Abstract
E1 enzymes activate ubiquitin (Ub) and ubiquitin-like (Ubl) proteins in two steps by carboxy-terminal adenylation and thioester bond formation to a conserved catalytic cysteine in the E1 Cys domain. The structural basis for these intermediates remains unknown. Here we report crystal structures for human SUMO E1 in complex with SUMO adenylate and tetrahedral intermediate analogs at 2.45 Å and 2.6 Å, respectively. These structures show that side chain contacts to ATP·Mg are released after adenylation to facilitate a 130 degree rotation of the Cys domain during thioester bond formation that is accompanied by remodeling of key structural elements including the helix that contains the E1 catalytic cysteine, the cross-over and re-entry loops, and refolding of two helices that are required for adenylation. These changes displace side chains required for adenylation with side chains required for thioester bond formation. Mutational and biochemical analyses suggest these mechanisms are conserved in other E1s.
Keywords: E1, SUMO, adenylation, thioester, x-ray, inhibitor
Post-translational modification by ubiquitin (Ub) and ubiquitin-like (Ubl) proteins such as SUMO and Nedd8 regulate signal transduction pathways that contribute to differentiation, apoptosis, the cell cycle, and response to stress1–4. The enzymes required for Ub/Ubl conjugation are conserved across evolution; E1s activate the Ub/Ubl for transfer to E2 conjugating enzymes; and E2s are combined with a wide array of E3 ligases to promote Ub/Ubl conjugation and to ensure substrate specificity3–6. E1s activate Ub/Ubl proteins in two steps7. E1s utilize ATP and magnesium to adenylate the C-terminal Ub/Ubl glycine, releasing pyrophosphate. The C-terminal Ub/Ubl adenylate is then attacked by a conserved E1 cysteine, resulting in release of AMP and formation of a thioester bond between the C-terminal Ub/Ubl glycine and E1 active site cysteine. E2s are then recruited to the E1 to transfer the E1~Ub/Ubl thioester adduct to a conserved E2 cysteine to form an E2~Ub/Ubl thioester adduct.
Crystal structures of SUMO, Nedd8 and ubiquitin E1s have been determined in complex with their respective Ub/Ubl proteins8–11. These studies revealed that E1s share a similar multi-domain architecture that includes two evolutionarily related adenylation domains that bind ATP·Mg and the respective Ub/Ubl, a C-terminal ubiquitin-fold domain (UFD) that recruits E2s for thioester transfer, and a catalytic Cys domain (Cys) that contains the active site cysteine. The Cys domain is linked to one of the two adenylation domains by a cross-over loop that passes over the C-terminal residues of the Ub/Ubl protein and by a re-entry loop. Although much has been learned from these studies, several pertinent issues remain with respect to E1 function. First, no Ub/Ubl adenylate has been observed in E1s capable of transferring Ub/Ubls to E2s, despite the presence of ATP·Mg and the respective Ub/Ubl. Second, all E1 structures determined thus far reveal the Cys domain and cross-over loop in similar conformations that position the catalytic cysteine more than 30 Å from the presumed site of adenylation.
To capture intermediates formed during Ub/Ubl adenylation and thioester bond formation, we used a chemical approach involving the synthesis of SUMO derivatives that mimic the adenylate intermediate or form a covalent adduct to mimic the tetrahedral intermediate generated during thioester bond formation12. Crystal structures of the E1/SUMO adenylate analog or E1~SUMO tetrahedral intermediate analog were determined at 2.45 Å and 2.6 Å, respectively. These structures revealed that the thioester bond formation half-reaction is accompanied by a 130 degree rotation of the Cys domain and remodeling of several structural elements of key functional significance, including the helix that contains the active site cysteine, the cross-over and re-entry loops, and two helices that comprise part of the adenylation active site. The net result of these conformational changes is replacement of nearly half of the active site residues required for adenylation with residues from the Cys domain that are required for thioester bond formation. Mutational and biochemical analyses reveal that molecular interactions important for achieving these conformational changes during transitions from the substrate complex to adenylate intermediate to tetrahedral intermediate are likely conserved in other E1 enzymes.
E1 intermediate analogs
A non-hydrolyzable mimic of the acyl-adenylate intermediate (AMSN) was made by linking a cysteylglycylglycyl tripeptide to 5′-(sulfamoylaminodeoxy)adenosine (CGG-AMSN)12. To trap a covalent adduct with the E1 active site cysteine, we synthesized a 5′-(vinylsulfonylaminodeoxy)adenosine tripeptide variant (CGG-AVSN)12 containing an electrophilic center at the position predicted to be attacked by the E1 active site cysteine during thioester bond formation. These compounds were then coupled to SUMO or ubiquitin, which lacked the corresponding three C-terminal amino acids, via intein-mediated ligation13 to generate human SUMO1-AMSN and SUMO1-AVSN, S. cerevisiae SMT3-AVSN, and ubiquitin-AVSN (Fig. 1a,b; Methods).
Figure 1. Analogs of the Ub/Ubl-adenylate and E1~Ub/Ubl tetrahedral intermediates.
Chemical structures of a, Ub/Ubl-adenylate (top) and Ub-AMSN adenylate analog12 (bottom), b, the E1~Ub/Ubl tetrahedral intermediate (top) during thioester bond formation, the Ub/Ubl-AVSN adduct12 (middle), and the E1~Ub/Ubl-AVSN tetrahedral intermediate analog (bottom). Red atoms indicate modifications in AMSN and AVSN that deviate from AMP. c, DTT sensitivity of Ub/Ubl thioester and thioether adducts for human and S. cerevisiae SUMO E1s and S. pombe ubiquitin E1. d, Cross-linking assay and time course for SUMO1-AVSN, S. cerevisiae SMT3-AVSN, and S. pombe Ub-AVSN adducts to their cognate E1s. See Methods for assay conditions.
Incubation of SUMO E1 with SUMO1-AVSN resulted in formation of a cross-linked species that migrated on SDS-PAGE at a similar position to the E1~SUMO1 thioester adduct (Fig. 1c,d). Consistent with irreversible formation of a thioether linkage, the E1~SUMO1-AVSN adduct was stable during incubation with the thiol DTT, whereas the E1~SUMO1 thioester adduct was not. Serine substitution of the E1 catalytic cysteine confirmed that cross-linking was dependent on the cysteine12. Incubation of the S. cerevisiae SMT3 E1 with SMT3-AVSN or the ubiquitin E1 with ubiquitin-AVSN resulted in similar cross-linked adducts (Fig. 1d). Cross-link formation between Ub E1 and ubiquitin-AVSN was dependent on its catalytic cysteine (not shown). Similar to the E1~SUMO1-AVSN adduct, the E1~SMT3-AVSN and E1~Ub-AVSN thioether adducts were also resistant to thiolysis by DTT (Fig. 1c). The ability to generate the cross-linked species with several E1s demonstrates that our approach for capturing the presumed tetrahedral intermediate analog may be applicable to other E1s.
