Abstract
C53/LZAP (also named as Cdk5rap3) is a putative tumor suppressor that plays important roles in multiple cell signaling pathways, including DNA damage response and NF-κB signaling. Yet how its function is regulated remains largely unclear. Here we report the isolation and characterization of two novel C53/LZAP-interacting proteins, RCAD (Regulator of C53/LZAP and DDRGK1) and DDRGK1 (DDRGK domain-containing protein 1). Our co-immunoprecipitation assays confirmed their interactions, while gel filtration assay indicated that C53/LZAP and RCAD may form a large protein complex. Intriguingly, we found that RCAD knockdown led to dramatic reduction of C53/LZAP and DDRGK1 proteins. We also found that C53/LZAP and DDRGK1 became more susceptible to the proteasome-mediated degradation in RCAD knockdown cells, whereas their ubiquitination was significantly attenuated by RCAD overexpression. In addition, we found that RCAD, like C53/LZAP, also plays an important role in regulation of NF-κB signaling and cell invasion. Taken together, our findings strongly suggest that RCAD is a novel regulator of C53/LZAP tumor suppressor and NF-κB signaling.
Keywords: NF-κB Transcription Factor, Protein Stability, Protein-Protein Interactions, Tumor Suppressor, Ubiquitination, C53/LZAP, DDRGK1/C20orf116, KIAA0776
Introduction
C53/LZAP is a highly conserved protein that has been shown to be involved in multiple cell signaling pathways. We originally found that C53/LZAP overexpression potentiated DNA damage-induced cell death by modulating the G2/M checkpoint (1). Furthermore, we demonstrated that by antagonizing checkpoint kinase 1 (Chk1), C53/LZAP modulated the activation of cyclin-dependent kinase 1 (Cdk1), thereby influencing the DNA damage response and mitotic entry during the cell cycle progression (2). Meanwhile, Wang et al. (3) elegantly demonstrated that C53/LZAP functions as a novel tumor suppressor in primary head and neck cancers by specifically inhibiting NF-κB signaling. They found that decreased C53/LZAP expression promoted cellular transformation, xenograft tumor growth, and xenograft tumor vascularity. Loss of C53/LZAP increased cellular invasion, and NF-κB transcriptional activity. At the molecular level, C53/LZAP directly bound to RelA, impaired serine 536 phosphorylation of RelA and increased HDAC association with RelA, thereby inhibiting basal and stimulated NF-κB transcriptional activity. Interestingly, they found that C53/LZAP protein level was markedly decreased in one third of primary human head and neck squamous cell carcinomas (HNSCCs) and decreased C53/LZAP level in primary HNSCC correlated with increased expression of the NF-κB-regulated genes IL-8 and IκBα.
C53/LZAP appears to play important roles in cell signaling pathways that are involved in tumorigenesis and cancer metastasis, yet the mechanism to regulate its activity remains completely unknown. To further elucidate its biological functions and the molecular mechanism that regulates its activity, we attempted to identify and characterize its interacting partners. Here we report the isolation and characterization of a novel C53/LZAP-interacting protein that regulates its protein stability.
EXPERIMENTAL PROCEDURES
Tissue Culture Cells and Reagents
U-2 OS osteosarcoma cell (from ATCC) were grown in MaCoy's 5A medium supplemented with 10% fetal bovine serum (FBS),2 while HeLa, MCF7, T47D, and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics. Cycloheximide, MG132, and TNFα were purchased from Sigma.
RCAD and DDRGK1 cDNA and Expression Constructs
The EST clones containing full-length RCAD and DDRGK1 cDNAs (human and murine) were purchased from Open Biosystems Inc. Murine RCAD cDNA was PCR-amplified using primers: 5′-GGGAATTCGGATGGCGGACGCCTGGGAG-3′ (forward) and 5′-AAGCGGCCGCTTAGGTACCCTCCTCTGTGACAGATGA-3′ (reverse), and verified by DNA sequencing. RCAD cDNA was further subcloned in-frame into pCMV-Myc (Clontech), and the resulting construct was used to overexpress Myc-tagged RCAD. Human DDRGK1 cDNA was PCR-amplified using primers: 5′-GGGAATTCGGATGGCGGAGTCTGTGGAGCGC-3′ (forward) and 5′-GGGCGGCCGCTCAGGCTGGGGCTTGGGCAGG-3′ (reverse), and further subcloned into pCMV-Myc or pCMV-Flag vectors. C53/LZAP constructs were described in our previous study (2).
siRNAs for Human C53/LZAP, RCAD, and DDRGK1 and Transfection
Silencer Select predesigned siRNAs were purchased from Ambion, Inc. The following are the sense sequences of siRNAs we used in this study: RCAD-1: GGAACUUGUUAAUAGCGGA; RCAD-2: GAGGAGUAAUUUUUACGGA; C53-2: GGCAGGAGAUUAUAGCUCU; C53-3: GGAUUGGCAGGAGAUUAUA; DDRGK1-1: GAAAAUUGGAGCUAAGAAA; DDRGK1-2: CCAUAAAUCGCAUCCAGGA. The negative control siRNAs were Silencer Select negative control 1 and 2 siRNAs that were purchased from Ambion.
Reverse transfections were performed using Hiperfect (Qiagen) following the manufacturer's instruction with minor modification. 10 nm siRNA was used as the standard concentration in our knockdown assays.
Generation of Rat Polyclonal Antibodies
Human RCAD fragment (residues 204–515) and DDRGK1 fragment (residues 1–150) were subcloned into pET-28C vector (Novagen), and His-tagged fusion proteins were purified from soluble fractions by nickel-NTA columns (Novagen). Purified proteins were injected into rats to make rat polyclonal antibodies according to the standard protocol (1). Polyclonal antibodies were affinity-purified using corresponding antigens.
Northern Blotting
The Ambion FristChoice Northern blot mouse blot I (purchased from Ambion) was used for our Northern blot assay. The DNA probes for murine RCAD and DDRGK1 were amplified from the EST clones containing their corresponding cDNAs and then labeled with [α-32P]dATP using DECAprime II kit (Ambion). Hybridization of the blot was carried out in ULTRAhyb hybridization buffer (Ambion) according to the procedure provided by the manufacturer.
