Abstract
Sac family phosphoinositide (PI) phosphatases are an essential family of CX5R(T/S)-based enzymes, involved in numerous aspects of cellular function such as PI homeostasis, cellular signalling, and membrane trafficking. Genetic deletions of several Sac family members result in lethality in animal models and mutations of the Sac3 gene have been found in human hereditary diseases. In this study, we report the crystal structure of a founding member of this family, the Sac phosphatase domain of yeast Sac1. The 2.0 Å resolution structure shows that the Sac domain comprises of two closely packed sub-domains, a novel N-terminal sub-domain and the PI phosphatase catalytic sub-domain. The structure further shows a striking conformation of the catalytic P-loop and a large positively charged groove at the catalytic site. These findings suggest an unusual mechanism for its dephosphorylation function. Homology structural modeling of human Fig4/Sac3 allows the mapping of several disease-related mutations and provides a framework for the understanding of the molecular mechanisms of human diseases.
Keywords: lipid metabolism, phosphatase, phosphoinositide, Sac1, Sac3/Fig4
Introduction
Phosphoinositides (PIs) control numerous cellular processes such as cell signalling, proliferation, cytoskeleton organization, membrane trafficking, ion channel activity, transcription, and mRNA trafficking (Odorizzi et al, 2000; De Matteis and Godi, 2004; Di Paolo and De Camilli, 2006). PIs localize at membrane-cytoplasmic interfaces and achieve their functions through the binding of proteins to their cytoplasmic-exposed phosphorylated inositol head groups, which can be reversibly phosphorylated at the 3′, 4′, and 5′ positions to generate an ensemble of seven biologically active derivatives. Each PI isoform has a specific intracellular distribution, which provides spatial and temporal cues within the cell for PI-binding proteins. The recruitment of specific PI-binding proteins promotes the assembly of macromolecular complexes to initiate a series of physiological events including membrane trafficking and cytoskeletal dynamics. Moreover, in response to extracellular stimuli, PIs also serve as precursors for second messangers to initiate downstream intracellular signalling pathways. To maintain the selective concentration of specific PI species, as well as to generate the dynamic PI composition changes in response to acute signalling inputs, eukaryotic cells encode a large number of PI-metabolizing enzymes: including PI kinases and PI phosphatases. Despite their physiological importance, the properties of many of these enzymes remain poorly characterized.
One class of these enzymes contains a conserved PI phosphatase module termed the Sac domain. The Sac phosphatase domain comprises ∼500 amino acids and contains seven highly conserved motifs including the conserved catalytic CX5R(T/S) motif, which is essential for catalytic activity (Hughes et al, 2000a). There are five Sac phosphatase domain-containing proteins in humans, which appear to fall into two subfamilies. Members of the first subfamily, including the trans-membrane protein Sac1, cytosolic proteins Sac2/INPP5f, and Sac3/Fig4, have an N-terminal Sac phosphatase and no other recognizable structural domains. The second subfamily comprises two synaptojanin homologues, which have a PI 5-phosphatase domain immediately after the N-terminal Sac phosphatase module (Supplementary Figure 1). In yeast, there are also five Sac domain-containing enzymes including Sac1, Fig4, and three synaptojanin like proteins (Sjl1,2,3).
Sac1 is a 67 kDa type II membrane protein that localizes to the endoplasmic reticulum (ER) and Golgi apparatus (Whitters et al, 1993; Nemoto et al, 2000). It was originally identified by two independent genetic screens searching for modifiers of actin cytoskeleton defects and of trans-Golgi network exocytic failure caused by inactivation of Sec14p, the major yeast phosphatidylinositol/phosphatidylcholine transfer protein (Cleves et al, 1989; Novick et al, 1989; Bankaitis et al, 1990). In yeast, loss of Sac1 function has been implicated in a broad range of cellular defects (Strahl and Thorner, 2007), such as disorganization of the actin cytoskeleton (Novick et al, 1989), inositol auxotrophy (Whitters et al, 1993), cold sensitivity for growth (Novick et al, 1989), multiple drug sensitivities (Hughes et al, 1999), abnormal vacuolar morphology (Foti et al, 2001; Tahirovic et al, 2005), cell wall defects (Schorr et al, 2001), and unbalanced sphingolipid synthesis (Brice et al, 2009). Sac1 mutants in Drosophila die as embryos with defects in dorsal closure (Wei et al, 2003). Mouse strains deficient in Sac1 are embryonic lethal (Liu et al, 2008, 2009). Knock down of Sac1 expression in mammalian cell lines results in disorganization of Golgi membranes and mitotic spindles (Liu et al, 2008, 2009). These findings suggest essential roles for Sac1-mediated PI metabolism in multicellular organisms.
In vitro studies have shown that Sac1 dephosphorylates a number of PIs, including PI(3)P, PI(4)P, and PI(3,5)P2 (Guo et al, 1999). In vivo, genetic ablation of Sac1 activity in yeast results in a 10-fold increase in the steady-state levels of PI(4)P and modest increases in the levels of the 3-OH PIP species (Guo et al, 1999; Rivas et al, 1999; Nemoto et al, 2000), suggesting that yeast Sac1 is a major pathway for PI(4)P dephosphorylation in vivo.
Although Sac1 primarily dephosphorylates PI(4)P, other Sac phosphatase members have been shown to regulate other PI isoforms in vivo. Fig4 (Sac3 in mammals) was originally identified in a screen for genes induced by mating pheromone in Saccharomyces cerevisiae (Erdman et al, 1998). Sac3 is a PI 5-phosphatase that specifically hydrolyses PI(3,5)P2 to generate PI(3)P both in vitro and in vivo (Rudge et al, 2004; Duex et al, 2006a, b). Interestingly, Sac3/Fig4p forms a complex with a PI(3)P-5-kinase Fab1p and a scaffold protein Vac14p. This complex is also conserved in mammals and is responsible for the acute regulation of subcellular levels of PI(3,5)P2 (Botelho et al, 2008; Jin et al, 2008; Sbrissa et al, 2008). Genetic mutations affecting the function of Sac3/Fig4 lead to neurological diseases, including Charcot–Marie–Tooth (CMT) disease Type 4J, a subset of amyotrophic lateral sclerosis (ALS) in humans, and neurodegeneration in the pale tremor mouse (Chow et al, 2007, 2009).
