GBF1, p115, and BIG1 confirmed accumulation of Arf3_1-GFP and Arf1_3-GFP on cis- and trans-elements of the Golgi complex, respectively (Figure 6B). These results identify the C-terminus as critical for targeting Arf3 to the trans-side of the Golgi complex.
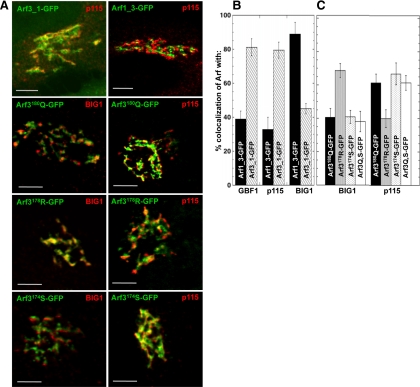
Figure 6.
Two residues in the C-terminal helix are critical for the specific TGN localization of Arf3. (A) NRK cells were transfected with the indicated Arf3_1-GFP, Arf1_3-GFP, Arf3174S-GFP, Arf3178R-GFP or Arf3180Q-GFP constructs. After 24 h, cells were fixed and stained for p115 or BIG1. Images were acquired using a confocal microscope. Bar, 5 μm. (B and C) Quantitative analysis of experiments similar to A showing signal overlap between the Arf3/Arf1 swap chimeras or single mutants and the specified markers. Error bars, mean ± SD (n ≥ 10 cells from two separate experiments).
To examine the role of specific residues, we constructed single mutants at the C-terminus in which Arf3 residues A174, K178, and K180Q were changed one by one to residues present in Arf1. NRK cells transfected with these constructs were then examined for a change from an Arf3-like trans-Golgi complex localization to a more Arf1-like distribution on the cis-side of the Golgi stack. Analysis of NRK cells expressing either Arf3A174S-GFP or Arf3K180Q-GFP revealed localization patterns similar to the cis-Golgi marker p115 and different from the trans-Golgi marker BIG1 (Figure 6A). Quantifying the extent of colocalization confirmed that membrane-bound Arf3A174S-GFP and Arf3K180Q-GFP indeed colocalize with p115 (∼70 and 65%, respectively) and overlap to a much smaller extent with BIG1 (∼40 and 35%, respectively; Figure 6C). A construct bearing both mutations (Arf3Q,S-GFP) localized similarly to cis-compartments (Figure 6C). Similar results were obtained in HeLa cells (Supplementary Figure 6). In sharp contrast, the conservative mutation K178R had minimal impact on accumulation of Arf3 on trans-compartments. These results suggest that both residues A174 and K180 participate in targeting Arf3 to the TGN.
Residues Identified as Critical for Temperature Sensitivity and TGN Recruitment Are Conserved and Unique to Arf3
The identification of specific residues at the N- and C-termini of human Arf3 critical to its membrane association and localization prompted us to examine their uniqueness within class I Arfs. To assess the evolutionary history of Arf3 and to obtain an appropriate sample of Arf homologues for sequence comparison, an in silico analysis was performed. Homology searching, using BLASTp against various metazoan genomes allowed the retrieval of Arf homologues of all three classes. Preliminary results using Bayesian and two methods of maximum-likelihood analysis readily distinguished class III from class I/II Arf homologues with Posterior probability and PhyML and RAxML bootstrap values of 1.00, 86, and 100, respectively (data not shown). Further analysis clearly distinguished the class I and class II homologues (Supplementary Figure 7), demonstrating that this gene duplication occurred earlier than previously thought (Li et al., 2004), after the separation of fungi and metazoa but before the evolution of multicellular animals.
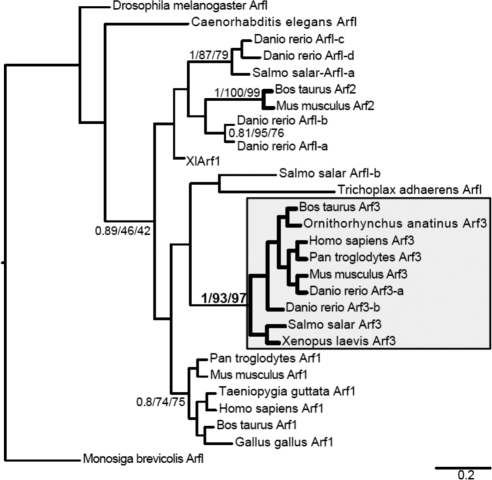
The final analysis (Figure 7), limited to class I homologues, robustly grouped Arf3 homologues from all vertebrates sampled (from fish to humans); it also allowed the identification of Arf1 and Arf2 homologues but with only moderate support and taxonomic range. T-his strongly suggests that single celled-animal ancestors, as well as invertebrates possess a single class I Arf homologue, but that Arf3 had already arisen before the emergence of vertebrates. Importantly, these analyses allowed us to clearly identify homologues of class I Arfs as well as specific homologues of Arf1, 2, and 3 for sequence comparison (Figure 8). This comparison revealed that residues identified as critical for temperature sensitivity and TGN recruitment are absolutely conserved and unique to Arf3. For example, L and I residues are present at positions 9 and 13 in all Arf3 sequences examined, whereas all other class I sequences displayed F residues at those positions. This is the case even for the single class I Arf expressed in invertebrates, in simple organisms such as Monosiga brevicolis, Nematostella vectensis, and Trichoplax adhaerens, or in the class I/II Arfs present in fungi. Similarly, A and K residues at positions 174 and 180 are uniquely present in Arf3 sequences.