Antibodies, Co-immunoprecipitation, Immunoblotting, and Immunofluorescence Staining
The procedures for immunoblotting, immunoprecipitation, and immunofluorescence staining were described previously (1, 2). The following antibodies at indicated dilutions were used for immunoblotting: GAPDH (Santa Cruz Biotechnology, 1:10,000), anti-Flag M2 (Sigma, 1:2,000), Myc (9E10, Santa Cruz Biotechnology, 1:3,000), HA monoclonal antibody (Sigma, 1:2,000), C53 rat polyclonal and monoclonal antibody (1:1,000 dilution), RCAD rat polyclonal antibody (affinity purified, 1:500), DDRGK1 rat polyclonal antibody (affinity purified, 1:500). Species-specific horseradish peroxidase- and fluorophore-conjugated secondary antibodies were obtained from Jackson ImmunoResearch.
Subcellular Fractionation
Subcellular fractionation was performed according to the protocol of differential sucrose gradient isolation of ER and mitochondria (17). HeLa or HepG2 cells (from three 75-cm flasks) that had reached 90% confluency were harvested and washed with ice-cold phosphate-buffered saline, and then lysed mechanically with sonication in MTE buffer (10 mm Tris-HCl, pH 7.4, 270 mm d-mannitol, 0.1 mm EDTA) plus protease inhibitors. The following procedures were performed at 4 °C. A low-speed centrifugation (700 × g) was used to remove large cellular debris. The supernatant from this step was collected as a total lysed protein fraction, and further centrifuged at 15,000 × g for 15 min to separate mitochondria from ER and other light membrane fractions. The supernatant from 15,000 × g centrifugation (containing crude ER) was loaded onto a three-layered discontinuous sucrose gradient (1.3, 1.5, and 2 m sucrose from the top to the bottom in MTE buffer) and centrifuged at 152,000 × g for 70 min. The top layer was collected as the fraction of cytosol, while the banded fraction at the interface of the 1.3 m sucrose layer was collected as the ER fraction. The subcellular fractions were subjected to SDS-PAGE and followed by immunoblotting analysis using specific antibodies. We used calnexin as the ER marker, and α-tubulin as the cytosol marker.
Real-time PCR
Cells (1 × 106) were collected and washed with ice-cold phosphate-buffered saline. mRNA was purified by RNeasy mini kit (Qiagen). The first-strand cDNA was synthesized by Superscript first-strand synthesis system for RT-PCR (Invitrogen), and used as the template for real-time PCR. QuantiTect SYBR Green PCR kit (Qiagen) was used for RT-PCR assays that were run on ABI 7500 RT-PCR system and analyzed by the relevant software. The primers were used for RT-PCR: human GAPDH: 5′-AAGGTGAAGGTCGGAGTCAA-3′ (forward); 5′-CCATGTAGTTGAGGTCAATGAAGG-3′ (reverse); human C53/LZAP: 5′-ATTTTTGGCCGATACTCTTCACA-3′ (forward); 5′-TCATAGTTGACATTCCGAACCAG-3′ (reverse); human RCAD: 5′-AGCAAACAGGCCTCAACTGT-3′ (forward); 5′-TTTCTGGTGCATCAGCTCAC-3′ (reverse); human DDRGK1: 5′-TGCTGGCTGAGGGGACTATAA-3′ (forward); 5′-CCGCTGTCGGATGAAGTTG-3′ (reverse).
NF-κB Reporter Assay
The NF-κB luciferase reporter plasmid (a gift from Dr. Junying Yuan) was transfected into HeLa or U2OS cells along with pRL-TK (Promega). At 24 h post-transfection, cells were treated with TNFα (10 ng/ml) for 8 h. Cells were lysed, and the luciferase activity was measured by Dual-Luciferase reporter assay kit (Promega).
Electron Microscopy
Cells were fixed with 2.5% glutaraldehyde in 0.1 m sodium phosphate buffer, pH 7. 4 at 4 °C for 30 min. Cells were then scraped from the tissue culture dish and harvested by centrifugation at 3000 rpm for 10 min. The cell pellet was further fixed in 2.5% glutaraldehyde for 2 h, and post-fixed with 1% osmium tetroxide for 2 h at 4 °C. The cell pellet was dehydrated in an ethanol gradient (50–100% for 10 min each) and embedded in Epon 812 at 60 °C for 2 days. Ultrathin sections were stained with uranyl acetate and lead citrate, and pictures were taken by a Zeiss 900 electron microscope.
Size Exclusion Chromatography
HeLa S100 extract was prepared as described by Dignam et al. (18). 0.5 ml of S100 extract was loaded on Superose 6 10/300 GL column (GE Healthcare) that was precalibrated with gel filtration calibration kit (GE Healthcare). The chromatography was performed using AKTApurifier (GE Healthcare) in the buffer of 20 mm HEPES, pH 7.4, 150 mm NaCl, 0.1 mm EDTA, 0.1 mm dithiothreitol plus protease inhibitors (0.3 ml/min flow rate). Fractions (0.5 ml) were collected, and aliquots of the fraction were subject to SDS-PAGE and immunoblotting with specific antibodies.
Invasion Assay
Cell invasion assays were performed in an invasion chamber, a 24-well tissue culture plate with 12 cell culture inserts. The inserts contain an 8-μm pore size polycarbonate membrane (Transwell Permeable Support, Corning Inc), coated with 60 μl of 1 mg/ml Matrigel (BD Sciences). U2OS cells were starved in serum-free medium overnight, trypsinized, and washed three times in DMEM containing 1% FBS. 2 × 104 cells in 1% FBS-DMEM were seeded into the upper chamber, and 600 μl of DMEM containing 10% or 1% FBS was placed in the lower chamber. After a 16-h incubation, matrigel and cells remaining in the chamber were removed using cotton swabs. Invasive cells which cling to the bottom of the membrane were stained with 0.5 μm Calcein AM (Sigma-Aldrich). Pictures of the stained cells were taken under a fluorescent microscope with×1 magnification. Cells were then counted using the Openlab software. All experiments were run in duplicate and were repeated three times.
Zymography
Zymography of MMP9 was performed using 10% SDS-PAGE with 0.1% gelatin. U2OS cells were starved in serum-free MaCoy's 5A medium overnight. An aliquot (10 μl) of conditional medium was mixed with 10 μl of 2× Laemmli buffer without β-mercaptoethanol, incubated at 50 °C for 5 min, and then subject to SDS-PAGE. The gel was washed twice for 15 min in 100 ml of Triton X-100 (2.5% in H2O) at room temperature, followed by incubation in 100 ml of development buffer (50 mm Tris-HCl, pH 8.0, 10 mm CaCl2). After agitation for 15 min at room temperature, the gel was transferred to a 37 °C incubator with slow agitation (50 rpm) overnight. After development, the gel was incubated in fixing/destaining solution (45% methanol and 10% acetic acid) for 15 min at room temperature, and then stained with Coomassie Blue R-250 (0.1% in 50% methanol and 10% acetic acid) for 2–3 h at room temperature, and destained in destaining solution for at least 2 h at room temperature.