Despite considerable attention on Sac domain-containing proteins, a lack of structural information of this unique Sac phosphatase domain has left large gaps in our knowledge of this protein family. For example, each Sac phosphatase prefers a specific subgroup of PIs as substrates, but how the substrate specificity is determined by the otherwise homologous phosphatase region is unresolved. Mutations have been mapped to Sac3/Fig4 in patients with CMT 4J and ALS, but how the function of the enzyme is impaired in disease mutants is still not understood. Hence, we performed structural studies on the Sac phosphatase. In this study, we report the first crystal structure of a Sac phosphatase, yeast Sac1. The structure shows a strikingly different configuration of the catalytic motif compared with protein tyrosine phosphatases and a large positively charged groove at the catalytic site that is surrounded by flexible loops enriched with hydrophobic and cationic residues. Our results further provide insights into the molecular mechanism for the enzymatic function of Sac phosphatases and suggest how mutations in Sac3/Fig4 affect its normal function in two human neuronal degeneration diseases (CMT4J and ALS).
Results
Structure determination and overall structure
The cytosolic portion of yeast Sac1 (residues 1–503) was expressed as an N-terminal His–sumo-tagged fusion protein. Crystals were obtained by using the hang drop vapour diffusion method with the reservoir solution containing 0.55 M Na citrate (pH 7.0) and 1.5 M NaCl. The crystals formed in space group I222 with unit cell dimensions of a=86.16 Å, b=94.73 Å, c=155.44 Å, and one molecule in the asymmetric unit. The structure was solved by selenomethionine single wavelength anomalous diffraction (SAD) method using the program HKL2MAP (Pape and Schneider, 2004). The final structure was refined against a 1.97 Å resolution data to a crystallographic Rwork=20.1% and Rfree=24.2% (Supplementary Table 1).
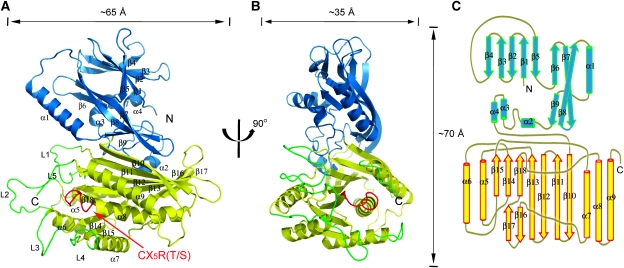
The crystal structure of the Sac domain of yeast Sac1p represents the first atomic structure of the approximate 500-residue Sac domain family. Figure 1 shows the ribbon diagram of the Sac phosphatase domain of yeast Sac1p. The overall structure has a dimension of about 65 Å × 35 Å × 70 Å, which can be divided into two domains, an N-terminal domain (residues 1–182 shown in blue and referred as the SacN domain thereafter) and a roughly 320-residue C-terminal catalytic domain (shown in yellow), which contains the CX5R(T/S) catalytic motif. It is also worth noting that in the Sac1 structure, there is a peptide stretch of about 50 residues at the COOH-terminus lacking any electron density, suggesting a flexible nature of this region.
Figure 1.
Overall crystal structure of Sac1. (A) Ribbon diagram of the Sac PI phosphatase. The SacN domain is coloured blue and the catalytic domain yellow. The secondary structure elements (strands β1–β18 and helices α1–α9) are labelled. The catalytic CX5R(T/S) motif (P-loop) is coloured red and labelled. Five protruding loops (L1–L5) surrounding the catalytic site are coloured green. (B) A 90° rotated view of (A). (C) Schematic diagram of the secondary structure topologies of Sac1.
The SacN domain of Sac1 has a unique fold
The SacN domain comprises three layers of β sheets and one long and three short α helices. The first layer of β sheet contains anti-parallel β1–β5. The second layer consists of β6 and half of the long and twisted β7–β8. The first and second β sheet layers stack on each other through extensive hydrophobic interactions. The third layer contains the other half of β7 and β8 and a short strand β9. This layer directly contacts with the catalytic domain. The α1 helix and its preceding loop between β8 and β9 are partially aligned with the region that was previously predicted to have a leucine zipper motif in mammalian Sac1 (Blagoveshchenskaya and Mayinger, 2009). The SacN domain of Sac1 has a novel fold, and structural homology search by DALI server (Holm and Sander, 1995) did not show any significant hits. The SacN domain is closely opposed to the catalytic domain, occluding a surface area of about 1300 Å2 on each domain. The function of the SacN domain is still unknown. A likely role of this domain is to control the enzymatic function through the interactions with other unknown factors.
The catalytic domain of Sac1
The catalytic domain of Sac1 consists of a nine-stranded and partially split β sheet that is flanked by five alpha helixes with two on one side and three on the other. The β sheet starts with four long anti-parallel strands (β10–β13) and then splits with three short parallel strands (β14, β15, and β18) in one branch and two anti-parallel strands (β16 and β17) in the other (Figure 1C). The P-loop (shown in red in Figure 1A and B), which contains the catalytic CX5R(T/S) motif (residues 392–399), extends from the end of β18, across the entire connecting loop, to the first turn of α8.
Interestingly, the P-loop is surrounded by five flexible loops (labelled from L1 to L5 and coloured in green in Figures 1A, B and 2). Three longer loops (L1, L2, and L5), side chains of which are partially invisible in our crystal structure, protrude out from the catalytic surface and form part of the edge of a large groove (see below). Although the primary sequences in these loop region are not as conserved as other regions across Sac domain family members, these loops are enriched with positively charged hydrophobic residues (Figure 2), the physicochemical nature of which is ideal to support interfacial membrane binding (Cho and Stahelin, 2005). These structural properties suggest that the catalytic site may be protected by the surrounding loops and on activation, the surrounding loops, particularly the three long protruding loops, may directly interact with or insert into the lipid bilayer.
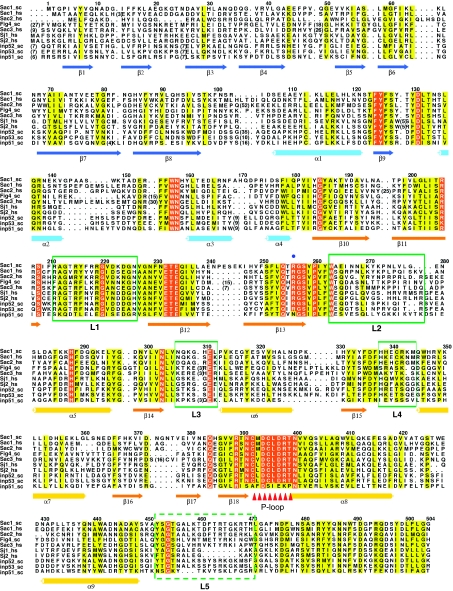
Figure 2.