Overall structures of the SUMO E1
A structure for E1/SUMO1-AMSN was determined by x-ray crystallography to 2.45 Å and refined to R/Rfree of 0.190/0.249 (Methods; Supplemental Table I). Electron density was evident for the covalent bond between SUMO1 and AMSN, thus, this adduct resembles the adenylate intermediate (Supplemental Fig. 1). This structure shares many overall similarities to structures of the SUMO E1 bound to SUMO1/ATP·Mg9, including the relative conformations of the UFD and Cys domains9 (Fig. 2). For discussion purposes, we term this the ‘open’ conformation. As expected, contacts between amino acid side chains that coordinate the magnesium ion and ATP β-γ phosphates, as observed in E1/SUMO1/ATP·Mg structures, were absent in the E1/SUMO1-AMSN structure. Another notable difference was that E1 amino acids 607–640 of UBA2 were observed in contacts with SUMO1 via a C-terminal SIM motif (aa 632–640; ELDDVIALD; Supplemental Fig. 2) although the functional significance of the SIM remains unclear because amino acids 550–640 are dispensible for human E1 activity in vitro and for yeast E1 function in vivo in S. cerevisiae9.
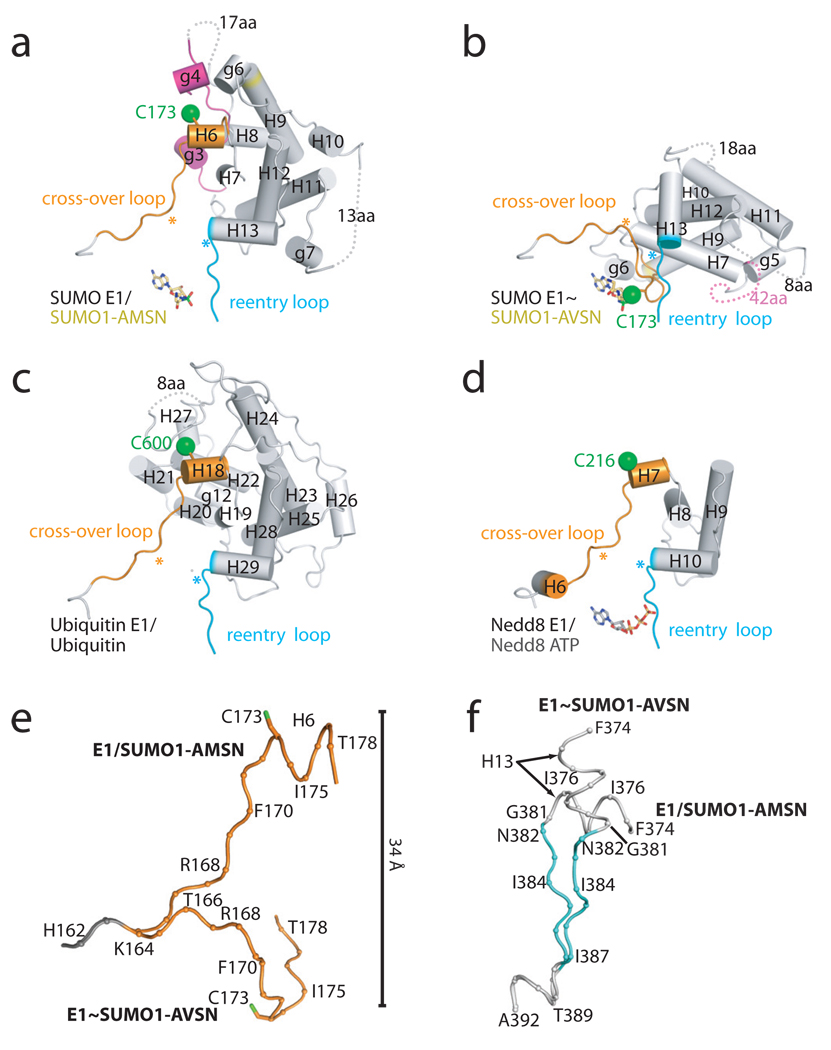
Figure 2. Structural changes in SUMO E1 accompany transitions from adenylate to tetrahedral intermediate.
a, Ribbon representation for the SUMO E1/SUMO1-AMSN adenylate analog (left) and SUMO E1~SUMO1-AVSN tetrahedral intermediate analog (right). Atoms for the catalytic cysteine (Cys173), AMSN and AVSN shown as spheres with E1 domains and SUMO color-coded and labeled. N term, N-terminus. b, Elements in SUMO E1 that undergo conformational changes are color-coded and labeled. N term, N-terminus. Similar regions in E1 structures are colored gray and SUMO1 is colored yellow. c, Cartoon representation of the structures color-coded and labeled as in a and b highlighting elements that undergo remodeling.
A structure for E1~SUMO1-AVSN was determined by x-ray crystallography to 2.6 Å and refined to R/Rfree values of 0.227/0.283 (Methods; Supplemental Table I). Electron density was evident for the covalent bond between the E1 cysteine and the sulfonamide β-carbon of SUMO1-AVSN, which is analogous to the carbonyl carbon of the C-terminal glycine residue in the native SUMO1 adenylate intermediate, thus, this adduct resembles the tetrahedral intermediate (Supplemental Fig. 1). This structure revealed a number of important differences, including a distinct conformation for the Cys domain that is related by a 130 degree rotation and 3 Å translation (center of mass) to that observed in other SUMO E1 structures (Fig. 2; Dyndom14). For discussion purposes, we term this the ‘closed’ conformation. In the open conformation, one Cys domain surface rests upon the SAE1 N-terminal helix and makes few contacts to UBA2, burying a total surface area of 2650 Å2. In the closed conformation, a surface on the opposite side of the Cys domain becomes buried, making fewer contacts to SAE1 but interacting more extensively with UBA2 surfaces (total surface area 3340 Å2). The two interfaces include distinct sets of amino acid residues in the Cys and E1 adenylation domains (Supplemental Fig. 3 and 4).
Rotation of the Cys domain is accompanied by a 125 degree change in the path of the cross-over loop and an orthogonal change in path for the re-entry loop. Analysis of the cross-over loop shows its trajectory is altered over several residues (aa 164–168) while changes in the re-entry loop are localized to Gly381 and Asn382 (Fig. 3; Supplemental Fig. 5). Furthermore, several elements that were structured in the Cys domain in the open conformation become disordered in the closed conformation, including helices g3 and g4 and the loop joining them, the loop between H10 and H11 (g7 is disordered), and the loop between H11 and H12 (Fig. 2c). Perhaps most relevant for thioester bond formation, the H6 helix that contains the active site cysteine (aa 172–178) melts in the closed conformation and the catalytic cysteine is now observed adjacent to the catalytic machinery of the adenylation pocket and the SUMO adenylate (Fig. 2 and 3).