RESULTS
Two Novel C53/LZAP-interacting Proteins, DDRGK1 and RCAD, Are Highly Conserved and Ubiquitously Expressed in Multiple Tissues and Cell Lines
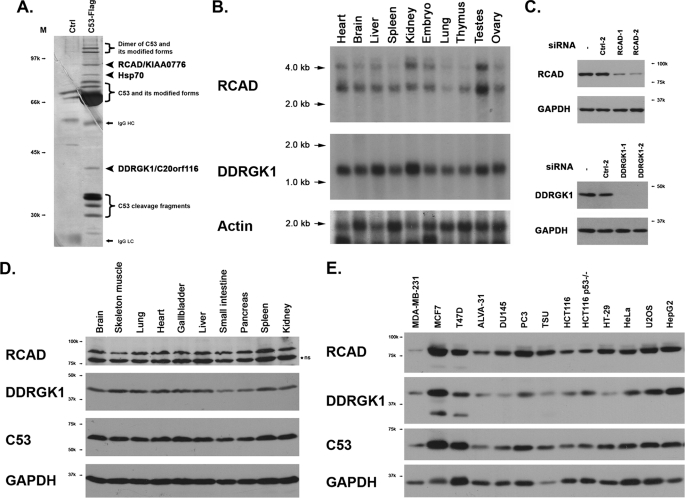
In an attempt to isolate C53/LZAP-interacting proteins, we performed co-immunoprecipitation using C-terminally Flag-tagged C53/LZAP as the bait, and identified potential interactors with mass spectrometry. As shown in Fig. 1A, there were at least three non-C53/LZAP proteins that were specifically immunoprecipitated with Flag-C53 protein, including heat shock protein 70 (Hsp70) and two uncharacterized proteins. A polypeptide with molecular mass of 40 kilodalton (kDa) was identified as DDRGK domain-containing protein 1 (DDRGK1, also named as C20orf116, CT116, and Dashurin) (4, 5), while a 90-kDa polypeptide was the uncharacterized protein KIAA0776 that was named hereafter as Regulator of C53/LZAP and DDRGK1 (RCAD) (Fig. 1A). Interestingly, our finding was in accordance with the data generated from a large-scale screening of protein-protein interactions (6). Both RCAD/KIAA0776 and DDRGK1/C20orf116 were among the potential C53 interactors in the screening (6) (also see the search result from the IntAct database using Cdk5rap3 as the keyword).
FIGURE 1.
Two novel C53/LZAP-interacting proteins: RCAD and DDRGK1. A, silver staining of the immunoprecipitate of C53-Flag fusion protein. C53-Flag fusion protein was overexpressed in 293T cells, and immunoprecipitated with M2-agarose beads (Sigma). An aliquot was subject to SDS-PAGE and sliver staining. Identities of the bands were determined by LC/MALDI mass spectrometry. B, mRNAs of RCAD and DDRGK1 in mouse tissues. FristChoice Northern blot mouse blot I (Ambion) was blotted with either mouse RCAD or DDRGK1 probes. C, specificity of siRNAs and antibodies. Endogenous RCAD or DDRGK1 was knockdown with specific siRNAs, and the cell lysates were subject to SDS-PAGE and immunoblotting with affinity-purified antibodies. D, RCAD and DDRGK1 proteins in rat tissues. Rat tissue lysates were subject to SDS-PAGE and immunoblotting with specific antibodies. Nonspecific band (ns) is marked by a star. E, RCAD and DDRGK1 expression in cancer cell lines. The total cell lysates of various cancer cells were subject to immunoblotting.
The human RCAD gene is located at chromosome 6q16.1. Similar to C53/LZAP, the human RCAD protein has 794 amino acid residues with no recognizable protein domains or motifs. Its orthologs can be found in the genomes of invertebrate, vertebrate, and plants, but not of yeast. Multiple sequence alignment indicates that its N-terminal portion is highly conserved (supplemental Fig. S1). RCAD is ubiquitously expressed in multiple tissues and cell lines. As shown in Fig. 1B, there are two major murine RCAD mRNA species with sizes of 2.8 and 4.2 kilobase (kb) in multiple mouse tissues. According to the EST data base, both mRNAs may give rise to the same protein with 793 amino acid residues. However, there are several human EST sequences with alternatively spliced exons that may encode RCAD isoforms with truncations. We generated an RCAD antibody that recognized an endogenous 90-kDa protein, while the antibody specificity was further confirmed by siRNA-mediated knockdown assay (Fig. 1C). Using this antibody, we examined RCAD expression in tissues and human cell lines. As illustrated in Fig. 1, D and E, RCAD was ubiquitously expressed in multiple tissues and cell lines. Interestingly, RCAD expression was remarkably lower in invasive breast cancer cell line MDA-MB-231 (Fig. 1E).
Human DDRGK1 protein has 314 amino acid residues with predicted molecular mass 35.6 kDa. Sequence analysis suggests that the first 1–28 residues are highly hydrophobic and may serve as a signal peptide. However, we could not detect any secretion of either endogenous or overexpressed DDRGK1 in tissue culture medium (data not shown). Furthermore, it contains a partial PCI (residues 229–273) domain that is frequently found in the subunits of the proteasome, COP-9 complex and translation initiation factors. DDRGK1 is a highly conserved protein, and its orthologs can be found in the genomes of vertebrates, invertebrates, and plants. Multisequence alignment suggests that its C-terminal portion contains several stretches of highly conserved sequences including the one with DDRGK pentapeptide (supplemental Fig. S2).
Northern blot analysis showed that murine DDRGK1 cDNA was about 1.3 kb, which is consistent with the size of both human and murine cDNA found in the full-length cDNA database (Fig. 1B). It was ubiquitously expressed in multiple tissues and organs, with relatively high-level expression in the liver, kidney, and testes. Our DDRGK1 antibody recognized a 42-kDa protein in SDS-PAGE, which is slightly larger than the predicted size (Fig. 1C). Similar to C53/LZAP and RCAD, DDRGK1 was expressed in multiple cancer cell lines with a relatively low expression in MDA-MB-231 cells.