Sequence alignment of the Sac domain region of all Sac phosphatases in human and yeast. The sequences corresponding to the Sac domain region were aligned by ClustalW 2 (Labarga et al, 2007) and coloured by ALSCRIPT (Barton, 1993). The conserved residues are shaded yellow and identical residues are shaded red. Secondary elements are drawn under the alignment. The catalytic site is marked by triangles. Regions that correspond to the five protruding loops (coloured green in Figure 1A and B) are indicated with green boxes. Entrez database accession numbers are as follow: Sac1_sc: gi: 486379; Inp51_sc, gi: 6322189; Fig4_sc, gi: 6324005; Inp52_sc, gi: 6324224; Inp53_sc, gi: 6324683; Sac3_hs, gi: 7662034; Sac2_hs, gi: 7662414; Sac1_hs, gi: 190014578; Sj1_hs, gi: 223460134; Sj2_hs, gi: 26190608.
Structure comparison of Sac1 with other phosphatases
Comparison of the structure of the catalytic domain of Sac1 with other CX5R(T/S)-based phosphatases shows that the topology of the catalytic core of Sac1 is conserved with other PTPs, including two lipid phosphatases MTMR2 and PTEN. They all share a common architecture of a central β sheet consisting of four parallel β strands flanked by anti-parallel ones. This sheet is sandwiched by a variable number of α helices on each side (Barford et al, 1994; Stuckey et al, 1994). We further searched the Protein Data Bank for structural similarities to the catalytic domain of Sac1 using the Dali server (Holm and Sander, 1995). The structure found to have the highest Z-score (7.4) is human PRL-1 (PDB ID: 1XM2) (Jeong et al, 2005). The overall structure of the two catalytic domains can be superimposed on that of PRL-1 with an r.m.s.d. of 3.5 Å over 128 Cα residues (Supplementary Figure 2A). Despite the limited similarity of the catalytic core topology, the catalytic domain of Sac1 is unique with its large and split central β sheet compared with other PTPs, including the low molecular weight phosphotyrosyl phosphatase and canonical PTP1B (Supplementary Figures 2A and B; and Figure 1C). Furthermore, structural comparison with the dual specific phosphatase, PTEN, shows the difference of the interactions between the catalytic domain and other regulatory domains. When the catalytic domain of Sac1 is superimposed with that of PTEN (Lee et al, 1999), the SacN domain of Sac1 ends up on the opposite side of the central β sheet in contrast to the C2 domain in PTEN (Supplementary Figure 3).
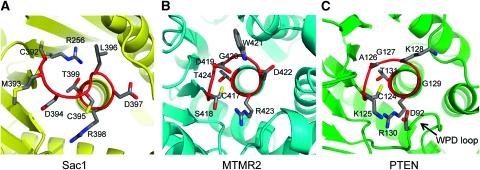
Another major difference is the unique conformation of the catalytic P-loop of the Sac phosphatase (Figure 3A). Among all PTPs, the P-loops in PTPs are strictly superimposable, and a consecutive seven main chain amide groups in this loop all face inward for the binding of a phosphate group of the substrate (Barford et al, 1998). The conformation of the P-loop is fairly stable, as evidenced by the fact that Cα tracing superimposition of P-loops from different PTPs (in the presence or absence of binding ligands) yields a minor standard deviation of <1 Å (Tabernero et al, 2008). However, this conserved P-loop configuration is disrupted in Sac1. Moreover, within the CX5R(T/S) motif, the distance between the γ-thiol group of C392 and the guanidinium group of R398 is 14 Å apart in Sac1, whereas the corresponding distance is only about 5 Å in other PTPs, including MTMR2 and PTEN (Figure 3B and C). Conformational deviation of the P-loop, resulting from the exposure of the enzyme to oxidants, has been reported (Salmeen et al, 2003; van Montfort et al, 2003; Yang et al, 2007). In PTP1B, formation of cyclic sulphenamide of the catalytic cysteine causes a significant change in conformation. However, oxidation is not likely the cause of the conformational variation in Sac1 as a high amount of reducing reagent, DTT, is present in the Sac1 protein stock and crystallization solutions. Furthermore, the electron density map around the P-loop does not show any extra density supporting the presence of oxidized forms of the thiol group of the catalytic C392 (Supplementary Figure 4). Structural comparison of the catalytic domain of Sac1 with reduced [PDB ID: 1AAX; (Puius et al, 1997)] and oxidized [in the cyclic sulphenamide state; PDB ID: 1OEM; (Salmeen et al, 2003)] forms of PTP1B shows that the P-loop conformation of Sac1 is strikingly different from that of either form of PTP1B (Supplementary Figure 5A and B). Particularly, the catalytic C392 and the conserved R398 residues in Sac1 are located on the opposite side of the P-loop, whereas in both reduced and oxidized PTP1Bs, these two residues are on the same side. Thus, this unique P-loop conformation of Sac1 may implicate an unusual mechanism for its catalytic function.
Figure 3.
Structural comparison of the catalytic site P-loop between Sac1, MTMR2, and PTEN. (A) The P-loop configuration in Sac1. This loop, which comprises the CX5R(T/S) motif, is coloured in red and residues in this loop are shown as sticks. This figure is generated in the same orientation as in Figure 2B. (B) The P-loop of MTMR2, and PTEN (C). The WPD loop in PTEN is indicated by an arrow.
Surface characteristics of Sac1
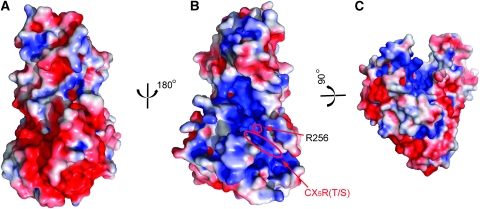
Interestingly, the Sac domain is an electrostatically bipolar protein with one surface negatively charged (Figure 4A) and the opposite surface enriched with positive charges (Figure 4B). The bipolar charge distribution has been observed in other membrane interacting protein modules, such as PH domains (Ferguson et al, 1994, 1995; DiNitto and Lambright, 2006). The positively charged face of the Sac domain is likely to interface with the lipid bilayer, which is concentrated with negatively charged phospholipids. In agreement with this notion, the catalytic CX5R(T/S) motif is localized on this positively charged surface.
Figure 4.
Surface representative of Sac1 phosphatase. The surfaces were coloured based on electrostatic potential with positively charged regions in blue (+3 kcal/electron) and negatively charged surface in red (−3 kcal/electron). (A) Rotated 180° about the perpendicular axis relative to (B). (B) The same orientation as in Figure 1B. The position of R256 is pinpointed by a red circle. (C) Rotated about ∼90° around the horizontal axis of (B) and then slightly tilted left for a view of the deep cleft. The catalytic CX5R(T/S) motif is highlighted with an oval shape. Note that the catalytic site is located at the bottom of the positively charged groove.
The other striking surface feature of the Sac domain is the presence of a deep canyon-like groove on the positively charged surface (Figure 4B and C). This groove is formed by both the SacN domain and the catalytic domain and travels a distance about 40 Å at an average depth of more than 10 Å. There is a highly conserved positively charged residue, R256 (labelled in Figure 4B and marked by a blue dot in Figure 2) aligned at the bottom of the groove. This arginine residue, together with some other positively charged residues, contributes to the electrostatic surface potential property in the groove.