Figure 7.
Arf3 evolved at least before the emergence of vertebrates. Phylogenetic analysis of class I Arf homologues rooted by the Monosiga brevicolis ArfI sequence. This tree shows the best Bayesian topology and support values for nodes with greater support than 0.80 posterior probability. Node values are given in the order of posterior probability values/PhyML bootstraps and RAxML bootstraps. Classification of Arfs with Roman numerals may reflect either the lack of resolution into an Arabic-numbered Arf clade or that they diverged before the duplications giving rise to that clade. The node supporting monophyly of proposed Arf3 homologues is shown in bold, and the clade is enclosed in the shaded box.
Figure 8.
Sequence alignment of human, bovine, and other nonmammalian class I Arfs. Sequence alignment of the amino and carboxy termini of class I Arfs from representative species. These include species that express multiple class I Arfs such as H. sapiens (Hs), Mus musculus (Mm), Xenopus laevis (Xl), Danio rerio (Dr), Salmo salar (Ss), Gallus gallus (Gg), and Taeniopygia guttata (Tg). Also included are species that express a single class I Arf such as Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce), Monosiga brevis (Mb), and N. vectensis (Nv). Widely used fungal model organisms such as Saccharomyces cerevisiae (Sc) and Schizosaccharomyces pombe (Sp) were included.
DISCUSSION
We previously established that Arf3 was much less potent than Arf1 in blocking an in vitro intra-Golgi assay that measures traffic between early Golgi compartments (Taylor et al., 1992). We speculated then “…that GGBF [Arf1] and GGBF* [Arf3] may direct assembly of coats from different organelles such as the Golgi and the trans-Golgi network.” Tagged Arf3 chimeras allowed us to overcome the lack of Arf3-specific antibodies for immunolocalization and finally revisit our hypothesis. Here, we established that Arf3 localizes preferentially to the trans-side of the Golgi complex, where it overlaps significantly with the trans-markers BIG1 and GalT-GFP and shows good separation from the cis-markers p115 and GBF1. Three complementary approaches established a functional link between Arf3 and the only Arf-GEFs localized at the TGN, BIG1, and BIG2. These included overexpression and knockdown of Golgi-localized Arf-GEFs, as well as brief BFA treatment of BFA-resistant BFY1 cell line. Our studies also uncovered a unique temperature sensitivity for Arf3 membrane recruitment upon shift to 20°C. This redistribution between the Golgi membranes and cytosol occurred slowly, with a half time of ∼10 min. Although temperature shift to 20°C clearly blocked VSVG trafficking at the Golgi complex, Arf3 knockdown did not affect either VSVG trafficking to the plasma membrane or the localization of several markers of the Golgi stack, TGN, or clathrin adaptors. Analysis of swap chimeras and point mutants demonstrated that temperature sensitivity and localization are readily separated and depend on residues present at opposite ends of Arf3. Residues at positions 9 and 13 of the variable N-terminal helix appear to determine temperature in/sensitivity of the membrane recruitment of Arf1 or Arf3. On the other hand, residues A174 and K180 appear to direct Arf3 to trans-compartments of the Golgi complex. Comparison of vertebrate class I Arf sequences confirmed that critical residues at positions 9, 13, 174, and 180 are both absolutely conserved and unique to Arf3.
Arf3 Is Uniquely Recruited to the TGN
Initial characterization of Arf3-GFP distribution first suggested that Arf3 localizes separately from the cis-Golgi marker GBF1 (J. Chun, unpublished observation). We extended and strengthened this observation by confirming Arf3 localization to the trans-side of the Golgi complex using both confocal and immunoelectron microscopy. This conclusion is based on a large number of complementary observations that involved multiple tagged forms in several cell lines costained for a wide variety of markers.