Figure 3. Conformational changes within the Cys domain.
The Cys domains of a, SUMO E1/SUMO1-AMSN, b, SUMO E1~SUMO1-AVSN, c, Ub E1/Ub complex10 and d, NEDD8 E1~NEDD8(t)/NEDD8(a)/Ubc12/ATP11 with helices labeled and depicted as tubes. Elements that undergo conformational changes colored as in Fig. 2b. Hinge points indicated by asterisks in the cross-over and re-entry loops. c, Superposition of cross-over and d, re-entry loops for E1/SUMO1-AMSN and E1~SUMO1-AVSN colored as in Fig. 2b. The catalytic cysteine (stick representation with sulfur colored green) is displaced by 34 Å during transitions between open and closed conformations. aa, amino acids
There are also major conformational rearrangements in the adenylation domains of SAE1 and UBA2 subunits, respectively (Fig. 2b,c). In SAE1, the N-terminal helices H1 and H2 that lie beneath the Cys domain and UBA2 helix 13 in the open conformation move out of the adenylation active site and become disordered in the closed conformation. In UBA2, the g1 helix (residues 53–57) that forms one side of the adenylation active site in the open conformation unfolds into a loop in the closed conformation that protrudes into space that was occupied by SAE1 helices H1 and H2 in the open conformation. Elements that move away from the adenylation active site in the closed conformation involve several residues known to be required for adenylation.
Cys domain elements are conserved in E1s
Many elements in the Cys domain are conserved at the level of sequence and structure in open forms of the Ub/Ubl E1s (Supplemental Fig. 4) including H6, H7, H12 and H13 in the SUMO E1 and respective helices in the Ub E1 and Nedd8 E1 (Fig. 3a,c,d). Also conserved are positions of the cross-over and re-entry loops that precede or follow the first or last of the conserved helices in the Cys domain. While distal from the active site in the open conformation, rotation of the Cys domain brings many of these conserved elements proximal to or in contact with the adenylate pocket in the closed conformation (Fig. 3b). In addition, the cross-over and re-entry loops that were once separated by 10 Å or more in the open conformation come together in a parallel β-sheet in the closed conformation.
Also noteworthy are structural elements that cover the helix that contains the active site cysteine in the open conformation, as they must move to uncover the cysteine for thioester bond formation. Helix g4 plays this role in the SUMO E1 while residues 778–783 in the loop between helices H26 and H27 cover the active site cysteine in the Ub E1. No corresponding element exists in the Nedd8 E1 Cys domain, but the large insertion domain in the Nedd8 APPBP1 subunit (specifically helices 10, 13, and 18) is proximal to its active site cysteine (not shown). Each element contains acidic motifs that are directed toward the active site cysteine and we envision this arrangement creates an acidic environment that could raise the cysteine pKa, maintaining its protonated form to protect it from oxidative or chemical damage. These elements could also protect the E1~Ub/Ubl thioester adduct from attack by unwanted nucleophiles as evident in the structure of the doubly loaded and activated form of the Nedd8 E1 (ref. 11). Elements that cover the SUMO E1 cysteine in the open conformation face away from the adenylate pocket or become disordered in the closed conformation.
Adenylation and thioester bond formation
Structures of E1 active sites that catalyze adenylation identified residues that contact ATP·Mg and the Ub/Ubl C-terminal glycine8,9,10,15. These structures all reveal the catalytic machinery poised to facilitate adenylation because basic residues stabilize the pyrophosphate leaving group, magnesium stabilizes the α phosphate that likely undergoes inversion during expulsion of the leaving group, and the C-terminal carboxylate nucleophile is coordinated for catalysis through hydrogen bonding and charge complementarity via interactions with the N-terminal end of UBA2 helix H2.
The structure of the E1/SUMO1-AMSN adenylate mimic revealed hydrogen bonding interactions between the SUMO1 C-terminal carbonyl oxygen and backbone amide of Ile28. Interactions were also observed between the backbone amide of Gly27 and one oxygen atom of the sulfamide moiety in SUMO1-AMSN, the presumed mimic of a phosphate non-bridging oxygen atom of the SUMO1-adenylate (Fig. 4a). The C-terminal carbonyl oxygen is pointed directly at the N-terminal end of helix H2, suggesting that helix H2 is ideally positioned to constitute the oxyanion hole by providing complementary positive electrostatic potential for stabilization of the transition state and tetrahedral intermediate during thioester bond formation.
Figure 4. Active sites in E1/SUMO1-AMSN and E1~SUMO1-AVSN.
a, Stereo representation (left) and schematic (right) of E1/SUMO1-AMSN depicting residues that contact the adenylate intermediate analog. b, Stereo representation (left) and schematic (right) of SUMO E1~SUMO1-AVSN depicting residues that contact the tetrahedral intermediate analog. The position analogous to SUMO1 G97 in E1~SUMO1-AVSN is denoted G97* to indicate the electrophilic center. Residues in stick representation labeled by single letter amino acid code with select waters as red spheres. Atoms colored as follows: UBA2 carbon (pink), SUMO1 carbon (yellow), oxygen (red), nitrogen (blue), and sulfur (green). Potential hydrogen bonds indicated by dashed lines.
What mechanism underlies activation of the catalytic cysteine as a nucleophile for attack at the SUMO1 carbonyl carbon? The E1~SUMO1-AVSN structure revealed that two oxygen atoms of the sulfonamide group, presumed to mimic the phosphate non-bridging oxygen atoms of the native leaving group (AMP), were coordinated by hydrogen bonding interactions with the UBA2 Thr174 hydroxyl and the backbone amide atoms of Gly27 and Ile28 (Fig. 4b). We also noted the position of His184, a potential general acid/base catalyst, because it moved closer to the catalytic Cys173 in the closed conformation in comparison to its position in the open conformation (Supplemental Fig. 6). While Thr174 and His184 are conserved in other E1s, mutational and biochemical analysis revealed that neither is essential for activity (Supplemental Fig. 4 and 6). Because no other side chains capable of acid/base catalysis were observed close enough to the active site to suggest a role in catalysis, we posit that the catalytic machinery of the adenylate pocket, namely the oxyanion hole provided by helix H2, is sufficient to stabilize transition states during adenylation and thioester bond formation.