RCAD Interacts with C53/LZAP and Forms a Large Complex
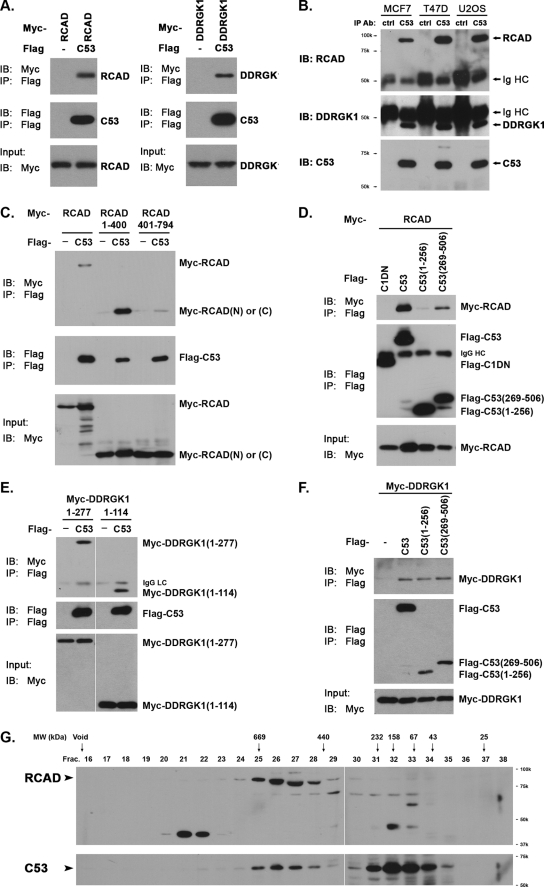
To further confirm our initial observation of the interactions among C53/LZAP, RCAD, and DDRGK1, we first performed co-immunoprecipitation (co-IP) assays using overexpressed proteins. As shown in Fig. 2A, both Myc-tagged RCAD and DDRGK1 were present in the Flag-tagged C53 immunoprecipitate. Endogenous RCAD and DDRGK1 were also co-immunoprecipitated with endogenous C53/LZAP in various cancer cells (Fig. 2B). Using co-IP assays, we briefly mapped the domains of both proteins for their interaction. It appeared that the N-terminal portion of RCAD (residues 1–400) was important for RCAD-C53 interaction, while the C (residues 269–506) terminal portion of C53/LZAP had a higher affinity to RCAD (Fig. 2, C and D). Additionally, we further mapped the interacting domains for the interaction between C53/LZAP and DDRGK1. As shown in Fig. 2, E and F, the N-terminal portion of DDRGK1 (1–114) was capable of binding to C53/LZAP, while both N- and C-fragments of C53/LZAP bound DDRGK1. It is not clear whether the C53-DDRGK1 interaction is direct or is mediated by RCAD.
FIGURE 2.
Protein interactions among C53/LZAP, RCAD, and DDRGK1. A, co-immunoprecipitation of overexpressed proteins. The proteins indicated in the figure panel were overexpressed in 293T cells. Co-immunoprecipitation was performed as described using M2 anti-Flag EZ beads (Sigma). B, co-immunoprecipitation of endogenous proteins. Endogenous C53/LZAP was immunoprecipitated with C53 polyclonal antibody, and the presence of RCAD and DDRGK1 was detected with corresponding antibodies. C, RCAD domain responsible for RCAD-C53 interaction. D, C53 domain for C53-RACD interaction. Caspase-1 dominant negative (C1DN) was used as the negative control. E, DDRGK1 domain for DDRGK1-C53 interaction. F, C53 domain for C53-DDRGK1 interaction. G, size exclusion chromatography. HeLa S100 cell lysate was subject to Superose 6 gel filtration. Aliquots of the fractions were subject to SDS-PAGE and immunoblotting. The Superose 6 column was precalibrated with molecular weight markers (GE Healthcare).
To investigate the interaction of endogenous proteins, we performed a size exclusion assay to examine if they exist in a large protein complex. HeLa cell S100 fraction was subject to gel filtration chromatography, and the collected fractions were immunoblotted with specific antibodies. As shown in Fig. 2G, RCAD protein was exclusively present in a large complex that peaked around 550 kDa. Intriguingly, a small fraction of C53/LZAP was also present in a complex with similar size, while the remaining C53/LZAP appeared in the fractions ranging between 60 and 200 kDa. The co-peaking of endogenous RCAD and C53/LZAP in the same fractions of gel filtration strongly suggests that they may be the components of a large protein complex. In contrast to RCAD and C53/LZAP, DDRGK1 was absent in the S100 fraction but present in the membrane fraction (see below).
DDRGK1 Is an ER Protein Anchored by Its N-terminal Signal Peptide
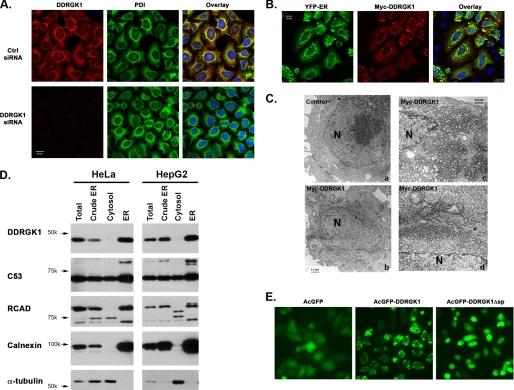
Unlike RCAD and C53/LZAP, DDRGK1 has a putative N-terminal signal peptide, indicating that it may be an ER or secreted protein. Our efforts failed to detect any secreted DDRGK1 in culture medium. Meanwhile, immunofluorescence staining with our DDRGK1 antibody showed that DDRGK1 co-localized with the ER marker PDI (Protein Disulfide Isomerase), and the specificity of our DDRGK1 antibody was confirmed in DDRGK1 knockdown cells (Fig. 3A). Furthermore, overexpressed Myc-tagged DDRGK1 co-localized with another ER marker YFP-ER (Fig. 3B), and it appeared to be on the cytoplasmic side of the ER membrane (supplemental Fig. S3). Interestingly, DDRGK1 overexpression caused abnormal ER aggregation around the nucleus (Fig. 3B). Low-magnification electron micrographs showed that electron-dense “clouds” surrounded the nuclei of the cells transfected with DDRGK1, whereas such “clouds” were absent in the control cells. By examining the EM pictures, we found that 75% of DDRGK1-overexpressing cells contained abnormal ER aggregation, whereas none of the control cells displayed any abnormality (Fig. 3C). Examination at a high magnification revealed that the electron-dense structures corresponded to massive proliferation and aggregation of the ER network in DDRGK1-overexpressing cells (Fig. 3C). We also performed subcellular fractionation to biochemically confirm its ER localization. As shown in Fig. 3D, DDRGK1 was exclusively present in the enriched ER fraction, while C53/LZAP and RCAD were present in both cytosolic and ER fractions. Of note, RCAD appeared to be cleaved in the cytosolic fraction, and the possible cause remains unclear.