Mutagenesis of catalytic site residues
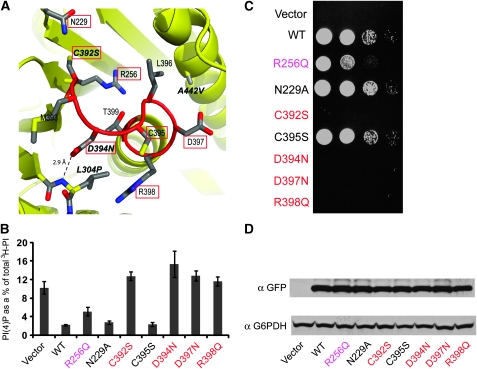
Previous studies have showed several interesting Sac1 mutant alleles. The catalytic cysteine to serine mutation (C392S) has been shown to be catalytically dead and fails to complement the sac1Δ phenotype in yeast (Rohde et al, 2003). The yeast D394N and A445V mutants (the product of the sac1-8 and sac1-10 alleles, respectively) have been shown to confer phenotypes consistent with the loss of phosphatase activity as reported earlier (Kearns et al, 1997; Nemoto et al, 2000; Liu et al, 2008). The L304P mutation has some puzzling effects in that it has normal in vivo functions when the cells were grown on YPD medium, but showed sac1 mutant phenotypes including elevated PI(4)P levels when grown on synthetic minimal medium (Hughes et al, 2000b). Interestingly, all these mutations are either located within or adjacent to the P-loop as showed by the Sac1 structure (Figure 5A). The carboxyl group of D394 is 2.9 Å away from the amide group of L304 and forms a hydrogen bond with the main chain amide proton of L304. This hydrogen bond would then be disrupted in the L304P mutant because of the lack of an amide proton in the proline residue, and thus may cause conformational changes of the P-loop that may affect the catalytic activity in the end.
Figure 5.
In vivo characterization of Sac1 mutants. (A) A close view of the catalytic P-loop of Sac1. Reported Sac1 mutant alleles are labelled in italic. Residues tested in this study are highlighted with red squares. (B) Quantitative analysis of PI(4)P is shown from sac1Δ cells transformed with empty vector control, N-term GFP-tagged wild type (WT) Sac1, and Sac1 mutants. The error bars represent ±s.e.m. of three independent experiments. (C) In vivo growth rescue assay of sac1 cold sensitivity. Sac1Δ cells were transformed with indicated Sac1 constructs or vector control. The cells were spotted onto plates with three serial of dilutions from left to right, and grown at restrictive temperature at 15°C for 6 days. (D) Western blots of samples prepared from the transformed cell show that the exogenously introduced Sac1 WT or mutants are expressed as stable proteins and at a comparable level (G6PDH is shown as a loading control).
To gain insight into the role of other conserved residues near the catalytic site, we generated several GFP-tagged Sac1 mutants in the vicinity of the catalytic site and introduced them into sac1Δ yeast cells. Sac1Δ yeast cells are viable when grown at permissive temperature, but with elevated PI(4)P level (6–8-fold greater than wild-type cells) and fail to grow at 15°C (Foti et al, 2001). These defects can be rescued by the re-introduction of functional Sac1. On the basis of these observations, we tested the ability of our mutants to rescue the growth defects of sac1Δ cells at 15°C and analysed the intracellular PI levels by HPLC (Figure 5B and C). Immunoblotting experiments showed that GFP-tagged wild type and mutant forms of Sac1 were expressed as stable proteins at a roughly similar level (Figure 5D). R256 in Sac1 is conserved among the Sac family members except for the catalytically inactive yeast Sjl1 (Figure 2). This conserved arginine contributes significantly to the positive electrostatic potentials at the catalytic site. In sac1Δ cells transformed with the R256Q mutant, the 3H incorporated PI(4)P represents ∼5% of total newly synthesized PI, which is nearly doubled compared to that of cells transformed with wild-type Sac1 (Figure 5B). In agreement with this result, the R256Q mutant can only partially rescue the growth defect of sac1Δ cell at 15°C (Figure 5C). We also tested several P-loop residues. As reported earlier, the catalytically dead C392S or D394N mutants completely fail to rescue the growth phenotype and the PI(4)P level is about 6–8-fold greater than that of sac1Δ cells transformed with wild-type Sac1. Similar to MTMR2, the Sac domain does not have the conserved aspartic acid residue (D92 in PTEN, Figure 4C) on its corresponding WPD loop. It has been suggested that in MTMR2, D422 within the P-loop may serve as the general acid and base during catalysis (Begley et al, 2003). This aspartic acid residue (D397 in yeast Sac1) is also conserved in Sac family phosphatases (Figure 2) and the D397N mutation has similar phenotypes as catalytically dead mutants. The only conserved basic residue within the catalytic P-loop, R398, seems also critical for the phosphatase function, as the R398Q mutant also has similar effects as catalytically dead mutants. Residues N229 is conserved among Sac phosphatases and is in the vicinity to the catalytic cysteine (C392). However, N229A has no apparent effect on the in vivo function of Sac1. Mutation of another conserved P-loop reside C395S also behaves like wild-type Sac1 (Figure 5B and C).
Collectively, these observations suggest that selective substitutions of some anionic residues near the catalytic site impair the normal function of Sac1, which may be due to direct impact on phospholipid binding. Both aspartic acid residues at the catalytic site are critical for the catalytic function by either serving as the general acid or maintaining proper P-loop conformation.