Remodeling is required for E1 activity
The E1/SUMO1-AMSN structure revealed that side chains contacting the Mg ion or ATP β-γ phosphates in E1/SUMO1/ATP·Mg were no longer involved in contacts to the adenylate analog (Fig. 5a,b). Furthermore, the E1~SUMO1-AVSN structure showed that many of these residues were fully displaced from the active site and replaced with residues from the Cys domain during thioester bond formation (Fig. 5c). These data suggest that residues required for adenylation should be dispensible for the thioester formation half reaction. The reverse should also hold true. To test this hypothesis, we mutated residues in the SUMO and Ub E1 and assayed these mutant E1s for their ability to form the adenylate, thioester adduct, or tetrahedral intermediate via cross-linking to the Ub/Ubl-AVSN adduct.
Figure 5. Side chains required for adenylation are dispensible for formation of the tetrahedral intermediate analog.
Amino acid contacts that contribute to E1 adenylation activity shown for a, E1/SUMO1/ATP·Mg9, b, E1/SUMO1-AMSN adenylate analog and c, E1~SUMO1-AVSN tetrahedral intermediate analog color-coded as in Fig. 2b. Water (red) and Mg (cyan) as spheres. Dashed lines indicate potential hydrogen bonds. d, Structure–function analysis of E1 side chains depicted in a–c in assays for E1~SUMO1-AVSN cross-linking (top), E1~SUMO1 thioester formation (middle), and SUMO1-adenylate formation (bottom). Assay conditions in Methods. e, Structure–function analysis of residues in S. pombe UBA1 in assays for UBA1~Ub-AVSN cross-linking (top) and UBA1-Ub thioester formation (bottom). f, Structure-based sequence alignment of regions for human SUMO E1/SUMO1-AMSN and E1~SUMO1-AVSN, S. cerevisiae UBA1/Ub10, and human NEDD8 E1/NEDD8/ATP·Mg8. Gaps indicated periods. Boxes indicate conservation. Secondary structure for E1/SUMO1-AMSN and E1~SUMO1-AVSN above alignment with dashed lines indicating disorder. Conformational changes are color-coded as in Fig. 2b. Asterisks above the alignment indicate residues participating in unique interactions in the respective structures. Residues probed by mutational analysis are indicated above the alignment color-coded by activity.
N-terminal SAE1 helices H1 and H2 are adjacent to the adenylation active site in E1 structures with SUMO1/ATP·Mg and SUMO1-AMSN (Fig. 5a,b). SAE1 Arg21 in helix H2 contacts the ATP γ phosphate and is important for adenylation in other E1s8,10. A RLW motif in SAE1 composed of Arg 24, Leu 25, and Trp 26 side chains seems to stabilize the positions of the SAE1 H1 and H2 helices through hydrophobic interactions with UBA2 Pro 385, Ile 387, and Tyr 144 side chains. Mutation of Arg21, the RLW motif, or deletion of the N-terminal 27 amino acids of SAE1, which removes helices H1 and H2, abrogated adenylation but had no affect on achieving the closed conformation during thioester bond formation, as evidenced by the mutant E1’s ability to form a cross-link with SUMO1-AVSN (Fig. 5d). Thus, side chains within SAE1 amino acids 1–27 are required for adenylation but dispensible for achieving the closed conformation for cross-linking to SUMO1-AVSN, a result consonant with our structure because these elements are fully displaced from the active site in the SUMO1-AVSN structure (Fig. 5c).
The UBA2 g1 helix and Lys72 form another surface of the adenylation pocket and residues therein contact the ATP and adenylate ligands in the open conformation (Fig. 5a,b). Individual alanine substitutions for UBA2 Asn56, Leu57, Arg59 and Lys72 resulted in mutants defective for adenylation (Fig. 5d). While N56A and L57A mutant E1s readily formed cross-links with SUMO1-AVSN, R59A and K72A mutants lost about half of their cross-linking activity. These results are again consistent with our model because the g1 helix melts and is displaced from the active site in the E1/SUMO1-AVSN structure (Fig. 5c). Diminished cross-linking activity for R59A and K72A can be explained by their dual roles in coordinating ATP in the open conformation and in stabilizing interactions in the closed conformation. Specifically, Arg59 contacts the ATP γ phosphate in the open conformation but stabilizes the g1 loop in the closed conformation through hydrogen bond and van der Waals interactions with Asn56 and Leu57 (Fig. 5c). Lys72 contacts the ribose 3′-OH, a non-bridging oxygen of β-phosphate and is proximal to the oxygen atoms of both Asn56 and Gln60 side chains in the open conformation but maintains hydrogen bonding interactions with the ribose 3′-OH and Asp50 side carboxylate (3.4 Å) in the closed conformation.
Amino acid residues within the SAE1 N-terminal helix and UBA2 g1 helix are highly conserved across evolution in Ub and Nedd8 E1 enzymes (Fig. 5f). To test if mutations of the analogous residues in the Ub E1 would block adenylation while maintaining the ability to achieve the closed conformation to form a cross-link with the Ub-AVSN analog, we deleted the N-terminal 27 amino acids from the Ub E1 and made individual alanine substitutions of Leu472 and Arg474 (correspond to UBA2 Leu57 and Arg59, respectively). As predicted, each mutant isoform was unable to catalyze Ub adenylation as evidenced by the inability to form an E1~Ub thioester (Fig. 5e, top). In contrast, each mutant was active in cross-linking assays with Ub-AVSN, suggesting these mutations do not prevent the Ub E1 from achieving the closed conformation during thioester bond formation (Fig. 5e, bottom).
We next turned our attention to residues conserved in SUMO, Ub and Nedd8 E1s that appeared important for achieving or stabilizing the closed conformation during thioester formation. The loop that contains the active site cysteine is coordinated by hydrogen bond interactions between the UBA2 Asp50 side chain and backbone amide atoms of Asn177 and Thr178 (Fig. 6c). Asp50 is conserved across evolution but is exposed to solvent in E1 structures in the open conformation (Fig. 6a,b). Alanine substitution of Asp50 in SUMO E1 abrogated cross-linking activity with SUMO1-AVSN and thioester bond formation with SUMO1 but had no detectable effect on adenylation. The conservative glutamate substitution (D50E) had no effect on adenylation activity and retained minimal thioester formation and cross-linking activity (Fig. 6d). Alanine substitution of the analogous aspartic acid in the Ub E1 also blocked thioester formation and cross-linking to Ub-AVSN (Fig. 6e). Thus, Asp50 is essential for maintaining a productive closed conformation during thioester bond formation.
Figure 6. Side chains required for formation of the thioester bond or the tetrahedral intermediate analog are dispensible for adenylation.