The lack of specific antibodies that will selectively recognize endogenous Arf3 forced us to use alternative means to examine the intracellular localization of Arf3. We took advantage of either tagged Arf3 chimeras or stained for overexpressed untagged Arf3 using a pan-specific Arf antibody (clone 1D9). To address concerns that arise from the use of C-terminal tags (Jian et al., 2010), we compared the localization of Arf3 tagged with the large GFP or the smaller HA epitope. Importantly, we also confirmed that tagged Arf3 chimeras yielded the same localization pattern as overexpressed untagged Arf3 (Figure 1). Finally, we examined Arf3 distribution in both NRK cells and HeLa cells and used multiple markers for both the cis- and the trans-side of the Golgi complex.
As in any experiments involving overexpression, we monitored closely the level of expression for each of the chimera tested. To avoid interference with normal function and proper localization, we selected for analysis transfected cells expressing low levels that displayed relatively weak cytoplasmic staining. Quantification was performed primarily with NRK cells because this cell line displays better separation of cis- and trans-Golgi compartments than HeLa cells. The Arf3-GFP chimera usually yielded lower expression level than Arf3-HA and was favored for analysis. The degree of separation of Arf3 with GBF1 and p115 appears similar to that observed for cis- and trans-Golgi markers in previous studies (Zhao et al., 2002; Schaub et al., 2006). In combination with the colocalization of Arf3 with GalT-GFP and BIG1, as well as complementary immuno-EM studies, these observations strongly suggests that Arf3 localizes at the TGN and most likely also at the trans-Golgi cisternae.
Arf3 Is Most Likely Activated by BIGs at the TGN
A series of three complementary experiments firmly established the functional link between BIGs and Arf3 suggested initially by their very similar localization patterns. First, the overexpression of BIG1 but not GBF1 protected Arf3 localization to the Golgi complex from a short BFA treatment. This result agrees with previous studies that demonstrated selective effects of GBF1 or BIG1 overexpression on early and late compartments (Manolea et al., 2008). These results apparently contradict a previous report that overexpression of GBF1 can protect not only Arf1, 4, and 5 but also Arf3 from BFA-induced redistribution (Kawamoto et al., 2002). We suggest that previous results linking Arf3 to GBF1/COPI (Kawamoto et al., 2002; Volpicelli-Daley et al., 2005) may have resulted from overexpression of WT and mutant Arf3 to extremely high levels.
Second, knockdown of BIGs redistributed Arf3 from the membranes but had no significant impact on Arf1 Golgi localization (Figure 2, B and E). Lastly, a brief BFA treatment of BFY1 cells, a cell line with a BFA-resistant GBF1 system but a BFA-sensitive BIGs system, redistributed Arf3 from the Golgi membranes, whereas Arf1 was only marginally affected (Figure 2C and Supplementary Figure 2). This last result provides strong confirmation of the unique link between Arf3 and BIGs. The mechanism responsible for the selective maintenance of an active GBF1/COPI system in BFY1 cells remains unknown but dramatically illustrates our previous observation that the Golgi stack assembles and functions relatively independently of BIGs and the TGN (Manolea et al., 2008).
The C-Terminus Determines the Unexpected Localization of Arf3 to Golgi trans-Compartments
To tease out the region in Arf3 important for its specific localization, we constructed swap chimeras between Arf1 and Arf3. This approach was greatly facilitated by the fact that Arf1 and Arf3 are 96% identical in sequence and that variations are limited to the N- and C-terminal helices. We initially expected that the same N-terminal region important for temperature sensitivity and for insertion into the lipid membrane (Antonny et al., 1997) would also be the one responsible for targeting Arf3. The presence of the N-terminus at the membrane provides obvious opportunities to either make contact with a specific lipid environment or to interact with a specific protein. However, quantitative analysis demonstrated unambiguously that the region important for Arf3 specific localization is not the N-terminus but rather the C-terminus. Colocalization of chimeras with either GBF1, p115, or BIG1 suggested that Arf3_1-GFP localized toward the cis-side of the Golgi complex, whereas Arf1_3-GFP localized like Arf3 toward the trans-side. Furthermore, analysis of single point mutants revealed that mutation of either A174 or K180 into the corresponding Arf1 residues caused the construct to lose trans-localization specific to Arf3 and localize instead to the cis-side of the Golgi stack. These results identify both residues A174 and K180 at the C-terminus as critical for targeting Arf3 to the trans-side of the Golgi complex.