Missense disease mutations in human Sac3/Fig4
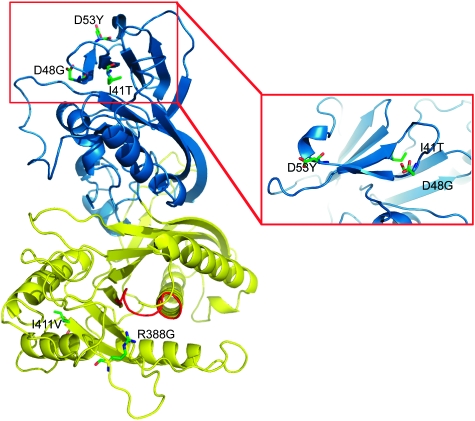
Several missense mutations of the human Sac3 are responsible for the genetic neurological disorders CMT4J and ALS. To understand how these single amino-acid substitutions can affect the function of the protein, we computationally modelled the structure of the Sac phosphatase domain of human Sac3 based on the crystal structure of Sac1 (Figure 6). We then mapped five known missense mutations to the modelled structure of the Sac domain of Sac3. The I41T is a recessive mutation found in patients with CMT4J. This residue localizes at the C-terminal end of β3 in the SacN domain and is about 40 Å away from the catalytic site. The hydrophobic side chain of this isoleucine is buried in a hydrophobic core between the first two layers of β sheets in the SacN domain. Thus, the I41T substitution may not directly affect the catalytic activity of the Sac3, but may affect the local folding or the stability of the protein. In agreement with this prediction, yeast Fig4 carrying with a corresponding I59T mutation was able to produce normal levels of PI(3,5)P2 and corrected the vacuolar enlargement phenotype when this mutant was expressed in fig4Δ cells (Chow et al, 2007). Two other mutations, D48G and D53Y, which have been predicted to be responsible for a subset of ALS disease, are located at the two ends of the β4 strand (Figure 6 inset). These two mutations are likely to affect protein folding or stability because both of these residues are surface exposed and located in flexible loop regions. Interestingly, these two residues, together with I41, are clustered in a surface area on the top of the SacN domain. This finding indicates that the SacN domain may be involved in protein–protein interaction through a surface area on the first layer of the β sheet. Human disease mutations in Sac3 may displace the binding with other proteins, which may cause defective regulation on its enzymatic activity. The last two mutations, R388G and I411V, are located in the catalytic domain. The R388G is in the close vicinity of the catalytic P-loop, thus it may have a direct impact on its enzymatic function. The I441V is away from the catalytic P-loop and on the opposite side of the catalytic domain. Although I441 forms part of a hydrophobic core, substitution by a hydrophobic valine residue at this position may not be deleterious. Consistent with this prediction, the I441V variant has an activity close to the wild type (Chow et al, 2009).
Figure 6.
Homology structural modelling and missense disease mutations in human Sac3. The modelled Sac domain of Sac3 has a similar two-domain structure: the SacN domain (blue) and the catalytic domain (yellow). The catalytic P-loop is coloured red. Genetic mutations that have been implicated in CMT4J or ALS are labelled and illustrated with sticks. The inset is a zoomed view of a region with a clustering of three mutations in the SacN domain.
Discussion
The structure of the Sac domain of yeast Sac1, the first atomic structure of the Sac PI phosphatase family, shows that the Sac domain contains two modules, the SacN and the catalytic sub-domains. Sequence and structure homology analysis suggests that the SacN domain has a unique sequence and a novel structural fold, both of which are only conserved in the Sac phosphatase family. Interestingly, several disease-related mutations found in Sac3/Fig4 form a cluster on the SacN domain, indicating that the SacN domain may mediate interactions with other unknown factors. Recent results also show that mammalian Sac1 orthologs contain a leucine zipper motif within the SacN domain region and that this motif is required for Sac1 oligomerization. The predicted leucine zipper region is not conserved in yeast Sac1. However, this region partially overlaps with the α1 helix in the SacN domain (Figure 2). It has been further shown that the leucine zipper-dependent oligomerization of Sac1 is required for its shuttling to the Golgi complex (Blagoveshchenskaya et al, 2008; Blagoveshchenskaya and Mayinger, 2009). These results suggest that the regulatory function of the SacN domain might also be mediated by Sac1 oligomerization and translocation.
The CX5R(T/S) catalytic motif of Sac1 is located between the C-terminus of a β-strand and the first turn of an α-helix. Despite the sequence variability of the X5 segment, the conformation of the P-loop is strictly superimposable among different PTPs. Strikingly, seven successive main chain amide groups in this loop all face inward, together with the guanidinium group of the CX5R(T/S) arginine, to coordinate the phosphate group of the substrate in place for nucleophilic attack by the Sγ atom of the catalytic cysteine (Figure 3B and C). Surprisingly, the conformation of the P-loop of the catalytic domain of Sac1 is different from all known PTP structures. These dissimilarities between Sac and other phosphatases suggest that the Sac domain family phosphatases use an unusual mechanism for PI dephosphorylation. However, it may also be possible that the structure we crystallized is in an inactive conformation; it requires a conformational change that flips the P-loop into the consensus P-loop confirmation for its full activity. A complex structure of Sac1 with its substrate analogues would resolve this ambiguity. We have intensively tried the co-crystallization of wild-type or catalytically dead C392S mutant Sac domains with inositol head groups [Ins(1,4)P2 and Ins(1,3)P2], but without success so far. Further structural studies are warranted for the delineation of the catalytic mechanisms and substrate specificity determinants of Sac phosphatases.
As the substrates of lipid phosphatases are embedded in membrane bilayers, Sac1 must bind to the membrane–water interface to hydrolyse its substrates on membranes. Unlike enzymatic reactions that occur in solution, interfacial enzymatic reactions have their own unique kinetic features. The enzyme first needs to be adsorbed to the lipid interface for activation. Once the enzyme is tightly bound to the membrane surface, it may catalyse the reaction with high processivity in a so called ‘scooting' mode (Jain et al, 1986). Under the ‘scooting' model, the enzyme hydrolyses many PI lipids without dissociating from the membrane surface. The crystal structure of Sac1 shows that the catalytic P-loop is surrounded by five flexible loops. Interestingly, some of these loops are enriched with cationic and hydrophobic residues, the physicochemical nature of which is ideal to support interfacial membrane binding (Gelb et al, 1999; Cho and Stahelin, 2005). The structure further shows a deep long groove on the catalytic surface of Sac1 (Figure 4B and C). This groove has its both ends open, which makes it possible for the enzyme to load its substrates from one end of the groove to the catalytic site for hydrolysis and to release the products through the other opening of the groove while the Sac domain remains ‘scooting' on the membrane. Thus, Sac1 may function in an efficient processive manner at the membrane–water interface.
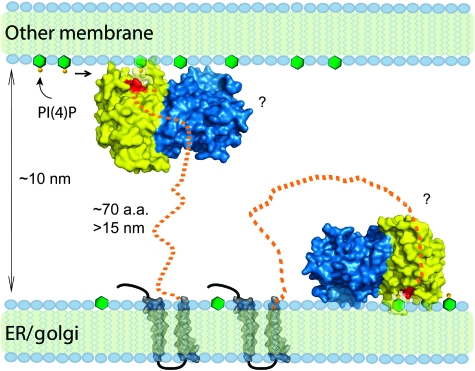
Sac1 is a type II membrane protein, which localizes to both the ER and Golgi apparatus (Whitters et al, 1993; Nemoto et al, 2000). A question that remains is whether its substrate is localized on the same membrane where Sac1 resides (cis) or on a different membrane (trans). Studies in yeast have shown that PI(4)P is concentrated on the Golgi and the plasma membrane. Inactivation of Sac1p leads to a specific increase in the cellular level of PI(4)P and the bulk of accumulated PI(4)P is generated by the phosphoinositol-4 kinase Stt4 at the plasma membrane (Foti et al, 2001; Tahirovic et al, 2005). These findings suggest that Sac1 has the ability to hydrolyse its substrate in trans. Thus, Sac1, and hence the ER, must be in close proximity to the plasma membrane. Indeed, ER membranes in close proximity to the PM (<100 Å) are often seen in ER images of both yeast and mammalian cells. In the Sac domain structure, there is a peptide stretch of 50 residues (residues 453–503) at the C-terminus lacking any electron density, suggesting a flexible nature of this region. Given an additional nearly 20-residue long spacer (residues 504–522, between the Sac domain and the first trans-membrane motif) that presumably also adopts a linear unfolded state, the Sac domain could extend far away from the membrane (a completely extended peptide of about 70 residues can reach a distance of 20 nm). This structural feature of Sac1 certainly allows it to overcome the space restriction to act in trans on the pool of PI(4)P generated by Stt4 at the plasma membrane, although we cannot rule out that Sac1 may also function in a cis mode.