Amino acid contacts deemed important for achieving the closed conformation are shown for a, E1/SUMO1/ATP·Mg9, b, E1/SUMO1-AMSN adenylate analog and c, E1~SUMO1-AVSN tetrahedral intermediate analog colored as in Fig. 2b. Potential hydrogen bonds indicated by dashed lines. d, Structure–function analysis of E1 side chains depicted in a–c in assays for E1~SUMO1-AVSN cross-linking (top), E1-SUMO thioester formation (middle), and SUMO-adenylate formation (bottom) assays. Assays conditions described in Methods. e, Structure–function analysis of select residues in S. pombe UBA1 in assays for UBA1~Ub-AVSN cross-linking (top) and UBA1-Ub thioester formation (bottom).
The UBA2 Arg176 side chain projects into the active site in the closed conformation where it participates in bipartite salt-bridging interactions with Asp117 (2.7 Å and 3.1 Å; Fig. 6c). In the open conformation, Arg176 interacts with the g4 helix in the Cys domain while Asp117 plays an essential role in adenylation by coordinating the magnesium ion in the ATP·Mg complex (Fig. 6a). Alanine substitution of UBA2 Arg176 resulted in a slight defect in cross-linking activity and thioester bond formation while maintaining nearly wild-type adenylation activity (Fig. 6d), while mutation of Ub E1 Lys596 resulted in no apparent defect in cross-linking or thioester formation (Fig. 6e). In contrast, UBA2 D117A and Ub E1 D537A blocked adenylation and abrogated cross-linking activity to low levels (Fig. 6d,e). We hypothesized that the remaining charged side chain in either single mutant might be rescued by eliminating the unpaired charge in the double mutant. As predicted, both UBA2 D117A/R176A or UBA1 D537A/K596A double mutants rescued wild-type activity in the cross-linking assay (Fig. 6d,e).
The cross-over loop and re-entry loops alter conformations in the open and closed forms of the E1 by 90 degrees or more (Fig. 3e,f). To determine if conformational changes in the cross-over loop impacted E1’s ability to catalyze adenylation or thioester formation, we made UBA2 K164A, P165G, T166V, R168P, F170A, P171A, I175A, and N177D substitutions. No single point mutant exhibited significant defects in adenylation, thioester formation, or cross-linking (not shown), consistent with conformational changes occurring over several residues in the cross-over loop. In contrast, conformational changes are localized between two amino acids, Gly381 and Asn382, in the re-entry loop. N382P and G381P/N382P diminished or abrogated thioester formation with SUMO1 or cross-linking to SUMO1-AVSN, respectively, without reduction in adenylation activity (Fig. 6d). The single point mutation corresponding to UBA2 N382P in the Ub E1 (K850P) also abrogated cross-linking activity and thioester formation (Fig. 6e). These data are consistent with our structure and suggest that conformational changes observed for the closed conformation of the SUMO E1 are important for E1 activity.
Conclusions
Structures of the SUMO E1 in complex with mimics of the SUMO adenylate and tetrahedral intermediates have revealed dramatic conformational changes that accompany adenylation and thioester bond formation. Domain alternation of the Cys domain is not sufficient to catalyze thioester bond formation as a number of other conserved structural elements of key functional significance in the Cys and adenylation domains undergo remodeling to allow the cysteine access to the adenylate pocket. The structural changes that accompany the transition between the open and closed forms of the E1 are remarkable in their complexity and are, to the best of our knowledge, without precedent in the literature. That is not to say that domain alternations have not been observed; in fact, rigid body domain alternations of similar magnitude (~140 degrees) have been described for ANL family of enzymes, so named for its constituent family members that include acyl-CoA synthetases, adenylation domains of nonribosomal peptide synthetases, and firefly luciferase16. While structurally unrelated to the Ub/Ubl E1 family, domain alteration in the ANL family has been proposed to create distinct active sites which are uniquely equipped to push the reaction forward during each catalytic step. We believe this also holds true for members of the Ub/Ubl E1 family.
What catalytic advantage is gained by active site remodeling in the Ub/Ubl E1s? Although Ub/Ubl E1 and ANL family members catalyze adenylation of a carboxylate to form an acyl adenylate intermediate followed by a second half-reaction that involves formation of a thioester, domain alternation is sufficient for catalysis in ANL enzymes without active site remodeling, presumably because these enzymes catalyze adenylation and thioester formation on diffusible ligands. We posit that domain alternation in Ub/Ubl E1s must be combined with active site remodeling because adenylation and thioester formation reactions result in a product that cannot diffuse away due to the covalent thioester bond between the Ub/Ubl and E1. So how does active site remodeling push the E1 reaction forward? If adenylation is catalyzed in the open configuration, conformational changes prior to thioester bond formation would dismantle the active site and favor adenylation by releasing pyrophosphate to prevent the reverse reaction, namely attack of the adenylate by pyrophosphate to reform ATP (a distinct possibility since this step is rate limiting17 and because all Ub/Ubl E1 enzymes characterized thus far in the open configuration are bound to ATP, despite the presence of the Mg cofactor and Ub/Ubl substrate). Displacement of residues required for adenylation would also remove steric impediments that could block the active site cysteine from coming in direct contact with the Ub/Ubl C-terminal carbonyl carbon during thioester bond formation. Once the thioester bond is formed, the Cys domain rotates away from the active site, releasing AMP and allowing the adenylation active site to reform to bind ATP·Mg and the next Ub/Ubl substrate, once again favoring the forward reaction by blocking the reverse reaction. Loading a second Ub/Ubl to block the reverse reaction provides one additional reason for why E1s are more efficient at transferring the Ub/Ubl to an E2 when doubly loaded11,18.
Structures of the SUMO E1 described herein provide unique insight to the chemical mechanisms used for adenylation and thioester bond formation. Our structures suggest that helix H2 contributes to transition state stabilization by providing an oxyanion hole to promote transfer between acyl phosphate and thioester intermediates. No other amino acid side chains capable of general acid/base chemistry were observed in the vicinity of the active site in the closed conformation, suggesting that thioester bond formation is facilitated by transition state stabilization and by placing the cysteine nucleophile proximal to the adenylate. The notable absence of general acid/base catalysis in the E1 active site is, perhaps, reminiscent of E2s, which catalyze isopeptide bond formation by pKa suppression of the nucleophile and transition state stabilization19–21.