Accumulation of Arf3 at the TGN could arise either from the relative substrate specificity of the cis-Golgi and TGN-localized GEFs or from active recruitment of the substrate to TGN membranes by an Arf3 receptor. However, we consider it unlikely that residues A174 or K180 determine GEF specificity because they lie on the surface opposite to that interacting with the Sec7 domain (Goldberg, 1998; Renault et al., 2003). Furthermore, the fact that all Arf3 chimeras and mutants are recruited to Golgi membranes in a BFA-sensitive manner clearly demonstrate that Arf3 can be activated by GBF1 when mis-targeted to the cis-Golgi. Instead, we consider it more likely that residues A174 or K180 act as part of a targeting sequence that binds directly the putative receptor. We cannot rule out the possibility that they function indirectly through changes in the tertiary structure that regulate binding to a putative receptor or lipid domain. The available crystal structures of Arf1, Arf4, and Arf5 reveal that those residues lie on the protein surface near the N-terminal helix and should be available for interaction with a putative receptor. The fact that those residues are unique to Arf3 (Figure 8) strongly suggests that no other Arfs use that mechanism for preferential recruitment to the TGN.
Hydrophobic Residues in the N-Terminal Helix of Arf1 and Arf3 Dictate Temperature Insensitivity/Sensitivity of Membrane Recruitment
Arf3 differs from Arf1 not only by its restricted pattern within the Golgi complex but by its unique temperature sensitivity. Temperature shift from 37 to 20°C caused release of Arf3 from the membrane, whereas Arf1 remained largely unaffected. Importantly, this effect was fully reversible as Arf3 returned to Golgi membranes with similar kinetics after a shift from 20 to 37°C. Examination of Arf3/Arf1 chimeras demonstrated that the temperature sensitivity determinant lies within the N-terminus. More detailed analysis of single and double mutants identified residues 9 and 13 as being important in directing both the extent of binding as well as its temperature sensitivity.
Of the four residues different between the Arf3 and Arf1 N-termini, several lines of evidence made us focus initially on hydrophobic residues at positions 9 and 13. In particular, the myristoylated N-terminal helix has been implicated in Arf-membrane association, and aromatic residues in Arf1 appear to interact directly with the membrane (Antonny et al., 1997). It is important to note that long aliphatic L and I residues at 9 and 13 appear absolutely conserved and unique to Arf3 (Figure 8). This observation suggests that the temperature sensitivity of Arf3 is conserved throughout all vertebrate species and might be an important feature of Arf3 function. Also, the presence of F9 and F13 in all the other Arfs that localize at the Golgi complex suggests that their membrane recruitment might be temperature insensitive, as observed with Arf1. The reason for the impact of F/I substitution remains unknown, but may result from the fact that aromatic residues such as F are more polarizable than aliphatic ones such as L and I and may better interact at the interface between the polar head groups and hydrophobic core of the membrane. This property accounts for their relative abundance at this interface in the transmembrane helices of single and multispanning membrane proteins and may explain the higher-temperature–insensitive binding of Arf1. Elucidation of the mechanism responsible for the correlation between of L/I residues and reduced-temperature–sensitive recruitment will require a more detailed mutagenesis analysis.
The unusual temperature sensitivity of Arf3 membrane association may provide insights into the unique mechanism of Arf3 recruitment. The delay in the redistribution to and from the Golgi complex and cytosol suggests either a rearrangement of the membrane composition, most probably lipids, or posttranslational changes in a putative membrane receptor. We examined three obvious possibilities, displacement of BIGs, loss of Golgi polarization, or reduction in Golgi-associated PtdIns(4)P levels and found that none of them could account for our observation. We should point out that the temperature sensitivity does not appear to be a property of an Arf localized only to the TGN, because the Arf3_1-GFP construct redistributed from the cis-Golgi membranes upon shift to 20°C (Figures 6, A and B, and Supplementary Figure 5A). We conclude that the release mechanism of Arf3 and chimeras involves a global change on the Golgi complex because it occurs slowly and irrespective of whether the chimeras localize on the cis- or trans-Golgi.
Arf3 Function at the TGN
The presence of multiple class I Arfs in vertebrates begs explanation. Previous work suggests that complexity within the membrane trafficking system arose from an evolutionary mechanism that involves cycles of gene duplication of organelle identity proteins (e.g., rabs, SNAREs, coats, and more) and coevolution with their interacting partners (Dacks and Field, 2007; Dacks et al., 2008); this process ultimately yields specialized protein machineries for differentiated endomembrane organelles. Phylogenetic analysis suggests that Arf diversity is guided by the same evolutionary mechanism (Logdson and Kahn, 2003; Li et al., 2004). As shown in Figure 8, class I Arfs underwent at least two rounds of recent gene duplication, and some functional redundancy is to be expected. For example, the fact that knockdown of Arf3 had no apparent effect on the localization of either AP-1 or PtdIns(4)P to the TGN suggests that Arf3 functions redundantly with Arf1, Arf4 or Arf5 in AP-1 assembly and/or PtdIns(4)P synthesis at the TGN. However, our observation of differential localization and response to temperature shift clearly suggests that the process of specialization has already begun. Unique functions for Arf3 remain unknown but could include targeting a unique set of effectors or regulation of BIG recruitment, as reported for Arf6 and ARNO (Cohen et al., 2007). Our observations provide new avenues to uncover the nature of these Arf3 specializations and their implications for regulation of cargo sorting at the TGN.