The structural properties of Sac1 led us to propose a membrane interaction model for this PI phosphatase (Figure 7). In this model, the cationic surface mediates membrane association. On this surface, the catalytic P-loop is surrounded by several flexible loops that are enriched with cationic and hydrophobic residues. Once the Sac domain gets adsorbed to membrane bilayer, the deep open groove at the catalytic site would allow Sac1 to hydrolyse PIs in a processive manner. The long linker region between the catalytic domain and the first trans-membrane motif allows the ER-anchored Sac1 phosphatase to reach its substrates embedded in the plasma membrane.
Figure 7.
Functional model of Sac1 at the membrane interface. The Sac domain of Sac1 is shown by molecular surfaces coloured cyan (the SacN sub-domain) and yellow (the catalytic sub-domain). The catalytic site P-loop is coloured red. The polarized charge distribution controls the membrane docking of the Sac domain by orienting the catalytic site to its membrane-embedded substrates. Several loops surrounding the catalytic site may directly interact or insert into the lipid bilayer and may facilitate the processive hydrolysis of PIs. The long flexible linker between the catalytic domain and the membrane anchors allows Sac1 to hydrolyse its substrates either in a trans or cis mode.
In conclusion, the structure of the Sac domain of yeast Sac1 provides the first representative structure of the Sac phosphatase family. The structure shows a large positively charged groove at the catalytic site and a strikingly different configuration of the catalytic P-loop of the Sac domain. These findings suggest a novel catalytic mechanism for Sac1. The structure also provides a three-dimensional framework for the understanding of disease mechanisms of the missense mutations found in human Sac3. Further structural and functional studies of enzyme substrate complexes and of other Sac family members will lead to a more complete understanding of the structural determinants for substrate recognition and specificity.
Materials and methods
Protein expression and purification
The DNA sequence encoding a variety portions of the cytoplasmic Sac domain of wild-type yeast Sac1 were PCR amplified and subcloned into a pET28-based bacterial expression vector in frame with an N-terminal His–sumo tag. The constructs were confirmed by DNA sequencing. Recombinant proteins were overexpressed in Escherichia coli BL21 cells. The selenomethionine substituted proteins were expressed in M9 minimum media supplied with Drop-out mix synthetic minus methionine powder (USBiological) and selenomethionine powder was added on induction. Recombinant proteins were purified with metal affinity beads (Clontech). The His–sumo tag was removed by the sumo-specific protease Ulp1 during overnight dialysis in a buffer of 20 mM hepes, pH 7.3, 20 mM NaCl, 5 mM β-mercaptoethanol. The cutting mixture was further purified by Hi-trap Q column followed by gel filtration. The peak corresponding to Sac1 was pooled and concentrated to 10 mg/ml in a buffer containing 20 mM Hepes, pH 7.3, 20 mM NaCl, 10 mM DTT.
Crystallization and preliminary X-ray crystallographic analysis
Crystallization trials were set up with a PHENIX liquid handling system (Art Robbins Instruments). The construct containing residues 1–503 of ySac1 yielded crystals and was thus pursued for further structural analysis. Sac1 crystals used for native data set were grown by hanging drop vapour diffusion at 4°C by mixing protein (10 mg/ml) with reservoir solution containing 1.5 M NaCl, 0.1 M Na Citrate pH 7.5, 0.45 M Na citrate in 1:1 ratio (v/v). Crystals were quite sensitive to pH and obtained in the pH range of 5.5–5.9. SeMet-Sac1 crystals were grown under similar conditions. Crystals were transferred into the same solution supplemented with 20% glycerol before cooling to 100 K in a nitrogen stream. The crystal diffracted up to 1.97 Å at the Cornell synchrotron light source, MacCHESS beam line A1. The crystal belongs to space group I222 with a=86.16 Å, b=94.7 Å 3, c=155.44 Å, α=β=γ=90 with the calculated Matthews coefficient Vm=2.88 and with 57.3% of solvent in the crystal and one protein molecule in an asymmetric unit (Matthews, 1968). All the X-ray diffraction data were processed by using hkl2000 (Otwinowski and Minor, 1997).
Structure determination and refinement
A complete three wavelength data set was collected at wavelengths around the selenium K edge. However, because of crystal decay during data collection, only the data collected at the peak wavelength (0.9794 Å) was used for phasing. The initial phase was calculated by SAD method using the program HKL2MAP (Pape and Schneider, 2004). As there are only a total of four methionine sites (including the NH2-terminal starting codon) in the 503-residue Sac1 domain, the anomalous signal contributed by selenium atoms was relatively weak and the initial electron density is not of quality expected for a 2.2 Å resolution anomalous data set. The ab initio protein model was built manually in Xtalview (McRee, 1999) with the aid of automatic modelling building by using program ARP/wARP 7.0 (Perrakis et al, 1999) in the CCP4 program suite (Collaborative Computational Project, 1994). The model was completed by iterative model building with COOT (Emsley and Cowtan, 2004) refinement with CNS (Brunger et al, 1998). The amino acids beyond 456 were not visible in electron density maps and presumed to be disordered. The final Sac1 structure consists of 1–456 amino acids with excellent stereochemistry and good crystallographic statistics (Supplementary Table 1).
Homology structure modelling
For Sac3 homology modelling, the Sac1 crystal structure was used as the template for modelling. Secondary structure prediction on the Sac3 sequence was performed using the PSIPRED program (McGuffin et al, 2000). The CLUSATLW2 program (Labarga et al, 2007) was used to get the best sequence alignment between Sac3 and Sac1 using secondary structures as constraints. The Sac3 homology model was built using the computational Modeler 9V4 program (Sali et al, 2008). The Sac3 structure was further refined and validated using Ramachandran plot analysis indicating no residues in disallowed regions.
