Abstract
Collagen IV networks are ancient proteins of basement membranes that underlie epithelia in metazoa from sponge to human. The networks provide structural integrity to tissues and serve as ligands for integrin cell-surface receptors. They are assembled by oligomerization of triple-helical protomers and are covalently cross-linked, a key reinforcement that stabilizes networks. We used Fourier-transform ion cyclotron resonance mass spectrometry and nuclear magnetic resonance spectroscopy to show that a sulfilimine bond (-S=N-) crosslinks hydroxylysine-211 and methionine-93 of adjoining protomers, a bond not previously found in biomolecules. This bond, the nitrogen analog of a sulfoxide, appears to have arisen at the divergence of sponge and cnidaria, an adaptation of the extracellular matrix in response to mechanical stress in metazoan evolution.
Collagen IV networks are ancient proteins of basement membranes, a specialized form of extracellular matrix, that underlie epithelia in metazoa from sponge to human. The networks confer structural integrity to tissues, serve as scaffolds for the assembly of other macromolecular components, and serve as ligands for integrin cell-surface receptors that mediate cell adhesion, migration, growth and differentiation (1-3). The networks participate in signaling events in Drosophila development, in the clustering of receptors in the development of mammalian neuromuscular junction (4), and they are involved in autoimmune and genetic diseases (5-7). The networks are assembled by oligomerization of triple-helical protomers by end-to-end associations and by intertwining of triple helices (8, 9). At the C-terminus, two protomers associate through their trimeric non-collagenous (NC1) domains forming a hexamer structure. The protomer-protomer interface is covalently cross-linked, a key reinforcement that strengthens the structural integrity of networks. In the case of humans, the cross-link also confers immune privilege to the collagen IV antigen of Goodpasture autoimmune disease (10, 11).
The chemical nature of these cross-links has been the subject of numerous investigations for two decades; yet, the identity of the covalent bond has remained unknown. Initially, the cross-links were identified as disulfide bonds (12), which were subsequently ruled out by the x-ray crystal structure of NC1 hexamers (13, 14). Electron density maps suggested connectivity between methionine-93 (Met93) and lysine-211 (Lys211) at the interface of adjoining protomers (14); however, the connectivity is gradually degraded by x-rays, rendering precise characterization a challenge for structural analysis by crystallography (15, 16). Using mass spectrometry (MS) analyses of crosslinked tryptic (Tp) peptides and a smaller crosslinked post-proline endopeptidase (PPE) peptides, both derived from the α1α2α1 collagen IV network of placenta, we found that Lys211 is modified to hydroxylysine (Hyl211) and that Hyl211 is covalently linked to Met93 forming a S-hydroxylysyl-methionine (sHM) cross-link (17). In the α3α4α5 network (10), we found that the sHM crosslink connects the α3 and α5 NC1 domains, but the α4 NC1 domains are connected by a S-lysyl-methionine cross-link, involving Lys211 instead of Hyl211, indicating that this post-translational modification is not a requirement for cross-link formation (10). The nature of the bond linking Met93 and Hyl211 could not be determined at that time because the observed difference of one mass unit between the uncross-linked and cross-linked peptides fell within experimental error.
Herein, we deduce the chemical nature of the bond using Fourier transform ion cyclotron resonance (FTICR) (18), which can achieve very high mass accuracy (e.g. < 2 ppm, approximately ±0.001 mass units for a peptide with a mass of ~5,000), and NMR spectroscopy to analyze cross-linked Tp-peptides, derived from the α1NC1-α1NC1 dimer. Table 1 shows the mass values for the multiple-charge states (+4, +5, and +6) of the cross-linked Tp-peptide. For each charge state, ten consecutive scans were averaged to obtain the observed monoisotopic mass value of 5010.471 ± 0.022 (Table 1’). This value is 2.017 mass units lower than the theoretical total mass (5012.486) of the two constituent tryptic peptides, a Met93-containing peptide (T-3599.688) and a Hyl211-containing peptide (T-1412.777), which were previously described (17).
Table 1.