Supplementary Material
ACKNOWLEDGMENTS
We thank Z. Schapovalova for construction of several plasmids encoding multiple tagged forms of Arf3, as well as H. Chan for technical help with confocal microscopy. We thank Drs. B. Antonny (CNRS, Université de Nice-Sophia Antipolis, France), R. McElheny, and M. Glover and D. Quilty (University of Alberta) for helpful discussions. We thank Dr. A. De Matteis for the gift of affinity-purified anti-GM-130 antibodies, Dr, L. Berthiaume for affinity-purified antibody to GFP, Dr. R. Kahn for Arf3 antibody, and W. J. Cho (University of Alberta) for performing the immunogold electron microscopy. This study was supported by a grant to P.M. from the Canadian Institutes of Health Research and to J.B.D. from the Natural Sciences and Engineering Research Council of Canada.
Abbreviations used:
- AP
Adaptor protein
- Arf
ADP-ribosylation factor
- BFA
brefeldin A
- BIG
brefeldin A–inhibited guanine nucleotide exchange factor
- COP
coat protein
- FAPP
four-phosphate-adaptor protein
- GalT
galactosidase T
- GBF
Golgi-specific brefeldin A resistance factor
- GEF
guanine nucleotide exchange factor
- GGA
gamma ear Golgi-localized Arf-binding protein
- IF
immunofluorescence
- NRK
normal rat kidney
- VSVG
vesicular stomatitis virus glycoprotein
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-01-0016) on March 31, 2010.
REFERENCES
- Abascal F., Zardoya R., Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Allan V. J., Kreis T. E. A microtubule-binding protein associated with membranes of the Golgi apparatus. J. Cell Biol. 1986;103:2229–2239. doi: 10.1083/jcb.103.6.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny B., Beraud-Dufour S., Chardin P., Chabre M. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 1997;36:4675–4684. doi: 10.1021/bi962252b. [DOI] [PubMed] [Google Scholar]
- Bard F., Malhotra V. The formation of TGN-to-plasma-membrane transport carriers. Annu. Rev. Cell Dev. Biol. 2006;22:439–455. doi: 10.1146/annurev.cellbio.21.012704.133126. [DOI] [PubMed] [Google Scholar]
- Berger S. J., Claude A., Melançon P. Analysis of recombitant human ADP-ribosylation factors by reversed phase HPLC and Electrospray MS. Analyt. Biochem. 1998;264:53–65. doi: 10.1006/abio.1998.2821. [DOI] [PubMed] [Google Scholar]
- Boehm M., Aguilar R. C., Bonifacino J. S. Functional and physical interactions of the adaptor protein complex AP- 4 with ADP-ribosylation factors (ARFs) EMBO J. 2001;20:6265–6276. doi: 10.1093/emboj/20.22.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman A. L., Kuai J., Zhu X., Chen J., Kuriyama R., Kahn R. A. Arf proteins bind to mitotic kinesin-like protein 1 (MKLP1) in a GTP-dependent fashion. Cell Motil. Cytoskelet. 1999;44:119–132. doi: 10.1002/(SICI)1097-0169(199910)44:2<119::AID-CM4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Boman A. L., Salo P. D., Hauglund M. J., Strand N. L., Rensink S. J., Zhdankina O. ADP-ribosylation factor (ARF) interaction is not sufficient for yeast GGA protein function or localization. Mol. Biol. Cell. 2002;13:3078–3095. doi: 10.1091/mbc.E02-02-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J. S., Glick B. S. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Braulke T., Bonifacino J. S. Sorting of lysosomal proteins. Biochim. Biophys. Acta. 2008;1793:605–614. doi: 10.1016/j.bbamcr.2008.10.016. [DOI] [PubMed] [Google Scholar]
- Bui Q. T., Golinelli-Cohen M. P., Jackson C. L. Large Arf1 guanine nucleotide exchange factors: evolution, domain structure, and roles in membrane trafficking and human disease. Mol. Genet. Genomics. 2009;282:329–350. doi: 10.1007/s00438-009-0473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavrier P., Goud B. The role of ARF and Rab GTPases in membrane transport. Curr. Opin. Cell Biol. 1999;11:466–475. doi: 10.1016/S0955-0674(99)80067-2. [DOI] [PubMed] [Google Scholar]
- Chun J., Shapovalova Z., Dejgaard S. Y., Presley J. F., Melancon P. Characterization of class I and II ADP-ribosylation factors (Arfs) in live cells: GDP-bound Class II Arfs associate with the ER-Golgi intermediate compartment independently of GBF1. Mol. Biol. Cell. 2008;19:3488–3500. doi: 10.1091/mbc.E08-04-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claude A., Zhao B. P., Kuziemsky C. E., Dahan S., Berger S. J., Yan J. P., Armold A. D., Sullivan E. M., Melançon P. GBF1, a novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J. Cell Biol. 1999;146:71–84. [PMC free article] [PubMed] [Google Scholar]
