Abstract
The major immediate-early promoter (MIEP) region of human cytomegalovirus (HCMV) plays a critical role in the regulation of lytic and latent infections by integrating multiple signals supplied by the infecting virus, the type and physiological state of the host cell, and its extracellular surroundings. The interaction of cellular transcription factors with their cognate binding sites, which are present at high densities within the enhancer upstream from the MIEP core promoter, regulate the rate of IE gene transcription and thus affect the outcome of HCMV infection. We have shown previously that the NF-κB binding sites within the MIEP enhancer and cellular NF-κB activity induced by HCMV infection are required for efficient MIEP activity and viral replication in quiescent cells (P. Caposio, A. Luganini, G. Hahn, S. Landolfo, and G. Gribaudo, Cell. Microbiol. 9:2040-2054, 2007). We now show that the inactivation of either the Elk-1 or serum response factor (SRF) binding site within the enhancer also reduces MIEP activation and viral replication of recombinant HCMV viruses in quiescent fibroblasts. In these cells, we show that the expression of either Elk-1 or SRF is required for optimal IE gene expression, and that the HCMV-stimulated activation of the MEK1/2-ERK1/2 signaling axis leads to Elk-1 transcriptional competency. Furthermore, the replication kinetics of recombinant viruses in which NF-κB, Elk-1, and SRF binding sites all are inactivated demonstrate that the higher levels of Elk-1 and SRF binding to MIEP in proliferating cells can compensate even for a lack of HCMV-induced NF-κB-mediated MIEP transactivation. These observations highlight the importance of the combination of different MIEP binding sites to optimize IE gene expression in cells in different physiological states.
A hallmark of human cytomegalovirus (HCMV) pathogenesis is its ability to productively replicate in a remarkably broad range of different cell types, including epithelial, fibroblast, endothelial, macrophage, dendritic, smooth-muscle, and neuronal cells, as well as hepatocytes (2, 5, 19, 23, 29, 38, 39). Macrophages and neurons are terminally differentiated and do not undergo cellular division, whereas endothelial, epithelial, and smooth-muscle cells remain predominantly within the stationary phase of the cell cycle and divide only when specific circumstances are presented, such as tissue injury. It therefore seems that the coevolution of HCMV with its host has resulted in HCMV developing mechanisms that enable it to manipulate the host cell's regulatory systems in nonproliferating cell types that present intracellular environments that are unfavorable for high levels of viral DNA replication and virus production. Numerous studies have indeed demonstrated the ability of HCMV to modulate (both activate and repress) various cellular signaling pathways and transcription factor systems that positively regulate viral gene expression. However, despite the fact that most of the cell types susceptible to HCMV infection in vivo are in a growth-arrested state, the vast majority of these studies have utilized rapidly dividing cell cultures rather than cultures of quiescent cells (29, 48).
A critical step in HCMV replication is the synthesis of the major immediate-early (MIE) proteins IE1 and IE2, which regulate subsequent early (E) and late (L) gene expression during lytic infections and manipulate a variety of cellular functions to optimize the cellular environment for viral replication (28, 29, 42). Failure to express a robust IE program, as observed in undifferentiated cell types, results in a latent outcome of HCMV infection (37). The expression of MIE genes is under the control of the major immediate-early promoter (MIEP) region, which contains one of the most powerful transcriptional enhancers identified to date (28, 29, 41). The MIEP enhancer spans the region between approximately −550 and −39 relative to the MIE transcription start sites at +1, and it displays a characteristic array of repetitive binding sites for various cellular transcription factors that may act as positive or negative regulatory factors of MIEP activity (28, 29, 41). The MIEP enhancer has two components that are proximal (−39 to −300) and distal (−300 to −550) relative to the transcription start site of the MIE promoter. The proximal and distal enhancer portions both are required for efficient MIE gene expression and viral replication (17, 27, 41). However, the reason why the MIEP enhancer should contain a multitude of diverse transcription binding sites is unclear. Comparative analyses have shown that the arrangement and number of binding sites of the MIEP enhancer vary among different species-specific viruses (41). Thus, it can be hypothesized that present-day MIEPs result from evolutionary molecular negotiations between the virus and its specific hosts, generating viral transcriptional regulatory elements that, in turn, ensure adequate levels of MIEP activation and IE gene expression across a variety of cell types susceptible to infection and a variety of conditions under which such cells become infected (23, 28, 29, 39).
Of the MIEP transcription factor binding sites that are common across the various CMVs and thought to positively contribute to IE gene regulation, the four NF-κB binding sites and their cognate signaling pathway have received the most attention, despite the controversial results that have been obtained regarding their role during viral replication (1, 7, 8, 11, 15). For example, while the pharmacological inhibition of NF-κB activity was shown to impair HCMV replication, suggesting that the NF-κB signaling axis is important for the life cycle of HCMV (7, 11), the deletion of the four MIEP NF-κB sites in the context of the virus cycle did not significantly affect viral replication in cycling human fibroblasts (1, 8, 15). More recently, we found that the integrity of the NF-κB signaling and MIEP binding sites is essential for IE gene expression and viral replication in growth-arrested but not in proliferating fibroblasts and endothelial cells (8). Thus, these observations suggest that the requirement of NF-κB signaling for MIEP activity and HCMV replication depends on the proliferative state of the host cell, and that proliferating but not quiescent cells may contain transcription factors that bind to MIEP and compensate for NF-κB inactivation.
In fact, the MIEP contains binding sites that interact with transcription factors whose activity may be stimulated by growth factors, such as the activator protein 1 (AP-1), the serum response factor (SRF), and the ETS domain transcription factor ets-like gene-1 (Elk-1) (28, 40). Although AP-1 activity in HCMV-infected fibroblasts has been shown to bind to a sequence within the MIEP located between −168 and −174, the role of this protein in MIEP activation still is unclear (3, 4, 34). On the other hand, previous work with growth factor-regulated cellular c-fos and egr-1 promoters has shown that Elk-1 does not bind directly to the serum response element (SRE) (a combination of ETS and SRF binding sites) but instead forms a serum-inducible ternary protein complex with SRF named the ternary complex factor (TCF) (6, 30, 36, 43). The TCF complex thus contains the 67-kDa constitutively expressed SRF protein that binds to the core motif CCATATTAGG and the growth factor-activated 62-kDa Elk-1 protein that binds to the adjacent ETS motif CAGGAT, located 2 bp 5′ of the SRF motif (6, 36). The phosphorylation of Elk-1 within its regulatory domain by the mitogen-activated protein kinase/extracellular signal regulated kinase (MAPK/ERK) pathway triggers an important series of conformational changes that enhance its recruitment to the SRE and potentiate the transcriptional activity of TCF (6, 14, 36). A sequence similar to that of the SRE and referred to as the SRF/Elk-1 element (SEE) is located between −538 and −523 relative to the transcription start site at +1 in the HCMV MIEP. The SEE was shown to promote the formation of TCF in nuclear extracts from uninfected human tumor cell lines (9); however, its role in the context of HCMV replication is unknown, as is whether all or only a subset of the TCF factors are necessary for MIEP activation and IE gene expression.
The aim of this study was to investigate the role of the TCF partners in the context of HCMV productive replication in growth-arrested and proliferating cells. These two cell conditions (i.e., quiescent and proliferating) were adopted as two distinct experimental models to consider at least two of the wide range of cell contexts under which the MIEP is forced to operate during infection in the natural host. In quiescent cells, we show that Elk-1 is rapidly phosphorylated by the HCMV-activated MEK-ERK signaling pathway, and that MEK activation is required for optimal IE and E gene expression. Furthermore, we show that the expression of both Elk-1 and SRF and their binding to the SEE is critical for HCMV replication in quiescent cells, and that high levels of TCF activity in proliferating cells are able to compensate for the lack of virus-induced NF-κB activity. Since most of the cells productively infected by HCMV in the natural host exist in a nondividing growth state, the HCMV-induced NF-κB and TCF activation observed in the quiescent cell condition might play a major role in the regulation of MIE gene expression and the initiation of the viral replication cycle.
MATERIALS AND METHODS
Oligonucleotides.
All oligonucleotides used for PCR, mutagenesis, or sequencing were obtained from Invitrogen (Table 1 shows all oligonucleotide sequences).
TABLE 1.
Oligonucleotides used for cloning, BAC mutagenesis, shRNA expression, and IE mRNA analysis
| Primer designation | Sequence (5′ to 3′)a |
|---|---|
| MIEP forward | GTTTGCGTCAATGGGG |
| MIEP reverse | GCATATGTTGTATCCCCACATATC |
| AP-1-mut-EcoRI | ACTTGGAAATCCCCGTgaattcAACCGCTATCCACGG |
| NF-κB-2-AP-1-mut-KpnI-EcoRI | GGAGACTTGCAAggtaccGTgaattcAACCGCTATCCA |
| Elk-1-mut-EcoRI | CGTAAGTTATGTAACGCgaattcCCATATATGGGCTATGAAC |
| SRF-mut-KpnI | ATGTAACGCGGAACTCCAggtaccTGCTATGAACTAATGACC |
| SEE-mut-EcoRI-KpnI | TTATGTAACGCgaattcCCAggtaccTGCTATGAACTAATGACC |
| NF-κB-1-mut-StuI | GTTGTTACGACATTTTCCAAaggcctGTTGATTTTGGTGCCA |
| NF-κB-2-mut-KpnI | TGGGGTGGAGACTTGCAAggtaccGTGAGTCAAACCGCTAT |
| NF-κB-3-mut-BglII | ATGTACTGCCAAGTAGCCagatctCCATAAGGTCATGTACT |
| NF-κB-4-mut-BglII | CACCCATTGACGTCAATGCCagatctCTATTGGCGTTACTA |
| MIEP-galK forward | GCCCATTTGCGTCAATGGGGCGGAGTTATTACGACATTTTGGAAAGTCCCCCTGTTGACAATTAATCATCGGCA |
| MIEP-galK reverse | GCATATGTTGTATCCATATCATAATATGTACATTTATATTGGCTCATGTCTCAGCACTGTCCTGCTCCTT |
| shElk-1 A | TCCGCAAGGTGAGCGGCCAGAAGTTCGTC |
| shElk-1 B | CTCGCTGCCTCCTAGCATTCACTTCTGGA |
| shSRF C | CACTGATTCAGACCTGCCTCAACTCGCCA |
| shSRF D | GCAACTGACTTCATTTGTGCCACACGCAT |
| shRNA GFP | CACAAGCTGGAGTACAACTACAACAGCCA |
| IE1 forward | CAAGTGACCGAGGATTGCAA |
| IE1 reverse | CACCATGTCCACTCGAACCTT |
| IE2 forward | TGACCGAGGATTGCAACGA |
| IE2 reverse | CGGCATGATTGACAGCCTG |
| β-Actin forward | CAAAAGCCTTCATACATCTC |
| β-Actin reverse | TCATGTTTGAGACCTTCAA |
Lowercase boldface letters indicate restriction enzyme sites in oligonucleotide sequences.