Mass values for the cross-linked Tp-peptides from the α1NC1-α1NC1 dimer
| Z | Theoretical mass |
Observed ion (m/z) |
Observed peptide mass |
Difference (Theo – Obs) |
|---|---|---|---|---|
| +4 | 1253.6204 | 5010.473 ± 0.008 | ||
| +5 | 1003.1014 | 5010.483 ± 0.022 | ||
| +6 | 836.0806 | 5010.459 ± 0.009 | ||
|
| ||||
| 5012.488 | 5010.471 ± 0.022* | 2.017 | ||
The average of the observed peptide mass is indicated with an asterisk
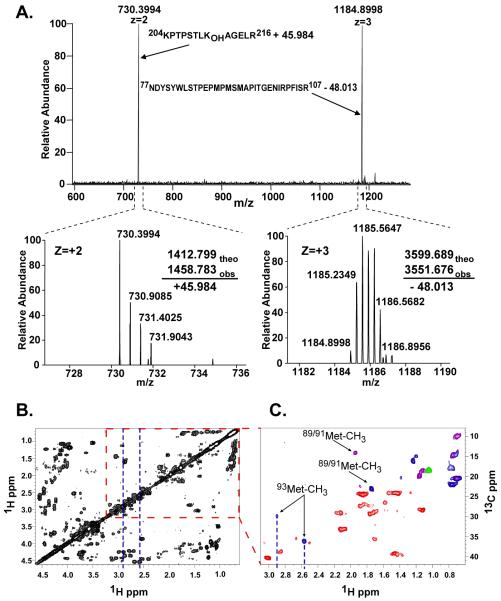
To determine the location of modifications that lead to the loss of 2.017 mass units, the tryptic complex was analyzed by collision-induced dissociation (CID) MS3 fragmentation analysis. The mass-to-charge ratio m/z of 1003.1014 (+5) ion was selected for MS2 fragmentation, which generated a m/z 730.399 ion, corresponding to the T-1412.799 peptide plus 45.984 mass units and a m/z 1184.900 ion, corresponding to the T-3599.689 peptide that lost 48.013 mass units (Fig. 1A). To locate these mass changes to specific residues, the m/z 730.3994 and 1184.900 ions were selected for further CID (MS3). The y- and b-series of the spectra confirm not only the sequence of the T-1412.799 peptide, but also that the location of the mass change of +45.984 corresponds to the side chain of Hyl211 (Fig. S1A). The MS3 fragmentation profile for the m/z 1184.900 ion also verified the peptide sequence, and localized the loss of 48.013 mass units to Met91 or Met93 (Fig. S1B). A smaller cross-linked PPE-peptide complex derived from the cross-linked Tp-peptides (Fig. S2) confirmed that the loss of 48.013 mass units is localized to the side chain of Met93. An analogous fragmentation has been observed in methionine sulfoxide containing peptides that undergo concomitant neutral loss of methane sulfenic acid (CH3SOH) (19, 20). In this case, however, this group remains attached to the side chain of Hyl211, demonstrating the covalent nature of the interaction between Met93 and Hyl211. These findings for the crosslink in the α1NC1-α1NC1 dimer are identical to that for the α2NC1-α2NC1 dimer (Fig. S3)
Fig. 1.
Mass spectrometric and NMR analyses of the cross-linked Tp-peptides from the α1NC1-α1NC1 dimer. A) MS2 spectrum showing the fragmentation by collision-induced dissociation of the m/z 1003.1014 (+5) ion (Table 1). The bottom panel shows the isotopic envelope for each ion and the mass difference of each fragment with respect to the uncross-linked peptides. B) NMR studies showing the 1H-1H correlated spectroscopy COSY spectrum in 50 mM phosphate buffer, pH=7.0, 20°C. C) Edited 1H-13C HSQC spectrum of an expanded portion of panel B in which the methyl groups of Val, Leu, Thr, Iso, and Met are expected. Correlation peaks are color-coded according to multiplicity edited HSQC spectra, optimized for correlation selection of CH2 (red), CH/CH3 (blue) and CH (green) only. Several peaks contain overlapping signals from CH3 and CH groups are indicated in purple.