- Cockcroft S., Thomas G. M., Fensome A., Geny B., Cunningham E., Gout I., Hiles I., Totty N. F., Truong O., Hsuan J. J. Phospholipase D: a downstream effector of ARF in granulocytes. Science. 1994;263:523–526. doi: 10.1126/science.8290961. [DOI] [PubMed] [Google Scholar]
- Cohen L. A., Honda A., Varnai P., Brown F. D., Balla T., Donaldson J. G. Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol. Biol. Cell. 2007;18:2244–2253. doi: 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R., Mason-Gamer R. J., Jackson C. L., Segev N. Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol. Biol. Cell. 2004;15:1487–1505. doi: 10.1091/mbc.E03-06-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza-Schorey C., Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey C., Li G., Colombo M. I., Stahl P. D. A regulatory role for ARF6 in receptor-mediated endocytosis. Science. 1995;267:1175–1178. doi: 10.1126/science.7855600. [DOI] [PubMed] [Google Scholar]
- Dacks J. B., Field M. C. Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J. Cell Sci. 2007;120:2977–2985. doi: 10.1242/jcs.013250. [DOI] [PubMed] [Google Scholar]
- Dacks J. B., Poon P. P., Field M. C. Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc. Natl. Acad. Sci. USA. 2008;105:588–593. doi: 10.1073/pnas.0707318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis M. A., Luini A. Exiting the Golgi complex. Nat. Rev. Mol. Cell Biol. 2008;9:273–284. doi: 10.1038/nrm2378. [DOI] [PubMed] [Google Scholar]
- Donaldson J. G. Multiple roles for Arf6, sorting, structuring, and signaling at the plasma membrane. J. Biol. Chem. 2003;278:41573–41576. doi: 10.1074/jbc.R300026200. [DOI] [PubMed] [Google Scholar]
- Donaldson J. G., Cassel D., Kahn R. A., Klausner R. D. ADP-ribosylation factor, a small GTP-binding protein, is required for binding of the coatomer protein beta-COP to Golgi membranes. Proc. Natl. Acad. Sci. USA. 1992;89:6408–6412. doi: 10.1073/pnas.89.14.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duden R. ER-to-Golgi transport: COP I and COP II function (Review) Mol. Membr. Biol. 2003;20:197–207. doi: 10.1080/0968768031000122548. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S. M., Harborth J., Weber K., Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- Fischer K. D., Helms J. B., Zhao L., Wieland F. T. Site-specific photocrosslinking to probe interactions of Arf1 with proteins involved in budding of COPI vesicles. Methods. 2000;20:455–464. doi: 10.1006/meth.2000.0958. [DOI] [PubMed] [Google Scholar]
- Garcia-Mata R., Szul T., Alvarez C., Sztul E. ADP-ribosylation factor/COPI-dependent events at the endoplasmic reticulum-Golgi interface are regulated by the guanine nucleotide exchange factor GBF1. Mol. Biol. Cell. 2003;14:2250–2261. doi: 10.1091/mbc.E02-11-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B. S., Nakano A. Membrane traffic within the Golgi stack. Annu. Rev. Cell Dev. Biol. 2009;25:113–132. doi: 10.1146/annurev.cellbio.24.110707.175421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A., Di Campli A., Konstantakopoulos A., Di Tullio G., Alessi D. R., Kular G. S., Daniele T., Marra P., Lucocq J. M., De Matteis M. A. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Goldberg J. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- Griffiths G., Fuller S. D., Back R., Hollinshead M., Pfeiffer S., Simons K. The dynamic nature of the Golgi complex. J. Cell Biol. 1989;108:277–297. doi: 10.1083/jcb.108.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Harborth J., Elbashir S. M., Bechert K., Tuschl T., Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. p. 726. [Google Scholar]
- Honda A., Al-Awar O. S., Hay J. C., Donaldson J. G. Targeting of Arf-1 to the early Golgi by membrin, an ER-Golgi SNARE. J. Cell Biol. 2005;168:1039–1051. doi: 10.1083/jcb.200409138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam A., Shen X., Hiroi T., Moss J., Vaughan M., Levine S. J. The brefeldin A-inhibited guanine nucleotide-exchange protein, BIG2, regulates the constitutive release of TNFR1 exosome-like vesicles. J. Biol. Chem. 2007;282:9591–9599. doi: 10.1074/jbc.M607122200. [DOI] [PubMed] [Google Scholar]