FIGURE 3.
DDRGK1 is an ER protein anchored by its N-terminal signal peptide. A, immunofluorescence staining of endogenous Myc-DDRGK1. HeLa cells were transfected with either negative control siRNA or DDRGK1 siRNA 1. At 48 h post-transfection, the cells were fixed and stained with DDRGK1 antibody along with an ER marker PDI. The confocal pictures were captured by Zeiss 510 Meta microscope. B, immunostaining of Myc-DDRGK1. HeLa cells were transfected with Myc-DDRGK1 construct along with YFP-ER marker. At 24 h post-transfection, the cells were fixed and stained with Myc antibody, and the pictures were taken by Zeiss 510 confocal microscope. C, electron micrographs of HeLa cells overexpressing Myc-DDRGK1. The left panels are the images with low magnification (×4,400), whereas the right panels are the images with high magnification (×20,000). The nuclei are marked by N. The electron dense “cloud” around the nucleus in DDRGK1-overexpressing cells indicated massive amplification of the ER network, which is better illustrated in the images in the right panels. D, subcellular fractionation. The cytosolic and ER fractions were isolated by discontinuous sucrose gradient, and the aliquots were subject to SDS-PAGE and immunoblotting. Calnexin was used as the ER marker, whereas α-tubulin was used as the cytosolic marker. E, deletion of DDRGK1 signal peptide. Monomeric AcGFP-DDRGK1 fusion protein and its mutant without the N-terminal signal peptide were transfected into HeLa cells, and the images were taken with OpenLab software at 24 h post-transfection.
Except for its N-terminal putative signal peptide, DDRGK1 does not have any other recognizable transmembrane domains. We therefore speculated that this putative signal peptide may be responsible for its ER anchorage. As shown in Fig. 3E, deletion of the N-terminal 28 residues resulted in nuclear localization of AcGFP-DDRGK1 fusion protein, suggesting that the N-terminal signal peptide of DDRGK1 is indeed responsible for its ER localization.
RCAD Knockdown Causes Dramatic Reduction of C53/LZAP and DDRGK1 Proteins
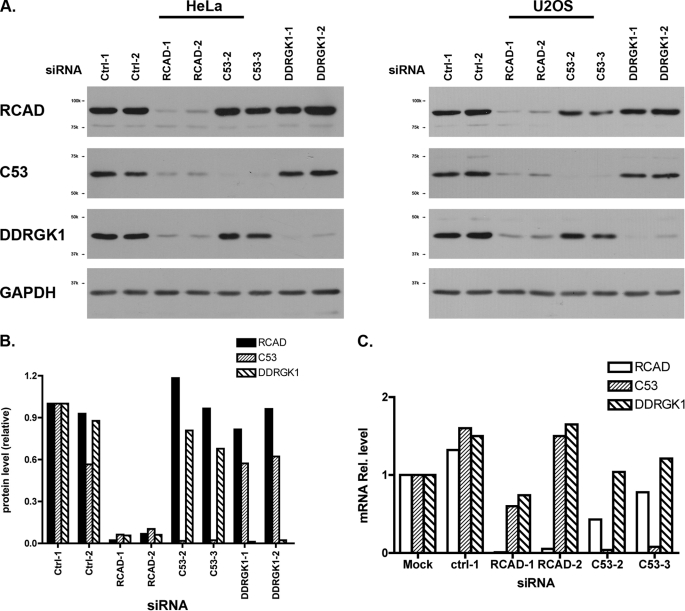
To elucidate the biological function of RCAD, we examined the effect of RCAD depletion. As shown in Fig. 4A, two siRNAs effectively knocked down endogenous RCAD (more than 90%) in both HeLa and U2OS cells, and they did not significantly affect cell morphology and proliferation. However, RCAD knockdown in HeLa cells caused remarkable reduction of C53/LZAP (94% for RCAD siRNA-1 and 90% for RCAD siRNA-2, respectively) and DDRGK1 (94% for RCAD siRNA-1 and 2) (Fig. 4, A, left panel, and B). The similar phenotype was also observed in U2OS cells (Fig. 4A, right panel). More importantly, the reduction of protein levels was not caused by the change of mRNA levels (Fig. 4C). In comparison to the mock, transfection of either negative control or RCAD siRNAs modestly altered the mRNA levels of C53/LZAP and DDRGK1, and the alterations may be due to the nonspecific effect of siRNA transfection. Yet the variations on mRNA levels appeared random and were limited within the range of 2-fold change (Fig. 4C). Therefore, the dramatic drop of C53/LZAP and DDRGK1 protein levels in RCAD knockdown cells was unlikely to be caused by transcriptional change. Additionally, we examined if C53/LZAP can affect RCAD and DDRGK1 levels. In HeLa cells, C53/LZAP knockdown exerted a modest effect on the level of DDRGK1 (80% reduction of DDRGK1 in C53/LZAP knockdown cells) and minimal effect on RCAD (Fig. 4, A and B). In contrast, C53/LZAP knockdown in U2OS cells appeared to have more significant effect on both RCAD and DDRGK1, leading to reduction of both proteins. This result indicates that unlike RCAD, the effect of C53/LZAP appears to be cell type-specific.
FIGURE 4.
RCAD knockdown results in dramatic reduction of C53 and DDRGK1 proteins. A, effect of RCAD knockdown on C53 and DDRGK1 proteins. The endogenous proteins were knockdown by indicated siRNAs, and the total lysates were subject to immunoblotting with specific antibody. B, quantitation of the immunoblots in A. The immunoblots of HeLa cells was quantified by OpenLab software. C, mRNA levels in siRNA-mediated knockdown cells. Real-time PCR was performed to evaluate the relative mRNA levels in specific knockdown cells.