Cells and culture conditions.
Low-passage human embryonic lung fibroblasts (HELFs) were grown as monolayers in minimum essential medium (MEM) (Gibco-BRL) supplemented with 10% fetal calf serum (FCS) (Gibco-BRL), 2 mM glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate (high-serum medium). Quiescent HELFs (arrested in G0/G1 phase) were obtained by culturing subconfluent cultures for 96 h in minimum essential medium (MEM) containing 0.5% FCS (low-serum medium). Flow cytometry demonstrated that more than 90% of the cells were growth arrested.
Virus preparation and infections.
HCMV strain AD169 (VR-538) was purchased from the ATCC. Viral stocks were prepared by infecting HELF cells at a virus-to-cell ratio of 0.01. Cells were incubated in MEM supplemented with 1% heat-inactivated FCS and cultured until a marked cytopathic effect was observed. Stocks then were prepared from sonicated cells, subjected to centrifugal clarification, and frozen at −80°C. Mock infection fluid was prepared from uninfected cells by the same procedure. Virus titers were determined by plaque assay on HELFs. For the determination of viral replication kinetics, quiescent or proliferating HELFs were infected with fusion-inducing factor X (FIX)-bacterial artificial chromosome (BAC)-derived viruses at a multiplicity of infection (MOI) of 0.1. Mock-infected control cultures were exposed to an equal volume of mock-infecting fluid. Virus adsorptions were carried out for 2 h at 37°C. For all experiments, the time at which virus first was added to the cells is considered time zero. Following infection, cultures were maintained in low-serum medium or 10% serum medium for various times postinfection (p.i.). Thereafter, the cells and supernatants were harvested and disrupted by sonication. Viral titers then were measured by an indirect immunoperoxidase staining procedure on HELF cells (13). The HCMV strain used in the control experiments for the growth kinetics analysis was reconstituted from the FIX-BAC containing the genome of HCMV clinical isolate VR1814 (16, 32).
BAC mutagenesis.
The HCMV IE enhancer sequence (positions −52 to −667 relative to the IE1/IE2 transcription start site) was amplified out of the purified HCMV VR1814 genome by PCR using the primers shown in Table 1. The HCMV VR1814 IE enhancer fragment then was ligated between restriction sites HindIII and NheI of the pGL3-basic vector (Promega) to obtain the pMIEPGL3 construct. The correctness of the amplified viral sequences was confirmed by sequencing. To derive MIEP sequences with inactivated binding sites for AP-1, Elk-1, SRF, SEE (Elk-1 and SRF), and all four NF-κB sites (designated 4NF-κB), a Stratagene QuikChange XL site-directed mutagenesis kit was used. The AP-1 (at −169 with respect to the IE1/IE2 transcription start site), Elk-1 (at −522), SRF (at −532), SEE (Elk-1 plus SRF; from −521 to −539), and four NF-κB (at −98, −161, −265, and −412) sites of pMIEPGL3 were changed to unique restriction sites (−169 to −174, EcoRI; −522 to −528, EcoRI; −532 to −538, KpnI; −98 to −103, StuI; −161 to −166, −265 to −270, and −412 to −421, BglII) using the oligonucleotides AP-1-mut-EcoRI, Elk-1-mut-EcoRI, SRF-mut-KpnI, SEE-mut-EcoRI-KpnI, NF-κB-1-mut-StuI, NF-κB-2-mut-KpnI, NF-κB-3-mut-BglII, and NF-κB-4-mut-BglII (Table 1 lists all sequences) and their complementary oligonucleotides. To generate the mutation in the AP-1 site when the NF-κB site at −161 already was mutated, the primer NF-κB-2-AP-1-mut-KpnI-EcoRI (Table 1) and its complement were used. The correctness of the introduced mutations in the resulting plasmids pMIEP ΔAP-1, pMIEP ΔElk-1, pMIEP ΔSRF, pMIEP ΔSEE, pMIEP Δ4NF-κB, pMIEP Δ4NF-κB-AP-1, and pMIEP Δ4NF-κB-SEE was confirmed by sequencing.
VR1814 mutants with mutations in the MIEP AP-1, Elk-1, SRF, SEE, and 4NF-κB response elements were generated by a two-step replacement strategy using the galk positive/negative selection method described by Warming et al. (44). Briefly, the FIX-BAC (a gift from T. Shenk) was electroporated into Escherichia coli SW102 (a gift from N. Copeland). In the first step, the galK open reading frame (ORF) was amplified from pgalK (a gift from N. Copeland) by PCR using the MIEP-galk primer set. At the 3′ ends of the forward and reverse primers, specific sequences (of 24 and 20 bp, respectively) dictate the amplification of the galK cassette, and their 5′-end 50-bp tails are homologous to the HCMV VR1814 IE enhancer sequences between nucleotides −52498 and −53104 of the FIX-BAC complete sequence (GenBank accession no. AC146907). Following PCR, the 1,241-bp PCR product was digested with DpnI to remove any plasmid template and gel purified. To accomplish the homologous recombination, approximately 50 ng of DNA was electroporated into SW102 bacteria harboring FIX-BAC. Cells then were plated on minimal medium (M63) agar plates containing 0.2% galactose and chloramphenicol and incubated at 32°C for 5 days. The colonies that appeared were streaked twice on MacConkey agar plates containing 0.2% galactose and chloramphenicol that produced bright red bacterial colonies. One of the single Gal+ FIX ΔMIEP-BAC colonies was further characterized for MIEP replacement by PCR and used to initiate the counterselection step. To replace the galK cassette in the second step, the VR1814 MIEP enhancer sequences, each containing one of the different binding site mutations (AP-1, Elk-1, SRF, SEE, or 4NF-κB binding sites), were amplified by PCR from the pMIEP ΔAP-1, pMIEP ΔElk-1, pMIEP ΔSRF, pMIEP ΔSEE, pMIEP Δ4NF-κB, pMIEP Δ4NF-κB-AP-1, and pMIEP Δ4NF-κB-SEE plasmids using the MIEP primer set (Table 1) corresponding to the 3′ ends of the homology arms used in the first selection step. The PCR products were digested with DpnI and gel purified, and 200 ng was electroporated in SW102 cells harboring the FIX ΔMIEP-BAC clone. To select for bacteria with the loss of the galK gene, the transformed bacteria were plated on M63 agar plates containing 0.2% 2-deoxygalactose (DOG) with glycerol as the sole carbon source and chloramphenicol (44). Gal− colonies were characterized for the replacement of galK sequences with the mutated MIEP versions by the PCR amplification of the whole segment, followed by restriction enzyme analysis and sequencing. Two FIX ΔAP-1, ΔElk-1, ΔSRF, ΔSEE, Δ4NF-κB, Δ4NF-κB-AP-1, and Δ4NF-κB-SEE BAC clones were selected and produced. To rescue the mutations of FIX ΔSEE and FIX Δ4NF-κB-SEE, the wild-type (wt) IE enhancer sequence was amplified by PCR from pMIEPGL3 with the same primers set as that used for the amplification of the 4NF-κB- and SEE-mutated sequences. The substitution of the mutated MIEP element in FIX ΔSEE-BAC and in FIX Δ4NF-κB-SEE-BAC then was performed using the two-step replacement strategy and the galK positive/negative selection method as described above. Recombinant FIX ΔSEE REV-BAC and FIX Δ4NF-κB-SEE REV-BAC were characterized for the substitution of the 4NF-κB- and SEE-mutated IE enhancer with the wt MIEP using PCR amplification followed by restriction enzyme analysis and sequencing. Recombinant FIX ΔAP-1, FIX ΔElk-1, ΔSRF, FIX ΔSEE, FIX Δ4NF-κB, FIX Δ4NF-κB-AP-1, FIX Δ4NF-κB-SEE, FIX ΔSEE REV, and FIX Δ4NF-κB-SEE REV viruses, as well as the wt FIX virus, were reconstituted by the electroporation of the corresponding BACs into HELF cells. A plasmid expressing HCMV pp71, the pCMV-pp71 (a gift from T. Shenk), was cotransfected with BAC DNAs.
Nuclear extract isolation and electrophoretic mobility shift assay (EMSA).
HELFs were grown to subconfluence, serum starved, and infected with HCMV AD169 (MOI, 5 PFU/cell). Where indicated, growth-arrested HELFs were treated with U0126 (20 μM; Sigma) for 1 h prior to infection. U0126, a selective inhibitor of MEK1/2 (12), also was present during infection and subsequent incubation periods. At the indicated times p.i., cells were washed in cold PBS, harvested, and centrifuged to collect the pellet. The pellets were incubated for 20 min on ice with a cytoplasmic isolation buffer (10 mM HEPES [pH 7.6], 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.5% NP-40, protease inhibitor cocktail [Sigma]). Samples were centrifuged, and the nuclear pellets were collected by removing the supernatant containing the cytoplasmic extract, washed in cytoplasmic isolation buffer without NP-40, centrifuged, and incubated for 30 min on ice with a nuclear isolation buffer (20 mM Tris-HCl [pH 8.0], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM PMSF, 25% glycerol, protease inhibitor cocktail [Sigma]). After centrifugation, supernatants containing the nuclear extracts were collected and stored at −70°C.