An overlay of correlation spectroscopy (COSY) and heteronuclear multiple quantum correlation (1H-13C HMQC) spectra of the cross-linked Tp-peptides was used to evaluate the chemical shift of the methyl group of Met93 (Fig 1B and Fig S4A). The sequences of cross-linked Tp-peptides have a total of 25 methyl groups, three of which belong to Met89, Met91, and Met93. Typical proton chemical shift for methyl groups in peptides of Leu, Thr, Iso, and Ala is between 0 and 1.5 ppm, and that of Met is about 2.1 ppm. Eighteen out of 22 total methyl groups of Leu, Thr, Ile, and Ala were identified within the expected chemical shift range in the HMQC spectrum (Fig S4B). An edited HSQC analysis was carried out to identify the methyl groups of Met89, Met91, and Met93. In the chemical shift range expected for Met, two signals were observed; one at 1H 1.9 ppm/13C 14 ppm and the other at 1H 1.7/13C 24 ppm, presumably correspond to Met89 and/or Met91, (Fig 1C). In addition, two downfield-shifted methyl resonances were observed that had atypical chemical shifts at 1H 2.9 ppm/13C 30 ppm and 1H 2.6ppm/13C 36 ppm. These features likely correspond to the methyl group of Met93 in two different chemical environments such as cyclic or alternate chiral forms of the crosslink (Fig S5).
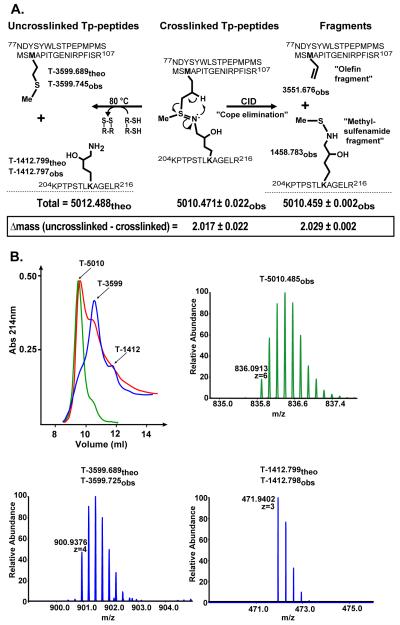
The collective evidence from the MS and NMR indicates the existence of a sulfilimine bond in the cross-linked Tp-peptides (Fig 2A). Mass spectrometry analysis revealed that the linkage between Met93 and Hyl211 is characterized by the loss of two hydrogen atoms, which is consistent with oxidation resulting in a double bond connecting the sulfur atom of Met93 and the nitrogen atom of Hyl211 (Fig 2A). The MS2 fragmentation of the tryptic complex can be explained by a concerted process in which a β-hydrogen is taken up by the nitrogen, which leads to the cleavage of the bond between sulfur and γ-carbon of Met93 (Cope elimination) (21). This syn elimination is also observed in the fragmentation of methionine sulfoxide and is analogous to the Cope elimination of amine oxides. The resulting products, an olefin fragment and a methylsulfenamide fragment, are shown in Fig 2A. The sulfilimine structure is further supported by a down-shifted methyl peak in the NMR spectrum of the cross-linked Tp-peptides, which is consistent with the shift of the methyl group of S-methylisothiazolidinium nitrate (1H 2.78 ppm).
Fig. 2.
Summary of the MS analyses of the crosslinked Tp-peptides before and after reduction with DTT. A) The uncross-linked tryptic peptides, T-3599.689 and T-1412.799, derived from the α1-NC1 domain, display the side chains of Met93 and Hyl211, respectively. T-5012.488 corresponds to the total theoretical mass of both peptides. The sulfilimine double bond crosslinking the tryptic peptides is shown. The difference between the theoretical (theo) and observed (obs) mass reveals that two hydrogen atoms are lost upon sHM crosslink formation. Fragmentation of the sulfilimine bond by CID produces peptide fragments containing an olefin fragment derived from Met93 and a methylsulfenamide fragment derived from Hyl211 as a result of the “Cope elimination” event in the gas phase. However, chemical reduction with DTT, formally involving addition of 2 H-atoms, severs the sulfilimine link and recovers Met93 and Hyl211, as indicated below. B) Cross-linked Tp-peptides were separated by gel filtration chromatography before (green) and after incubation in 100 mM DTT at room temperature (red) and 80 °C (blue). The arrows indicate the identity of each chromatographic peak as revealed by MS analysis.