RCAD Interferes with Ubiquitination of C53/LZAP and DDRGK1
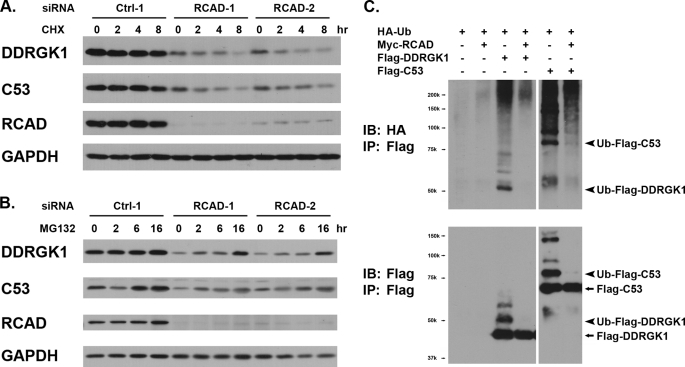
To understand how RCAD influences the protein levels of C53/LZAP and DDRGK1, we first examined the kinetics of protein degradation. As shown in Fig. 5A, treatment of control HeLa cells with cycloheximide, a commonly used protein translation inhibitor, did not cause much degradation of either C53/LZAP or DDRGK1 during the time course of treatment. In contrast, cycloheximide treatment on RCAD knockdown cells led to more rapid degradation of C53/LZAP and DDRGK1 (Fig. 5A). Although we could not exclude the possibility that RCAD may affect microRNA-mediated pathways, our result provided indirect evidence that RCAD may influence protein stability of C53/LZAP and DDGRK1.
FIGURE 5.
RCAD regulates protein stability and ubiquitination of C53/LZAP and DDRGK1. A, RCAD knockdown promoted protein degradation of C53/LZAP and DDRGK1. Control and RCAD knockdown HeLa cells were treated with cycloheximide (100 μg/ml) for periods of indicated time. Total cell lysates were subject to immunoblotting. B, proteasome inhibitor MG132 prevented degradation of C53/LZAP and DDRGK1 in RCAD knockdown cells. Control and RCAD knockdown HeLa cells were treated with MG132 (20 μm) for the indicated time, and the protein levels were evaluated by immunoblotting. C, RCAD overexpression blocked ubiquitination of C53/LZAP and DDRGK1. HeLa cells were transfected with indicated constructs. After 24 h of transfection, Flag-tagged C53/LZAP (or DDGRK1) was immunoprecipitated with Flag antibody, and then immunoblotted with hemagglutinin antibody for detection of ubiquitinated proteins.
The ubiquitin/proteasome system (UPS) is the major cellular pathway for protein degradation. To further understand the molecular mechanism underlying RCAD function, we attempted to examine whether the UPS is involved in degradation of C53/LZAP and DDRGK1. HeLa cells were transfected with either control or RCAD siRNAs, and RCAD knockdown were confirmed by immunoblotting at 60 h post-transfection (Fig. 5B). The cells were then treated with proteasome inhibitor MG132 for various periods of time. As shown in Fig. 5B, in the cells transfected with negative control siRNA, MG132 treatment did not significantly affect the levels of three proteins. This result indicates that in the control cells, all three proteins are fairly stable, and the proteasome-mediated degradation of these proteins is not very robust. In contrast, protein levels of C53/LZAP and DDRGK1 were significantly elevated by MG132 treatment in RCAD knockdown cells, and the increase became obvious even at 2 h after treatment (Fig. 5B). This result strongly suggests that RCAD knockdown renders both C53/LZAP and DDRGK1 more susceptible to the UPS-mediated protein degradation. We further tested whether RCAD can affect ubiquitination of C53/LZAP and DDRGK1. As shown in Fig. 5C, both C53/LZAP and DDRGK1 can be ubiquitinated by HA-Ubiquitin, but the modification was dramatically inhibited by RCAD overexpression. This result strongly suggests that RCAD regulates degradation of C53/LZAP and DDRGK1 by affecting their ubiquitination.
RCAD Knockdown Causes Elevated NF-κB Activity and Cell Invasion
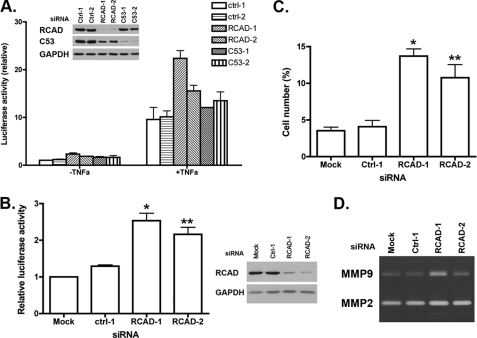
Wang et al. (3) have demonstrated that C53/LZAP acts as a suppressor of NF-κB signaling. Loss of C53/LZAP leads to the increase of NF-κB transcriptional activity and expression of its downstream targets such as MMP9. As shown above, RCAD knockdown resulted in a dramatic reduction of C53/LZAP level. Therefore, we speculated that RCAD may also influence the NF-κB activity. To test this hypothesis, we first used a luciferase reporter assay to measure the NF-κB activity in RCAD knockdown cells. In HeLa cells, knockdown of either RCAD or C53/LZAP led to elevation of both basal and TNFα-stimulated NF-κB activity (Fig. 6A). A similar result was obtained in U2OS cells (Fig. 6B). We consistently observed about a 2-fold increase of basal NF-κB activity (Fig. 6, A and B). Furthermore, like C53/LZAP, RCAD knockdown promoted cell invasion of U2OS cells (Fig. 6C), possibly due to the increased expression of matrix metalloprotease 9 (MMP9) (Fig. 6D). Taken together, our results suggest that RCAD plays an important role in NF-κB signaling.
FIGURE 6.
RCAD is involved in regulation of NF-κB signaling. A, RCAD knockdown caused elevated basal and stimulated NF-κB activity in HeLa cells. Control and RCAD knockdown HeLa cells were transfected with NF-κB luciferase reporter, and then treated with TNFα for 8 h. The NF-κB activity was scored with dual luciferase assay. B, RCAD knockdown led to elevated basal NF-κB activity in U2OS cells. *, p = 0.004; **, p = 0.03. C, RCAD knockdown promoted cell invasion of U2OS cells. *, p = 0.02; **, p = 0.04. D, zymography of conditional medium of U2OS cells.