The unique structure of the Cys domain in the closed conformation may also have implications for thioester transfer between the E1 and E2. The landmark structural work on the doubly loaded Nedd8 E1 revealed one Nedd8 bound in the adenylation pocket, one Nedd8 linked to the Nedd8 E1 Cys domain by a thioester bond, and the E2 Ubc12 bound to the UFD in a conformation that brought the E1 and E2 catalytic cysteine residues to within 20 Å (ref. 11). However, it is interesting to note that because the Nedd8 E1 was in the fully open conformation, the E1 active site cysteine pointed away from the E2. We modeled a path for the Cys domain during its alternation and were able to bring the E1 catalytic cysteine to within 3.5 Å of the Ubc12 active site suggesting that thioester transfer between the E1 and E2 may occur when the Cys domain achieves an intermediate state between closed and open conformations. Domain alternation has also been observed in HECT E3 ligases22 and we speculate that active site remodeling could also play a role in thioester transfer between E2s and HECT E3 ligases because the mechanism underlying this process remains elusive. E1s are now validated targets for small molecule inhibitors23 and our structural data suggests that several distinct conformations of the E1 along the path through adenylation and thioesterification could be targeted for therapeutic intervention. In a final note, domain alternation and active site remodeling as observed for the SUMO E1 may provide a broader conceptual framework to better understand other multistep biochemical processes.
Methods Summary
Proteins were expressed in E. coli and purified to homogeneity9,24. Ub/Ubl-AMSN and Ub/Ubl-AVSN adducts were generated using intein-mediated ligation12,13. SUMO E1/SUMO1-AMSN and E1~SUMO1-AVSN were purified, crystallized, and their structures determined by molecular replacement using the structure of E1/SUMO1/ATP·Mg as the search model9. SUMO E1 and Ub E1 mutants were generated by PCR-based mutagenesis and proteins purified as described above. E1~Ub/Ubl thioester formation and cross-linking assays were performed as described9 using native Ub/Ubl or Ub/Ubl-AVSN, respectively. Adenylation assays were performed in a reactions containing 2.5 µM hE1, 5 µM SUMO1, 5 mM MgCl2, 20 mM Hepes pH 7.5, 50 mM NaCl and 200 µM ATP, incubated for 5 minutes at 25 °C, desalted to remove excess ATP·Mg and pyrophosphate, and analyzed by SDS-PAGE.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature
Methods
Cloning, expression, and protein purification
Human SUMO E1 (hE1) was prepared as described9. Briefly, the DNA fragment encoding full-length human SAE1 (hSAE1; residues 1–349) was cloned into vector pET-11c for the expression of native polypeptide. The DNA fragments encoding residues 1–640 or 1–550 of human UBA2 (hUBA2FL and hUBA2ΔCT), respectively were cloned into vector pET-28b to introduce a thrombin cleavable histidine tag to the N-terminus of UBA2. The same DNA fragments were also cloned into vector pSMT3 (ref. 24) so as to introduce a Ulp1-cleavable His6-SMT3 tag to the N-terminus of UBA2. The DNA fragment encoding full-length S. cerevisiae AOS1 (ScAOS1; residues 1–346) was cloned into vector pET-15b. The DNA fragment encoding residues 1–637 of S. cerevisiae UBA2 (ScUBA2) was cloned into vector pET-28b and pSMT3. To generate ubiquitin E1, the DNA fragment encoding residues 1-1012 of S. pombe UBA1 (SpUBA1) was cloned from an S. pombe cDNA library into vector pSMT3. All mutations were generated using PCR-based mutagenesis.
hSAE1/hUBA2 or ScAOS1/ScUBA2 constructs were co-transformed and the SpUBA1 construct was transformed into Escherichia coli strain BL21 (DE3) Codon Plus RIL (Novagen). Large-scale cultures were grown at 37°C in baffled flasks to an A600 of 0.8. For hE1 and yE1 cultures, the temperature was reduced to 30°C and protein co-expression was induced by the addition of isopropyl-β-D-thiogalactoside to 1 mM for 4 hours whereas the SpUBA1 cultures were incubated at 18°C for 18 hours post-induction. Cells were harvested by centrifugation and re-suspended in buffer containing 20 mM Tris HCl (pH 8.0), 20% w/v sucrose, 350 mM NaCl and 20 mM imidazole. Cells were lysed by sonication and lysates cleared by centrifugation. hE1, yE1, and SpUBA1 were purified to homogeneity by metal-affinity (Ni NTA; Qiagen), gel filtration (Superdex 200; Amersham), and anion-exchange (MonoQ 10/10; Amersham) chromatography. His6-SMT3 tags were removed using Ulp1 protease (ref 24) before gel filtration to generate tagless proteins, whereas proteins containing thrombin cleavable histidine tags were purified with the tag intact.
DNA fragments encoding residues 1–94 of human SUMO1, residues 1–95 of S. cerevisiae SMT3, or residues 1–73 of S. pombe ubiquitin (from S. pombe genomic DNA) were cloned into vector pTXB1 to introduce an intein-chitin binding domain to the C-terminus of the Ub/Ubl. Each of these constructs lacked the three C-terminal amino acid residues of the mature Ub/Ubl protein that included the conserved di-glycine motif. The resulting fusion constructs were placed into pET-28b to add an N-terminal hexahistidine tag. A C52A mutant of human SUMO1 was generated in which a non-conserved, partially solvent exposed cysteine residue (Cys52) was substituted to alanine to minimize nonspecific disulfide bond formation or cross-linking to the vinyl group of AVSN. This mutant was used in cross-linking assays with the SUMO E1. Plasmids were transformed into E. coli BL21 (DE3) Codon Plus RIL (Novagen) and cultures were prepared and harvested using procedures outlined for hE1. Ub/Ubl-intein-CBD fusion proteins were applied to a column containing 20 ml chitin beads (New England Biolabs). The intein-CBD tag was removed and a reactive Ub/Ubl~thioester generated by incubating the Ub/Ubl-bound chitin beads in buffer containing 20 mM Tris pH 8.0, 50 mM NaCl, and 50m M 2-mercaptoethanesulfonate (MESNa) for 12 hours at 4°C. Following elution from the chitin beads in the same buffer, Ub/Ubl~MESNa thioester was purified using gel filtration chromatography (Superdex 75; Amersham). The histidine tagged Ub/Ubl-intein-CBD and histidine tagged SUMO1-intein-CBD with the C52A mutation were prepared using the same protocol.
Protein ligation
Ub/Ubl~MESNa thioester was mixed with CGG-AMSN or CGG-AVSN at an approximate 1:1 molar ratio (250 µM) in 20 mM Tris pH 8.0, 50 mM NaCl and 15 mM MESNa. The mixture was incubated at 4 °C for 12–18 hours and the progress of the ligation reaction was monitored as a shift in the mobility of Ub/Ubl protein on an SDS-PAGE gel. The ligation mixture was subsequently applied to a Superdex 75 column to separate un-reacted AMSN or AVSN tripeptides from the respective Ub/Ubl adducts. Fractions containing Ub/Ubl-AMSN or Ub/Ubl-AVSN constructs were pooled and concentrated to 400 µM. The diglycine motifs of the Ub/Ubls are regenerated upon ligation to AMSN/AVSN, and cysteine substitution of the residue immediately N-terminal to the diglycine motif results in T95C for human SUMO1, I95C for S. cerevisiae SMT3, and R74C for S. pombe ubiquitin. Mutation of this non-conserved residue was previously shown to have no effect on the specificity or activity of SUMO1 (ref. 25).