Electrophoretic mobility shift assays were carried out as previously described (8). Briefly, nuclear extracts (10 μg of proteins) were incubated in a binding buffer (10 mM Tris-HCl [pH 7.5], 50 mM NaCl, 0.5 mM EDTA, 0.5 mM dithiothreitol, 1 mM MgCl2, 5% glycerol) containing 1 μg of poly(dI-dC) (Pharmacia) and the 32P oligonucleotide probe representing the wild-type HCMV SEE motif (−518 to −547 with respect to the IE1/2 transcription start site). Complexes were analyzed by nondenaturating 4% polyacrylamide gel electrophoresis, dried, and detected by autoradiography. For supershift experiments, 1 μg of rabbit polyclonal antibodies recognizing SRF (sc-335X; Santa Cruz Biotechnology) or 1 μg of a mouse monoclonal against p-Elk-1 (Ser383; sc-8406X; Santa Cruz Biotechnology) was added to the solutions after protein-DNA incubation for 30 min at 20°C, and the DNA-protein complexes were resolved in nondenaturing 4% polyacrylamide gel. Unlabeled 30-bp annealed SEE oligonucleotide was added as the competitor DNA in 300-fold molar excess above the level of the SEE probe.
Immunoblotting.
For immunoblotting, fibroblasts were grown to subconfluence, serum starved, and infected with HCMV AD169 (MOI, 5 PFU/cell). Where indicated, they were treated with U0126 (20 μM; Sigma) for 1 h prior to infection. U0126 also was present during infection and subsequent incubation periods. At the indicated times p.i., whole-cell protein extracts were prepared by resuspending pelleted cells in lysis buffer containing 50 mM Tris-Cl (pH 6.8), 1% sodium dodecyl sulfate (SDS), 1× protease inhibitor cocktail (P8340; Sigma), 1× phosphatase inhibitor cocktail 1 (P2850; Sigma), 1× phosphatase inhibitor cocktail 2 (P5726; Sigma). After being boiled at 95°C for 5 min, soluble proteins were collected by centrifugation at 15,000 × g for 10 min. Supernatants were analyzed for protein concentration with a Bio-Rad Dc protein assay kit (Bio-Rad Laboratories) and stored at −80°C. Proteins were separated by SDS-12% polyacrylamide gel electrophoresis (50 μg of protein per lane) and then transferred to Immobilon-P membranes (Millipore). The filters were blocked in a solution of 5% nonfat dry milk, 10 mM Tris-Cl (pH 7.5), 100 mM NaCl, and 0.1% Tween 20 and then immunostained with rabbit polyclonal antibodies against phosphorylated MEK1/2 (p-MEK1/2) (Ser217/221; 9121; Cell Signaling Technology) (diluted 1:500), p-ERK1/2 (Thr202/Tyr204; 9101; Cell Signaling Technology) (diluted 1:1,000), ERK1/2 (9102; Cell Signaling Technology) (diluted 1:1,000), Elk-1 (9182; Cell Signaling Technology) (diluted 1:1,000), SRF (sc-335X; Santa Cruz Biotechnology) (diluted 1:500), or mouse monoclonal antibodies against p-Elk-1 (Ser383; sc-8406X; Santa Cruz Biotechnology), IEA (IE1 plus IE2) (11003; Argene) (diluted 1:250), and UL44 (clone 1202; Goodwin Institute, Plantation, FL) (diluted 1:1,000); mouse monoclonal antibody (MAb) against actin (MAB1501R; Chemicon, Temecula, CA) (diluted 1:2,000) was used as the control for protein loading. Immunocomplexes were detected with sheep anti-mouse or donkey anti-rabbit immunoglobulin antibodies conjugated to horseradish peroxidase (Amersham) and visualized by enhanced chemiluminescence (Super Signal; Pierce).
Inhibition of Elk-1 and SRF protein expression.
HELFs cells were transiently transfected using TransIT-LT1 transfection reagent (OriGene Technologies, Inc.) with 5 μg of either a pRS short hairpin RNA (shRNA) expression plasmid containing a 29-nucleotide sequence insert specific for either Elk-1 (shElk-1 A or shElk-1 B) or SRF (shSRF C or shSRF D) or a pRS plasmid containing a noneffective shRNA cassette against green fluorescent protein (GFP) as a negative control for specific gene downregulation (NCS) (Origene Technologies, Inc.) (Table 1 lists the sequences) and then incubated in high- or low-serum medium for 72 h. Thereafter, total cell extracts were prepared and analyzed by immunoblotting with rabbit anti-Elk-1 or anti-SRF antibodies. To investigate the effects of shRNAs on HCMV IE gene expression, transiently transfected cells were infected with HCMV AD169 (MOI of 3 PFU/cell), and at 24 h p.i. total RNA was isolated and reverse transcribed; real-time reverse transcription-PCR (RT-PCR) was carried out for IE1, IE2, and β-actin (see below).
RT-PCR analysis.
Real-time RT-PCR analysis was performed on an Mx 3000 P (Stratagene) using SYBR green as a nonspecific PCR product fluorescent label. After HCMV infection and cell treatment, total cellular RNA was isolated using Eurozol reagent. RNA (1 μg) then was retrotranscribed at 42°C for 60 min in PCR buffer (1.5 mM MgCl2) containing 5 mM random primers, 0.5 mM deoxynucleoside triphosphate (dNTP), and 100 U of Moloney murine leukemia virus reverse transcriptase (Ambion) in a final volume of 20 μl. cDNAs (2 μl) (or water, as a control) were amplified in duplicate by real-time RT-PCR using the Brilliant SYBR Green QPCR Master Mix (Stratagene) in a final volume of 25 μl. Primer sequences for assessing IE1, IE2, and β-actin mRNA levels are listed in Table 1. Following an initial denaturing step at 95°C for 2 min to activate 0.75 U of Platinum TaqDNA polymerase (Invitrogen), the cDNAs were amplified for 30 cycles (95°C for 1 min, 58°C for 1 min, and 72°C for 1 min). For quantitative analysis, semilogarithmic plots were constructed of the change in fluorescence versus the cycle number, and a threshold was set for the changes in fluorescence at a point in the linear PCR amplification phase (CT). The CT values for each gene were normalized to the CT values for β-actin using the ΔCT equation. The level of target RNA, normalized to the endogenous β-actin reference and relative to results for the cells infected for 12 h, was calculated by the comparative CT method and the 2−ΔΔCt equation.
RESULTS
HCMV infection stimulates TCF formation in quiescent cells and Elk-1 activation through the MEK1/2-ERK1/2 pathway.
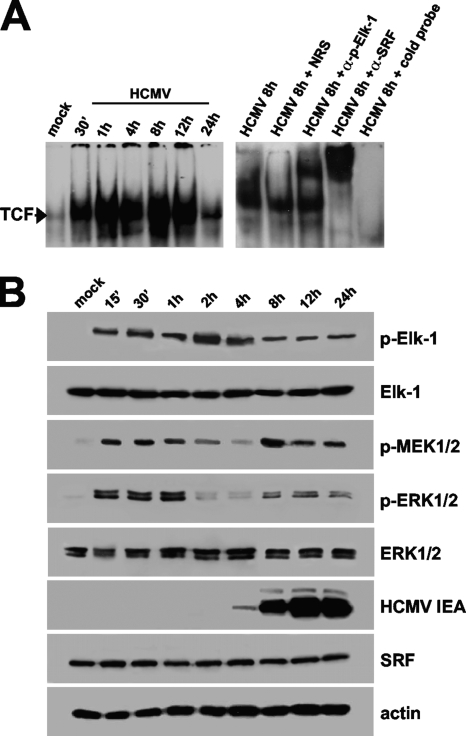
Elk-1 can be activated through multiple pathways, including the protein kinase C (PKC), mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK), and the p38/MAPK pathways (14, 26, 31, 35, 46). Although HCMV infection has been reported to stimulate the activity of both the MAPK/ERK1/2 and p38/MAPK pathways (10, 20, 21), it is unclear whether Elk-1 becomes activated. In addition, it is not known whether this activation results in the ability of Elk-1 to stimulate the formation of transcriptionally competent TCF on the SEE of the MIEP in the context of HCMV infection. To address these issues, EMSA analyses were performed using a probe containing the SEE nucleotide sequence (from −518 to −547 relative to IE1/2 transcription start site) and a nuclear extract prepared from quiescent HELFs that had been infected with HCMV for the indicated times (p.i.). As shown in Fig. 1A (left), HCMV infection stimulated a rapid and sustained binding of a protein complex to the SEE probe. Since the transcriptional activation of TCF is known to require the interaction of phosphorylated Elk-1 with both SRF and its cognate SRE (6, 36), supershift analyses using anti-SRF and anti-phospho-Elk-1 antibodies were performed to investigate whether the protein complex formed on the SEE probe was indeed composed of these two transcription factors. The results confirm that the HCMV-induced complex contains both SRF and phosphorylated Elk-1 (Fig. 1A, right). Moreover, the mutation of the SRF binding site within the SEE probe abolished TCF formation in extracts from quiescent HELFs infected with HCMV (data not shown), thus suggesting a role of SRF as the constitutive scaffold protein required to initiate TCF assembly by binding to the SEE and recruiting phosphorylated Elk-1.
FIG. 1.