DISCUSSION
C53/LZAP is a putative tumor suppressor that has been implicated in multiple cell signaling pathways, including DNA damage response and NF-κB signaling (1–3). Here we report the isolation and characterization of two novel C53/LZAP-interacting proteins, RCAD and DDRGK1. In this study, we confirmed the interactions among these three proteins (Figs. 1 and 2). The result of size exclusion chromatography further indicated that RCAD and C53/LZAP may form a protein complex larger than 440 kDa (Fig. 2C). Interestingly, we found that RCAD knockdown led to remarkable reduction of C53/LZAP and DDRGK1 proteins (Fig. 4). Furthermore, we found that C53/LZAP and DDRGK1 were more susceptible to the proteasome-mediated degradation in RCAD knockdown cells, while their ubiquitination was significantly attenuated by RCAD overexpression (Fig. 5). In addition, we found that like C53/LZAP, RCAD also plays an important role in regulation of NF-κB signaling (Fig. 6). Taken together, our findings strongly suggest that RCAD is a novel regulator of C53/LZAP tumor suppressor and NF-κB signaling pathway.
By searching public databases and our own observations, we have found a few interesting features (or lack of features) shared by these three relatively novel proteins. First, like C53/LZAP, both RCAD and DDRGK1 are highly conserved during evolution. The orthologs of three proteins are readily found in the genomes of vertebrates, invertebrates, and plants, but not of yeast. Second, extensive protein domain/motif search failed to reveal any significant known domains/motifs except a signal peptide and a partial PCI domain in DDRGK1. Third, all of them are ubiquitously expressed in multiple tissues, organs, and cell lines even though the protein levels may vary. These observations indicate that these three proteins may co-evolve and play pivotal roles in fundamental cellular processes. Further structural and functional studies will provide more insight into biochemical mechanisms of these proteins.
As shown in our study, DDRGK1 is an ER protein that is anchored by a putative N-terminal signal peptide (Fig. 3), a result that is in agreement with the recently published reports (4, 5). Although we cannot exclude the possibility that its signal peptide is cleaved under certain circumstances, so far we have not detected any cleavage product or secretion of this protein. Interestingly, overexpression of DDRGK1 leads to significant amplification and re-organization of the ER network in HeLa and other cell lines (Fig. 3C).3 It has been reported that overexpression of certain ER proteins can cause ER amplification and formation of so-called “organized smooth ER” (OSER) with stacked ER membrane arrays (7). For example, overexpression of ER enzyme b (5) and its GFP fusion protein induced formation of karmellae, whorls, and cyrstalloid OSER structures, and the formation of OSER involved homotypic interactions between cytoplasmic domains of those ER-anchored proteins (7). OSER structures were also reported in a variety of cells under physiological conditions (8–10). Examination of the electron micrographs of DDRGK1-expressing cells at a high-magnification suggests that DDRGK1-induced ER re-organization appeared less symmetric and organized than the one induced by GFP-b(5) (Fig. 3C). More studies will be performed to address whether DDRGK1-induced ER structures are OSER, and whether DDRGK1 play a role in ER re-organization under physiological conditions. Interestingly, we also found that a large fraction of C53/LZAP and RCAD are also contained in the ER fraction (Fig. 3D), indicating a possible role of these three proteins in ER-associated cellular processes.
RCAD is another novel C53/LZAP-interacting protein, and plays a pivotal role in regulation of protein levels of both C53/LZAP and DDRGK1. Our results showed that RCAD-mediated regulation occurred on a post-transcriptional level. C53/LZAP and DDRGK1 are relatively stable under normal condition, while they become more susceptible to the UPS-mediated protein degradation in the absence of RCAD (Fig. 4). Ubiquitination of both C53/LZAP and DDRGK1 is significantly blocked by RCAD overexpression, indicating that RCAD may regulate ubiquitination of its downstream targets (Fig. 4). The remaining question is how RCAD acts. As the biochemical mechanism of C53/LZAP (or DDRGK1) ubiquitination remains completely unknown, we can only speculate some possible scenarios. First, direct binding of RCAD to C53/LZAP (or DDRGK1) may sterically hinder ubiquitination process. Nonetheless, our study so far indicates that this scenario may be unlikely. Despite the fact that the exact stoichiometry of the putative RCAD/C53 complex remains to be determined, our gel filtration assay demonstrated that the majority of cytosolic C53/LZAP was free of RCAD (Fig. 2G). In contrast, RCAD knockdown resulted in more than 90% reduction of C53/LZAP protein (Fig. 4). These observations prompt us to postulate that RCAD may function in a catalytic way. Very recently, Tatsumi et al. (4, 11) reported that RCAD (designated as Ufl1 in their report) functions as a novel E3-ligase in the Ufm1 system, a newly identified ubiquitin-like system. Intriguingly, DDRGK1/C20orf116 is a target of RCAD/Ufl1-mediated ufmylation (4). It would be of great interest to test whether RCAD/Ufl1-mediated modification by Ufm1 affects protein stability of C53/LZAP and DDRGK1.
Our results strongly suggest that RCAD and C53/LZAP may form a larger protein complex in vivo. First, our co-immunoprecipitation and in vitro interaction assays showed that RCAD and C53/LZAP interacted with each other. Second, cytosolic RCAD and C53/LZAP co-peaked in the same fractions of size exclusion chromatography. Interestingly, nearly all RCAD was in a complex form, while only a small fraction of C53/LZAP remained in a complex form. Third, knockdown of one protein affects the level of another one, although the effects are not equivalent. RCAD knockdown led to a dramatic reduction of C53/LZAP (more than 90%), while C53/LZAP knockdown resulted in an only approximate two-third reduction of RCAD in U2OS cells, and had a minimal effect in HeLa cells. This inequality of their influences on each other may reflect different molecular mechanisms of regulation. As discussed above, we speculate that RCAD may regulate C53/LZAP and its other targets in a catalytic manner, such as acting as an E3 ligase for Ufm1. In contrast, C53/LZAP may regulate RCAD in a stoichiometric fashion, and therefore, RCAD is stable only while complexing with C53/LZAP. This may provide an explanation why C53/LZAP's effect on RCAD differs in various cells. It is possible that more RCAD and C53/LZAP are in the complex form in U2OS cells than in HeLa cells, whereas HeLa cells may contain more free C53/LZAP that compensates for siRNA-mediated depletion, and thereby stabilizes RCAD. Therefore, C53/LZAP knockdown in U2OS cells would have more significant effect on RCAD level than in HeLa cells.