- Jian X., Cavenagh M., Gruschus J. M., Randazzo P. A., Kahn R. A. Modifications to the C-terminus of Arf1 alter cell functions and protein interactions. Traffic. 2010 doi: 10.1111/j.1600-0854.2010.01054.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh H., Williger B. T., Exton J. H. Arfaptin 1, a putative cytosolic target protein of ADP-ribosylation factor, is recruited to Golgi membranes. J. Biol. Chem. 1997;272:5421–5429. doi: 10.1074/jbc.272.9.5421. [DOI] [PubMed] [Google Scholar]
- Kawamoto K., Yoshida Y., Tamaki H., Torii S., Shinotsuka C., Yamashina S., Nakayama K. GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factors, is localized to the cis-Golgi and involved in membrane association of the COPI coat. Traffic. 2002;3:483–495. doi: 10.1034/j.1600-0854.2002.30705.x. [DOI] [PubMed] [Google Scholar]
- Levine T. P., Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr. Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- Li Y., Kelly W. G., Logsdon J. M., Jr, Schurko A. M., Harfe B. D., Hill-Harfe K. L., Kahn R. A. Functional genomic analysis of the ADP-ribosylation factor family of GTPases: phylogeny among diverse eukaryotes and function in C. elegans. FASEB J. 2004;18:1834–1850. doi: 10.1096/fj.04-2273com. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Cole N., Presley J. Unravelling Golgi membrane traffic with green fluorescent protein chimeras. Trends Cell Biol. 1998;8:16–20. doi: 10.1016/s0962-8924(97)01199-9. [DOI] [PubMed] [Google Scholar]
- Logdson J.M.J., Kahn R. A. The Arf family tree. In: Kahn R. A., editor. Arf Family GTPases. Dordrecht: Kluwer Academic; 2003. pp. 1–21. [Google Scholar]
- Manolea F., Claude A., Chun J., Rosas J., Melançon P. Distinct functions for Arf guanine nucleotide exchange factors at the Golgi complex: GBF1 and BIGs are required for assembly and maintenance of the Golgi stack and trans-Golgi network, respectively. Mol. Biol. Cell. 2008;19:523–535. doi: 10.1091/mbc.E07-04-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S. J., Skaug J., Zhao X. H., Giordano J., Scherer S. W., Melançon P. p200 ARF-GEP1, a Golgi-localized guanine nucleotide exchange protein whose Sec7 domain is targeted by the drug brefeldin A. Proc. Natl. Acad. Sci. USA. 1999;96:7968–7973. doi: 10.1073/pnas.96.14.7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra P., Salvatore L., Mironov A., Jr, Di Campli A., Di Tullio G., Trucco A., Beznoussenko G., Mironov A., De Matteis M. A. The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Mol. Biol. Cell. 2007;18:1595–1608. doi: 10.1091/mbc.E06-10-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin K. S., Simons K. Reduced temperature prevents transfer of a membrane glycoprotein to the cell surface but does not prevent terminal glycosylation. Cell. 1983;34:233–243. doi: 10.1016/0092-8674(83)90154-x. [DOI] [PubMed] [Google Scholar]
- Mogelsvang S., Marsh B. J., Ladinsky M. S., Howell K. E. Predicting function from structure: 3D structure studies of the mammalian Golgi complex. Traffic. 2004;5:338–345. doi: 10.1111/j.1398-9219.2004.00186.x. [DOI] [PubMed] [Google Scholar]
- Mouratou B., Biou V., Joubert A., Cohen J., Shields D. J., Geldner N., Jurgens G., Melançon P., Cherfils J. The domain architecture of large guanine nucleotide exchange factors for the small GTP-binding protein Arf. BMC Genom. 2005;6:20. doi: 10.1186/1471-2164-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z., Randazzo P. A. Arf GAPs and membrane traffic. J. Cell Sci. 2006;119:1203–1211. doi: 10.1242/jcs.02924. [DOI] [PubMed] [Google Scholar]
- Ooi C. E., Dell'Angelica E. C., Bonifacino J. S. ADP-Ribosylation factor 1 (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J. Cell Biol. 1998;142:391–402. doi: 10.1083/jcb.142.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L., Glick B. S., Rothman J. E. A new type of coated vesicular carrier that appears not to contain clathrin: its possible role in protein transport within the Golgi stack. Cell. 1986;46:171–184. doi: 10.1016/0092-8674(86)90734-8. [DOI] [PubMed] [Google Scholar]