HCMV infection in quiescent cells induces TCF assembly and stimulates Elk-1 activation through the MAPK MEK1/2-ERK1/2 pathway. (A) HCMV infection rapidly induces TCF formation on the SEE of MIEP. HELF cells were growth arrested for 96 h in low-serum medium and then infected with HCMV AD169 (MOI of 5 PFU/cell) or mock infected. Nuclear extracts then were prepared at the indicated times p.i. and assayed for TCF complex formation by EMSA using the SEE probe. Autoradiographs demonstrate the nucleoprotein complex formed with the 32P-labeled wild-type MIEP SEE (from −518 to −547 with respect to the IE1/2 transcription start site). Supershift experiments were performed by adding 1 μg of rabbit polyclonal antibodies raised against SRF, 1 μg of a mouse monoclonal raised against p-Elk-1, or normal rabbit serum (NRS) as a control. An unlabeled 30-bp annealed SEE oligonucleotide was added as competitor DNA in a 300-fold molar excess above the level of the SEE probe. (B) Activation of MEK1/2, ERK1/2, and Elk-1 following HCMV infection in quiescent cells. Fibroblasts were grown to subconfluence, serum starved for 96 h, and then infected with HCMV AD169 (MOI, 5 PFU/cell). At the indicated times p.i., total protein cell extracts were prepared and analyzed by immunoblotting with rabbit anti-SRF, anti-Elk-1, anti-p-MEK1/2, anti-ERK1/2, or anti-p-ERK1/2 polyclonal antibody and with an anti-p-Elk-1 MAb. Actin immunodetected with a MAb served as an internal control.
To investigate further the mechanisms of Elk-1 activation following HCMV infection, immunoblot analyses were performed on extracts prepared from quiescent cells infected with HCMV at different times p.i. As seen in Fig. 1B, the HCMV infection of quiescent cells stimulated the phosphorylation of Elk-1 within 15 min, and prolonged levels of phospho-Elk-1 were detected up to 24 h p.i. The virus also induced the activation of the upstream ERK1/2 and MEK1/2 kinases with the same two-phases kinetics as those previously described (21), confirming this MAPK cascade to be a virally regulated transduction pathway leading to Elk-1 activation.
Taken together, these results demonstrate that HCMV infection in growth-arrested cells induces a rapid and sustained formation of genuine TCF containing the phosphorylated form of Elk-1 as well as the constitutive SRF protein, and that Elk-1 is regulated by the virus-activated MEK1/2-ERK1/2 pathway.
HCMV-activated MEK1/2 signaling is required for efficient TCF formation and IE and E gene expression in quiescent cells.
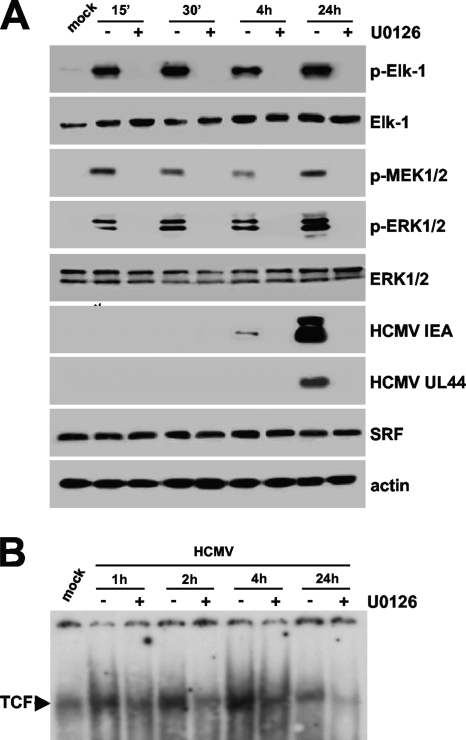
The observation that the MEK1/2-ERK1/2 pathway was activated by virus infection within the first hour p.i. suggests that it plays a role in TCF activation and therefore in MIEP activation and the initiation of viral gene expression in quiescent cells. To examine the importance of this HCMV-activated signaling for efficient MIEP activation in quiescent cells, EMSA analyses were performed with the SEE probe and the nuclear extracts prepared from quiescent HELFs infected in the presence of UO126, a potent and specific inhibitor of both MEK1 and MEK2 kinase activity (12). Immunoblot analyses demonstrated that treatment with UO126 (20 μM) for 1 h prior to infection was able to abolish HCMV-induced MEK1/2, ERK1/2, and Elk-1 phosphorylation throughout infection (Fig. 2A). Moreover, as can be seen in the EMSA autoradiograph reported in Fig. 2B, UO126 significantly reduced the formation of virus-mediated TCF, indicating that the HCMV-mediated activation of MEK1/2 is required for optimal TCF assembly above the SEE.
FIG. 2.
Effects of the inhibition of HCMV-induced MEK1/2 activation on TCF formation and viral IE and E gene expression. (A) UO126 inhibits HCMV IE and E gene expression. Fibroblasts were grown to subconfluence and serum starved for 96 h, and then they were infected with HCMV AD169 (MOI, 5 PFU/cell). Where indicated, serum-starved HELFs were treated with U0126 (20 μM) for 1 h prior to infection. U0126 also was present during infection and subsequent incubation periods. At the indicated times p.i., total cell extracts were prepared and analyzed by immunoblotting with rabbit anti-SRF, anti-Elk-1, anti-p-MEK1/2, anti-ERK1/2, or anti-p-ERK1/2 polyclonal antibody and with anti-p-Elk-1, anti-IEA, or anti-UL44 MAb. Actin immunodetected with a MAb served as an internal control. (B) Virus-mediated TCF induction is reduced by UO126. HELF cells were growth arrested for 96 h in low-serum medium and then infected with HCMV AD169 (MOI of 5 PFU/cell) or mock infected. Where indicated, serum-starved HELFs were treated with U0126 (20 μM) for 1 h prior to infection. U0126 also was present during infection and subsequent incubation periods. Nuclear extracts then were prepared at the indicated times and assayed for TCF complex formation by EMSA analysis using the 32P-labeled wild-type MIEP SEE motif (from −518 to −547 with respect to the IE1/2 transcription start site).
Having established that the inhibition of MEK1/2 by UO126 inhibited both Elk-1 phosphorylation and HCMV-mediated TCF induction, we next examined the consequence of this inhibition upon the IE and E program of HCMV gene expression. In order to do so, the extracts of quiescent HELFs that had been treated with UO126 and infected with HCMV were analyzed for their content of IE and E proteins by immunoblotting with specific antibodies. The expression of IEA (IE1 and IE2) and UL44 was assessed as a control for IE and E proteins. As shown in Fig. 2A, UO126 inhibited the expression of these HCMV proteins. Taken together, these results demonstrate the requirement of virus-activated MEK1/2 signaling for the assembly and activation of the TCF complex and, in turn, for optimal HCMV IE and E gene expression.
Elk-1 and SRF both are required for optimal MIEP activity and IE gene expression in quiescent cells.
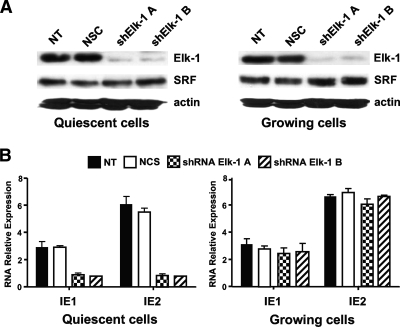
Since Elk-1 and SRF constitute the TCF protein partners required for both DNA binding and transcriptional activation, we next investigated the role that these two proteins play in the regulation of MIEP activity in quiescent and proliferating cells. pRS plasmids containing specific 29mer short hairpin RNAs (shRNAs) directed against either Elk-1 or SRF were transfected into HELFs that subsequently were incubated in high- or low-serum medium for 72 h. Total protein extracts then were prepared, and the Elk-1 and SRF protein contents were measured by immunoblotting. As shown in Fig. 3A, the two distinct shRNA sequences directed against Elk-1 (shElk-1 A and shElk-1 B) significantly reduced Elk-1 expression in both quiescent and growing cells without affecting the SRF protein levels. To determine the effects of Elk-1 silencing on the expression of HCMV IE genes, IE1 and IE2 mRNA levels were measured in quiescent and in proliferating HELFs that had been transfected with Elk-1 shRNAs for 72 h and then further infected with HCMV for a further 24 h. As shown in Fig. 3B, both IE1 and IE2 expression levels were significantly reduced in quiescent HELF cells. Since this reduction was not observed in Elk-1-silenced proliferating HELFs, it is clear that Elk-1 activity plays a role in growth-arrested cells for optimal IE gene expression.
FIG. 3.
Elk-1 silencing reduces MIEP activity in quiescent cells. (A) Inhibition of cellular Elk-1 protein expression by short hairpin RNAs (shRNAs). HELFs cells were transiently transfected with 5 μg of either a pRS shRNA expression plasmid specific for Elk-1 (shElk-1 A or shElk-1 B) or a pRS plasmid containing a noneffective shRNA cassette against GFP as a negative control for specific gene downregulation (NCS). Cells then were incubated in high- or low-serum medium for 72 h. Total cell extracts subsequently were prepared and analyzed by immunoblotting with rabbit anti-Elk-1 antibodies. The immunodetection of SRF served as a control for the specificity of shRNA-mediated Elk-1 silencing. NT, nontransfected HELF cells. (B) Silencing of cellular Elk-1 expression reduces HCMV IE1 and IE2 expression in quiescent cells. HELF cells were transiently transfected with 5 μg of either a pRS shRNA expression plasmid specific for Elk-1 (shElk-1 A or shElk-1 B) or a pRS plasmid containing a noneffective shRNA cassette against GFP (NCS) and then incubated in high- or low-serum medium for 72 h. Quiescent or proliferating shRNA Elk-1-expressing HELFs then were infected with HCMV AD169 (MOI of 3 PFU/cell). At 24 h p.i., total RNA was isolated and reverse transcribed. Real-time RT-PCR then was carried out with the appropriate IE1, IE2, and β-actin primers to quantify the expression levels of IE1 and IE2 mRNA. The results were analyzed using a standard-curve model. The levels of IE1 and IE2 mRNA were normalized to levels of endogenous β-actin mRNA. The data shown are the averages of three experiments ± standard errors of the means (error bars). NT, nontransfected HELF cells that were infected with HCMV as described above.