One interesting observation from the study of Tatsumi et al. (4) is that RCAD/Ufl1 lacks typical signature domains of E3 ligases, such as ring finger and HECT. Our finding that C53/LZAP and RCAD may form a stable large protein complex raises an intriguing possibility that this putative Ufm1 E3 ligase may consist of multiple components. Interestingly enough, both Ufm1 and Ufc1 are among the list of C53/LZAP-interacting proteins (6). Therefore, it is plausible that C53/LZAP may also play a role in ufmylation, probably through sequestering multiple components of the Ufm1 system. It would be of great importance to fully identify the components of this putative C53/LZAP and RCAD complex, and investigate their biochemical role in Ufm1-mediated modification.
In addition to its role in regulation of protein stability of C53/LZAP and DDRGK1, RCAD also plays a role in regulation of NF-κB signaling. RCAD knockdown leads to elevated NF-κB activity and cell invasion. However, it remains unclear whether RCAD-mediated regulation of NF-κB signaling is C53/LZAP-dependent or not. In contrast to C53/LZAP, overexpression of RCAD did not suppress NF-κB activity,3 suggesting that RCAD may not directly inhibit NF-κB activity. In addition, we constantly observed that RCAD knockdown resulted in a slightly higher NF-κB activity than C53/LZAP knockdown in luciferase report assays even though C53/LZAP level in RCAD knockdown cells remained slightly higher than the one in C53/LZAP knockdown cells (Fig. 6). This observation indicates that in addition to C53/LZAP, RCAD may also regulate other components of the NF-κB signaling pathway. Because RCAD modulates ubiquitination and protein stability of C53/LZAP and DDRGK1, it would be of great interest to examine the effect of RCAD on other components of the NF-κB pathway, which are also heavily regulated by ubiquitination and other ubiquitin like modification. As a note, we observed that RCAD knockdown led to slightly lower level of IκBα in U2OS cells, and more rapid degradation upon TNFα stimulation,3 even though IκBα mRNA level was higher (1.5–2-fold) in RCAD knockdown cells.3
Another intriguing question remains to be answered is whether RCAD, like C53/LZAP, functions as a tumor suppressor in tumorigenesis and metastasis. The human RCAD gene is located in chromosome 6q16.1, a region that was reported to be frequently lost in prostate and gastric cancers, as well as osteosarcoma, melanoma, gallbladder, and bile duct cancer cell lines (12–16). The RCAD level in invasive breast cancer cell MDA-MB-231 is remarkably lower than in other breast cancer cell lines (Fig. 1E). We also found that RCAD is involved in regulation of C53/LZAP stability and NF-κB signaling. Taken together, our study raises an interesting possibility for RCAD involvement in cancer biology, which will be fully addressed by future genetic and cancer model studies.
Acknowledgments
We thank Dr. Fei Sun (University of Pittsburg) for the HA-ubiquitin construct and Dr. Erik Snapp (Albert Einstein College of Medicine) for insightful advice on OSER.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM081776 (to H. L.), the Children's Memorial Research Center, and the Clarke family.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
H. Li, unpublished observations.
- FBS
- fetal bovine serum
- DMEM
- Dulbecco's modified Eagle's medium
- ER
- endoplasmic reticulum
- EST
- expressed sequence tag
- RCAD
- regulator of C53/LZAP and DDRGK1.
REFERENCES
- 1.Jiang H., Luo S., Li H. (2005) J. Biol. Chem. 280, 20651–20659 [DOI] [PubMed] [Google Scholar]
- 2.Jiang H., Wu J., He C., Yang W., Li H. (2009) Cell Res. 19, 458–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J., An H., Mayo M. W., Baldwin A. S., Yarbrough W. G. (2007) Cancer Cell 12, 239–251 [DOI] [PubMed] [Google Scholar]
- 4.Tatsumi K., Sou Y. S., Tada N., Nakamura E., Iemura S. I., Natsume T., Kang S. H., Chung C. H., Kasahara M., Kominami E., Yamamoto M., Tanaka K., Komatsu M. (2010) J. Biol. Chem. 285, 5417–5427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neziri D., Ilhan A., Maj M., Majdic O., Baumgartner-Parzer S., Cohen G., Base W., Wagner L. (2009) Biochim. Biophys. Acta 1800, 430–438 [DOI] [PubMed] [Google Scholar]
- 6.Ewing R. M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M. D., O'Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y. V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J. P., Duewel H. S., Stewart, Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S. L., Moran M. F., Morin G. B., Topaloglou T., Figeys D. (2007) Mol. Syst. Biol. 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snapp E. L., Hegde R. S., Francolini M., Lombardo F., Colombo S., Pedrazzini E., Borgese N., Lippincott-Schwartz J. (2003) J. Cell Biol. 163, 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yorke M. A., Dickson D. H. (1985) Cell Tissue Res. 241, 629–637 [DOI] [PubMed] [Google Scholar]
- 9.Abran D., Dickson D. H. (1992) Exp. Eye Res. 54, 737–745 [DOI] [PubMed] [Google Scholar]
- 10.Gong F. C., Giddings T. H., Meehl J. B., Staehelin L. A., Galbraith D. W. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2219–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komatsu M., Chiba T., Tatsumi K., Iemura S., Tanida I., Okazaki N., Ueno T., Kominami E., Natsume T., Tanaka K. (2004) EMBO J. 23, 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu T., Hano H. (2008) Prostate Cancer Prostatic Dis. 11, 357–361 [DOI] [PubMed] [Google Scholar]
- 13.van Gils W., Kilic E., Brüggenwirth H. T., Vaarwater J., Verbiest M. M., Beverloo B., van Til-Berg M. E., Paridaens D., Luyten G. P., de Klein A. (2008) Melanoma Res. 18, 10–15 [DOI] [PubMed] [Google Scholar]
- 14.Sun M., Srikantan V., Ma L., Li J., Zhang W., Petrovics G., Makarem M., Strovel J. W., Horrigan S. G., Augustus M., Sesterhenn I. A., Moul J. W., Chandrasekharappa S., Zou Z., Srivastava S. (2006) DNA Cell Biol. 25, 597–607 [DOI] [PubMed] [Google Scholar]
- 15.Saito S., Ghosh M., Morita K., Hirano T., Miwa M., Todoroki T. (2006) Oncol. Rep. 16, 949–956 [PubMed] [Google Scholar]
- 16.Li B. C., Chan W. Y., Li C. Y., Chow C., Ng E. K., Chung S. C. (2003) Diagn. Mol. Pathol. 12, 193–200 [DOI] [PubMed] [Google Scholar]
- 17.Bozidis P., Williamson C. D., Colberg-Poley A. M. (2007) Current Protocols in Cell Biology, Chapter 3, Unit 3.27, John Wiley & Sons, Inc., Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 18.Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]