Although a sulfilimine bond has not been reported in any native biomolecule, they occur in other molecules (22). They are the nitrogen analog of sulfoxides, S(IV), and are also known as sulfur-nitrogen ylides. Most sulfilimines that have been reported are stabilized by strong electron withdrawing groups on the nitrogen end of the sulfilimine linkage (23). However, cyclic sulfilimines derived from the oxidation of methionine analogs by I2 have been characterized in detail as their isothiazolidinium salts following protonation of the nitrogen (24, 25). These studies showed direct bonding of the nitrogen and sulfur atoms and a new stereogenic center at the sulfur atom, consistent with the two methyl resonances seen by NMR. In the case of collagen IV, sulfilimine crosslinks are likely to be formed through a 2 electron, heteroatom transfer class oxidation (26). For example, the Met93 sulfide could react with a chemical or enzymatic oxidant to form a transient, sulfur-(IV) sulfonium intermediate, which is captured by the Hyl/Lys211 amine to form the sulfilimine. The latter Sn2-like step entails inversion at sulfur as the S-X bond breaks heterolytically, and is analogous to the known oxidation of free methionine to dehydromethionine mediated by iodine (25). This sulfilimine linkage may not occur only in collagen IV but in other proteins as well.
Sulfilimines are reduced by thiols to yield the parent amine and thioether groups (23). Thus, the susceptibility of the cross-linked Tp-peptide to dithiothreitol (DTT) reduction was evaluated (Fig. 2B). Partial reduction was achieved with 100 mM DTT at room temperature and complete reduction at 80 °C at pH 7.8. Mass spectrometry analyses revealed that DTT breaks the cross-link with the concomitant generation of the T-3599 and T-1412 peptides with complete recovery of both Met93 and Hyl211, respectively (Fig 2, A and B). The susceptibility of the cross-link to reduction is comparable to that of disulfide bonds of insulin (Fig S6). Because the cross-linked Tp-peptides do not contain cysteines (Fig. 2A), these results provide further support for the existence of a sulfilimine bond between Hyl211 and Met93.
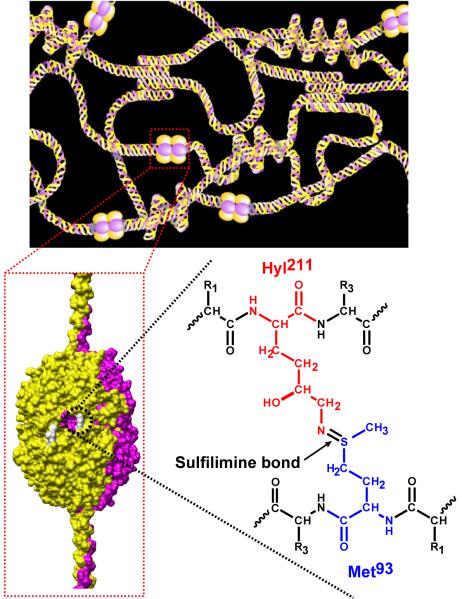
The location of the sulfilimine linkage within the α1α2α1 collagen IV network is shown in Fig 3. Up to six sulfilimine bonds fasten the interface of the trimeric NC1 domains of two adjoining protomers, reinforcing the quaternary structure of the networks. Furthermore, the sulfilimine bond also occurs in the α3α4α5 collagen IV network (Fig S7) because fragmentation pattern of its cross-linked tryptic peptides (10) is identical to that of the α1α2α1 network described herein.
Fig. 3.
Proposed chemical structure of the sHM cross-link. Schematic of the α1α2α1 collagen IV network (top panel) illustrating the interaction between the NC1 domain of triple-helical protomers. A space-filling model of the NC1 hexamer quaternary structure (bottom left) shows the location of the sHM cross-links (white). The sulfilimine bond that constitutes the sHM crosslink connecting the side chains of Met93 and Hyl211 is shown for the α1NC1-α1NC1 dimer. The same bond connects Met93 and Hyl211 in the α2NC1-α2NC1 dimer.