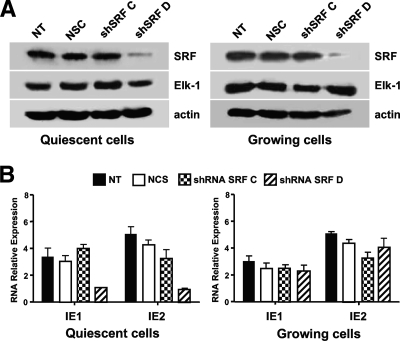
When SRF protein expression was reduced by transfecting the shRNA sequence shSRF D (Fig. 4A), a significant decrease in IE1 and IE2 mRNA expression was seen once again in the quiescent but not in the proliferating cells (Fig. 4B). This result again suggests that SRF plays a positive role in the optimization of MIEP activity in growth-arrested cells. This reduction in IE mRNA levels was not observed in HELFs transfected with the shSRF C sequence that was ineffective in reducing SRF protein expression (Fig. 4A), thus supporting the specificity of silencing by shSRF D.
FIG. 4.
SRF expression is required for efficient IE gene expression in quiescent cells. (A) Silencing SRF protein expression by shRNA. HELFs were transiently transfected with 5 μg of either a pRS shRNA expression plasmid with a 29-nucleotide SRF-specific sequence insert (shSRF C or shSRF D) or a pRS plasmid containing a noneffective shRNA cassette against GFP as a negative control for specific gene downregulation (NCS) and then incubated in high- or low-serum medium for 72 h. Thereafter, total cell extracts were prepared and analyzed by immunoblotting with rabbit anti-SRF. The immunodetection of Elk-1 served as a control for the specificity of shRNA-mediated SRF silencing. NT, nontransfected HELF cells infected with HCMV as described above. (B) SRF expression is required for optimal IE1 and IE2 mRNA expression in quiescent cells. HELF cells were transiently transfected with 5 μg of either a pRS shRNA expression plasmid specific for SRF (shSRF C or shSRF D) or a pRS plasmid containing a noneffective shRNA cassette against GFP (NCS) and then incubated in high- or low-serum medium for 72 h. Quiescent or proliferating shRNA Elk-1-expressing HELFs then were infected with HCMV AD169 (MOI of 3 PFU/cell). At 24 h p.i., total RNA was isolated and reverse transcribed, and the levels of IE1 and IE2 mRNA were determined as described above. The data shown are the averages of two experiments ± standard errors of the means (error bars).
Taken together, these results indicate that the expression of both Elk-1 and SRF is required for optimal MIEP activity and IE gene expression in quiescent cells.
Mutagenesis of SRF and Elk-1 binding sites in the MIEP impairs HCMV replication in quiescent cells.
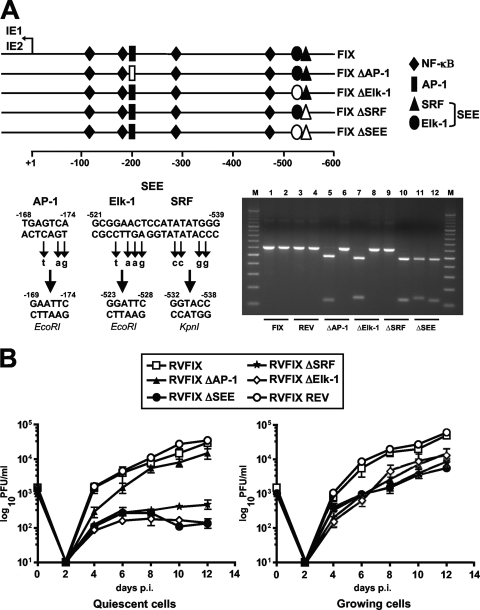
To investigate the contribution of the TCF complex partners (Elk-1, SRF, and cognate SEE) to HCMV replication in quiescent and proliferating cells, HMCV mutants were produced that contained point mutations in either the Elk-1 or the SRF binding site or in both sites to generate a total inactivation of the SEE (Fig. 5A). Mutations also were introduced into the unique AP-1 site (at −168 relative to the +1 transcription start site) to eliminate its binding to a distinct MIEP element that interacts with growth factor-activated transcription factors (28, 34). To generate these mutants, the MIEP region from HCMV VR1814 DNA was amplified and subjected to site-directed mutagenesis by introducing the restriction site EcoRI into the AP-1 site, EcoRI into the Elk-1 site, KpnI into the SRF site, and EcoRI and KpnI into the SEE (Fig. 5A). These mutations abolished the ability of the transcription factors to bind to their respective sites within the SEE (data not shown). MIEPs, each containing a single mutation (AP-1, Elk-1, SRF, or SEE site), then were used to generate BAC plasmids by using a two-step replacement strategy (44) and the FIX-BAC derived from the HCMV VR1814 strain. Infectious recombinant viruses (RVs) subsequently were reconstituted by the electroporation of the BAC plasmids into HELFs and analyzed for their growth properties.
FIG. 5.
Mutagenesis of the MIEP SEE negatively affects HCMV replication in quiescent cells. (A) A schematic representation of the HCMV MIEP region and the mutations that were introduced to inactivate AP-1, Elk-1, SRF, and SEE (SRF and Elk-1) binding sites. The position of the AP-1 (at −168), Elk-1 (at −521), SRF (at −529), and SEE (between −521 and −539) sites are numbered with respect to the IE1/2 transcription start site. The point mutations and unique restriction sites introduced into each specific element are indicated below the wild-type sequence (−168 to −174, EcoRI; −521 to −528, EcoRI; −532 to −538, KpnI). To confirm the successful disruption of the AP-1, Elk-1, SRF, and SEE sites, the MIEP enhancer sequences between nucleotides −52498 and −53104 were PCR amplified from FIX (lanes 1 and 2), FIX ΔSEE REV (lanes 3 and 4), FIX ΔAP-1 (lanes 5 and 6), FIX ΔElk-1 (lanes 7 and 8), FIX ΔSRF (lanes 9 and 10), and FIX ΔSEE (lanes 11 and 12) reconstituted viruses (RVs). The amplified products were restriction digested with either EcoRI (lanes 1, 3, 5, 7, 9, and 11) or KpnI (lanes 2, 4, 6, 8, 10, and 12), and the DNA fragments were separated by gel electrophoresis. (B) Growth kinetics of ΔAP-1, ΔElk-1, ΔSRF, and ΔSEE viruses in growing and quiescent cells. Growth-arrested or proliferating HELF cells were infected with the parental RVFIX, RVFIX ΔAP-1, RVFIX ΔElk-1, RVFIX ΔSRF, RVFIX ΔSEE, or RVFIX SEE REV (MOI of 0.1 PFU/cell). The extent of viral replication was measured at the indicated days p.i. by titrating the infectivity of cell suspension supernatants on HELFs using the IE antigen indirect immunoperoxidase staining technique (13). The data shown are the averages of three experiments ± standard errors of the means (error bars).
To determine the viral growth kinetics of the mutant viruses, HELFs in either quiescent or proliferating states were infected with one of the following RVs: RVFIX (parental), RVFIX ΔAP-1, RVFIX ΔElk-1, RVFIX ΔSRF, RVFIX ΔSEE, or the revertant RVFIX ΔSEE REV at an MOI of 0.1 PFU/cell, and titers were determined for infectious virus at 0, 2, 4, 6, 8, 10, and 12 days p.i. As can be seen in Fig. 5B, the replication kinetics of RVFIX ΔSEE in proliferating cells displayed only a minimal defect compared to the kinetics of RVFIX, RVFIX ΔAP-1, and RVFIX ΔSEE REV. In contrast, the RVFIX ΔSEE virus in quiescent cells exhibited a greater-than 2-log decrease in titers compared to those of RVFIX, RVFIX ΔAP-1, and RVFIX ΔSEE REV. Recombinant viruses with either an Elk-1 (RVFIX ΔElk-1) or SRF (RVFIX ΔSRF) mutation exhibited replication defects in quiescent cells similar to those observed with the SEE double mutant (Fig. 5B), indicating the individual contributions of SRF and Elk-1 sites to HCMV replication in growth-arrested cells. Moreover, the reconstitution of the wild-type SEE, as in RVFIX ΔSEE REV, clearly indicates that the replication defect of the ΔSEE virus in quiescent cells was due to the specific disruption of the SRF and Elk-1 sites rather than to alterations in other regions of the HCMV genome.
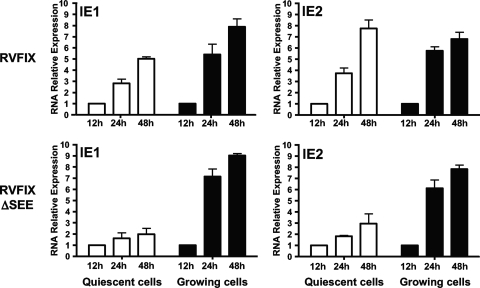
To gain further insight into the growth defect of the ΔSEE virus in quiescent cells, the expression of IE genes was measured in growing and quiescent cells infected with either RVFIX or RVFIX ΔSEE. As indicated by the real-time RT-PCR analysis shown in Fig. 6, no significant differences in IE1 and IE2 mRNA levels were found in growing cells between the parental RVFIX and the mutant RVFIX ΔSEE at any of the time points analyzed. In contrast, in quiescent cells infected with the ΔSEE virus, IE1 and IE2 mRNA levels were significantly reduced, suggesting that IE gene transcription is impaired in quiescent cells by mutations in the SEE site. Thus, the inefficient replication of the ΔSEE virus in quiescent cells stems from its incapacity to express adequate amounts of pivotal IE proteins. Taken together, these data indicate that the SEE binding element in the MIEP is required for both the transcription of IE genes and productive virus replication in quiescent cells.