Yeast in vivo experiments and PI analysis
SAC1 was cloned into pGOGFP N-terminal GFP vector (Odorizzi et al, 1998) with BglII and Sal1. Point mutants were constructed from the pGOGFP-SAC1 template by site-directed mutagenesis and confirmed by sequencing. The resultant constructs were transformed into sac1Δ yeast cells (strain MFY62) (Foti et al, 2001). For the cold sensitive growth assay, transformants were grown to mid-log (OD600=0.5), adjusted to 1 OD/ml, serially diluted 1:10 3 times, spotted onto selection plates, and left at 15°C for 6 days. For western blot, 5 OD600 of midlog cells were collected, precipitated in 10% trichloroacetic acid, washed 2 × in acetone, and bead beated in sample buffer; 1 OD600 equivalent of the samples was run on an 8% SDS PAGE and immunoblotted against GFP (Santa Cruz Biotechnology sc-9996) and G6PDH (Sigma A-0251). Intracellular PI level analysis was performed as described earlier (Botelho et al, 2008). Briefly, cells transformed with control empty vector or vector harbouring wild type or mutant Sac1 were grown to log phase. Cells (5 OD600) were collected, washed in media lacking inositol, and labelled with myo-[2-3H] inositol for 1 h. Lipids were deacylated, glycerophospholipids were extracted, and 107 c.p.m. of samples were separated by HPLC.
Accession code
The coordinates and structure factors have been deposited in the RCSB with the accession code 3LWT.
Supplementary Material
Acknowledgments
We thank Dr Anthony Bretscher, Linda Nicholson and Volker Vogt for critical discussion and suggestions, Adam Brady for comments on the paper. This work is supported by the start up fund for YM from Weill Institute for Cell and Molecular Biology, Cornell University. The X-ray data were collected at MacChess beamline A1 and F2. CHESS is supported by the NSF & NIH/NIGMS via NSF award DMR-0225180, and the MacCHESS resource is supported by NIH/NCRR award RR-01646.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bankaitis VA, Aitken JR, Cleves AE, Dowhan W (1990) An essential role for a phospholipid transfer protein in yeast Golgi function. Nature 347: 561–562 [DOI] [PubMed] [Google Scholar]
- Barford D, Das AK, Egloff MP (1998) The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct 27: 133–164 [DOI] [PubMed] [Google Scholar]
- Barford D, Flint AJ, Tonks NK (1994) Crystal structure of human protein tyrosine phosphatase 1B. Science 263: 1397–1404 [PubMed] [Google Scholar]
- Barton GJ (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng 6: 37–40 [DOI] [PubMed] [Google Scholar]
- Begley MJ, Taylor GS, Kim SA, Veine DM, Dixon JE, Stuckey JA (2003) Crystal structure of a phosphoinositide phosphatase, MTMR2: insights into myotubular myopathy and Charcot-Marie-Tooth syndrome. Mol Cell 12: 1391–1402 [DOI] [PubMed] [Google Scholar]
- Blagoveshchenskaya A, Cheong FY, Rohde HM, Glover G, Knodler A, Nicolson T, Boehmelt G, Mayinger P (2008) Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. J Cell Biol 180: 803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoveshchenskaya A, Mayinger P (2009) SAC1 lipid phosphatase and growth control of the secretory pathway. Mol Biosyst 5: 36–42 [DOI] [PubMed] [Google Scholar]
- Botelho RJ, Efe JA, Teis D, Emr SD (2008) Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol Biol Cell 19: 4273–4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brice SE, Alford CW, Cowart LA (2009) Modulation of sphingolipid metabolism by the phosphatidylinositol-4-phosphate phosphatase Sac1p through regulation of phosphatidylinositol in Saccharomyces cerevisiae. J Biol Chem 284: 7588–7596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Cho W, Stahelin RV (2005) Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct 34: 119–151 [DOI] [PubMed] [Google Scholar]
- Chow CY, Landers JE, Bergren SK, Sapp PC, Grant AE, Jones JM, Everett L, Lenk GM, McKenna-Yasek DM, Weisman LS, Figlewicz D, Brown RH, Meisler MH (2009) Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet 84: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, Szigeti K, Shy ME, Li J, Zhang X, Lupski JR, Weisman LS, Meisler MH (2007) Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature 448: 68–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves AE, Novick PJ, Bankaitis VA (1989) Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J Cell Biol 109: 2939–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project N (1994) The CCP4 suite: programs for protein crystallography. Acta Cryst D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- De Matteis MA, Godi A (2004) PI-loting membrane traffic. Nat Cell Biol 6: 487–492 [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P (2006) Phosphoinositids in cell regulation and membrane dynamics. Nature 443: 651–657 [DOI] [PubMed] [Google Scholar]
- DiNitto JP, Lambright DG (2006) Membrane and juxtamembrane targeting by PH and PTB domains. Biochim Biophys Acta 1761: 850–867 [DOI] [PubMed] [Google Scholar]
- Duex JE, Nau JJ, Kauffman EJ, Weisman LS (2006a) Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot Cell 5: 723–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex JE, Tang F, Weisman LS (2006b) The Vac14p-Fig4p complex acts independently of Vac7p and couples PI3,5P2 synthesis and turnover. J Cell Biol 172: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Erdman S, Lin L, Malczynski M, Snyder M (1998) Pheromone-regulated genes required for yeast mating differentiation. J Cell Biol 140: 461–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB (1994) Crystal structure at 2.2 A resolution of the pleckstrin homology domain from human dynamin. Cell 79: 199–209 [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB (1995) Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell 83: 1037–1046 [DOI] [PubMed] [Google Scholar]
- Foti M, Audhya A, Emr SD (2001) Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell 12: 2396–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb MH, Cho W, Wilton DC (1999) Interfacial binding of secreted phospholipases A(2): more than electrostatics and a major role for tryptophan. Curr Opin Struct Biol 9: 428–432 [DOI] [PubMed] [Google Scholar]
- Guo S, Stolz LE, Lemrow SM, York JD (1999) SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem 274: 12990–12995 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1995) Dali: a network tool for protein structure comparison. Trends Biochem Sci 20: 478–480 [DOI] [PubMed] [Google Scholar]
- Hughes WE, Cooke FT, Parker PJ (2000a) Sac phosphatase domain proteins. Biochem J 350: 337–352 [PMC free article] [PubMed] [Google Scholar]
- Hughes WE, Pocklington MJ, Orr E, Paddon CJ (1999) Mutations in the Saccharomyces cerevisiae gene SAC1 cause multiple drug sensitivity. Yeast 15: 1111–1124 [DOI] [PubMed] [Google Scholar]