The sulfilimine bond likely occurs in diverse metazoan species. NC1 dimer subunits, a signature structural feature indicative of cross-links, have been identified in collagenase digests of basement membranes including human (27), bovine (27), dog (28), and mouse (27). Furthermore, a phylogenetic analysis of the Lys211 and Met93 residues, based on a multiple sequence alignment of the NC1 domain across the metazoan phylum (Fig. 4 and (29)), revealed that the sulfilimine bond may occur in many metazoans, except in hydra, flatworm, sponge, and placozoa. A further comparison of the sequence motif (X-K-A/S/G) that confers hydroxylation of lysyl residues by lysyl hydroxylase (30) occurs in the NC1 domains of all metazoa except hydra, sponge and placozoa. The motif is also absent in the α4 NC1 domain of human, mouse, bovine and chick, which in the case of bovine Lys211 does not undergo hydroxylation and leads to the formation of sKM cross-link (10). In one species of the phylum cnidaria, Nematostella vectensis, both Met93 and Lys211 and the hydroxylation motif of Lys are conserved (Fig 4), suggesting that the sHM cross-link appeared at the time of the divergence of sponge and cnidaria, an apparent evolutionary adaptation that arose in response to mechanical stress on organisms.
Fig. 4.
Multiple sequence alignment of collagen IV NC1 domain sequences encompassing Met93 and Lys211. Met93 and Lys211 are shown in bold. Conserved amino acid residues are indicated with an asterisk (*). Semi-conserved residues are indicated with a colon (:). The hydroxylation motif for lysyl hydroxylase, X-K-(A/S/G) is shown at the bottom of the alignment. Abbreviations are as follow: Homo sapiens (Hsa; A1, NP_001836.2; A2, NM_001846.2; A3, ABX71213.1; A4, X81053.1; A5, ABW24668.1; A6, AL136080.6), Ciona intestinalis (Cintestinalis, A1, XP_2120982.1; A2, XP_2119477.1), Strongylocentrotus purpuratus (Spu, A2, NP_999676.1; A3, NP_999631.1), Drosophila melanogaster (Dme, A1, AAA28404.1; A2, AAB64082.1), Caenorhabditis elegans (Cel, A1, AAB59179.1; A2, AAA27989.1), Brugia malayi (Bmalayi, XP_1902932.1), Ascaris Suum (Asc_suum, AAA18014.1), Nematostella vectensis (Nvectensis, XP_1626265.1), Hydra magnipapillata (Hydra_mag, XP_2157001.1), Hydra vulgaris (Hydra_vul, AAG40729.1), Schistosoma japonicum (Sjaponcum, AAX25734.2), Trichoplax adhaerens (Tadhaerens, A1, EDV21329.1; A2, EDV21231.1). The alignments were generated with ClustalW.
One sentence summary.
A novel sulfilimine bond was identified as a key reinforcement of an extracellular matrix protein, collagen IV, an apparent adaptation crucial for metazoan evolution.
Supplementary Material
Acknowledgments
We thank D. Liebler for facilitating the initial contact with T. D. Veenstra from SAIC-Frederick, Inc. The technical assistance of P. Todd and Rafi Mohamed is greatly ppreciated. We would also like to thank S. Hill and H. McDonald, from the Proteomics Laboratory at the Mass Spectrometry Research Center of Vanderbilt University, for their assistance with the Orbitrap-LTQ instrument. This work was supported in part by National Institutes of Health Grants DK065123 and DK18381 (to B. G. H.), and the W.M. Keck Foundation (K.B.S.) and the Skaggs Institute for Chemical Biology (K.B.S.).