FIG. 6.
MIEP SEE binding site is required for efficient viral IE gene expression in quiescent cells. Growing or quiescent HELFs were infected with the parental RVFIX or RVFIX ΔSEE (MOI of 0.1 PFU/cell). Total RNA was isolated at the indicated time p.i. and reverse transcribed. Real-time RT-PCR was carried out with the appropriate IE1, IE2, and β-actin primers to quantify the expression levels of IE1 and IE2 mRNA. The results then were analyzed using a standard-curve model, and the levels of IE1 and IE2 mRNA were normalized to levels of endogenous β-actin mRNA. The data shown are the averages of three experiments ± standard errors of the means (error bars). The value at each time point then was normalized to the value observed with cells infected for 12 h, which was set at 1.
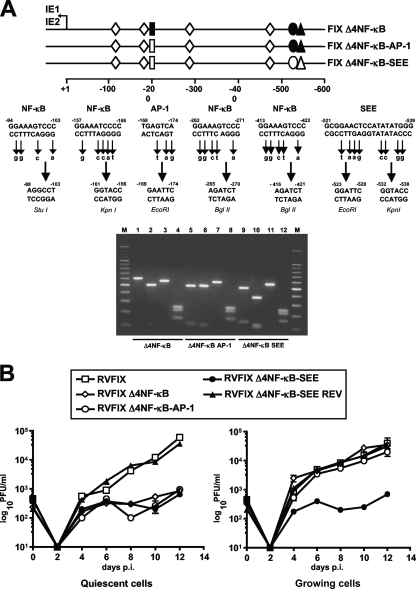
SEE-interacting factors compensate for inactivation of the NF-κB sites during viral replication in proliferating cells.
We previously found that the disruption of the four MIEP NF-κB binding sites does not hamper viral replication in growing cells (8). We thus hypothesized that other transcription factors that bind to the MIEP and whose activity is sustained in actively cycling cells could compensate for the inhibition of NF-κB (8). To investigate the potential role of SRF and Elk-1 as compensators, inactivating mutations were introduced into either the SEE or AP-1 site in a MIEP in which the four NF-κB sites also were disrupted (Fig. 7A). To generate these mutants, the MIEP region was amplified from RVFIX Δ4NF-κB DNA (8) and subjected to site-directed mutagenesis by introducing the restriction site EcoRI into the AP-1 site and EcoRI and KpnI into the SEE (Fig. 7A). Recombinant HCMV infectious viruses containing these mutations then were generated and examined for their growth properties. As can be seen in Fig. 7B, the growth of recombinant viruses RVFIX Δ4NF-κB, RVFIX Δ4NF-κB-AP-1, and RVFIX Δ4NF-κB-SEE exhibited, as expected, a severe growth defect when examined in quiescent cells. In contrast, only the RVFIX Δ4NF-κB-SEE virus displayed more than a 2-log decrease in titers in proliferating cells compared to those of RVFIX Δ4NF-κB, RVFIX Δ4NF-κB-AP-1, the revertant RVFIX Δ4ΝF-κΒ-SEE REV, and the parental RVFIX viruses (Fig. 7B).
FIG. 7.
SEE of HCMV MIEP compensates for the lack of NF-κB-mediated MIEP transactivation in proliferating cells. (A) A schematic representation of the HCMV MIEP region and the mutations that were introduced to inactivate the AP-1 binding site, SEE (SRF and Elk-1 binding sites), and the four NF-κB binding sites. The AP-1, SEE, and NF-κB sites were defined with respect to the IE1/2 transcription start site. The point mutations and unique restriction sites introduced into each specific element are indicated below the wild-type sequence (−98 to −103, StuI; −161 to −166, KpnI; −168 to −174, EcoRI; −265 to −270 and −412 to −421, BglII; −522 to −528, EcoRI; and −532 to −538, KpnI). To confirm the successful disruption of AP-1, SEE, and all four NF-κB sites, the MIEP enhancer sequences between nucleotides −52498 and −53104 were PCR amplified from FIX Δ4NK-κB (lanes 1, 2, 3, and 4), FIX Δ4NK-κB-AP-1 (lanes 5, 6, 7, and 8), and FIX Δ4NK-κB-ΔSEE (lanes 9, 10, 11, and 12) reconstituted viruses (RVs). The amplified products were restriction digested using EcoRI (lanes 1, 5, and 9), KpnI (lanes 2, 6 and 10), StuI (lanes 3, 7 and 11), or BglII (lanes 4, 8 and 12), and the DNA fragments were separated by gel electrophoresis. (B) The NF-κB and SEE binding sites of MIEP both are required for optimal viral replication in quiescent cells. Growth-arrested or proliferating HELF cells were infected with the parental RVFIX, RVFIX Δ4NF-κB, RVFIX Δ4NF-κB-AP-1, RVFIX Δ4NF-κB-SEE, or RVFIX Δ4NF-κB-SEE REV (MOI of 0.1 PFU/cell). The extent of viral replication then was assessed at the indicated days p.i. by titrating the infectivity of the cell suspension supernatants on HELFs using the IE antigen indirect immunoperoxidase staining technique (13). The data shown are the averages of three experiments ± standard errors of the means (error bars).
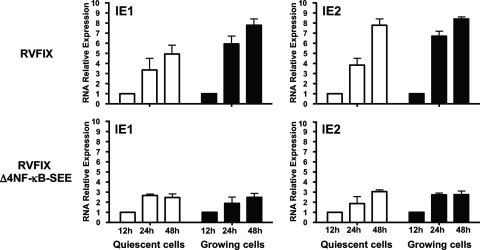
Finally, to determine whether the growth defect of the Δ4NF-κB-SEE virus in proliferating cells was related to a decrease in MIEP activity, mRNA was extracted from proliferating HELF cells that had been infected with either the RVFIX or RVFIX Δ4NF-κB-SEE viruses and analyzed by real-time RT-PCR for IE1 and IE2 mRNA content. Figure 8 shows that in proliferating cells at 24 h p.i., RVFIX Δ4NF-κB-SEE displayed a 4- and 4.5-fold reduction in IE1 and IE2 mRNA expression levels, respectively, compared to that of parental RVFIX. Taken together, these results indicate that the increased activities of Elk-1 and SRF and their sustained binding to the SEE in cells stimulated by growth factors compensate for the effects of the disruption of the MIEP NF-κB sites.
FIG. 8.
Reduced replication rate of the RVFIX Δ4NF-κB-SEE virus in proliferating cells stems from reduced levels of IE gene expression. Growing or quiescent HELFs were infected with the parental RVFIX or RVFIX Δ4NF-κB-SEE (MOI of 0.1 PFU/cell). Total RNA was isolated at the indicated time p.i. and reverse transcribed. Real-time RT-PCR was carried out using the appropriate IE1, IE2, and β-actin primers to quantify the expression levels of IE1 and IE2 mRNA. The results then were analyzed using a standard-curve model, and the levels of IE1 and IE2 mRNA were normalized to the endogenous levels of β-actin mRNA. The data shown are the averages of three experiments ± standard errors of the means (error bars). The value at each time point then was normalized to the value observed with cells infected for 12 h, which was set at 1.
DISCUSSION
This study was undertaken to investigate further the regulation of the MIEP in different cell growth states. We have shown previously that the progressive disruption of the NF-κB response elements within the MIEP in recombinant HCMV viruses did not significantly affect virus replication in proliferating cells (8). However, the ΔNF-κB mutant viruses displayed a severe growth defect in quiescent cells, thus indicating that the requirement of NF-κB signaling for MIEP activity and HCMV replication depends upon the particular physiological condition of the host cell, such as its exit from the cell cycle. These observations also suggested that the importance of transcription factor(s) other than NF-κB for optimizing MIEP activity during HCMV infection differs depending on whether cells are in a proliferating or quiescent state. The activity of these factors may be increased in growing cells, and thus their sustained binding to the MIEP may have masked the effects of the disruption of the NF-κB binding sites. To verify this hypothesis, we have performed a functional analysis of the MIEP binding sites thought to bind cellular transcription factors, such as AP-1, SRF, and Elk-1, which are stimulated by growth factors.
Here, we demonstrate that the nucleoprotein members of the TCF (Elk-1 and SRF) are required for the efficient transcription of IE genes and productive viral replication in quiescent cells. Moreover, a robust IE gene expression in quiescent cells was found to depend on the virus-induced activation of the MEK1/2-ERK1/2 signaling axis that ultimately led to rapid and prolonged Elk-1 phosphorylation. We also observed that the high level of SRF and Elk-1 binding to the SEE in proliferating cells compensates for the effects resulting from the absence of the virus-induced NF-κB binding upon overall MIEP activity. However, these results deserve further consideration. First, in a previous study that used transiently transfected SEE reporter constructs, a synergistic interaction between SRF and Elk-1 binding sites in the regulation of the basal MIEP activity and in its phorbol ester responsiveness was observed in transformed monocytes and T-lymphocyte cell lines (9). In the same study, EMSA analyses showed a substantial assembly of the TCF in extracts from tumor cells but not in extracts from uninfected human primary fibroblasts, although SRF was constitutively present in every cell type examined. The present work extends these observations by showing that during the HCMV infection of permissive fibroblasts, the binding of the virus-induced TCF complex to the SEE is critical for optimal IE gene expression and viral replication in quiescent cells. The formation of a transcriptionally competent TCF is thought to result from the MAPK-dependent phosphorylation of Elk-1 within its the regulatory domain (6, 33, 36, 46). This event is known to trigger conformational changes in Elk-1 that enhance the recruitment of coactivators (e.g., p300/CBP) and mediator subunits that ultimately leads to the stimulation of TCF transcriptional activity (6, 36, 47). Indeed, supershift experiments performed in the present study using an anti-phospho-Elk-1 MAb demonstrated the occurrence of the phosphorylated form of Elk-1 in the protein complex, which forms over the SEE in extracts from HCMV-infected HELFs (Fig. 1A).