- Peters P. J., Hsu V. W., Ooi C. E., Finazzi D., Teal S. B., Oorschot V., Donaldson J. G., Klausner R. D. Overexpression of wild-type and mutant ARF1 and ARF6, distinct perturbations of nonoverlapping membrane compartments. J. Cell Biol. 1995;128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R., Randazzo P. A., Presley J. F., Hartnell L. M., Bonifacino J. S. The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell. 2001;105:93–102. doi: 10.1016/s0092-8674(01)00299-9. [DOI] [PubMed] [Google Scholar]
- Rambourg A., Clermont Y. Three-dimensional electron microscopy: structure of the Golgi apparatus. Eur. J. Cell Biol. 1990;51:189–200. [PubMed] [Google Scholar]
- Renault L., Guibert B., Cherfils J. Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature. 2003;426:525–530. doi: 10.1038/nature02197. [DOI] [PubMed] [Google Scholar]
- Robinson M. S., Kreis T. E. Recruitment of coat proteins onto Golgi membranes in intact and permeabilized cells: effects of brefeldin A and G protein activators. Cell. 1992;69:129–138. doi: 10.1016/0092-8674(92)90124-u. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. MrBayes3, Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Saraste J., Palade G. E., Farquhar M. G. Temperature-sensitive steps in the transport of secretory proteins through the Golgi complex in exocrine pancreatic cells. Proc. Natl. Acad. Sci. USA. 1986;83:6425–6429. doi: 10.1073/pnas.83.17.6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub B. E., Berger B., Berger E. G., Rohrer J. Transition of galactosyltransferase 1 from trans-Golgi cisterna to the trans-Golgi network is signal mediated. Mol. Biol. Cell. 2006;17:5153–5162. doi: 10.1091/mbc.E06-08-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R., Mellman I. Does COPI go both ways? Cell. 1997;90:197–200. doi: 10.1016/s0092-8674(00)80326-8. [DOI] [PubMed] [Google Scholar]
- Shin H. W., Morinaga N., Noda M., Nakayama K. BIG2, a guanine nucleotide exchange factor for ADP-ribosylation factors: its localization to recycling endosomes and implication in the endosome integrity. Mol. Biol. Cell. 2004;15:5283–5294. doi: 10.1091/mbc.E04-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinotsuka C., Waguri S., Wakasugi M., Uchiyama Y., Nakayama K. Dominant-negative mutant of BIG2, an ARF-guanine nucleotide exchange factor, specifically affects membrane trafficking from the trans-Golgi network through inhibiting membrane association of AP-1 and GGA coat proteins. Biochem. Biophys. Res. Commun. 2002a;294:254–260. doi: 10.1016/S0006-291X(02)00456-4. [DOI] [PubMed] [Google Scholar]
- Shinotsuka C., Yoshida Y., Kawamoto K., Takatsu H., Nakayama K. Overexpression of an ADP-ribosylation factor-guanine nucleotide exchange factor, BIG2, uncouples brefeldin A-induced adaptor protein-1 coat dissociation and membrane tubulation. J. Biol. Chem. 2002b;277:9468–9473. doi: 10.1074/jbc.M112427200. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Taylor T. C., Kahn R. A., Melançon P. Two distinct members of the ADP-ribosylation factor family of GTP- binding proteins regulate cell-free intra-Golgi transport. Cell. 1992;70:69–79. doi: 10.1016/0092-8674(92)90534-j. [DOI] [PubMed] [Google Scholar]
- Volpicelli-Daley L.A., Li Y., Zhang C. J., Kahn R. A. Isoform-selective effects of the depletion of Arfs1–5 on membrane traffic. Mol. Biol. Cell. 2005;16:4495–4508. doi: 10.1091/mbc.E04-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. G., Clary D. O, Rothman J. E. A novel 115-kD peripheral membrane protein is required for intercisternal transport in the Golgi stack. J. Cell Biol. 1992;118:1015–1026. doi: 10.1083/jcb.118.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J. P., Colon M. E., Beebe L. A., Melancon P. Isolation and characterization of mutant CHO cell lines with compartment-specific resistance to brefeldin A. J. Cell Biol. 1994;126:65–75. doi: 10.1083/jcb.126.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Lasell T. K., Melançon P. Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: evidence for distinct functions in protein traffic. Mol. Biol. Cell. 2002;13:119–133. doi: 10.1091/mbc.01-08-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.