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full…
Materials and Methods
Figure legends
Figs. S1-S7
References and Notes
- 1.Moser M, Legate KR, Zent R, Fassler R. Science. 2009 May 15;324:895. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- 2.Hynes RO. Cell. 2002 Sep;110:673. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 3.Yurchenco PD, Furthmayr H. Biochemistry. 1984 Apr 10;23:1839. doi: 10.1021/bi00303a040. [DOI] [PubMed] [Google Scholar]
- 4.Fox MA, et al. Cell. 2007 Apr 6;129:179. [Google Scholar]
- 5.Gould DB, et al. N Engl J Med. 2006 Apr 6;354:1489. doi: 10.1056/NEJMoa053727. [DOI] [PubMed] [Google Scholar]
- 6.Gould DB, et al. Science. 2005 May 20;308:1167. [Google Scholar]
- 7.Hudson BG, Tryggvason K, Sundaramoorthy M, Neilson EG. N Engl J Med. 2003 Jun 19;348:2543. doi: 10.1056/NEJMra022296. [DOI] [PubMed] [Google Scholar]
- 8.Khoshnoodi J, Pedchenko V, Hudson BG. Microsc Res Tech. 2008 May;71:357. doi: 10.1002/jemt.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khoshnoodi J, et al. J Biol Chem. 2006 Mar 3;281:6058. doi: 10.1074/jbc.M506555200. [DOI] [PubMed] [Google Scholar]
- 10.Vanacore RM, et al. J Biol Chem. 2008 Aug 15;283:22737. doi: 10.1074/jbc.M803451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borza DB, et al. J Biol Chem. 2005 Jul 22;280:27147. doi: 10.1074/jbc.M504050200. [DOI] [PubMed] [Google Scholar]
- 12.Siebold B, Deutzmann R, Kuhn K. Eur J Biochem. 1988;176:617. doi: 10.1111/j.1432-1033.1988.tb14321.x. [DOI] [PubMed] [Google Scholar]
- 13.Sundaramoorthy M, Meiyappan M, Todd P, Hudson BG. J Biol Chem. 2002 Aug 23;277:31142. doi: 10.1074/jbc.M201740200. [DOI] [PubMed] [Google Scholar]
- 14.Than ME, et al. Proc. Natl. Acad. Sci. (USA) 2002;99:6607. [Google Scholar]
- 15.Than ME, Bourenkov GP, Henrich S, Mann K, Bode W. Biological Chemistry. 2005;386:759. doi: 10.1515/BC.2005.089. [DOI] [PubMed] [Google Scholar]
- 16.Vanacore RM, et al. J Biol Chem. 2004 Oct 22;279:44723. doi: 10.1074/jbc.M406344200. [DOI] [PubMed] [Google Scholar]
- 17.Vanacore RM, Friedman DB, Ham AJ, Sundaramoorthy M, Hudson BG. J Biol Chem. 2005 Jun 10;280:29300. doi: 10.1074/jbc.M502752200. [DOI] [PubMed] [Google Scholar]
- 18.Full description of the materials and methods are available as supporting materials on Science online.
- 19.Lagerwerf FM, van de Weert M, Heerma W, Haverkamp J. Rapid Commun Mass Spectrom. 1996;10:1905. doi: 10.1002/(SICI)1097-0231(199612)10:15<1905::AID-RCM755>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 20.Reid GE, Roberts KD, Kapp EA, Simpson RI. J Proteome Res. 2004 Jul-Aug;3:751. doi: 10.1021/pr0499646. [DOI] [PubMed] [Google Scholar]
- 21.Cope A, Foster T, Towle P. Journal of the American Chemical Society. 1949;71:3929. [Google Scholar]
- 22.Koval IV. Russian Chemical Reviews. 1990 Sep;59:1409. [Google Scholar]
- 23.Gilchrist TL, Moody CJ. Chemical Reviews. 1977;77:409. [Google Scholar]
- 24.Lambeth DO, Swank D. Federation Proceedings. 1979;38:830. [Google Scholar]
- 25.Glass RS, Duchek JR. Journal of the American Chemical Society. 1976;98:965. doi: 10.1021/ja00420a016. [DOI] [PubMed] [Google Scholar]
- 26.Johnson CR, Bacon CC, Kingsbur Wd. Tetrahedron Letters. 1972:501. [Google Scholar]
- 27.Weber S, Engel J, Wiedemann H, Glanville R, Timpl R. Eur J Biochem. 1984 Mar 1;139:401. doi: 10.1111/j.1432-1033.1984.tb08019.x. 1984. [DOI] [PubMed] [Google Scholar]
- 28.Thorner PS, Zheng K, Kalluri R, Jacobs R, Hudson BG. J Biol Chem. 1996;271:13821. doi: 10.1074/jbc.271.23.13821. [DOI] [PubMed] [Google Scholar]
- 29.Aouacheria A, et al. Mol Biol Evol. 2006 Dec;23:2288. doi: 10.1093/molbev/msl100. [DOI] [PubMed] [Google Scholar]
- 30.Kivirikko KI, Pihlajaniemi T. Adv Enzymol Relat Areas Mol Biol. 1998;72:325. doi: 10.1002/9780470123188.ch9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.