Previous studies of the prototypic immediate-early gene c-fos promoter found that Elk-1 binding to c-fos SRE could not be detected in the absence of SRF (6, 36), indicating that the prior assembly of the SRF-SRE binary complex is a prerequisite for Elk-1 recruitment and TCF formation. Thus, in the absence of mitogenic stimuli, the c-fos SRE forms a constitutive transcriptionally inactive binary complex with SRF dimers. Upon stimulation by growth factors, the activation of the MAPK pathway stimulates Elk-1 phosphorylation and thus promotes TCF activation (6, 36, 47). Similarly, in the HCMV MIEP, the SRF requirement for TCF formation was suggested by the finding that TCF assembly over the SEE was completely abolished in extracts from uninfected human U937 cells by a competitor oligonucleotide containing only the core SRF binding site (9). In agreement with this study, we have observed that a radiolabeled SEE probe mutated in the SRF binding site did not form any DNA-protein complexes in extracts from HCMV-infected HELFs (data not shown), suggesting that SRF and Elk-1 both are required for optimal TCF activation in quiescent cells infected with HCMV.
To address further the role played by Elk-1 and SRF in MIEP activation and to assess whether they are both essential for IE gene expression and whether they play different roles, we have reduced their levels within the cells by expressing specific shRNAs. The results clearly show that the silencing of either Elk-1 or SRF negatively influences IE gene expression in quiescent cells (Fig. 3 and 4), thus suggesting that the actions of both transcription factors are required to stimulate a robust MIEP transcriptional activity. The analysis of the growth kinetics of RVFIX ΔElk-1 and RVFIX ΔSRF viruses further supports this conclusion (Fig. 5B). Thus, the likely scenario in quiescent cells is that following HCMV infection, the MEK1/2-ERK1/2 signaling axis is rapidly activated by virion components and promotes the phosphorylation of Elk-1. Thus, when input viral DNA is transported into the nucleus, transcriptionally competent TCF containing SRF and the phosphorylated form of Elk-1 forms at the newly available SEE of MIEP. SRF thus may be regarded as a constitutive transcription factor acting as a scaffold protein that is required for the recruitment and the binding to the SEE of virally regulated Elk-1.
The SEE and its cognate factors Elk-1 and SRF also were found to compensate for the lack of NF-κB-mediated MIEP transactivation in proliferating cells (Fig. 7). The high extent of TCF formation on the SEE in cells stimulated by growth factors thus overcomes the negative consequences arising from the disactivation of the MIEP NF-κB sites. In contrast, the levels of TCF activity induced by HCMV infection in quiescent cells (Fig. 1) may not be able to compensate for the lack of HCMV-induced NF-κB activity and thus result in overall weak MIEP activity and IE gene expression. It also is clear that the virus-induced NF-κB and TCF activities only represent limiting factors for HCMV replication when the host cells reside in specific physiological states, including a cell's exit from the cell cycle. In such cell states, the signaling pathways that lead to NF-κB and TCF activation synergistically optimize MIEP activity, since the deactivation of either the NF-κB element or SEE negatively impacts upon IE gene expression and viral replication.
In vivo, HCMV is able to productively infect a wide range of nonproliferating cell types (2, 29, 39) that may be in either a reversible or an irreversible growth-arrested state. In these cells, the activity and/or relative concentrations of the host transcription factor networks that regulate MIEP activity may vary in response to the cell differentiation and growth states. Thus, the activation of a specific pathway may present only a limiting factor for HCMV replication when the host cell is in a particular physiological condition. Consequently, it is unlikely that the regulation of MIEP activity in proliferating fibroblasts in cell culture reflects that which occurs in nondividing cell types in the natural host, which are those predominantly infected by the virus during acute infection (2, 39). In vitro studies have indeed demonstrated that MIEP transcriptional activity varies according to cell type, stage of cellular differentiation, and activity of a particular transduction pathway (28, 37, 41, 48). Moreover, it has been reported that the disruption of the four CREB binding sites (19-bp repeat) within the HCMV MIEP of an infectious BAC (MIEP ΔCREB) also does not affect MIE gene expression or viral replication in proliferating cells (22, 28), as we and others observed for the four NF-κB sites (1, 8, 15). However, a more recent report has described that the disactivation of the CREB site within a 19-bp repeat at −137 relative to the IE1/2 transcription start site significantly affects viral replication depending on the infected cell type (24). Thus, these results suggest that, similarly to the inability of Δ4NF-κB recombinant HCMV to replicate in quiescent cells (8), different cell type-specific transcription factor networks compensate for the ΔCREB mutation (24). The ability of HCMV to exploit its exceptionally broad cellular tropism thus may be related to the high density of regulatory interactions of the MIEP and their apparent redundancy, which may confer functional flexibility and help the virus to kick start its transcriptional program under various cellular conditions. It therefore is conceivable that the differentiation and/or growth state of infected cells influences the importance of HCMV-induced NF-κB and TCF activation to start the viral gene expression program. Thus, the dynamic nature of the MIEP regulation allows the virus to successfully exploit the different transcriptional environments under which it is forced to operate during the infection of different cell types in different physiological states.
We also have observed a rapid and prolonged activation of the MEK1/2-ERK1/2 signaling axis in quiescent cells infected with HCMV (Fig. 1). This finding provides a further example of the virus’ ability to manipulate a cell's signaling pathways to facilitate its own gene expression and, thus, to promote its replication, survival, and persistence within the host in a wide array of cell types and under different cell conditions. As far as the MAPK pathway is concerned, evidence from a variety of studies indicates that both DNA and RNA viruses, including porcine circovirus (45), bovine papillomavirus (25), simian virus 40 (SV40) (40), and human immunodeficiency virus type 1 (HIV-1) (18), can stimulate the activation of MAPK-mediated signaling and exploit this pathway to regulate their own viral expression. In the case of HCMV, Johnson et al. (21) reported that viral infection can activate the MAPK ERK1/2 pathway with two-phases kinetics, whereby the first phase occurs between 10 and 30 min after infection and the second between 4 and 12 h p.i. Our results extend these observations by showing that the rapid HCMV-induced activation of ERK1/2 and its immediate upstream partner MEK results in the phosphorylation of Elk-1 within 15 min. Moreover, the reduced levels of IE1, IE2, and UL44 protein content measured in HCMV-infected cells treated with the MEK1/2 inhibitor U0126 (Fig. 2A) directly and further substantiate the requirement of the virus-induced MEK1/2-ERK1/2 signaling in the productive cycle of the virus in cultured cells.
In conclusion, this study is the first to demonstrate (i) a critical role of HCMV-induced TCF activation for the optimization of HCMV replication in postmitotic cells, and (ii) that TCF activity compensates for NF-κB deactivation and maintains MIEP activity in proliferating cells. These findings contribute to the deepening of our knowledge of the complexities of MIEP regulation and thus enable us to assess better the molecular mechanisms that regulate viral gene expression and replication across a wide range of infected cell types and the variety of conditions under which cells become infected. Since most cells productively infected with HCMV in the natural host are in a nonproliferating growth state (2, 29, 39), the HCMV-induced activation of both the MEK1/2-ERK1/2-Elk-1 and NF-κB pathways in these cells are expected to exert a greater effect upon the initiation of the viral cycle during the phases of productive viral replication and reactivation following latency. Future studies, using experimental models of HCMV latency, are needed to determine the importance of the TCF- and NF-κB-dependent MIEP regulation during HCMV reactivation.
Acknowledgments
We thank Tom Shenk for the FIX GFP BAC and pCGN71, Neal Copeland for the E. coli SW102 strain and pgalK, and Giuseppe Gerna for the HCMV VR1814 strain. We also thank Jay Nelson for his critical review of the manuscript.
This work was supported by grants from the Italian Ministry for University and Scientific Research (Research Programmes of Significant National Interest Projects, PRIN 2007) to G.G., the University of Turin, and the Piedmont Region (Ricerca Sanitaria Finalizzata 2007 and 2008) (to G.G. and S.L.).
Footnotes
Published ahead of print on 10 February 2010.