Preparation of E1~SUMO1-AVSN and E1/SUMO1-AMSN for crystallization
Complexes of hE1FL or hE1ΔCT and SUMO1-AMSN were generated by incubating 1.2 mg/mL hE1 and 515 µg/mL SUMO1-AMSN in buffer containing 20 mM Tris pH 8.0, 50 mM NaCl, and 1 mM β-mercaptoethanol (β-ME). This mixture was applied to a Superdex 200 gel filtration column that was pre-equilibrated in 20 mM Tris pH 8.0, 150 mM NaCl, 1 mM β-ME. Fractions containing hE1/SUMO1-AMSN were pooled, buffer exchanged into 20 mM Tris pH 8.0, 100 mM NaCl, 5 mM β-ME, concentrated to 10 mg/ml. The protein was used immediately for crystallization trials and the remaining sample was snap-frozen in liquid nitrogen and stored at −80 °C.
Cross-linking of hE1FL or hE1ΔCT to SUMO1-AVSN was performed in a reaction mixture containing 1 mg/ml hE1, 2.3 mg/ml SUMO1-AVSN, 20 mM Tris pH 8.0, 50 mM NaCl, and 0.5 mM tris (2-carboxyethyl) phosphine (TCEP). The cross-linking reactions were carried out at room temperature and formation of the UBA2-SUMO1-AVSN adduct was monitored via SDS-PAGE as a shift in the migration of UBA2 corresponding to ~15 kDa. Upon completion of cross-linking, the reaction mixture was subjected to gel filtration chromatography to separate excess SUMO1-AVSN from the hE1~SUMO1-AVSN adduct. Fractions containing hE1~SUMO1-AVSN adduct were pooled and subjected to nickel affinity chromatography. Binding of the hE1~SUMO1-AVSN adduct to the nickel resin via the histidine tag of SUMO1-AVSN allowed for the separation of any remaining un-reacted hE1. The adduct was eluted from the nickel resin, buffer exchanged into 20 mM Tris pH 8.0, 75 mM NaCl, 1 mM MgCl2, and 1 mM TCEP, and concentrated to 8 mg/mL. The protein was subsequently subjected to crystallization trials. The remaining sample was snap-frozen in liquid nitrogen and stored at −80 °C.
Crystallization and data collection
E1~SUMO1-AVSN and E1/SUMO1-AMSN samples were subjected to sparse-matrix screening in 96-well Greiner microplates (150 nl sitting drop vapor diffusion format) using a mosquito crystallization robot (Molecular Dimensions Ltd.). Sparse matrix screens were performed in duplicate and incubated at 6°C or 18°C. Diffraction quality crystals of the E1/SUMO1-AMSN complex were grown by mixing 1.5 µL protein (10 mg/mL, 20 mM Tris pH 8.0, 100 mM NaCl, 5 mM β-ME) with 1.5 µL crystallization buffer (0.1 M MES pH 6.4, 24% polyethylene glycol 3350, 1.3 M ammonium acetate, 10 mM ATP, 10 mM MgCl2, 10 mM TCEP, 2.5% polyethylene glycol 400) by hanging drop vapor diffusion at 6 °C. Diffraction quality crystals of the E1~SUMO1-AVSN adduct were grown by mixing 1.5 µL protein (8 mg/mL, 20 mM Tris pH 8.0, 75 mM NaCl, 1 mM MgCl2, 1 mM TCEP) with 1.5 µL crystallization buffer (22% polyethylene glycol 2000 [monomethyl ether], 0.2 M di-ammonium tartrate, 3% ethylene glycol) by hanging drop vapor diffusion at 6 °C.
E1/SUMO1-AMSN crystals did not require further cyro-protection and were flash-frozen in liquid nitrogen. E1/SUMO1-AMSN crystals belong to the orthorhombic space group P212121 with unit cell dimensions a=59.1 Å, b=133.8 Å, c= 159.3 Å. There is one E1/SUMO1-AMSN complex per asymmetric unit and the crystals have a solvent content of ~52%. E1~SUMO1-AVSN crystals were flash-frozen in liquid nitrogen in a final cryoprotectant solution comprised of mother liquor and 25% ethylene glycol. E1~SUMO1-AVSN crystals belong to the orthorhombic space group P21212 with unit cell dimensions a=101.3 Å, b= 115.2 Å, c= 90.8 Å. There is one E1~SUMO1-AVSN complex per asymmetric unit and the crystals have a solvent content of ~48%. Diffraction data were collected on an ADSC Quantum 315 detector at Advanced Photon Source (Argonne, IL), NE-CAT beamline 24-IDC. The data were indexed, integrated, and scaled using HKL2000 (ref. 26).
Structure determination and refinement
A data set was collected to a resolution of 2.45 Å for the E1/SUMO1-AMSN complex and the program PHASER26 was used to find a molecular replacement solution using the coordinates for SAE1/UBA2 (PDB entry 1Y8Q)9 as the search model (Supplemental Table 1 and Methods). The entire E1 structure was manually inspected and rebuilt prior to modeling SUMO1 or the AMSN motif. A model for SUMO1 was obtained from a complex between SUMO1 and Senp2 (PDB entry 1TGZ)28 and used to dock SUMO1 amino acids 20–94 into electron density manually. The CGG-AMSN motif was modeled last. The final model contained SUMO1(19–97)-AMSN, SAE1 amino acids (aa) 9–180 and 205–345, and UBA2 aa 5–217, 235–291, 305–548 and 608–640. As full-length proteins were used in crystallization, residues not observed in electron density were presumed disordered. This model was refined to R/Rfree values of 0.190/0.249 via iterative rounds of refinement and rebuilding using CNS29, CCP4’s REFMAC30 and the program O31. The model has good geometry32 with 87.1%, 11.8%, 1.0%, and 0.1% of residues in most favored, generous, allowed, and disallowed regions of Ramachandran space. The single residue (SAE1 Asp322) that falls outside of allowed Ramachandran space is well defined by electron density and was also observed in this configuration in previous SUMO E1 structures9.