- Hughes WE, Woscholski R, Cooke FT, Patrick RS, Dove SK, McDonald NQ, Parker PJ (2000b) SAC1 encodes a regulated lipid phosphoinositide phosphatase, defects in which can be suppressed by the homologous Inp52p and Inp53p phosphatases. J Biol Chem 275: 801–808 [DOI] [PubMed] [Google Scholar]
- Jain MK, Rogers J, Jahagirdar DV, Marecek JF, Ramirez F (1986) Kinetics of interfacial catalysis by phospholipase A2 in intravesicle scooting mode, and heterofusion of anionic and zwitterionic vesicles. Biochim Biophys Acta 860: 435–447 [DOI] [PubMed] [Google Scholar]
- Jeong DG, Kim SJ, Kim JH, Son JH, Park MR, Lim SM, Yoon TS, Ryu SE (2005) Trimeric structure of PRL-1 phosphatase reveals an active enzyme conformation and regulation mechanisms. J Mol Biol 345: 401–413 [DOI] [PubMed] [Google Scholar]
- Jin N, Chow CY, Liu L, Zolov SN, Bronson R, Davisson M, Petersen JL, Zhang Y, Park S, Duex JE, Goldowitz D, Meisler MH, Weisman LS (2008) VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J 27: 3221–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns BG, McGee TP, Mayinger P, Gedvilaite A, Phillips SE, Kagiwada S, Bankaitis VA (1997) Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature 387: 101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarga A, Valentin F, Anderson M, Lopez R (2007) Web services at the European bioinformatics institute. Nucleic Acids Res 35: W6–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP (1999) Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell 99: 323–334 [DOI] [PubMed] [Google Scholar]
- Liu Y, Boukhelifa M, Tribble E, Bankaitis VA (2009) Functional studies of the mammalian Sac1 phosphoinositide phosphatase. Adv Enzyme Regul 49: 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Boukhelifa M, Tribble E, Morin-Kensicki E, Uetrecht A, Bear JE, Bankaitis VA (2008) The Sac1 phosphoinositide phosphatase regulates Golgi membrane morphology and mitotic spindle organization in mammals. Mol Biol Cell 19: 3080–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BW (1968) Solvent content of protein crystals. J Mol Biol 33: 491–497 [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16: 404–405 [DOI] [PubMed] [Google Scholar]
- McRee DE (1999) XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J Struct Biol 125: 156–165 [DOI] [PubMed] [Google Scholar]
- Nemoto Y, Kearns BG, Wenk MR, Chen H, Mori K, Alb JG Jr, De Camilli P, Bankaitis VA (2000) Functional characterization of a mammalian Sac1 and mutants exhibiting substrate-specific defects in phosphoinositide phosphatase activity. J Biol Chem 275: 34293–34305 [DOI] [PubMed] [Google Scholar]
- Novick P, Osmond BC, Botstein D (1989) Suppressors of yeast actin mutations. Genetics 121: 659–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD (1998) Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95: 847–858 [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD (2000) Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem Sci 25: 229–235 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pape T, Schneider TR (2004) HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J Appl Crystallogr 37: 843–844 [Google Scholar]
- Perrakis A, Morris R, Lamzin VS (1999) Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6: 458–463 [DOI] [PubMed] [Google Scholar]
- Puius YA, Zhao Y, Sullivan M, Lawrence DS, Almo SC, Zhang ZY (1997) Identification of a second aryl phosphate-binding site in protein-tyrosine phosphatase 1B: a paradigm for inhibitor design. Proc Natl Acad Sci USA 94: 13420–13425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MP, Kearns BG, Xie Z, Guo S, Sekar MC, Hosaka K, Kagiwada S, York JD, Bankaitis VA (1999) Pleiotropic alterations in lipid metabolism in yeast sac1 mutants: relationship to ‘bypass Sec14p' and inositol auxotrophy. Mol Biol Cell 10: 2235–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde HM, Cheong FY, Konrad G, Paiha K, Mayinger P, Boehmelt G (2003) The human phosphatidylinositol phosphatase SAC1 interacts with the coatomer I complex. J Biol Chem 278: 52689–52699 [DOI] [PubMed] [Google Scholar]
- Rudge SA, Anderson DM, Emr SD (2004) Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol Biol Cell 15: 24–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Webb B, Madhusudhan MS, Shen M-Y, Marti-Renom MA, Eswar N, Alber F, Topf M, Oliva B, Fiser A, Snchez R, Yerkovich B, Badretdinov A, Melo F, Overington JP, Feyfant E (2008) MODELLER A Program for Protein Structure Modeling, Version 9v4. San Francisco, CA: University of California [Google Scholar]
- Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D (2003) Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature 423: 769–773 [DOI] [PubMed] [Google Scholar]
- Sbrissa D, Ikonomov OC, Fenner H, Shisheva A (2008) ArPIKfyve homomeric and heteromeric interactions scaffold PIKfyve and Sac3 in a complex to promote PIKfyve activity and functionality. J Mol Biol 384: 766–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorr M, Then A, Tahirovic S, Hug N, Mayinger P (2001) The phosphoinositide phosphatase Sac1p controls trafficking of the yeast Chs3p chitin synthase. Curr Biol 11: 1421–1426 [DOI] [PubMed] [Google Scholar]
- Strahl T, Thorner J (2007) Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim Biophys Acta 1771: 353–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey JA, Schubert HL, Fauman EB, Zhang ZY, Dixon JE, Saper MA (1994) Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 A and the complex with tungstate. Nature 370: 571–575 [DOI] [PubMed] [Google Scholar]
- Tabernero L, Aricescu AR, Jones EY, Szedlacsek SE (2008) Protein tyrosine phosphatases: structure-function relationships. FEBS J 275: 867–882 [DOI] [PubMed] [Google Scholar]
- Tahirovic S, Schorr M, Mayinger P (2005) Regulation of intracellular phosphatidylinositol-4-phosphate by the Sac1 lipid phosphatase. Traffic 6: 116–130 [DOI] [PubMed] [Google Scholar]
- van Montfort RL, Congreve M, Tisi D, Carr R, Jhoti H (2003) Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature 423: 773–777 [DOI] [PubMed] [Google Scholar]
- Wei HC, Sanny J, Shu H, Baillie DL, Brill JA, Price JV, Harden N (2003) The Sac1 lipid phosphatase regulates cell shape change and the JNK cascade during dorsal closure in Drosophila. Curr Biol 13: 1882–1887 [DOI] [PubMed] [Google Scholar]
- Whitters EA, Cleves AE, McGee TP, Skinner HB, Bankaitis VA (1993) SAC1p is an integral membrane protein that influences the cellular requirement for phospholipid transfer protein function and inositol in yeast. J Cell Biol 122: 79–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Groen A, Lemeer S, Jans A, Slijper M, Roe SM, den Hertog J, Barford D (2007) Reversible oxidation of the membrane distal domain of receptor PTPalpha is mediated by a cyclic sulfenamide. Biochemistry 46: 709–719 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.