REFERENCES
- 1.Benedict, C. A., A. Angulo, S. Ha, H. Huang, M. Messerle, C. F. Ware, and P. Ghazal. 2004. Neutrality of the canonical NF-κB pathway for human and murine cytomegalovirus transcription and replication in vitro. J. Virol. 78:741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bissinger, A. L., C. Sinzger, E. Kaiserling, and G. Jahn. 2002. Human cytomegalovirus as a direct pathogen: correlation of multiorgan involvement and cell distribution with clinical and pathological findings in a case of congenital inclusion disease. J. Med. Virol. 67:200-206. [DOI] [PubMed] [Google Scholar]
- 3.Boldogh, I., S. AbuBakar, C. Z. Deng, and T. Albrecht. 1991. Transcriptional activation of cellular oncogenes fos, jun, and myc by human cytomegalovirus. J. Virol. 65:1568-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boldogh, I., M. P. Fons, and T. Albrecht. 1993. Increased levels of sequence-specific DNA-binding proteins in human cytomegalovirus infected cells. Biochem. Biophys. Res. Commun. 197:1505-1510. [DOI] [PubMed] [Google Scholar]
- 5.Britt, W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr. Top. Microbiol. Immunol. 325:417-470. [DOI] [PubMed] [Google Scholar]
- 6.Buchwalter, G., C. Gross, and B. Wasylyk. 2004. Ets ternary complex transcription factors. Gene 324:1-14. [DOI] [PubMed] [Google Scholar]
- 7.Caposio, P., M. Dreano, G. Garotta, G. Gribaudo, and S. Landolfo. 2004. Human cytomegalovirus stimulates IKK2 activity and requires the enzyme for productive replication. J. Virol. 78:3190-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caposio, P., A. Luganini, G. Hahn, S. Landolfo, and G. Gribaudo. 2007. Activation of the virus-induced IKK/NF-κB signalling axis is critical for the replication of human cytomegalovirus in quiescent cells. Cell. Microbiol. 9:2040-2054. [DOI] [PubMed] [Google Scholar]
- 9.Chan, Y. J., C. J. Chiou, Q. Huang, and G. S. Hayward. 1996. Synergistic interactions between overlapping binding sites for the serum response factor and ELK-1 proteins mediates both basal enhancement and phorbol ester responsiveness of primate cytomegalovirus major immediate-early promoters in monocyte and T-lymphocyte cell types. J. Virol. 70:8590-8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J., and M. F. Stinski. 2002. Role of regulatory elements and the MAPK/ERK or p38 MAPK pathways for activation of human cytomegalovirus gene expression. J. Virol. 76:4873-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeMeritt, I. B., L. Milford, and A. D. Yurochko. 2004. Activation of the NF-κB pathway in human cytomegalovirus-infected cells is necessary for efficient transactivation of the major immediate-early promoter. J. Virol. 78:4498-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Favata, M. F., K. Y. Horiuchi, E. J. Manos, A. J. Daulerio, D. A. Stradley, W. S. Feeser, D. E. Van Dyk, W. J. Pitts, R. A. Earl, F. Hobbs, R. A. Copeland, R. L. Magolda, P. A. Scherle, and J. M. Trzaskos. 1998. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 273:18623-18632. [DOI] [PubMed] [Google Scholar]
- 13.Gerna, G., F. Baldanti, M. Zavattoni, A. Sarasini, E. Percivalle, and M. G. Revello. 1992. Monitoring of ganciclovir sensitivity of multiple human cytomegalovirus strains coinfecting blood of an AIDS patient by an immediate-early antigen plaque assay. Antivir. Res. 19:333-345. [DOI] [PubMed] [Google Scholar]
- 14.Gille, H., M. Kortenjann, O. Thomae, C. Moomaw, C. Slaughter, M. H. Cobb, and P. E. Shaw. 1995. ERK phosphorylation potentiates Elk-1 mediated ternary complex formation and transactivation. EMBO J. 14:951-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gustems, M., E. Borst, C. A. Benedict, C. Perez, M. Messerle, P. Ghazal, and A. Angulo. 2006. Regulation of the transcription and replication cycle of human cytomegalovirus is insensitive to genetic elimination of the cognate NF-κB binding sites in the enhancer. J. Virol. 80:9899-9904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn, G., H. Khan, F. Baldanti, U. H. Koszinowski, M. G. Revello, and G. Gerna. 2002. The human cytomegalovirus ribonucleotide reductase homolog UL45 is dispensable for growth in endothelial cells, as determined by a BAC-cloned clinical isolate of human cytomegalovirus with preserved wild-type characteristics. J. Virol. 76:9551-9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isomura, H., T. Tsurumi, and M. F. Stinski. 2004. Role of the proximal enhancer of the major immediate-early promoter in human cytomegalovirus replication. J. Virol. 78:12788-12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacqué, J. M., A. Mann, H. Enslen, N. Sharuva, B. Brichacek, R. J. Davis, and M. Stevenson. 1998. Modulation of HIV infectivity by MAPK, a virion-associated kinase. EMBO J. 17:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarvis, A. M., and J. A. Nelson. 2002. Human cytomegalovirus persistence and latency in endothelial cells and macrophage. Curr. Opin. Microbiol. 5:403-407. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, R. A., S.-M. Huong, and E.-S. Huang. 2000. Activation of the mitogen-activated protein kinase p38 by human cytomegalovirus infection through two distinct pathways: a novel mechanism for activation of p38. J. Virol. 74:1158-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, R. A., X. L. Ma, A. D. Yurochko, and E. S. Huang. 2001. The role of MKK1/2 kinase activity in human cytomegalovirus infection. J. Gen. Virol. 82:493-497. [DOI] [PubMed] [Google Scholar]
- 22.Keller, M. J., D. G. Wheeler, E. Cooper, and J. L. Meier. 2003. Role of the human cytomegalovirus major immediate-early promoter's 19-based-pair-repeat cAMP response element in acutely infected cells. J. Virol. 77:6666-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landolfo, S., M. Gariglio, G. Gribaudo, and D. Lembo. 2003. The human cytomegalovirus. Pharmacol. Ther. 98:269-297. [DOI] [PubMed] [Google Scholar]
- 24.Lashmit, P., S. Wang, H. Li, H. Isomura, and M. F. Stinski. 2009. The CREB site in the proximal enhancer is critical for cooperative interaction with the other transcription factor binding sites to enhance transcription of the MIE early genes in human cytomegalovirus infected cells. J. Virol. 83:8893-8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, P., W. C. Vass, J. T. Schiller, D. R. Lowry, and T. J. Velu. 1989. The bovine papillomavirus E5 transforming protein can stimulate the transforming activity of EGF and CSF receptors. Cell 59:21-32. [DOI] [PubMed] [Google Scholar]
- 26.Martín-Blanco, E. 2000. p38 MAPK signaling cascades: ancient roles and new functions. BioEssays 22:637-645. [DOI] [PubMed] [Google Scholar]
- 27.Meier, J. L., and J. A. Pruessner. 2000. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early expression. J. Virol. 74:1602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meier, J. L., and M. F. Stinski. Major immediate-early enhancer and its gene products, p. 151-156. In M. J. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister Academic Press, London, United Kingdom.
- 29.Mocarski, E. S., Jr., T. Shenk, and R. F. Pass. 2006. Cytomegaloviruses, p. 2701-2772. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 30.Oikawa, T., and T. Yamada. 2003. Molecular biology of the Ets family of transcription factors. Gene 303:11-34. [DOI] [PubMed] [Google Scholar]
- 31.Price, M. A., F. H. Cruzalegui, and R. Treisman. 1996. The p38 and ERK MAP kinase pathways cooperate to activate ternary complex factors and c-fos transcription in response to UV light. EMBO J. 15:6552-6563. [PMC free article] [PubMed] [Google Scholar]
- 32.Revello, M. G., F. Baldanti, E. Percivalle, A. Sarasini, L. De Giuli, E. Genini, D. Lilleri, N. Labo, and G. Gerna. 2001. In vitro selection of human cytomegalovirus variants unable to transfer virus and virus products from infected cells to polymorphonuclear leukocytes and to grow in endothelial cells. J. Gen. Virol. 82:1429-1438. [DOI] [PubMed] [Google Scholar]
- 33.Robinson, M. J., and M. H. Cobb. 1997. Mitogen-activated protein kinase pathway. Curr. Opin. Cell Biol. 9:180-186. [DOI] [PubMed] [Google Scholar]
- 34.Sambucetti, L. C., J. M. Cherrington, G. W. Wilkinson, and E. S. Mocarski. 1989. NF-kappa B activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 8:4251-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seger, R., and E. G. Krebs. 1995. The MAPK signaling cascade. FASEB J. 9:726-734. [PubMed] [Google Scholar]
- 36.Sharrocks, A. D. 2002. Complexities in ETS-domain transcription factor function and regulation: lessons from TCF (ternary complex factor) subfamily. Biochem. Soc. Trans. 30:1-9. [DOI] [PubMed] [Google Scholar]
- 37.Sinclair, J. 12 August 2009, posting date. Chromatin structure regulates human cytomegalovirus gene expression during latency, reactivation and lytic infection. Biochim. Biophys. Acta [Epub ahead of print.] doi: 10.1016/j.bbagrm.2009.08.001. [DOI] [PubMed]
- 38.Sinzger, C., A. Grefte, B. Plachter, A. S. H. Gouw, T. H. The, and G. Jahn. 1995. Fibroblasts, epithelial cells, endothelial cells and smooth muscle cells are the major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J. Gen. Virol. 76:741-750. [DOI] [PubMed] [Google Scholar]
- 39.Singzer, C., M. Digel, and G. Jahn. 2008. Cytomegalovirus cell tropism. Curr. Top. Microbiol. Immunol. 325:63-83. [DOI] [PubMed] [Google Scholar]
- 40.Sontag, E., S. Fedorov, C. Kamibayashi, D. Robbins, M. Cobb, and M. Mumby. 1993. The interaction of SV40 small tumor antigen with protein phosphatase 2A stimulates MAP kinase pathway and induces cell proliferation. Cell 75:887-897. [DOI] [PubMed] [Google Scholar]
- 41.Stinski, M. F., and H. Isomura. 2008. Role of the cytomegalovirus major immediate early enhancer in acute infection and reactivation from latency. Med. Microbiol. Immunol. 197:223-231. [DOI] [PubMed] [Google Scholar]
- 42.Stinski, M. F., and D. T. Petrick. 2008. Functional roles of the human cytomegalovirus essential IE86 protein. Curr. Top. Microbiol. Immunol. 325:133-152. [DOI] [PubMed] [Google Scholar]
- 43.Treisman, R. 1986. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell 46:567-574. [DOI] [PubMed] [Google Scholar]
- 44.Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei, L., and J. Liu. 2009. Porcine circovirus type 2 replication is impaired by inhibition of extracellular-regulated kinase (ERK) signaling pathway. Virology 386:203-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Widmann, C., S. Gibson, M. B. Jarpe, and G. L. Johnson. 1999. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79:236-244. [DOI] [PubMed] [Google Scholar]
- 47.Yang, S.-H., and A. D. Sharrocks. 2006. Convergence of the SUMO and MAPK pathways on the ETS-domain transcription factor Elk-1. Biochem. Soc. Symp. 73:121-129. [DOI] [PubMed] [Google Scholar]
- 48.Yurochko, A. D. 2008. Human cytomegalovirus modulation of signal transduction. Curr. Top. Microbiol. Immunol. 325:205-220. [DOI] [PubMed] [Google Scholar]