A data set was obtained to a resolution of 2.6 Å for the covalent adduct between SUMO E1 and SUMO1-AVSN by merging data obtained from two crystals. The structure was determined by molecular replacement using SUMO E1 coordinates; however the search model did not include the Cys or UFD domains. Electron density was manually inspected and elements in the adenylation domains of the E1 model were rebuilt or removed depending on the observed electron density. Electron density for SUMO1 and the UFD domain were observed and models were docked and refined prior to modeling the Cys domain. Electron density was evident for Cys domain throughout this process however it was clear that this domain was not in the same orientation as observed in other E1 structures. Three helices comprise the Cys domain core and these were placed into electron density first. The remaining elements of the Cys domain were manually built into 2fo-fc, fo-fc and simulated annealing omit map electron density (Supplemental Fig. 1). The covalent adduct between the catalytic cysteine and AVSN was modeled last. The final model includes SUMO1(20–97)-AVSN, SAE1 aa 25–183 and 204–345 and UBA2 aa 4–197, 240–290, 309–336 and 345–548. SUMO1(1–97)-AVSN, SAE1 and UBA2 (1–550) were crystallized, so residues not observed in electron density were presumed disordered. This model was refined to R/Rfree values of 0.227/0.283 using TLS refinement for SAE1, three UBA2 domains (adenylation, UFD, Cys) and SUMO. The model has good geometry32 with 90.1%, 9.5%, 0.3%, and 0.1% of residues in most favored, generous, allowed, and disallowed regions of Ramachandran space. The single residue (SAE1 Asn327) that falls outside of allowed Ramachandran space is well defined by electron density. All molecular graphics representations of the structure were generated using PYMOL33.
Biochemical assays
SUMO1 adenylation assays were performed in a reaction mixture containing 2.5 µM hE1FL, 5 µM SUMO1, 5 mM MgCl2, 20 mM Hepes pH 7.5, 50 mM NaCl and 200 µM ATP or 200 µM BODIPY FL ATP (Invitrogen). Reactions were incubated for 5 minutes at 25 °C and applied to Micro Bio-spin P-6 buffer exchange columns (BioRad) to remove excess Mg, ATP or BODIPY FL ATP and pyrophosphate. Samples were denatured in non-reducing SDS-PAGE buffer, subjected to SDS-PAGE and visualized with a Fujifilm FLA5000 imaging system using a 473 nm excitation laser with LPB (long pass blue) filter. Gels were subsequently stained with Sypro Ruby (BioRad) (Supplemental Fig. 7).
SUMO E1~SUMO1 thioester formation assays for wild type and mutant isoforms were performed in a reaction containing 1 µM hE1ΔCT, 2 µM SUMO1, 5 mM MgCl2, 20 mM Hepes pH 7.5, 50 mM NaCl, and 2 µM ATP. Reactions were incubated for 30 seconds at 25 °C, then denatured in non-reducing SDS-PAGE buffer, subjected to SDS-PAGE and visualized by staining with Sypro Ruby (Biorad). SpUBA1~ubiquitin thioester formation assays were performed using reactions conditions described for SUMO E1.
Cross-linking of wild type and mutant constructs of hE1FL to SUMO1(C52A)-AVSN was performed in a reaction mixture containing 1 µM hE1FL, 2 µM SUMO1(C52A)-AVSN, 20 mM Tris pH 8.0, 50 mM NaCl, and 0.5 mM TCEP. Reactions were incubated for 15 minutes at 25 °C and denatured in reducing SDS-PAGE buffer, subjected to SDS-PAGE and visualized by staining with Sypro Ruby. Cross-linking of SpUBA1 to Ub-AVSN was performed using the same conditions as described for SUMO E1.
Supplementary Material
Acknowledgements
We thank A. Armstrong for Ub and Ub E1 clones and N. Takeda for assistance in graphic art preparation. This work is based upon research conducted at the NE-CAT beamlines of the Advanced Photon Source, supported by RR-15301 from the NCRR at the NIH. Use of the APS is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. D.S.T. is an Alfred P. Sloan Research Fellow. X.L. and D.S.T. were supported by NIH R01 AI068038 and the NYSTAR Watson Investigator Program. S.K.O., A.D.C., and C.D.L. were supported by NIH R01 GM065872, F32 GM075695 (A.D.C.), and the Rita Allen Foundation (C.D.L.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions Experiments were performed by S.K.O., A.D.C., X.L., and C.D.L. Data were analyzed by S.K.O., A.D.C., X.L., D.S.T. and C.D.L. The manuscript was prepared by S.K.O., D.S.T., and C.D.L.
Author Information The atomic coordinates and structure factors have been deposited in the RCSB database under accession codes 3KYC and 3KYD. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
References
- 1.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 2.Melchior F. SUMO--nonclassical ubiquitin. Annu Rev Cell Dev Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 3.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 4.Laney JD, Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97(4):427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 5.Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 6.Capili AD, Lima CD. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr Opin Struct Biol. 2007;17(6):726–735. doi: 10.1016/j.sbi.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10(5):319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walden H, et al. The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol Cell. 2003;12(6):1427–1437. doi: 10.1016/s1097-2765(03)00452-0. [DOI] [PubMed] [Google Scholar]
- 9.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24(3):439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee I, Schindelin H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell. 2008;134(2):268–278. doi: 10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
- 11.Huang DT, et al. Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature. 2007;445(7126):394–398. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu X, et al. Designed semisynthetic protein inhibitors of Ub/Ubl E1 activating enzymes. J. Am. Chem. Soc. 2010;132(6):1748–1749. doi: 10.1021/ja9088549. [PubMed:20099854] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu Rev Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 14.Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins. 1998;30(2):144–154. [PubMed] [Google Scholar]
- 15.Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature. 2001;414(6861):325–329. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- 16.Gulick AM. Conformational Dynamics in the Acyl-CoA Synthetases, Adenylation Domains of Non-ribosomal Peptide Synthetases, and Firefly Luciferase. ACS Chem Biol. 2009 doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas AL, Rose IA. The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J Biol Chem. 1982;257(17):10329–10337. [PubMed] [Google Scholar]
- 18.Pickart CM, Kasperek EM, Beal R, Kim A. Substrate properties of site-specific mutant ubiquitin protein (G76A) reveal unexpected mechanistic features of ubiquitin-activating enzyme (E1) J Biol Chem. 1994;269(10):7115–7123. [PubMed] [Google Scholar]
- 19.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435(7042):687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu PY, et al. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003;22(19):5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13(6):491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 22.Verdecia MA, et al. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol Cell. 2003;11(1):249–259. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- 23.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 458(7239):732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 24.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5(5):865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 25.Knuesel M, Cheung HT, Hamady M, Barthel KK, Liu X. A method of mapping protein sumoylation sites by mass spectrometry using a modified small ubiquitin-like modifier 1 (SUMO-1) and a computational program. Mol Cell Proteomics. 2005;4(10):1626–1636. doi: 10.1074/mcp.T500011-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 3):432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 28.Reverter D, Lima CD. A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure. 2004;12(8):1519–1531. doi: 10.1016/j.str.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 29.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231(4):1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 33.Delano W. The PyMOL molecular graphics system. San Carlos CA, USA: DeLano Scientific; 2002. http://www.pymol.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.